Abstract
Many forms of long-lasting behavioral and synaptic plasticity require the synthesis of new proteins. For example, long-term potentiation (LTP) that endures for more than an hour requires both transcription and translation. The signal-transduction mechanisms that couple synaptic events to protein translational machinery during long-lasting synaptic plasticity, however, are not well understood. One signaling pathway that is stimulated by growth factors and results in the translation of specific mRNAs includes the rapamycin-sensitive kinase mammalian target of rapamycin (mTOR, also known as FRAP and RAFT-1). Several components of this translational signaling pathway, including mTOR, eukaryotic initiation factor-4E-binding proteins 1 and 2, and eukaryotic initiation factor-4E, are present in the rat hippocampus as shown by Western blot analysis, and these proteins are detected in the cell bodies and dendrites in the hippocampal slices by immunostaining studies. In cultured hippocampal neurons, these proteins are present in dendrites and are often found near the presynaptic protein, synapsin I. At synaptic sites, their distribution completely overlaps with a postsynaptic protein, PSD-95. These observations suggest the postsynaptic localization of these proteins. Disruption of mTOR signaling by rapamycin results in a reduction of late-phase LTP expression induced by high-frequency stimulation; the early phase of LTP is unaffected. Rapamycin also blocks the synaptic potentiation induced by brain-derived neurotrophic factor in hippocampal slices. These results demonstrate an essential role for rapamycin-sensitive signaling in the expression of two forms of synaptic plasticity that require new protein synthesis. The localization of this translational signaling pathway at postsynaptic sites may provide a mechanism that controls local protein synthesis at potentiated synapses.
Enduring changes in synaptic strength, such as long-term potentiation (LTP) in the mammalian brain or long-term facilitation (LTF) in Aplysia neurons, are one mechanism by which nervous systems can store information for long periods. Changes that underlie short-term plasticity can be accomplished by covalent modifications of preexisting proteins. New protein synthesis, however, is required for long-lasting LTP (L-LTP) (1, 2), neurotrophin-induced synaptic potentiation (3), and LTF in Aplysia (4), and for long-term behavioral memory in intact animals (5). Newly synthesized proteins are thought to deposit in activated synapses to facilitate the long-lasting structural modifications that encode memory. One mechanism that has been proposed for protein deposition is synaptic tagging (6), which hypothesizes that synaptic activity creates a tag at the activated synapse that directs the subsequent targeting of proteins synthesized in the soma. Another proposed mechanism is dendritic protein synthesis, which suggests that synaptic activity leads to protein synthesis at activated synaptic sites (e.g., refs/ 3, 7, and 8). Although it remains to be tested directly, the latter scenario is supported by observations that mRNA (9), polyribosomes, and translation factors are present in the postsynaptic regions of hippocampal neurons, and mRNAs can be translated in isolated dendrites (10–12). Despite the importance of new protein synthesis for LTP expression and memory retention, we know little about how synaptic activity initiates protein translation in neurons. Previous studies in other cell types have suggested that translation activation and repression can be mediated by modulating the activity of various translation factors (13). Such mechanisms may also be used by neurons, as suggested by results of recent experiments. For instance, Wu et al. reported that visual experience can stimulate polyadenylation as well as translation of CaMKIIα mRNA, although a causal relationship between these events has not been established (14). The activity of eukaryotic elongation factor 2 (eEF-2) is down-regulated by visual stimulation-induced phosphorylation. This phosphorylation is mediated by an N-methyl-D-aspartate-receptor-dependent activation of eEF-2 kinase (15, 16 ). A link between synaptic activity and translation activation has also been suggested recently by the observation that stimulation of metabotropic glutamate receptors induces the translocation of p90rsk kinase to polyribosomes (17). This translocation may be responsible for the phosphorylation and activation of multiple ribosome-binding proteins that are involved in translation control (18).
Recent studies have characterized a rapamycin-sensitive translational signaling pathway that regulates the translation of a specific subclass of mRNAs in mammalian cells (18). Various growth factors can stimulate this pathway, leading to the phosphorylation and activation of mammalian target of rapamycin (mTOR), a serine/threonine protein kinase that modulates the activity of several translation regulatory factors. mTOR activation can be specifically inhibited by rapamycin (19). Activation of mTOR can contribute to translational initiation by phosphorylating proteins that bind eukaryotic initiation factor-4E (eIF-4E); these proteins are known as eIF4E-binding proteins (4E-BPs). The association of 4E-BPs with eIF-4E inhibits the ability of eIF-4E to associate with eIF-4G and initiate translation. The phosphorylation of 4E-BPs by mTOR or other kinases results in their dissociation from eIF-4E and a subsequent initiation of translation (20, 21). mTOR kinase may also regulate translation by direct or indirect phosphorylation other translation-related protein factors such as p70S6K and eIF-4GI (22). p70S6K is present at synapses most likely by means of its association with the PDZ-domain-containing protein neurabin (23).
We have conducted experiments to assess the possible involvement of the rapamycin-sensitive pathway in translation-dependent synaptic plasticity. We show here that several key components of this signaling pathway are present in the hippocampus and are enriched at postsynaptic sites. Disruption of this signaling pathway with rapamycin inhibits the expression of both enduring LTP and brain-derived neurotrophic factor (BDNF)-induced synaptic potentiation in hippocampal slices. These results demonstrate the presence of this translational signaling pathway at synaptic sites and its essential role in long-term plasticity in the hippocampus. We propose that this pathway may provide a mechanism for the regulation of protein synthesis at synapses.
Materials and Methods
Antibodies.
Anti-eIF-4E mouse antibody was from Transduction Laboratories (Lexington, KY); anti-4E-BP1 and 4E-BP2 rabbit antibodies were made in-house (N.S.); anti-mTOR rabbit antibody was provided by R. T. Abraham (Mayo Clinic). All antibodies were affinity-purified. Antibody dilutions used in Western blot analysis: anti-eIF-4E, 1:250; anti-4E-BP1, 1:250; anti-4E-BP2, 1:250; anti-mTOR, 1:250. Antibody dilutions used in immunostaining: anti-eIF-4E, 1:100; anti-4E-BP1, 1:50; anti-4E-BP2, 1:50; anti-mTOR, 1:200.
Western Blot Analysis.
Adult hippocampi were homogenized in cold PBS containing 0.5% Triton X-100 and proteinase inhibitors at 4°C. The homogenate was boiled for 3 min, cooled on ice, and centrifuged for 10 min at 4°C. The supernatant was transferred to fresh tubes, and the protein concentration was determined by the Bradford method (Bio-Rad). Ten micrograms of total protein was mixed with 2× SDS gel-loading buffer (100 mM Tris⋅Cl/200 mM DTT/4% SDS/0.2% bromophenol blue/20% glycerol), heated for 2 min at 95°C, and run on SDS-4–15% polyacrylamide gel. Protein transfer and Western blot analysis were performed according to Harlow and Lane (24). Target proteins were detected by using enhanced chemiluminescence reagents (Amersham Pharmacia).
Immunostaining.
Slices (500 μm) were fixed on ice with 4% paraformaldehyde and 0.2% glutaraldehyde for 1 h and transferred to PBS on ice for overnight. Sections (50 μm) were then cut by using a Vibratome and a sapphire knife. The sections were incubated sequentially with 0.7% Triton X-100 in PBS at room temperature for 1 h followed by two 5-min washes with PBS; 0.1 M glycine in PBS at room temperature for 1 h followed by one 5-min wash with ddH2O; 1% sodium borohydride in ddH2O at room temperature for 20 min followed by one 5-min wash with ddH2O; preblock buffer (0.05% Triton-X, 5% goat serum in PBS) at 4°C for 1.5 h; primary antibodies in preblock buffer at 4°C overnight. After primary antibody incubation, the sections were washed for 30 min three times with preblock buffer and then incubated with FITC-conjugated secondary antibodies (Jackson ImmunoResearch; dilution, 1:200) at room temperature for 1 h followed by one 3-min wash with preblock buffer and two 30-min washes with PBS. Sections were mounted in Immuno Floure Mounting Medium (ICN). Cultured hippocampal cells (P2, 14 DIV) were fixed in cold 4% paraformaldehyde in PBS containing 4% sucrose on ice for 20 min and then in cold (−20°C) methanol on ice for 10 min. The fixed cells were incubated with 0.2% Triton X-100 in PBS for 10 min and then with preblock buffer at 4°C for 1 h. The cells were incubated with primary antibodies in preblock buffer overnight at 4°C, washed (for 15 min three times) with preblock buffer at room temperature, incubated with FITC- or Cy3-conjugated secondary antibodies for 1 h and washed for 15 min once with preblock buffer and for 15 min twice with PBS before mounted. Immunostained specimens were viewed with an Olympus (New Hyde Park, NY) confocal microscope. Images were recorded through standard emission filters at contrast settings for which the crossover between the two channels was negligible.
Electrophysiology.
Hippocampal slices were prepared by using a Stoelting tissue chopper from young (6- to 8-week-old) adult male Sprague–Dawley rats or young (5- to 7-week-old) adult male BALB/c57 l mice. Before electrophysiological recording, slices were stored for at least 1.5 h on a Millipore membrane placed over a tissue culture dish containing oxygenated artificial cerebrospinal fluid (ACSF) solution. The slice was exposed to 95% O2/5% CO2 circulating in an enclosed chamber. For electrophysiological recordings, slices were submerged in a stream of ACSF (119 mM NaCl/2.5 mM KCl/1.3 mM MgSO4/2.5 mM CaCl2/1.0 mM NaH2PO4/26.2 mM NaHCO3/11.0 mM glucose) maintained at room temperature (22–25°C) and gassed with 95% O2/5% CO2. Field excitatory postsynaptic potentials (EPSPs) measured in stratum radiatum were evoked by stimulation of the Schaffer collateral-commissural afferents (one stimulation every 30 sec). Extracellular recording electrodes were filled with 3 M NaCl. In L-LTP experiments, tetanic stimulation was delivered at the test intensity in 1-sec trains at 100 Hz, with four trains delivered 5 min apart; slices were maintained at 28°C.
Results
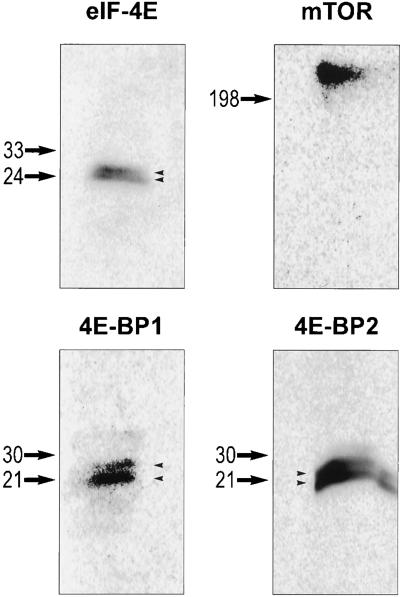
Previous studies have identified a rapamycin-sensitive signaling pathway that regulates protein translation in mammalian cells (20). We decided to test its involvement in hippocampal synaptic plasticity. We first examined whether the components of this pathway are present in the rat hippocampus. Western blot analyses identified mTOR and its downstream targets or effectors, including eIF-4E, 4E-BP1, and 4E-BP2 in hippocampal lysates (Fig. 1). eIF-4E, 4E-BP1, and 4E-BP2 were detected as multiple bands on Western blots, suggesting the existence of phosphorylated forms as indicated by studies of other cell types (25, 26).
Figure 1.
Western blot analysis of eIF-4E, 4E-BP1, 4E-BP2, and mTOR proteins in hippocampal lysates. Total lysates of the rat adult hippocampus were used for Western blot analysis, with antibodies specific for eIF-4E, 4E-BP1, 4E-BP2, and mTOR (see Material and Methods for details). The size of protein markers is labeled on the left of each blot. Note the doublet bands detected by anti-eIF-4E, 4E-BP1, and 4E-BP2 (arrowheads).
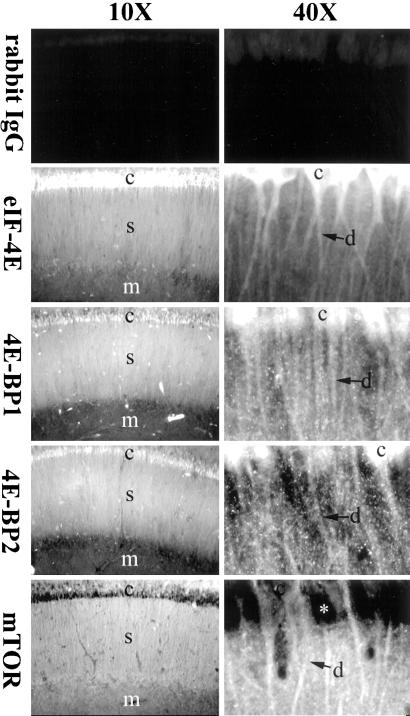
To determine the distribution patterns of these factors within the hippocampus, we performed fluorescent immunostaining with specific primary antibodies against eIF-4E, 4E-BP1, 4E-BP2, and mTOR and FITC-conjugated secondary antibodies. By confocal microscopy analysis, fluorescent signals from eIF-4E, 4E-BP1, 4E-BP2, and mTOR were observed in the CA1 region of hippocampal slices (Fig. 2). Immunostaining for all the proteins was clearly observed in both the cell body (stratum pyramidale) and synaptic (stratum radiatum) regions of the hippocampal slice. Immunopositive neurites radiating from immunopositive cell bodies strongly suggest dendritic localization of these proteins (Fig. 2). Staining for these proteins was also lower in the molecular layer (stratum lacunosum moleculare) than in the stratum radiatum (Fig. 2).
Figure 2.
Spatial distribution of eIF-4E, 4E-BP1, 4E-BP2, and mTOR in hippocampal slices. Shown are confocal images of hippocampal slices stained with primary antibodies against purified rabbit IgG (control staining), eIF4-E, 4E-BP1, 4E-BP2, or mTOR proteins, and FITC-conjugated secondary antibodies. Note the staining signals for eIF-4E, 4E-BP1, 4E-BP2, and mTOR in cell bodies and dendrites. Because mTOR is mainly a membrane protein, little signal for this protein is detected in the cytoplasm of the cell body (asterisk). c, cell body; d, dendrites; s, stratum radiatum; m, molecular layer.
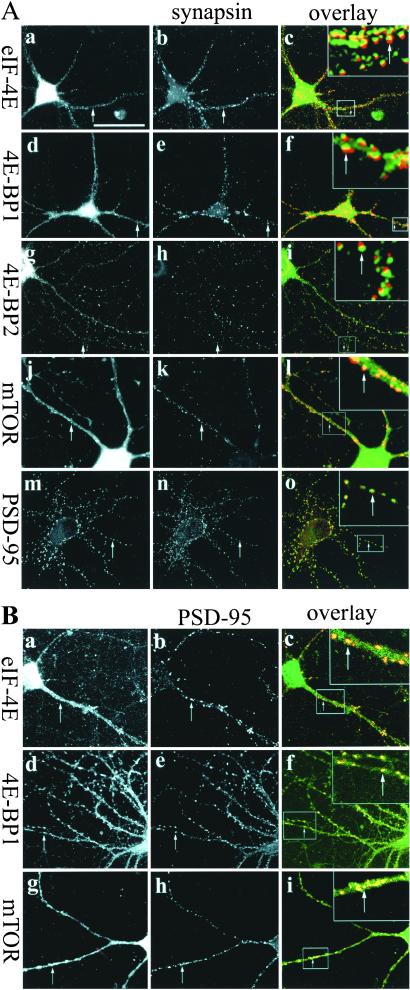
To characterize further the subcellular distribution of these factors in hippocampal neurons, we performed immunostaining on neurons (P2) cultured in vitro for 2 weeks. As shown in Fig. 3A, all the factors exhibited a somatodendritic pattern of localization. In the dendrites, eIF-4E, 4E-BP1, and 4E-BP2 were distributed in a punctate pattern. Some of the immunopositive puncta corresponded to synaptic regions as revealed by synapsin double labeling. At high magnification, the immunostaining signals for eIF-4E, 4E-BP1, and 4E-BP2 were seen to oppose and slightly overlap with the signals for synapsin-I (a presynaptic protein) staining, suggesting their localization on the postsynaptic side (see Fig. 3 Ac, Af, and Ai Insets). In particular, 4E-BP2 seemed to be significantly enriched in the postsynaptic region with a lower signal evident in the dendritic shaft, when compared with eIF-4E and 4E-BP1. The mTOR-positive regions were observed in the dendritic shaft in a diffuse pattern that extended into synaptic regions (Fig. 3Al). Consistent with the notion of postsynaptic localization of these proteins, double labeling with PSD-95 showed the signals from eIF-4E, 4E-BP1, and mTOR at synaptic regions almost completely overlapped with PSD-95 signals (Fig. 3B).
Figure 3.
Synaptic localization of eIF-4E, 4E-BP1, 4E-BP2, and mTOR. (A) Dissociated hippocampal neurons (P2; 14 DIV) were double-stained with components of the mTOR pathway (first column) including anti-eIF-4E (a), 4E-BP1 (d), 4E-BP2 (g), mTOR (j), or PSD-95 (m) antibodies and anti-synapsin I antibodies (second column) (b, e, h, and k). Signals from eIF-4E, 4E-BP1, 4E-BP2, mTOR, and PSD-95, and signals from the corresponding anti-synapsin staining on the same cells were overlaid in c, f, i, l, and o, respectively. In the overlaid images, green signals were generated by anti-eIF-4E, 4E-BP1, 4E-BP2, mTOR, or PSD-95 staining, red signals were generated by anti-synapsin staining, and yellow signals resulted from the overlapping of green and red signals. Insets in c, f, i, l, and o are the higher magnification images for the dendritic regions marked by boxes. (B) Double-labeling of eIF-4E (a), 4E-BP1 (d), mTOR (g) with PSD-95 (b, e, h). Signals from eIF-4E, 4E-BP1, and mTOR and signals from the corresponding PSD-95 staining on the same cells were overlaid in c, f, l, and i, respectively. Insets in c, f, and i are the higher magnification images for the dendritic regions marked by boxes (green signals resulted from eIF-4E, 4E-BP1, or mTOR staining; red signals from PSD-95 staining; yellow signals from the overlapping of green and red signals). Arrows indicate the synaptic regions that are double-stained. (Bar = 50 μm.)
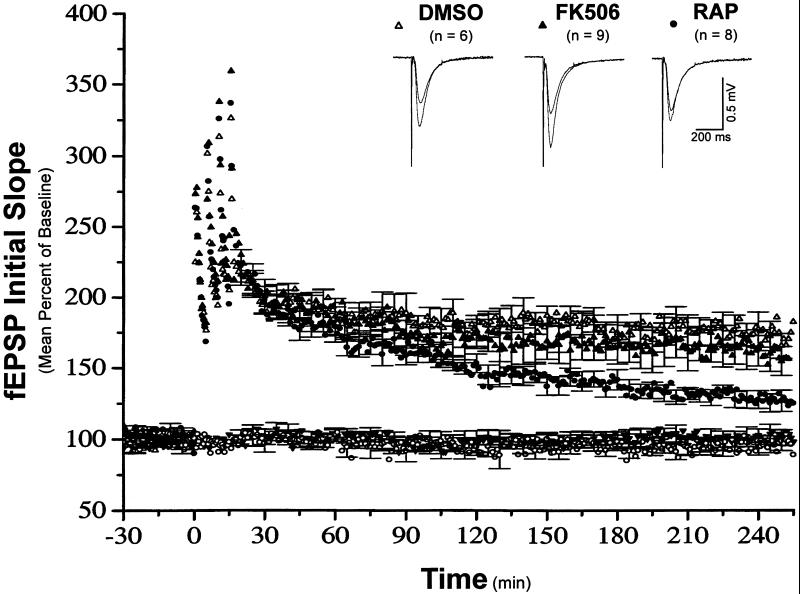
To determine the functional significance of mTOR-mediated signaling in LTP expression, we examined the sensitivity of both early- and late-phase LTP to inhibition by rapamycin. LTP was induced by four trains of 100-Hz stimulation (1 sec each) separated by 5-min intervals. As shown in Fig. 4, the presence of 200 nM rapamycin inhibited the magnitude of late-phase LTP (L-LTP, measured 230–240 min after the last tetanus), whereas the early phase of LTP (E-LTP, measured 50–60 min after the last tetanus) was unaffected [mean percent of baseline 50–60 min after tetanus, 164.9 ± 6.6; 230–240 min after tetanus, 127.5 ± 6.4 (n = 8)]. In contrast, application of the same concentration of FK506, a drug that has a similar structure and can bind to the same receptor (FKBP12) as rapamycin but does not inhibit mTOR activity (19), did not affect the magnitude of either early- or late-phase LTP [mean percent of baseline 50–60 min after tetanus, 183.6 ± 10.8; 220–240 min after tetanus, 158.6 ± 11.4 (n = 9)]. Furthermore, FK506-treated slices were indistinguishable from vehicle (DMSO)-control-treated slices [mean percent of baseline 50–60 min after tetanus, 184.0 ± 7.0; 230–240 min after tetanus, 173.4 ± 8.7 (n = 6)]. Rapamycin-treated slices exhibited significantly less L-LTP than either FK-506- (P ≤ 0.05) or DMSO-treated (P ≤ 0.05) slices. We also monitored the basal synaptic transmission from a second pathway in the same slices on which experiments were performed, and found rapamycin did not affect it during the process of recording (Fig. 4). These results demonstrate a role for mTOR in L-LTP expression.
Figure 4.
Rapamycin decreases the magnitude of late-phase LTP in hippocampal slices. Shown are ensemble averages for all DMSO, FK506, and rapamycin experiments (n = 6, 9, and 8, respectively). All drugs were applied in the ACSF at least 30 min before the first tetanus. Mean DMSO control fEPSP slope values were 0.20 ± 0.01 mV/msec before LTP induction, and 0.35 ± 0.03 mV/msec 220–240 min after LTP induction. Mean FK506-treated fEPSP slope values were 0.21 ± 0.02 mV/msec before LTP induction, and 0.30 ± 0.09 mV/msec 220–240 min after LTP induction. Mean rapamycin-treated fEPSP slope values were 0.24 ± 0.05 mV/msec before LTP induction, and 0.31 ± 0.07 mV/msec 220–240 min after LTP induction. The control pathways for FK506 and rapamycin experiments are symbolized by the filled inverted triangle and the open circles, respectively.
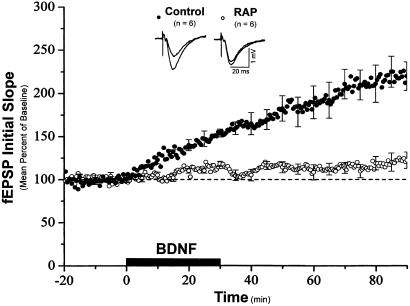
Previous studies have suggested that BDNF plays a role in L-LTP expression (27, 28). In addition, BDNF-induced synaptic enhancement requires new protein synthesis (3). Thus, we examined the effect of rapamycin on BDNF-induced synaptic potentiation. Slices were pretreated with either rapamycin or a vehicle control for 30 min before the addition of BDNF to the medium. As shown in Fig. 5, BDNF applied in the presence of control ACSF enhanced synaptic transmission as reported (3, 29) [mean percent of baseline 50–60 min after the onset of BDNF application: control, 186.2 ± 17.0% (n = 6)]. In the presence of rapamycin, however, BDNF failed to enhance synaptic strength [mean percent of baseline 50–60 min after the onset of BDNF application: rapamycin, 113.2 ± 5.5% (n = 6)]. Taken together, these results show that plasticity induced by two different means relies on rapamycin-sensitive signaling.
Figure 5.
Rapamycin prevents BDNF-induced synaptic potentiation in hippocampal slices. Shown are ensemble averages for all control and rapamycin experiments. BDNF (50 ng/ml) was applied in the ACSF at the time indicated by the bar. Mean control fEPSP slope values were 0.13 ± 0.03 mV/msec before BDNF application and 0.23 ± 0.04 mV/msec after BDNF application. Mean rapamycin-treated fEPSP slope values were 0.16 ± 0.01 mV/msec before BDNF application and 0.18 ± 0.01 mV/msec after BDNF application.
Discussion
Pharmacological studies with protein translation inhibitors have pointed to a requirement for new protein synthesis during BDNF-induced potentiation (3) and the late phase of LTP (1, 28). We have examined the potential role of a rapamycin-sensitive translational signaling pathway in hippocampal synaptic plasticity. Our Western blot analyses and immunostaining studies demonstrated the presence of several key components of this pathway, including mTOR, eIF-4E, 4E-BP1, and 4E-BP2 in the rat hippocampus; immunopositive labeling for all proteins was evident in both the cell body and dendritic layers of the hippocampal slice. Double-immunolabeling studies in cultured hippocampal neurons revealed that these proteins are also present at postsynaptic sites. This pattern of localization suggests that these proteins may mediate translational regulation at postsynaptic sites and couple synaptic events with the protein translation machinery. Consistent with this suggestion, our experiments demonstrate a requirement for this signaling pathway in both L-LTP- and BDNF-induced synaptic potentiation.
Pretreatment of hippocampal slices with rapamycin inhibited both BDNF- and high-frequency stimulation-induced synaptic plasticity, albeit with different temporal profiles. Rapamycin inhibited the early aspect BDNF-induced potentiation. In contrast, the early phase of LTP was unaffected by rapamycin, but the late phase (e.g., 2–3 h after tetanus) was significantly reduced. These temporal profiles of inhibition parallel the profiles observed with conventional protein synthesis inhibitors (1, 3, 28).
These experiments do not allow us to ascertain the location of the activity of the rapamycin-sensitive kinase during plasticity expression. Our immunostaining studies show the presence of mTOR in both the soma and the dendrites of hippocampal neurons. Rapamycin was present in the ACSF beginning 30 min before the initiation of potentiation and maintained at the same concentration for the duration of the experiment. The very early inhibition of the BDNF-induced potentiation is consistent with a local block of mTOR activity in dendrites or synaptic sites. The cell body layer is ≈500 μm from the recording making it implausible that rapamycin is inhibiting somatically synthesized proteins destined for synaptic sites. Consistent with this notion, the downstream substrates of mTOR, 4E-BP1, and 4E-BP2, are enriched at the postsynaptic sites, suggesting a local function of mTOR on the regulation of synaptic protein synthesis. In addition, recent work performed in cultured Aplysia sensory-motor neuron system have demonstrated a synaptic role of the rapamycin-sensitive signaling in LTF (30). In this experiment, the inhibition of late-, but not early-, phase LTP is consistent with either a somatic or dendritic location for the rapamycin-sensitive store. If rapamycin is acting at the soma, the long-latency inhibition could reflect either the transport time for synaptically generated signals to travel to the cell body and stimulate translation, or the transport time for newly synthesized proteins to be delivered to synaptic sites, or both. Alternatively, the long-latency inhibition by rapamycin could reflect the time required for mRNA transport to the dendrites. According to this idea, LTP-induced transcription is followed by mRNA transport to dendrites and then local, rapamycin-sensitive translation occurs, the protein products of which allow the synaptic potentiation to endure.
We currently do not know the mechanism by which synaptic activity activates this signaling pathway during LTP expression. Previous studies have identified protein kinase B (PKB, Akt) and phosphotidylinositol-3-kinase (PI3K) as the upstream regulators of mTOR (25, 31), and various growth factors have been shown to activate PI3K and PKB (32). As neurotrophins may be released during LTP (28, 33, 34), they may initiate mTOR signaling through PI3K and PKB activation. The second messenger cAMP produced after synaptic activation may also activate mTOR signaling by cAMP-dependent protein kinase A (PKA), because PKA is able to activate PKB (35). Future studies will establish a molecular connection between synaptic activity and mTOR translational signaling activation.
Dendritic protein synthesis at synaptic sites has been proposed as a mechanism that might contribute to local synaptic changes during LTP expression (7, 36, 37). Consistent with this hypothesis, various mRNAs and polyribosomes are present in these areas (9, 38). Recent studies have also demonstrated the existence of endoplasmic reticulum and Golgi apparatus in the postsynaptic site opposing synaptic boutons, suggesting the presence of translational and posttranslational modification machinery in the postsynaptic regions (39, 40). We showed here in cultured hippocampal neurons that eIF-4E, 4E-BP1, 4E-BP2, and mTOR are also present in the postsynaptic region. These findings put the mTOR translational signaling in a position to regulate local protein synthesis during long-term synaptic plasticity. This idea is supported by the recent observation that BDNF induces a rapamycin-sensitive phosphorylation of eIF4E and 4EBP1 (41).
Acknowledgments
We thank Michael Tsung and Holli Weld for making cultured hippocampal neurons. E.M.S. is an Assistant Investigator of the Howard Hughes Medical Institute.
Abbreviations
- LTP
long-term potentiation
- mTOR
mammalian target of rapamycin
- eIF-4E
eukaryotic initiation factor-4E
- 4E-BP
eIF-4E-binding protein
- BDNF
brain-derived neurotrophic factor
- ACSF
artificial cerebrospinal fluid
- fEPSP
field excitatory postsynaptic potential
References
- 1.Nguyen P V, Abel T, Kandel E R. Science. 1994;265:1104–1107. doi: 10.1126/science.8066450. [DOI] [PubMed] [Google Scholar]
- 2.Frey U, Morris R G. Trends Neurosci. 1998;21:181–188. doi: 10.1016/s0166-2236(97)01189-2. [DOI] [PubMed] [Google Scholar]
- 3.Kang H, Schuman E M. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 4.Montarolo P G, Goelet P, Castellucci V, Morgan J, Kandel E R, Schacher S. Science. 1986;234:1249–1254. doi: 10.1126/science.3775383. [DOI] [PubMed] [Google Scholar]
- 5.Davis H P, Squire L R. Psych Bull. 1984;96:518–559. [PubMed] [Google Scholar]
- 6.Frey U, Morris R G M. Nature (London) 1997;385:533–536. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- 7.Schuman E M. Neuron. 1997;18:339–342. doi: 10.1016/s0896-6273(00)81234-9. [DOI] [PubMed] [Google Scholar]
- 8.Martin K C, Casadio A, Zhu H, E, Y, Rose J C, Chen M, Bailey C H, Kandel E R. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- 9.Steward O. Neuron. 1997;18:9–12. doi: 10.1016/s0896-6273(01)80041-6. [DOI] [PubMed] [Google Scholar]
- 10.Torre E R, Steward O. J Neurosci. 1992;12:762–772. doi: 10.1523/JNEUROSCI.12-03-00762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crino P B, Eberwine J. Neuron. 1996;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- 12.Aakalu G, Smith W B, Nguyen N, Jiang C, Schuman E M. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 13.Mathews M B, Sonenberg N, Hershey J W B. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 1–29. [Google Scholar]
- 14.Wu L, Wells D, Tay J, Mendis D, Abbott M-A, Barnitt A, Quinlan E, Heynen A, Fallon J R, Richter J D. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 15.Scheetz A J, Nairn A C, Constantine-Patton M. Proc Natl Acad Sci USA. 1998;94:14770–14775. doi: 10.1073/pnas.94.26.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheetz A J, Narin A C, Constantine-Paton M. Nat Neurosci. 2000;3:211–216. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- 17.Angenstein F, Greenough W T, Weiler I J. Proc Natl Acad Sci USA. 1998;95:15078–15083. doi: 10.1073/pnas.95.25.15078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown E J, Schreiber S L. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 19.Sabatini D M, Lai M M, Snyder S H. Mol Neurobiol. 1997;15:223–239. doi: 10.1007/BF02740635. [DOI] [PubMed] [Google Scholar]
- 20.Beretta L, Gingras A C, Svitkin Y V, Hall M N, Sonenberg N. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 21.Brunn G J, Hudson C C, Sekulic A, Williams J M, Hosoi H. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 22.Gingras A-C, Raught B, Sonenberg N. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 23.Burnett P E, Blackshaw S, Lai MM, Quereshi I A, Burnett A F, Sabatini D M, Snyder S H. Proc Natl Acad Sci USA. 1998;95:8351–8356. doi: 10.1073/pnas.95.14.8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 25.Gingras A C, Kennedy S G, O'Leary M A, Sonenberg N, Hay N. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waskiewicz A J, Johnson J C, Penn B, Mahalingam M, Kimball S R, Cooper J A. Mol Cell Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korte M, Kang H, Bonhoeffer T, Schuman E M. Neuropharmacology. 1998;37:553–559. doi: 10.1016/s0028-3908(98)00035-5. [DOI] [PubMed] [Google Scholar]
- 28.Kang H, Shelton D, Welcher A, Schuman E M. Neuron. 1997;19:653–664. doi: 10.1016/s0896-6273(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 29.Kang H, Schuman E M. Science. 1995;267:1658–1662. doi: 10.1126/science.7886457. [DOI] [PubMed] [Google Scholar]
- 30.Casadio A, Martin K C, Giustetto M, Zhu H, Chen M, Bartsch D, Bailey C H, Kandel E R. Cell. 1999;99:221–237. doi: 10.1016/s0092-8674(00)81653-0. [DOI] [PubMed] [Google Scholar]
- 31.Nave B T, Ouwens D M, Withers D J, Aless D R, Shepherd P R. Biochem J. 1999;344:427–431. [PMC free article] [PubMed] [Google Scholar]
- 32.Hawkins P T, Welch H, McGregor A, Eguinoa A, Gobert S, Krugmann S, Anderson K, Stokoe D, Stephens L. Biochem Soc Trans. 1997;25:1147–1151. doi: 10.1042/bst0251147. [DOI] [PubMed] [Google Scholar]
- 33.Korte M, Griesbeck O, Gravel C, Carroll P, Staiger V, Thoenen H, Bonhoeffer T. Proc Natl Acad Sci USA. 1996;93:12547–12552. doi: 10.1073/pnas.93.22.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patterson S L, Abel T, Deuel TA S, Martin K C, Rose J C, Kandel E R. Neuron. 1996;16:1137–1145. doi: 10.1016/s0896-6273(00)80140-3. [DOI] [PubMed] [Google Scholar]
- 35.Filippa N, Sable C L, Filloux C, Hemmings B, Van Obberghen E. Mol Cell Biol. 1999;19:4989–5000. doi: 10.1128/mcb.19.7.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayford M, Baranes D, Podsypanina K, Kandel E R. Proc Natl Acad Sci USA. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuman Neuron. 1999;23:645–648. doi: 10.1016/s0896-6273(01)80023-4. [DOI] [PubMed] [Google Scholar]
- 38.Steward O, Levy W B. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardiol A, Racca C, Triller A. J Neurosci. 1999;19:168–179. doi: 10.1523/JNEUROSCI.19-01-00168.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pierce J P, van Leyen K, McCarthy J B. Nat Neurosci. 2000;3:311–313. doi: 10.1038/73868. [DOI] [PubMed] [Google Scholar]
- 41.Takei N, Kawamura M, Hara K, Yonezawa K, Nawa H. J Biol Chem. 2001;276:42818–42825. doi: 10.1074/jbc.M103237200. [DOI] [PubMed] [Google Scholar]