Abstract
Ancient Alu elements have been shown to be included in mature transcripts by point mutations that improve their 5′ or 3′ splice sites. We have examined requirements for exonization of a younger, disease-associated AluYa5 in intron 16 of the human ACE gene. A single G>C transversion in position −3 of the new Alu exon was insufficient for Alu exonization and a significant inclusion in mRNA was only observed when improving several potential splice donor sites in the presence of 3′ CAG. Since complete Alu exonization was not achieved by optimizing traditional splicing signals, including the branch site, we tested whether auxiliary elements in AluYa5 were required for constitutive inclusion. Exonization was promoted by a SELEX-predicted heptamer in Alu consensus sequence 222–228 and point mutations in highly conserved nucleotides of this heptamer decreased Alu inclusion. In addition, we show that Alu exonization was facilitated by a subset of serine/arginine-rich (SR) proteins through activation of the optimized 3′ splice site. Finally, the haplotype- and allele-specific ACE minigenes generated similar splicing patterns in both ACE-expressing and non-expressing cells, suggesting that previously reported allelic association with plasma ACE activity and cardiovascular disease is not attributable to differential splicing of introns 16 and 17.
INTRODUCTION
Splicing removes introns from eukaryotic precursor (pre-)mRNA and joins exons together to form mature transcripts. This step-wise process involves multiple and relatively weak interactions of the pre-mRNA substrate, several small nuclear ribonucleoprotein particles (snRNPs U1, U2, U4/U6 and U5) and a large number of non-snRNP proteins (1–3). In addition to numerous trans-acting factors, splicing requires cis-elements in the pre-mRNA that include the 5′ splice site (5′ss; consensus MAG/GURAGU), branchpoint sequence (BPS; YNCURAY), polypyrimidine tract (PPT) and the 3′ splice site (3′ss; YAG). These sequences are degenerate in higher eukaryotes, but are by no means sufficient for accurate splicing. Efficient intron removal often requires auxiliary sequences, known as exonic and intronic splicing enhancers (ESEs and ISEs) and silencers (ESSs and ISSs), that promote or inhibit splicing, respectively. Splicing enhancers have been identified through mutations or polymorphisms that alter splicing (4–6), through computational comparisons (7–9) and through selection of sequences that activate splicing or bind splicing regulatory factors (10,11), most notably a family of serine/arginine-rich (SR) proteins [reviewed in (12)]. Typically, SR proteins bind to ESEs in both constitutive and alternative exons through one or more N-terminal RNA recognition motifs (RRMs) (12). Unlike more extensively studied ESEs or ESSs, intronic auxiliary splicing signals have been studied in much less detail (13–19).
Human introns contain a large number of repetitive elements that may influence pre-mRNA splicing (20–27). The most abundant class of intronic repeats is represented by short interspersed elements that include Alu sequences. Alus are primate-specific, ∼300 nt repeats that constitute ∼10% of the human genome in over 106 copies. Alu insertions may cause genetic disease (28–30) and can contribute to protein diversity through exonization (31–34). About 5% of alternatively spliced internal exons in the human genome contain an Alu sequence and Alus have recently been shown to be exonized by single point mutations that improve their 5′ or 3′ss (32,33). Alus may therefore provide suitable systems for studying evolutionary aspects of both traditional and auxiliary splicing signals.
The human ACE gene encodes angiotensin converting enzyme (ACE; EC 3.4.15.1), which converts inactive angiotensin I into a critical vasoconstrictor and blood pressure regulator angiotensin II. Mice lacking ACE have low blood pressure, renal pathology and reduced male fertility (35,36). ACE has been identified as a major component of the genetic variance of highly heritable ACE plasma concentrations (37–41). Closely linked ACE polymorphisms have been associated with cardiovascular diseases, including arterial hypertension, myocardial infarction, left ventricular hypertrophy and coronary arteriosclerosis (42–47), and with Alzheimer's disease (48). In particular, associations of an insertion/deletion polymorphism (I/D) of antisense AluYa5 in ACE intron 16 with these phenotypes have been reported in a large number of independent studies, with meta-analyses supporting an association of a modest effect (47–51). Detailed linkage disequilibrium studies have attempted to map ACE levels as a quantitative trait locus to a particular polymorphism within ACE, supporting I/D and several intragenic single nucleotide polymorphisms (SNPs) as probable candidates for causal variants (47,52,53). However, genetic mapping of low penetrance genes in regions with high linkage disequilibrium has major limitations in complex traits and a biological mechanism for the proposed allelic and haplotype association needs to be demonstrated. Although several hypotheses have been considered, including a putative influence of I/D haplotypes on transcriptional regulation (54), no reliable evidence has been provided to date.
Here, we have tested if ACE I/D and closely linked SNPs influence pre-mRNA splicing and expression of ACE transcripts using minigene assays. In addition, we have examined the requirements for I/D Alu exonization by modifying both traditional and auxiliary splicing signals. We found that this Alu could not be included in mature transcripts by improving either 3′ or 5′ss and that the AluYa5 exonization required at least one mutation in both splice sites. This process was facilitated by a conserved ESE in the new Alu exon. Finally, we show that the Alu I/D inclusion in mRNA was promoted by a subset of SR proteins through increased utilization of the optimized 3′ss.
MATERIALS AND METHODS
Plasmid constructs
Minigenes A and B (Figure 1A) were cloned into HindIII/EcoRI of pCR3.1 (Invitrogen) using primers 1 (5′-gat caa gct taa gtg caa agg agt aca gct cat tg) and 2 (gat cga att cct cac tca cac tgt ggt ccg tct tt) and a DNA sample heterozygous for the I/D polymorphism. Constructs C–F were made using minigenes A and B as templates and a combination of vector primers PL1 (act cac tat agg gag acc) and PL2 (ggc tga tca gcg ggt tta) with ACE-specific primers 3 (ctg gat ccgccc agc tcc cac att aga acaat), 4 (tcg gat cca agg gca ttt cct ggg cac gct gac) and 5 (ctg gat cct ccc atc ctt tct ccc att tct ct, Figure 1A). Mutagenesis was carried out using a two-step overlap-extension PCR as described (6). ASF/SF2 cDNA that lacked the sequence cgg cgg ggg tgg agg tgg cgg encoding a heptaglycine motif between RRM1 and RRM2 was cloned as described (55). All constructs were validated by sequencing using automated sequencer ABI377 as described previously (56).
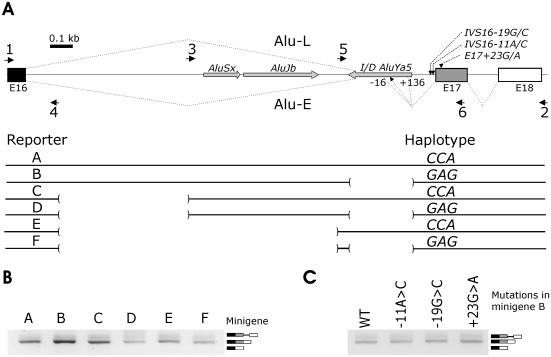
Figure 1.
A lack of a haplotype-specific pre-mRNA splicing pattern of ACE minigenes. (A) Construction of ACE minigenes. Minigenes A and B were amplified with primers 1 and 2 and a DNA sample with defined AluYa5 I/D genotype. Minigenes C and D were prepared by deleting a segment of intron 16 with a combination of vector primers and primers 3 and 4. Minigenes E and F were constructed by deleting tandemly arranged sense Alu repeats AluSx and AluJb using primers 4 and 5. Antisense AluYa5 was present in constructs A, C and E. Primers are shown as black arrows. Exons (E) are represented by boxes, introns by lines. The position and orientation of Alu sequences is denoted by grey arrows. Exons, introns and Alus are shown to scale; scale units are kilobases (kb). Exon and intron numbering is consensual as referred to in previous studies (52,54). Location of three tested SNPs around the splice acceptor site of intron 16 is shown near the 3′ss of intron 16 (the upper panel). Putative susceptibility (minigenes B, D and F) and protective (A, C and E) haplotypes in the indicated SNP are shown as 3 nt above each construct (the lower panel). Lines in the lower panel represent DNA sequences of the ACE gene that are present in the indicated minigene, whereas deleted sequences are highlighted with brackets. Alu-E (for Exonized Alu) and Alu-L (for a Long isoform retaining intron 17 and 3′ end of intron 16) represent RNA products generated by mutated constructs (see Figures 2–4). Alu-E isoforms spliced to the predicted 5′ss (black arrow) and positions −16 and +136 relative to the predicted 5′ss are shown below the I/D AluYa5. (B) Haplotype-specific minigenes A–F were transfected into 293T cells and their splicing pattern was examined by RT–PCR 48 h post-transfection. Exon 17 skipping was not observed after 50 PCR cycles (data not shown). (C) A splicing pattern of a truncated minigene individually mutated in the indicated SNPs. We mutated minigene B, which has unabridged intron 16 and lacks AluYa5 to study a putative effect of these mutations in the natural context and to facilitate mutagenesis.
Cell lines and transient transfection
HeLa cells (derived from human cervix epitheloid carcinoma), PANC1 cells (human pancreatic ductal carcinoma) and human embryonic kidney 293T cells were transfected in 6-well or 12-well plates as described (6). Transient transfection was with FuGENE 6 (Roche) according to the manufacturer's recommendations. Cells were harvested for RNA extraction 48 h later. The genotype of ACE-expressing 293T and PANC1 cells was I/D, whereas non-expressing HeLa cells had genotype D/D.
Detection of ACE isoforms
Total RNA was extracted using Tri Reagent (Sigma, USA) according to the manufacturer's recommendation. RNA was resuspended in DEPC-treated water and quantified on a spectrophotometer. Two micrograms of RNA from each sample were treated with DNase I (Ambion, USA) and reverse-transcribed using M-MLV (Promega, USA) in the presence of RNasin (Promega) at 42°C for 1 h. RT–PCR was carried out with vector-specific primers PL1/PL2 and a combination of vector primer PL1 and primer 6 (ggc agc ctg gtt gat gag tt) in exon 17 to validate the ratios of RNA products in independent amplifications (Figure 1A). The number of PCR cycles was 28 or lower to maintain approximately linear relationship between the RNA input and signal. RT–PCR products were separated on agarose or polyacrylamide gels and the identity of each isoform was confirmed by sequencing as described (56). The relative ratios of RNA products were measured with FluorImager 595 using FluorQuant and Phoretix software (Nonlinear Dynamics Inc., USA).
RESULTS
A lack of haplotype-specific splicing pattern of ACE minigenes
To test the influence of ACE variants and Alu sequences on pre-mRNA splicing, we prepared minigene constructs carrying predisposing and protective haplotypes (Figure 1A) that have been identified previously in independent human populations (57,58). The constructs contained either full (minigene A, B) or truncated (C, D) intron 16. Since extensive self-complementarity in intronic sequences may influence exon inclusion through modifying secondary RNA structure (20) and intron 16 Alus in the opposite orientation would be predicted to create stable stem–loop structures (data not shown), we also deleted tandemly arranged sense Alu elements in the latter constructs to create minigenes E and F (Figure 1A).
Examination of the splicing pattern after transfection of the wild-type minigenes with (A, C, E) and without (B, D, F) AluYa5 into 293T (Figure 1B), HeLa and PANC1 cells (data not shown) revealed a correctly spliced RNA product and a transcript retaining short intron 17, but no exon 17 skipping. Mutations in three SNPs located close to the 3′ss of intron 16 (Figure 1A), including IVS16-11C/A in PPT that may weaken 3′ss recognition, did not induce exon 17 skipping or intron 16 retention (Figure 1C).
ACE intron 16 I/D Alu is not exonized by a single mutation at the predicted splice acceptor site
ACE intron 16 I/D Alu belongs to a young, rapidly accumulating Ya5 subfamily (59–61), which tend to occupy less Alu-dense environment than older relatives of the Y subfamily or more ancient AluS/J subfamilies (62). Exonization of old and intermediate Alus could be achieved with a single mutation (33,34) and was facilitated by altering concentration of a protein involved in the 3′ss selection (33) or U1 snRNA (34). We therefore examined requirements of a polymorphic, recently integrated and disease-associated Alu for constitutive inclusion in ACE mRNA.
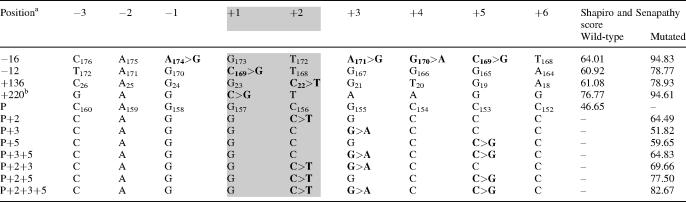
Table 1 shows alignments of the consensus sequences of AluJ/S subfamilies with ACE AluYa5 at the predicted 3′ss. Unlike the AluJ subfamily, most AluS-derived exons have been spliced via proximal AG as they contain inactivating guanosine in the distal AG (position +3 relative to the proximal AG or −2 relative to the distal AG, Table 1) (33). The proximal 3′ AG is repressed in the presence of guanosine −3 (33). Comparison of ACE AluYa5 with exonized Alus showed that this element was most similar to the AluS subfamily (Table 1), suggesting that optimizing position −3 relative to the proximal AG may exonize this sequence. We replaced guanosine −3 with cytosine, which is a preferred nucleotide in competing AG sites (63,64) and the most evolutionarily conserved nucleotide in vertebrates (65), and examined the expression of Alu-containing isoforms after transfection (Figures 1A and 2A). However, neither mRNA products transcribed from minigene A (Figure 1A, data not shown) nor mRNA products transcribed from truncated minigene E (lanes 1–2, Figure 2A) had any detectable exonization of ACE AluYa5.
Table 1.
Sequence alignment of the 3′ splice sites of ACE I/D AluYa5 and AluJ/S consensus sequences
aPosition is relative to the proximal 3′ AG. Sequence variations are in bold. Minigene mutation is indicated in italics. Position in the Alu consensus sequence (33) is indicated by a subscript. Proximal (Alu position 280 and 281) and distal (276 and 277) AGs are shaded. PGT Alu was significantly exonized following a single G282>C mutation (33).
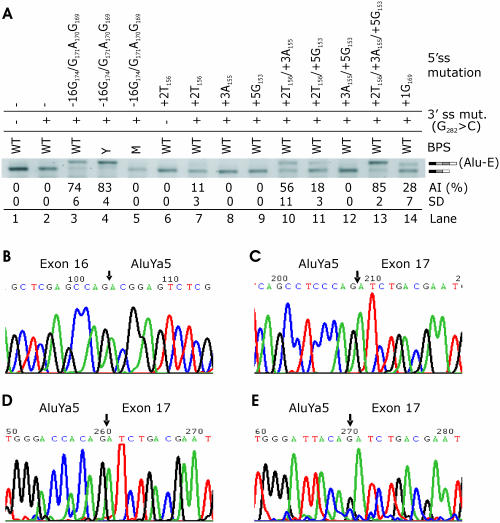
Figure 2.
Alu exonization generated by minigenes mutated in BPS and 3′ and 5′ss. Alu inclusion in mRNA was tested following an introduction of a series of mutations at the 3′ss, 5′ss and the BPS in a minigene transfected into 293T cells. (A) Inclusion of AluYa5 in mature transcripts following mutations in the 5′ss, 3′ss and the BPS in minigene E, which lacks most of intron 16 to simplify overlap-extension PCR and reduce the proportion of constructs with undesired mutations. AI, Alu inclusion levels as a mean (and SD) of triplicate transfections into 293T cells. Inclusion of AluYa5 in mRNA is indicated by a light grey rectangle on the right side. WT, wild-type BPS CAGUCA−19C. M, mutation in putative BP A−19>T; Y, yeast (S.cerevisiae) BPS UACUAA−19C. Adenosine −19 was also predicted as a putative BP (BPS score 3.25) through comparison of human and mouse introns (http://ast.bioinfo.tau.ac.il/). (B) Nucleotide sequence of the junction (arrow) between upstream exon and exonized Alu; (C–E) boundaries (arrow) between the exonized Alu and downstream exon generated by optimizing the 5′ss in position −16 (C; see lanes 3 and 4 in Figure 2A), in the predicted splice sites (D; lanes 7, 10, 11 and 13 in Figure 2A) and in position +136 (E).
Exonization of ACE AluYa5 by optimizing several 5′ splice sites in the presence of 3′CAG
To test whether optimized 5′ss can exonize this Alu in the presence of 3′ CAG or GAG, we mutated positions +2, +3 and +5 of the predicted 5′ss (34). In addition, we improved the match to the 5′ss consensus by introducing single nucleotide mutations in one upstream (position −12 relative to the predicted 5′ss) and two downstream (+136 and +220) potential splice donor sites (Table 2). Furthermore, we created a high Shapiro–Senapathy score 5′ss in position −16 by multiple mutations in both the full-length (A) and truncated (E) constructs. To further improve recognition of the 3′ss, we mutated the closest match (CAGUCA−19C) to the BPS consensus to the almost invariant BPS of Saccharomyces cerevisiae (UACUAA−19C), which is also an optimal mammalian BPS (66). Alu-containing isoforms (designated Alu-E for exonized Alu, Figure 1A) were quantified after transfection of mutated constructs into 293T cells. Although mutations at the 5′ss in the presence of 3′ GAG did not exonize Alu (data not shown), combination of a strong 5′ss at position −16 with the 3′ CAG was capable of significant Alu inclusion. The Alu inclusion was further improved in reporters with the optimized BPS (Figure 2A, lanes 3–4, Figure 2B and C). A point mutation of the putative BP [CAGUC(A−19>U)C], which is located in the optimal distance from the 3′ AG (67), eliminated Alu from the mRNA (Figure 2A, lane 5), suggesting that A−19 is the BP of the new Alu exon. Introduction of uridine at position +2 of the predicted 5′ss, which creates a highly conserved GU consensus, was insufficient for Alu inclusion (lane 6), but when combined with the 3′ CAG, we observed ∼11% exonization (lane 7 and Figure 2D). In contrast to position +2U, mutations designed to improve putative base-pairing interactions with U1 snRNA (+3, +5) did not produce any Alu inclusion in the absence of the 5′ GU (lanes 8–9 and 12), but further improved exonization when combined with +2U, with adenosine +3 being more efficient than guanosine +5 (Figure 2A, lanes 10–11 and Figure 2D). Triple mutations at the predicted 5′ss producing a good match to the 5′ consensus sequence resulted in Alu inclusion levels comparable to those produced by optimal BPS in constructs with 5′ss in position −16 (cf. lanes 13 and 4 in Figure 2A). A C>G transversion in position −12 (Table 2) led to activation of the 5′ss in position −16 (lane 14), presumably through improved match in position +5 of the 5′ss. Sequencing of the RNA product generated by the +136C>T mutation showed that ∼30% of mature transcripts contained a longer, 256 nt Alu exon (Figure 2E). Finally, a construct mutated in position +220 (Table 2) did not produce any Alu inclusion despite a high-score 5′ss consensus (data not shown), most probably due to a shortened distance between this and downstream 3′ss and/or poor exon definition of a predicted 341 nt exon, which is more than twice the size of an average human exon.
Table 2.
Minigene mutations at the 5′ splice sites
aPosition is relative to the predicted 5′ss. Position in the consensus Alu sequence (33) is subscripted.
bPosition +220 is downstream of Alu I/D. The 5′ GT is shaded. P, clones mutated in predicted 5′ss.
Correlation of the Shapiro and Senapathy scores with Alu inclusion levels produced by minigenes mutated in the predicted 5′ss was significant (r = 0.73, p = 0.02, Figure 2A and Table 2). However, a slightly lower Alu inclusion observed for a clone with a high-score 5′ss at position −16 than for an optimized predicted 5′ss (cf. lanes 3 and 13, Figure 2A and Table 2) was indicative of the presence of auxiliary splicing signals outside a 9 nt 5′ss consensus. Thus, despite limitations of the Shapiro–Senapathy scores, such as independence of each position, they provided a reasonably good indicator of splicing outcomes, consistent with their high predictive values for utilization of competing alternative, mutated and cryptic donor sites (68,69).
Identification of splicing regulatory sequences in exonized ACE AluYa5
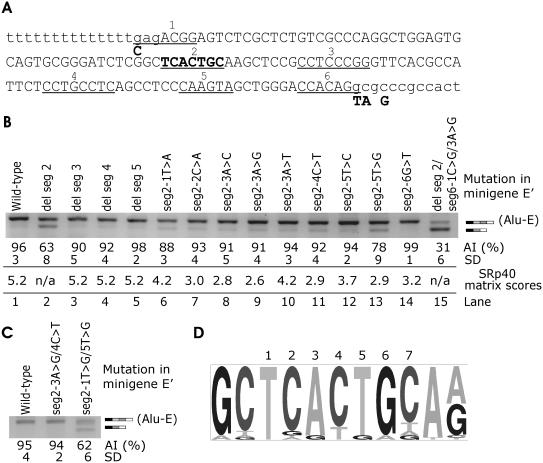
Strong traditional splicing signals did not result in complete exonization of the new Alu exon (Figure 2A, lane 4), suggesting that a constitutive inclusion requires auxiliary elements in the new Alu exon or in flanking introns. We first used ESE prediction tools that may facilitate identification of putative ESEs in the exonized Alu sequence (Figure 3A), including RESCUE-ESE (7), octamer ESE/ESS (9) and functional systematic evolution of ligands by exponential enrichment (SELEX) implemented in the ESE Finder facility (8). Putative ESEs (shown as segments 1–6, Figure 3A) that did not overlap splice sites were individually deleted in minigene E that was optimized at the 3′ and 5′ss (designated E′, Figure 3A). Deletion of segment 2 substantially reduced exonization (Figure 3B, lane 2), whereas deletions of segments 3–5 failed to do so (Figure 3B, lanes 3–5). Mutagenesis of each position in this heptamer to reduce Alu inclusion showed that a T>G transversion in position 5 had the strongest effect (Figure 3B, lanes 6–14). This alteration had one of the lowest SRp40 matrix scores (8), although no overall correlation was found between matrix scores and Alu inclusion levels (P > 0.2, Figure 3B). Mutation of the last heptamer position, which increased the SRp40 matrix score, did not alter exonization (data not shown). To test whether double mutations with minimized matrix scores can further reduce Alu inclusion, we created constructs containing either mutations 3G/4T (matrix score 0.24) or mutations 1G/5G (matrix score 0.42) in segment 2 (Figure 3C). Mutations 3G/4T had no effect, whereas mutations 1G/5G reduced Alu exonization to a level observed for the segment 2 deletion. Finally, since segments bridging the splice sites could not be deleted, two mutations were introduced at positions 1 and 3 of segment 6 (CCACAGg to GCGCAGg, Figure 3A). Alu exonization was reduced to 31% (lane 15, Figure 3B).
Figure 3.
Identification of a splicing enhancer element in ACE intron 16 AluYa5 Influence of segments 2–5 deletions and segment 2 point mutations on Alu exonization. (A) Exonized Alu segment (upper case) with putative ESEs (underlined and numbered 1–6). Segments 1, 2 and 6 were predicted by the ESE Finder (8), segments 1 (C allele only), 3 and 4 were significant ESEs identified through octamer sequences (9) and segment 5 was predicted by the RESCUE-ESE (7). Intronic sequence is in lower case. To facilitate mutagenesis, segments 2 through 5 were individually deleted in minigene E′. This minigene contained intron 16 truncation (Figure 1A) and four point mutations optimizing the splice sites (shown below the nucleotide sequence in bold). A segment 2, which was mutated individually in each position, is in the middle of the sequence in bold. (B) Alu inclusion levels following deletions of segments 2–5 and point mutations in segment 2. Transfection of all splicing reporters was into 293T cells. AI, Alu inclusion. (C) Alu inclusion levels in double mutants of minigene E. (D) Consensus sequence of segment 2 heptamer in alternatively spliced exons containing right arms of antisense Alus as identified previously (34). Representation of each heptamer position (numbered above) and flanking sequences was visualized using a pictogram utility available at http://genes.mit.edu/pictogram.html.
Comparison of segment 2 sequence with exonized Alus (34) showed that this heptamer was invariant (n = 18) or had only a single nucleotide change (n = 12) in 35 exons that contained the right arms of exonized Alus in antisense orientation. The last heptamer position was least conserved. In contrast, positions 1 and 5, which were most efficient in reducing Alu exonization (Figure 3B), were virtually invariant (Figure 3D), suggesting that they are critical for the splicing enhancement observed for segment 2. Variability of the heptamer was greater in Alu exons derived from Jb subfamilies (13 changes in 12 exons) than those derived from S subfamilies (11 changes in 19 exons). In addition, the heptamer sequence was invariant in most of over 8000 Alus that were identified as amenable to exonization [(34), data not shown]. Alignment of consensus sequences of major Alu subfamilies (62,70) with ACE AluYa5 also showed a high degree of conservation of segment 2 in all Alu subfamilies, except for AluSp. This subfamily typically contains cytosine in position 5, which decreased the SRp40 matrix score, but not Alu inclusion (lane 12, Figure 3B). Interestingly, among 61 Alu-derived exons (34), we found only a single AluSp entry.
SR-mediated inclusion of ACE I/D Alu in mature transcripts
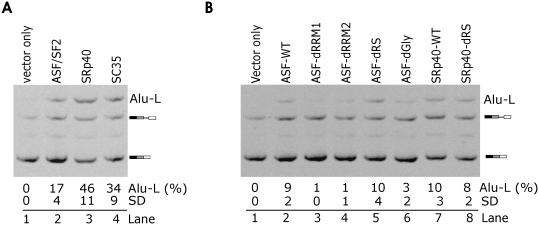
To test whether Alu exonization is influenced by factors known to control RNA processing, we co-transfected the wild-type reporters (data not shown) and minigenes optimized at the 3′ss with plasmids expressing SR proteins. Unlike wild-type constructs, minigenes containing 3′ CAG co-transfected with ASF/SF2, SRp40 and SC35 generated a larger, 1158 bp fragment (Figure 4, lanes 2–4). Sequencing of this RNA product revealed the presence of complete AluYa5, intron 17 and the 3′ end of intron 16 in mature transcripts. This isoform was designated Alu-L (Figure 1A). Minigene A with 3′CAG generated higher amounts of Alu-L as compared to minigene E containing the same 3′CAG mutation (cf. lanes 2 of Figure 4A and B), suggesting that the deleted segment of intron 16 contains sequences that promote utilization of the predicted 3′ss. Cells that were co-transfected with splicing reporters and identical empty vectors lacked Alu-L (lanes 1 in Figure 4A and B). These results are consistent with promotion of proximal splicing by SR proteins, first observed for ASF/SF2 (71,72).
Figure 4.
SR-mediated activation of the 3′ss of ACE AluYa5. Minigenes A and E were co-transfected with plasmids expressing a subset of SR proteins. (A) Inclusion of ACE IVS16 AluYa5 in mature transcripts in 293T cells transfected with 0.5 μg of minigene A containing 3′ CAG and 1 μg of plasmids expressing the indicated proteins. (B) RRMs are critical ASF/SF2 domains for the Alu-L formation. ASF-dRRM1, ASF-dRRM2, ASF-dRS and ASF-dGly denote ASF/SF2 lacking RRM1, RRM2, the RS domain and a heptaglycine stretch, respectively. WT, wild-type. Inclusion levels are mean values (SD) of two independent transfection experiments.
SR proteins contain one or two N-terminal RRMs and a C-terminal SR domain (73). To test their requirements for the Alu-L activation, we co-transfected the splicing reporters mutated at the 3′CAG with the wild-type and mutated ASF/SF2 plasmids that lacked sequences encoding RRM1, RRM2 and RS domains. In addition, we co-transfected this reporter construct with ASF/SF2 lacking a heptaglycine tract between RRM1 and RRM2 as described (55). Deletion of RRMs virtually eliminated the Alu-L formation (Figure 4B, lanes 3–4), whereas deletion of the remaining segments was insufficient to prevent the ASF/SF2-induced Alu-L activation, including the RS domain (Figure 4B, lanes 5 and 6). Similarly, Alu-L could still be induced upon deletion of the RS domain of SRp40 (lanes 7 and 8). Together, these results suggested that interaction of ASF/SF2 RRMs with the pre-mRNA was essential for Alu-L formation and that this process was RS-independent.
DISCUSSION
Antisense Alus are amenable to exonization since they contain strong PPTs and several potential 5′ and 3′ss (32–34). A single, 3′ GAG to 3′ CAG mutation in an Alu in the PGT gene, which has the 3′ss identical to the ACE AluYa5 (Table 1), resulted in almost constitutive inclusion in mRNA (33). In contrast, an identical mutation in ACE AluYa5 did not result in any inclusion in mature transcripts (Figure 2A). Differential effects of this mutation on Alu inclusion could explain a less significant contribution of the younger AluY subfamily to protein diversity as there are only few examples of AluY-containing exons (32).
Proximal 3′ AG (positions 280/281, Table 1) is selected in most exonized Alus that were derived from S subfamilies, whereas the distal AG (positions 276/277) is preferred by exonized Alu J sequences (33). The proximal 3′CAG was essential for successful exonization of this element, as we observed no Alu inclusion with the 3′ GAG minigenes carrying strong 5′ss at position −16 (Table 2, data not shown). This is consistent with an unproductive use of the proximal 3′ GAG in most Alu-derived exons (33) and with a general C ∼ T > A > G hierarchy in the 3′ss utilization (63,64). An apparently inefficient use of the 3′ CAG (Figure 2A, lane 2) could be due to a shorter PPT in our constructs (14 uninterrupted Ts) as compared with, on average, 19 ± 3 Ts in exonized Alus (33). The mean size of poly(A) tail of Alu S/J and the younger Ya5 was ∼21 and ∼26 nt, respectively, whereas the size range of recent, disease-causing Alu insertions was between 40 and 97 nt (74), suggesting that the size of poly(A) tails determines retropositional capability. If in antisense orientation, the 3′ss that contain longer tracts may be recognized more efficiently by the spliceosome than Alus with shorter tracts.
Since the BPS of the new Alu exon is located in a flanking intronic sequence, branch site variability may contribute to diverse exonization potential of similar Alu elements. This is supported by the observed increase of Alu inclusion in minigenes with optimized BPS consensus (lane 4, Figure 2A) and its elimination in a construct with A−19>T mutation (lane 5), suggesting that a poor BPS consensus limits exonization of weakly included Alus. Although we did not test if improvement of the 5′ss coupled with the optimal BPS would suffice for Alu inclusion in constructs containing 3′GAG, the enhancement mediated by the S.cerevisiae BPS was only minor (Figure 2A, lane 4).
Although the predicted 5′ss in ACE Alu (Table 2) was not used in the presence of the 3′ CAG, creation of the 5′ss GT consensus resulted in 11% Alu inclusion, which was further improved by optimizing 5′ss in positions 3 and 5 (Figure 2, lanes 7, 10, 11 and 13). The GT dinucleotide was essential for Alu inclusion, because mutations in these positions did not exonize Alus in constructs lacking the GT consensus (lanes 8 and 9, Figure 2A). The enhancement of Alu inclusion is likely to be mediated by optimized pairing with U1 snRNA, as shown earlier (34).
However, the presence of strong traditional splicing signals, including a long PPT, the preferred BPS and optimized 3′ and 5′ss, did not result in complete inclusion of the new Alu exon (lane 4 in Figure 2A). This suggested that auxiliary splicing signals, such as the presence of enhancers or a lack of silencers, or both (7,8,75), were essential for constitutive Alu derepression. Consistent with this assumption, deletion of a predicted enhancer element in Alu positions 222–228 diminished exonization in the presence of optimized 3′ and 5′ss and mutations in positions conserved in exonized Alus contributed most to Alu exclusion (Figure 3B). This segment was predicted by the ESE Finder (8) as a putative SRp40 binding site with a significant matrix score of 5.2, suggesting that it might be a binding site for this protein, although novel regulatory features in sequences created by deletions could not be excluded (Figure 3A). Although testing of putative ESEs in the exonized Alu of the ADAR2 gene did not identify any splicing auxiliary sequences (34), this heptamer is present in the ADAR2 Alu exon and it will be interesting to examine its activity in other exonized Alus and in a heterologous context. The presence of sequences that strongly repress splicing in sense Alus (55) may contribute to a low exonization potential of these elements. Since antisense Alus also contain sequences that repress splicing (14), it will be interesting to compare their inhibitory activities in future studies.
In addition to exonization of ACE Alu by optimizing both 3′ and 5′ss, we have shown that this element can be included in mature transcripts in cells over-expressing three SR proteins in the presence of a single point mutation G282>C. This effect, which depends critically on the interaction between ASF/SF2 RRMs with the pre-mRNA (Figure 4B), can be explained by promotion of proximal splicing (6,71,72,76) or could be attributed to altered mRNA export or translation in co-transfected cells. Future studies should test to what extent, if any, the promotion of proximal splicing depends on the sequences contributed by the I/D Alu. Together, these results suggest that ∼238 000 antisense Alus in human introns (33), which may represent a reservoir of ready-to-use segments expanding protein diversity (31), have a differential exonization potential and that trans-acting factors controlling coupled processes of transcription, splicing and mRNA export play an important role in this process.
Altered splicing contributes to aberrant expression of genes that may predispose to cardiovascular diseases, including LDLR, LPL, LIPA, LCAT, APOB and APOAII [reviewed in (77)]. The variability of ACE mRNA expression, its sensitivity to external signals and inaccessibility of relevant patient tissues make it difficult to study the importance of ACE SNPs in functional assays. Here, we have tested the hypothesis that ACE haplotypes and several SNPs flanking the 3′ss of intron 16, including IVS16-11C/A, result in differential exon 17 skipping, alter the amounts of natural transcripts and explain putative allelic association with disease. Purine residues around position IVS-11 show a great depletion in the PPTs of several mammals, zebrafish, fugu, ciona and other species (65), suggesting that this conserved position is important in evolution. As the IVS16-11C>A transversion may weaken the PPT by reducing interaction with the 65 kDa subunit of auxiliary factor of U2 snRNP and other polypyrimidine-binding proteins, carriers of the A allele would be predicted to show increased exon skipping and/or lower expression of natural transcripts. However, the absence of ACE AluYa5, which is in tight linkage disequilibrium with the IVS16-11A allele (Figure 1A), has been associated with a higher ACE activity in plasma and a higher risk of coronary disease (47,48,58). Intron 16 has a strong 5′ss (the Shapiro and Senapathy score 89.3) and was spliced out efficiently in our reporter assay (Figure 1B). A small reduction in the strength of the 3′ss scores (71.0 in minigenes A, C and E down to 68.5 in minigenes B, D and F, Figure 1A) due to IVS16-11A may not lead to recognizable differences in ACE expression. Although neither the exonic SNP nor PPT variants altered splicing (Figure 1C) and minigene assays generally show good reproducibility of the splicing patterns in vivo, we cannot formally exclude that the tested SNPs exert differential effect on mRNA expression in relevant tissues or distinct developmental stages. Taken together, our results do not support a role of allele-specific pre-mRNA splicing patterns generated by putative predisposing and protective ACE haplotypes in the analyzed gene segment. Future studies should address putative involvement of other ACE polymorphisms in mRNA processing, such as the recently implicated SNP rs4362 in a downstream exon (47).
Acknowledgments
We wish to thank Dr G. Screaton, University of Oxford, and Dr J. Cáceres, Medical Research Council Human Genetics Unit, Edinburgh, for reagents. We thank Dr J. Kralovicova for helpful discussions. This study was supported by the grants from the Wessex Medical Trust (Hope), the University of Southampton School of Medicine, and by the Value in People Award from the Wellcome Trust. Funding to pay the Open Access publication charges for this article was provided by JISC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Burge C.B., Tuschl T., Sharp P.A. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland R.F., Cech T.R., Atkins J.F., editors. The RNA World. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 2.Black D.L. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Z., Licklider L.J., Gygl S.P., Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 4.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 5.Pagani F., Stuani C., Tzetis M., Kanavakis E., Efthymiadou A., Doudounakis S., Casals T., Baralle F.E. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 2003;12:1111–1120. doi: 10.1093/hmg/ddg131. [DOI] [PubMed] [Google Scholar]
- 6.Královičová J., Houngninou-Molango S., Krämer A., Vořechovský I. Branch sites haplotypes that control alternative splicing. Hum. Mol. Genet. 2004;13:3189–3202. doi: 10.1093/hmg/ddh334. [DOI] [PubMed] [Google Scholar]
- 7.Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 8.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X.H., Chasin L.A. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H.X., Zhang M., Krainer A.R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tacke R., Manley J.L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graveley B.R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min H., Turck C.W., Nikolic J.M., Black D.L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 14.Sun H., Chasin L.A. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo N., Kawamoto S. An intronic downstream enhancer promotes 3′ splice site usage of a neural cell-specific exon. J. Biol. Chem. 2000;275:33641–33649. doi: 10.1074/jbc.M005597200. [DOI] [PubMed] [Google Scholar]
- 16.Deguillien M., Huang S.C., Moriniere M., Dreumont N., Benz E.J., Jr, Baklouti F. Multiple cis elements regulate an alternative splicing event at 4.1R pre-mRNA during erythroid differentiation. Blood. 2001;98:3809–3816. doi: 10.1182/blood.v98.13.3809. [DOI] [PubMed] [Google Scholar]
- 17.Expert-Bezancon A., Le Caer J.P., Marie J. Heterogeneous nuclear ribonucleoprotein (hnRNP) K is a component of an intronic splicing enhancer complex that activates the splicing of the alternative exon 6A from chicken beta-tropomyosin pre-mRNA. J. Biol. Chem. 2002;277:16614–16623. doi: 10.1074/jbc.M201083200. [DOI] [PubMed] [Google Scholar]
- 18.Hastings M.L., Wilson C.M., Munroe S.H. A purine-rich intronic element enhances alternative splicing of thyroid hormone receptor mRNA. RNA. 2001;7:859–874. doi: 10.1017/s1355838201002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCullough A.J., Berget S.M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell. Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howe K.J., Ares M., Jr Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc. Natl Acad. Sci. USA. 1997;94:12467–12472. doi: 10.1073/pnas.94.23.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grover A., Houlden H., Baker M., Adamson J., Lewis J., Prihar G., Pickering-Brown S., Duff K., Hutton M. 5′ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem–loop structure that regulates alternative splicing of exon 10. J. Biol. Chem. 1999;274:15134–15143. doi: 10.1074/jbc.274.21.15134. [DOI] [PubMed] [Google Scholar]
- 22.Gabellini N. A polymorphic GT repeat from the human cardiac Na+/Ca2+ exchanger intron 2 activates splicing. Eur. J. Biochem. 2001;268:1076–1083. doi: 10.1046/j.1432-1327.2001.01974.x. [DOI] [PubMed] [Google Scholar]
- 23.Pagani F., Buratti E., Stuani C., Romano M., Zuccato E., Niksic M., Giglio L., Faraguna D., Baralle F.E. Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 2000;275:21041–21047. doi: 10.1074/jbc.M910165199. [DOI] [PubMed] [Google Scholar]
- 24.Hui J., Stangl K., Lane W.S., Bindereif A. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nature Struct. Biol. 2003;10:33–37. doi: 10.1038/nsb875. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y., Stephan W. Compensatory evolution of a precursor messenger RNA secondary structure in the Drosophila melanogaster Adh gene. Proc. Natl Acad. Sci. USA. 2003;100:11499–11504. doi: 10.1073/pnas.1932834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hefferon T.W., Groman J.D., Yurk C.E., Cutting G.R. A variable dinucleotide repeat in the CFTR gene contributes to phenotype diversity by forming RNA secondary structures that alter splicing. Proc. Natl Acad. Sci. USA. 2004;101:3504–3509. doi: 10.1073/pnas.0400182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buratti E., Muro A.F., Giombi M., Gherbassi D., Iaconcig A., Baralle F.E. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol. Cell. Biol. 2004;24:1387–1400. doi: 10.1128/MCB.24.3.1387-1400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell G.A., Labuda D., Fontaine G., Saudubray J.M., Bonnefont J.P., Lyonnet S., Brody L.C., Steel G., Obie C., Valle D. Splice-mediated insertion of an Alu sequence inactivates ornithine delta-aminotransferase: a role for Alu elements in human mutation. Proc. Natl Acad. Sci. USA. 1991;88:815–819. doi: 10.1073/pnas.88.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knebelmann B., Forestier L., Drouot L., Quinones S., Chuet C., Benessy F., Saus J., Antignac C. Splice-mediated insertion of an Alu sequence in the COL4A3 mRNA causing autosomal recessive Alport syndrome. Hum. Mol. Genet. 1995;4:675–679. doi: 10.1093/hmg/4.4.675. [DOI] [PubMed] [Google Scholar]
- 30.Vervoort R., Gitzelmann R., Lissens W., Liebaers I. A mutation (IVS8+0.6kbdelTC) creating a new donor splice site activates a cryptic exon in an Alu-element in intron 8 of the human beta-glucuronidase gene. Hum. Genet. 1998;103:686–693. doi: 10.1007/pl00008709. [DOI] [PubMed] [Google Scholar]
- 31.Makalowski W., Mitchell G.A., Labuda D. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 32.Sorek R., Ast G., Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lev-Maor G., Sorek R., Shomron N., Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 34.Sorek R., Lev-Maor G., Reznik M., Dagan T., Belinky F., Graur D., Ast G. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in alu exons. Mol. Cell. 2004;14:221–231. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- 35.Esther C.R., Jr, Howard T.E., Marino E.M., Goddard J.M., Capecchi M.R., Bernstein K.E. Mice lacking angiotensin-converting enzyme have low blood pressure, renal pathology, and reduced male fertility. Lab. Invest. 1996;74:953–965. [PubMed] [Google Scholar]
- 36.Esther C.R., Marino E.M., Howard T.E., Machaud A., Corvol P., Capecchi M.R., Bernstein K.E. The critical role of tissue angiotensin-converting enzyme as revealed by gene targeting in mice. J. Clin. Invest. 1997;99:2375–2385. doi: 10.1172/JCI119419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cambien F., Alhenc-Gelas F., Herbeth B., Andre J.L., Rakotovao R., Gonzales M.F., Allegrini J., Bloch C. Familial resemblance of plasma angiotensin-converting enzyme level: the Nancy Study. Am. J. Hum. Genet. 1988;43:774–780. [PMC free article] [PubMed] [Google Scholar]
- 38.McKenzie C.A., Abecasis G.R., Keavney B., Forrester T., Ratcliffe P.J., Julier C., Connell J.M., Bennett F., McFarlane-Anderson N., Lathrop G.M., Cardon L.R. trans-ethnic fine mapping of a quantitative trait locus for circulating angiotensin I-converting enzyme (ACE) Hum. Mol. Genet. 2001;10:1077–1084. doi: 10.1093/hmg/10.10.1077. [DOI] [PubMed] [Google Scholar]
- 39.McKenzie C.A., Julier C., Forrester T., McFarlane-Anderson N., Keavney B., Lathrop G.M., Ratcliffe P.J., Farrall M. Segregation and linkage analysis of serum angiotensin I-converting enzyme levels: evidence for two quantitative-trait loci. Am. J. Hum. Genet. 1995;57:1426–1435. [PMC free article] [PubMed] [Google Scholar]
- 40.Tiret L., Rigat B., Visvikis S., Breda C., Corvol P., Cambien F., Soubrier F. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am. J. Hum. Genet. 1992;51:197–205. [PMC free article] [PubMed] [Google Scholar]
- 41.Tsukada K., Ishimitsu T., Tsuchiya N., Horinaka S., Matsuoka H. Angiotensin-converting enzyme gene polymorphism and cardiovascular endocrine system in coronary angiography patients. Jpn. Heart J. 1997;38:799–810. doi: 10.1536/ihj.38.799. [DOI] [PubMed] [Google Scholar]
- 42.Rigat B., Hubert C., Alhenc-Gelas F., Cambien F., Corvol P., Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soubrier F., Martin S., Alonso A., Visvikis S., Tiret L., Matsuda F., Lathrop G.M., Farrall M. High-resolution genetic mapping of the ACE-linked QTL influencing circulating ACE activity. Eur. J. Hum. Genet. 2002;10:553–561. doi: 10.1038/sj.ejhg.5200847. [DOI] [PubMed] [Google Scholar]
- 44.Soubrier F., Nadaud S., Williams T.A. Angiotensin I converting enzyme gene: regulation, polymorphism and implications in cardiovascular diseases. Eur. Heart J. 1994;15:24–29. doi: 10.1093/eurheartj/15.suppl_d.24. [DOI] [PubMed] [Google Scholar]
- 45.Ye S., Dhillon S., Seear R., Dunleavey L., Day L.B., Bannister W., Day I.N., Simpson I. Epistatic interaction between variations in the angiotensin I converting enzyme and angiotensin II type 1 receptor genes in relation to extent of coronary atherosclerosis. Heart. 2003;89:1195–1199. doi: 10.1136/heart.89.10.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uemura K., Nakura J., Kohara K., Miki T. Association of ACE I/D polymorphism with cardiovascular risk factors. Hum. Genet. 2000;107:239–242. doi: 10.1007/s004390000358. [DOI] [PubMed] [Google Scholar]
- 47.Katzov H., Bennet A.M., Kehoe P., Wiman B., Gatz M., Blennow K., Lenhard B., Pedersen N.L., de Faire U., Prince J.A. A cladistic model of ACE sequence variation with implications for myocardial infarction, Alzheimer disease and obesity. Hum. Mol. Genet. 2004;13:2647–2657. doi: 10.1093/hmg/ddh286. [DOI] [PubMed] [Google Scholar]
- 48.Kehoe P.G., Katzov H., Feuk L., Bennet A.M., Johansson B., Wiman B., de Faire U., Cairns N.J., Wilcock G.K., Brookes A.J., et al. Haplotypes extending across ACE are associated with Alzheimer's disease. Hum. Mol. Genet. 2003;12:859–867. doi: 10.1093/hmg/ddg094. [DOI] [PubMed] [Google Scholar]
- 49.Samani N.J. Molecular biology of the renin-angiotensin system: implications for hypertension and beyond. J. Cardiovasc. Pharmacol. 1991;18(Suppl 2):S1–S6. [PubMed] [Google Scholar]
- 50.Sayed-Tabatabaei F.A., Houwing-Duistermaat J.J., van Duijn C.M., Witteman J.C. Angiotensin-converting enzyme gene polymorphism and carotid artery wall thickness: a meta-analysis. Stroke. 2003;34:1634–1639. doi: 10.1161/01.STR.0000077926.49330.64. [DOI] [PubMed] [Google Scholar]
- 51.Keavney B., McKenzie C., Parish S., Palmer A., Clark S., Youngman L., Delepine M., Lathrop M., Peto R., Collins R. Large-scale test of hypothesised associations between the angiotensin-converting-enzyme insertion/deletion polymorphism and myocardial infarction in about 5000 cases and 6000 controls. International studies of infarct survival (ISIS) collaborators. Lancet. 2000;355:434–442. doi: 10.1016/s0140-6736(00)82009-7. [DOI] [PubMed] [Google Scholar]
- 52.Keavney B., McKenzie C.A., Connell J.M., Julier C., Ratcliffe P.J., Sobel E., Lathrop M., Farrall M. Measured haplotype analysis of the angiotensin-I converting enzyme gene. Hum. Mol. Genet. 1998;7:1745–1751. doi: 10.1093/hmg/7.11.1745. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W., Collins A., Abecasis G.R., Cardon L.R., Morton N.E. Mapping quantitative effects of oligogenes by allelic association. Ann. Hum. Genet. 2002;66:211–221. doi: 10.1017/S0003480002001112. [DOI] [PubMed] [Google Scholar]
- 54.Rosatto N., Pontremoli R., De Ferrari G., Ravazzolo R. Intron 16 insertion of the angiotensin converting enzyme gene and transcriptional regulation. Nephrol. Dial. Transplant. 1999;14:868–871. doi: 10.1093/ndt/14.4.868. [DOI] [PubMed] [Google Scholar]
- 55.Lei H., Vořechovský I. Identification of splicing silencers and enhancers in sense Alus: a role for pseudo-acceptors in splice site repression. Mol. Cell. Biol. 2005;25 doi: 10.1128/MCB.25.16.6912-6920.2005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vořechovský I., Luo L., Dyer M.J., Catovsky D., Amlot P.L., Yaxley J.C., Foroni L., Hammarstrom L., Webster A.D., Yuille M.A. Clustering of missense mutations in the ataxia-telangiectasia gene in a sporadic T-cell leukaemia. Nature Genet. 1997;17:96–99. doi: 10.1038/ng0997-96. [DOI] [PubMed] [Google Scholar]
- 57.Rieder M.J., Taylor S.L., Clark A.G., Nickerson D.A. Sequence variation in the human angiotensin converting enzyme. Nature Genet. 1999;22:59–62. doi: 10.1038/8760. [DOI] [PubMed] [Google Scholar]
- 58.Cox R., Bouzekri N., Martin S., Southam L., Hugill A., Golamaully M., Cooper R., Adeyemo A., Soubrier F., Ward R., et al. Angiotensin-1-converting enzyme (ACE) plasma concentration is influenced by multiple ACE-linked quantitative trait nucleotides. Hum. Mol. Genet. 2002;11:2969–2977. doi: 10.1093/hmg/11.23.2969. [DOI] [PubMed] [Google Scholar]
- 59.Batzer M.A., Kilroy G.E., Richard P.E., Shaikh T.H., Desselle T.D., Hoppens C.L., Deininger P.L. Structure and variability of recently inserted Alu family members. Nucleic Acids Res. 1990;18:6793–6798. doi: 10.1093/nar/18.23.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matera A.G., Hellmann U., Hintz M.F., Schmid C.W. Recently transposed Alu repeats result from multiple source genes. Nucleic Acids Res. 1990;18:6019–6023. doi: 10.1093/nar/18.20.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Batzer M.A., Deininger P.L., Hellmann-Blumberg U., Jurka J., Labuda D., Rubin C.M., Schmid C.W., Zietkiewicz E., Zuckerkandl E. Standardized nomenclature for Alu repeats. J. Mol. Evol. 1996;42:3–6. doi: 10.1007/BF00163204. [DOI] [PubMed] [Google Scholar]
- 62.Jurka J., Kohany O., Pavlicek A., Kapitonov V.V., Jurka M.V. Duplication, coclustering, and selection of human Alu retrotransposons. Proc. Natl Acad. Sci. USA. 2004;101:1268–1272. doi: 10.1073/pnas.0308084100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith C.W., Chu T.T., Nadal-Ginard B. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol. Cell. Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- 65.Yeo G., Hoon S., Venkatesh B., Burge C.B. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl Acad. Sci. USA. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhuang Y.A., Goldstein A.M., Weiner A.M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc. Natl Acad. Sci. USA. 1989;86:2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chua K., Reed R. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol. Cell. Biol. 2001;21:1509–1514. doi: 10.1128/MCB.21.5.1509-1514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roca X., Sachidanandam R., Krainer A.R. Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res. 2003;31:6321–6333. doi: 10.1093/nar/gkg830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roca X., Sachidanandam R., Krainer A.R. Determinants of the inherent strength of human 5′ splice sites. RNA. 2005;11:683–698. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kapitonov V., Jurka J. The age of Alu subfamilies. J. Mol. Evol. 1996;42:59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- 71.Krainer A.R., Conway G.C., Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 72.Fu X.D., Mayeda A., Maniatis T., Krainer A.R. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc. Natl Acad. Sci. USA. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caceres J.F., Krainer A.R. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roy-Engel A.M., Salem A.H., Oyeniran O.O., Deininger L., Hedges D.J., Kilroy G.E., Batzer M.A., Deininger P.L. Active Alu element ‘A-tails’: size does matter. Genome Res. 2002;12:1333–1344. doi: 10.1101/gr.384802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 76.Cáceres J.F., Stamm S., Helfman D.M., Krainer A.R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 77.von Kodolitsch Y., Pyeritz R.E., Rogan P.K. Splice-site mutations in atherosclerosis candidate genes: relating individual information to phenotype. Circulation. 1999;100:693–699. doi: 10.1161/01.cir.100.7.693. [DOI] [PubMed] [Google Scholar]