Abstract
Mutations in the gene encoding the peripheral myelin protein 22 (PMP22), a tetraspan protein in compact peripheral myelin, are one of the causes of inherited demyelinating peripheral neuropathy. Most PMP22 mutations alter the trafficking of the PMP22 protein in Schwann cells, and this different trafficking has been proposed as the underlying mechanism of the disease. To explore this problem further, we compared the aggregation of wild-type Pmp22 with those of the two Pmp22 mutations found in Trembler (Tr) and Trembler J (TrJ) mice. All three Pmp22s can be crosslinked readily as homodimers in transfected cells. Wild-type Pmp22 also forms heterodimers with Tr and TrJ Pmp22, and these heterodimers traffic with their respective mutant Pmp22 homodimers. All three Pmp22s form complexes larger than dimers with Tr Pmp22 especially prone to aggregate into high molecular weight complexes. Despite the differences in aggregation of Tr and TrJ Pmp22, these two mutant Pmp22s sequester the same amount of wild-type Pmp22 in heterodimers and heterooligomers. Thus, the differences in the phenotypes of Tr and TrJ mice may depend more on the ability of the mutant protein to aggregate than on the dominant-negative effect of the mutant Pmp22 on wild-type Pmp22 trafficking.
Peripheral myelin protein 22 (PMP22) is a tetraspan integral membrane protein mainly expressed by myelinating Schwann cells and is found with the major peripheral myelin protein Po in compact myelin (1). Duplication, deletion, or point mutations in the PMP22 gene underlie the most common human peripheral neuropathies, Charcot–Marie–Tooth (CMT)1A disease and hereditary neuropathy with pressure palsies (2). PMP22 is known as growth arrest-specific protein-3 because of its involvement in the control of fibroblast cell growth (2–4). Multiple point mutations in PMP22 have been identified in CMT1A and Dejerine-Sottas disease, a more severe demyelinating neuropathy, although CMT1A disease usually arises from PMP22 duplication (2). Well described Pmp22 point mutations are the Trembler (Tr, G150D) and Trembler J (TrJ, L16P) mutations, originally discovered in mice but later found in humans (2, 5–8). The Tr and TrJ mice are appropriate models in which to explore how mutations cause disease, especially because the two phenotypes are not identical. In mice, the Tr mutation is dominant, and the TrJ is semidominant over the wild-type allele (wt) (8, 9). The TrJ/TrJ genotype leads to a more severe peripheral myelin deficiency and a much shorter lifespan compared with the long-living Tr/Tr mice (8). Moreover, because the heterozygous Tr and TrJ mice display a more severe disease phenotype than the heterozygous Pmp22 knockout mice, both mutant alleles act via gain-of-function or dominant-negative mechanisms (10, 11).
Impaired protein trafficking in Schwann cells and other cell lines has been described for several Pmp22 mutant proteins, suggesting a common mechanism behind the demyelinating disease (12–15). Most of the mutant Pmp22 proteins do not reach the plasma membrane that could be interpreted as evidence that the mutant allele has a loss of function (16). However, the Tr and TrJ Pmp22 mutant proteins also exhibit a dominant-negative effect on the wt Pmp22, altering its trafficking pattern (11, 17). The Tr Pmp22 protein remains localized mainly in the endoplasmic reticulum (ER) and reduces the amount of wt Pmp22 that reaches the plasma membrane (17). The TrJ Pmp22 protein accumulates in the intermediate compartment (IC) between the ER and the Golgi (13). In this last instance a possible mechanism behind the dominant-negative effect has been described. Pmp22 forms dimers, and when coexpressed with TrJ Pmp22 it also forms wt-TrJ heterodimers (13). Thus, the altered trafficking of TrJ Pmp22 prevents a fraction of the wt Pmp22 from reaching the plasma membrane; this fraction accumulates with the TrJ Pmp22 in the IC.
The degree to which the trafficking of wt Pmp22 is altered in the presence of a mutant Pmp22 likely will depend on the formation and stability of the wt-mutant Pmp22 complexes. The formation of the complexes also may depend on the specific Pmp22 mutation. For these reasons we have compared the interaction of wt Pmp22 with Tr and TrJ Pmp22 to determine whether differences in the trafficking of wt Pmp22 might explain the differences in Tr and TrJ phenotypes. Because the aggregation of mutant proteins also contributes to their pathogenicity (23), we also compared the degree to which the Tr and TrJ Pmp22 proteins aggregate.
Materials and Methods
Molecular Cloning.
We used the c-Myc- (Myc) or hemagglutinin- (HA) tagged Pmp22 constructs as described earlier (13) for the transfection of COS7 cells (American Type Culture Collection). The Tr mutation (G150D) was introduced by PCR into the ORF of Pmp22 in the same way that the TrJ mutation (L16P) was introduced (13). A rat Po cDNA fragment was cut from the plasmid (a generous gift from Steven Scherer) with BamHI and NciI, and the protruding ends were filled to create blunt ends. The eukaryotic expression vector pCMX was cut with XbaI and NciI, and the ends were also blunted. The Po cDNA fragment then was religated with the pCMX fragment to make the construct. The Po-pCMX plasmid was sequenced to make sure that the Po cDNA was inserted in the right orientation.
Cell Culture and Transfection.
COS7 cells were maintained at 37°C in 5% CO2 on untreated tissue-culture dishes in DMEM containing 6% bovine calf serum, 6% equine serum (HyClone), and penicillin/streptomycin. HeLa cells (American Type Culture Collection) were cultured in DMEM containing 10% FBS and penicillin/streptomycin. Plates at 80% confluence were used for transfection as described earlier (13).
Cell Lysis and Immunoprecipitation.
After washing in Tris-buffered saline (25 mM Tris, pH 7.5/136 mM NaCl/2.6 mM KCl), cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl/0.2% SDS/1% Nonidet P-40/1% deoxycholate/50 mM Tris, pH 7.4), or Nonidet P-40 buffer (150 mM NaCl/0.1% Nonidet P-40/50 mM Tris, pH 7.5) supplemented with protease inhibitors (Complete, Roche Molecular Biochemicals). The lysates were rotated in a microcentrifuge tube for 30 min at 4°C and centrifuged (100,000 × g, 4°C) for 30 min. For immunoprecipitation, 500 μl of clarified lysates were rotated for 1 h at 4°C with 4 μl of polyclonal rabbit anti-c-Myc antibody, and then 2 h with 10 μl of protein G agarose beads (Amersham Pharmacia). The agarose beads were washed four times in RIPA buffer and then incubated for 5 min at 95°C in 1× SDS sample buffer (0.05 M Tris, pH 6.8/1% SDS/1% DTT/30 μM bromophenol blue/6% glycerol) before separation on SDS/PAGE (12.5% polyacrylamide/0.375 M Tris⋅HCl/0.1% SDS, pH 8.8, Bio-Rad MiniPROTEAN II system). Western blot analysis with mono- or polyclonal anti-c-Myc or mouse anti-HA antibodies was performed as described (13). The monoclonal antibody against Po was a gift from Juan Jose Archellos (Karl-Franzen-Universitat, Graz, Austria). For coculture experiments, individually transfected cells were mixed, lysed, and analyzed by the same procedure.
Gel Filtration Chromatography and Western Blot Analysis.
Cell lysates (50 μl) were made in Nonidet P-40 or RIPA buffer and separated on an Amersham Pharmacia Superdex 200 HR 10/30 column at 4°C in the same buffer. Twenty-eight 280-μl column fractions (34-61) were collected. Western blots of individual column fractions with anti-c-Myc or anti-HA antibodies were performed as described above. To disaggregate Pmp22 before column chromatography in Nonidet P-40 or RIPA buffer, 2% SDS was added to lysate, and the mixture was incubated for 5 min at 95°C.
Chemical Crosslinking.
COS7 cells, 36 h after transfection, were washed twice in reaction buffer (150 mM NaCl/50 mM Na2HPO4, pH 7.5) and cooled on ice. Dithiobis(succinimidyl propionate) (DSP, 50 mM, Pierce) freshly diluted in DMSO was added to the reaction buffer (final concentration, 1 mM DSP/2% DMSO), and the cells were incubated on ice for 30 min. Cells then were washed once in Tris-buffered saline and lysed in 400 μl of Nonidet P-40 or RIPA buffer. The samples made in RIPA buffer were immunoprecipitated with a polyclonal rabbit anti-c-Myc antibody, separated in a 12.5% acrylamide/SDS/PAGE gel, and analyzed by Western blot using monoclonal mouse anti-c-Myc antibody. To check for specificity, 0.1 M DTT was added to samples to cleave DSP. The samples made in Nonidet P-40 buffer were separated by gel filtration and analyzed by Western blot.
Results
Tr Pmp22 as Well as wt and TrJ Pmp22 Forms Dimers.
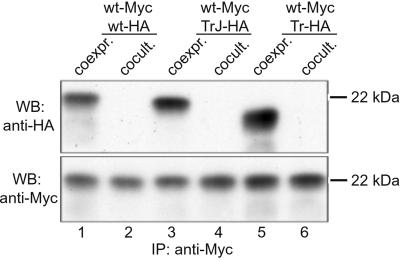
To determine whether the two different phenotypes of TrJ and Tr mice are influenced by the ability of the mutant Pmp22 to aggregate, we compared complex formation of wt, TrJ, and Tr Pmp22 with each other. Because Tr Pmp22 prevents ≈90% of the wt Pmp22 from being inserted into the plasma membrane (17), we assumed that wt and Tr Pmp22 would interact directly with each other. This result was confirmed by immunoprecipitating coexpressed Myc-tagged wt Pmp22 (wt-Myc) and HA-tagged Tr Pmp22 (Tr-HA) in COS7 cells. Immunoprecipitation of cell lysates with a polyclonal anti-c-Myc antibody revealed not only the precipitation of wt-Myc (Fig. 1, lane 5) but also, after immunoblotting with anti-HA antibody, of Tr-HA (lane 5). Similar results were obtained with coexpression of Myc- and HA-tagged wt Pmp22 (lane 1) and of wt-Myc and TrJ-HA (lane 3) as described previously (13). To exclude the possibility that Pmp22 proteins form nonspecific aggregates after membrane disruption, we cocultured cells expressing wt-Myc with cells expressing wt-HA, TrJ-HA, or Tr-HA and performed the same immunoprecipitation procedure. If cell lysis induces nonspecific aggregation, then HA-tagged Pmp22s should be immunoprecipitated together with Pmp22-Myc in each instance. However, no wt-HA, TrJ-HA, or Tr-HA were coimmunoprecipitated by anti-c-Myc antibody from cocultured cells (Fig. 1, lanes 2, 4, and 6). This result indicates that Pmp22 proteins have to be coexpressed in the same cell to interact with each other.
Figure 1.
Coimmunoprecipitation of wt Pmp22 with wt, TrJ, and Tr Pmp22. Myc-tagged wt Pmp22 (wt-Myc) and HA-tagged wt (wt-HA), TrJ (TrJ-HA), or Tr Pmp22 (Tr-HA) were either coexpressed in COS7 cells (coexpr.) or expressed separately but cultured on the same tissue-culture plate (cocult.). Cells were lysed in RIPA buffer and immunoprecipitated (IP) with a polyclonal anti-Myc antibody. After SDS/PAGE, Pmp22 was visualized on Western blot (WB) membrane by anti-HA (Upper) or anti-c-Myc (Lower) antibodies. Wt Pmp22 interacts with wt, TrJ, or Tr Pmp22 only when coexpressed in the same cell (lanes 1, 3, and 5).
The migration of the immunoprecipitated wt-myc band was constant in each of the coculture experiments (Fig. 1 Lower). However, the migration of the wt-HA, TrJ-HA, and Tr-HA species varied, with TrJ-HA and Tr-HA migrating further than wt-HA (see also Fig. 3). Such differences seem to be characteristic for each mutation. It has been shown previously that the differences in migration between TrJ and wt Pmp22 are not caused by degradation (13), and it is likely that the same applies to Tr Pmp22.
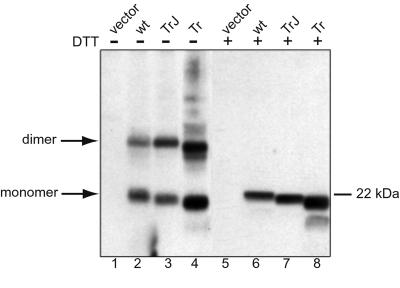
Figure 3.
Crosslinking of wt and mutant Pmp22 in transfected COS7 cells. Cells were transfected with Myc-tagged wt, TrJ, or Tr Pmp22 or the expression vector without DNA insert (vector). After crosslinking with DSP, the Myc-tagged Pmp22 proteins were immunoprecipitated with anti-c-Myc antibody. The immunoprecipitates were treated with sample buffer (lanes 1–4) or sample buffer with DTT (lanes 5–8) followed by separation by SDS/PAGE. Pmp22 was detected by Western blot using anti-Myc antibody.
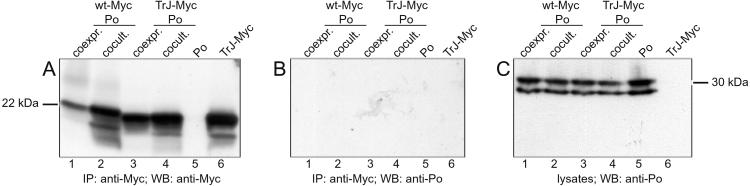
As a further test of the specificity of the Pmp22 interaction, the potential interaction of Pmp22 with the other compact myelin protein Po was tested by coimmunoprecipitation. In a previous study, the expression of Pmp22 in cultured rat Schwann cells had no effect on the distribution of endogenous Po, indicating a lack of interaction between these two proteins (13). Immunoprecipitation with an anti-c-Myc-antibody of lysates from COS7 cells coexpressing either wt-Myc or TrJ-Myc together with Po showed that only the Myc-tagged forms of Pmp22 were precipitated (Fig. 2 A and B). Similarly, in cells separately expressing wt-Myc and Po only the Myc-tagged Pmp22 was precipitated (Fig. 2A, lanes 1–4). When the anti-c-Myc immunoprecipitates were probed with an anti-Po antibody, no Po was precipitated in either of these situations (Fig. 2B, lanes 1–4), indicating that Po does not interact with wt or TrJ Pmp22 when coexpressed in the same cell. That the anti-Po antibody was capable of detecting Po was evident from the precipitation of Po from the lysates of the coexpression or coculture cells before immunoprecipitation (Fig. 2C, lanes 1–4) and of the Po-expressing control cells (Fig. 2C, lane 5). Exactly the same results were obtained when HeLa cells were used instead of COS7 cells (data not shown). Moreover, immunoprecipitation of lysates from radiolabeled Schwann cells with a polyclonal Pmp22 antibody precipitated only Pmp22 and not Po (data not shown). These results indicate that Pmp22 does not interact with Po in transfected cells even though both are found in compact myelin. These results are at variance with previous data on coimmunoprecipitation of Po and PMP22 (18), and the reasons for this discrepancy are unclear.
Figure 2.
Failure of Po to coimmunoprecipitate with wt and TrJ Pmp22. Myc-tagged wt Pmp22 (wt-Myc, lanes 1 and 2) or TrJ Pmp22 (TrJ-Myc, lanes 3 and 4) were coexpressed (coexpr., lanes 1 and 3) or cocultured (cocult., lanes 2 and 4) with Po in COS7 cells. Cells expressing only Po (lane 5) or Myc-tagged TrJ Pmp22 (lane 6) served as controls. Cells were lysed in RIPA buffer and immunoprecipitated (IP) with a polyclonal anti-c-Myc antibody. The immunoprecipitates and cell lysates were separated by SDS/PAGE. Pmp22 was visualized on Western blot (WB) membrane by anti-c-Myc antibody (A), and Po was probed with anti-Po antibody (B and C). Po did not coimmunoprecipitate with wt or TrJ Pmp22 in COS7 cells.
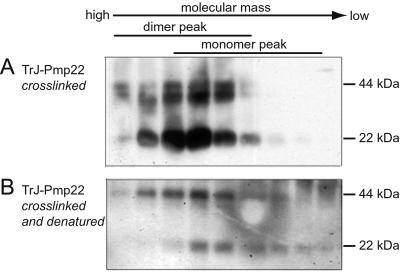
Coimmunoprecipitation experiments only indicate that proteins interact. To examine the stoichiometry of Pmp22 complexes in intact cells, proteins were crosslinked in transfected COS7 cells with a cell-permeable thiol-cleavable crosslinker, DSP, and Pmp22 was immunoprecipitated with polyclonal mouse anti-c-Myc antibody. To reverse the crosslinking reaction, a fraction of the sample was treated with DTT before SDS/PAGE to cleave the crosslinker. In untreated samples of wt, TrJ, and Tr Pmp22, two major bands migrated at the expected position of monomers and dimers (Fig. 3, lanes 3 and 4). After treatment with DTT only monomer bands were detected (lanes 6–8). It therefore is likely that a fraction of Pmp22 protein in the cell, whether wt or mutant, exists as dimers in the cell. Whereas wt and TrJ Pmp22 could be detected only as monomers or dimers, under nonreducing conditions various forms of high molecular weight Tr Pmp22 aggregates were detected above the mass of 44 kDa. Also a dimer band of slightly smaller size is clearly visible below the main dimer band. Under reducing conditions, a majority of Tr Pmp22 migrated as a full-length protein of 22 kDa, whereas a small fraction was degraded and migrated at a position of ≈18 kDa (Fig. 3, lanes 4 and 8). After complete deglycosylation of Tr Pmp22 with N-glycosidase F (13), both the full-length and the 18-kDa protein bands shifted by 4 kDa, showing that the latter protein is not a deglycosylated full-length protein but rather a glycosylated and partially degraded protein (data not shown).
The Characteristics of Pmp22 Complexes Are Altered in Mutant Pmp22s.
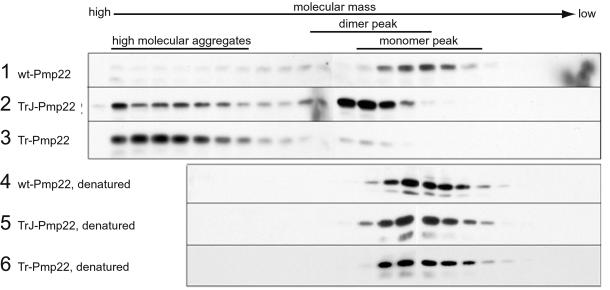
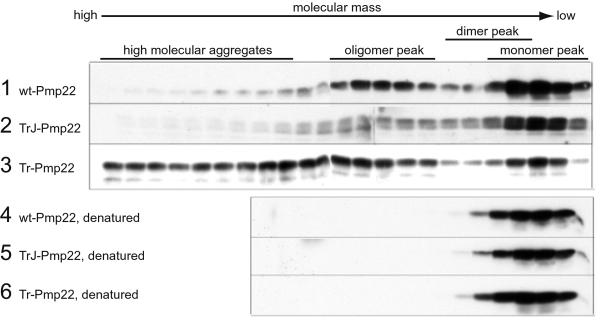
To characterize further the nature of the various Pmp22 complexes and to determine whether complexes larger than dimers occur, we separated lysates of Myc-tagged Pmp22-transfected COS7 cells on a Superdex 200 gel filtration column. Two different lysis conditions were used, one in Nonidet P-40 buffer and the other in RIPA buffer. Although these analyses do not necessary reflect the state of Pmp22 in the cell, they do reveal the potential complex forms that Pmp22 could adopt. The column fractions were analyzed by Western immunoblot using anti-c-Myc antibody. Because the electrophoretic step in this procedure is carried out in a denaturing gel, all complexes are reduced and migrate as the Pmp22 monomer. A lysis buffer containing 0.1% Nonidet P-40 was chosen initially because Pmp22 eluted from the column mainly in the position of monomer (Fig. 4, lane 1). The monomeric nature of wt Pmp22 under these conditions was deduced from the fact that denaturation of the sample before chromatography in 2% SDS at 95°C had no effect on the position at which wt Pmp22 eluted (Fig. 4, lane 4). The apparent molecular mass of Pmp22, as judged by soluble protein markers, was ≈158 kDa, although the actual mass of the Pmp22 monomer is 22 kDa. Presumably, hydrophobic interactions between Pmp22 and the material of the column account for this difference. Another tetraspan protein, the gap junction protein connexin 32, of monomer molecular mass 32 kDa eluted as a single peak with an apparent molecular mass of ≈100 kDa (data not shown).
Figure 4.
Gel filtration chromatography demonstrating aggregation of wt, TrJ, and Tr Pmp22. COS7 cells transiently transfected with Myc-tagged wt, TrJ, or Tr Pmp22 were lysed in Nonidet P-40 buffer. The lysates were separated on an Amersham Pharmacia Superdex 200 sizing column (rows 1–3) or incubated for 10 min at 95°C in the presence of 2% SDS before filtration chromatography (rows 4–6). Fractions were analyzed by Western blot using anti-c-Myc antibody. Most wt Pmp22 eluted in the monomer peak (row 1). A significant amount of TrJ Pmp22 (row 2) and most of the Tr Pmp22 (row 3) eluted from the column as high molecular aggregates. A major portion of the TrJ Pmp22 and a small amount of the Tr Pmp22 also eluted at a slightly higher molecular mass than the Pmp22 monomer in the dimer peak (see Fig. 5). Denatured wt, TrJ, and Tr Pmp22 all eluted only in the position of the Pmp22 monomer peak (rows 4–6).
Both mutations altered the aggregation of Pmp22. TrJ Pmp22 eluted at a slightly higher molecular mass than monomeric wt Pmp22 and formed complexes of high molecular mass (Fig. 4, lane 2), whereas most of the Tr Pmp22 protein formed the same high molecular mass complexes with only a minor fraction of low molecular mass material (Fig. 4, lane 3). The tendency of Tr Pmp22 to aggregate in complexes larger than dimers, noted in the crosslinking experiments (Fig. 3), is accentuated in the Nonidet P-40-containing buffer. When the TrJ and Tr Pmp22 samples were exposed to 2% SDS before chromatography, all proteins eluted in the position of monomer (Fig. 4, lanes 5 and 6).
A series of crosslinking experiments showed that the major fraction eluted first from the column in the analysis of TrJ Pmp22 was a dimer. Cell lysates of COS7 cells expressing TrJ Pmp22 in the Nonidet P-40-containing buffer were crosslinked with DSP, and the mixture was analyzed by chromatography (Fig. 5A). The TrJ Pmp22-containing fractions all eluted in a position several fractions ahead of the wt Pmp22 monomer peak. This observation was true not only of the crosslinked TrJ Pmp22 dimer migrating at ≈44 kDa but also of the noncrosslinked TrJ Pmp22 that appeared as monomers on the Western blot. The latter could only have come from noncrosslinked dimers. This conclusion was confirmed by the analysis of the anti-c-Myc immunoprecipitate of the TrJ Pmp22 lysate. In this procedure the immunoprecipitate was released from the protein G agarose beads in 1% SDS at 95°C before chromatography, a process that dissociates the uncrosslinked but not the crosslinked TrJ Pmp22 dimers. In this analysis (Fig. 5B) the crosslinked TrJ Pmp22 still eluted as dimers, whereas the noncrosslinked TrJ Pmp22 eluted as monomers.
Figure 5.
TrJ Pmp22 exists mainly as dimers, not monomers, in Nonidet P-40 buffer. COS7 cells expressing Myc-tagged TrJ Pmp22 were crosslinked with DSP and lysed in Nonidet P-40 buffer. The lysates were separated directly on the gel filtration column in the same buffer (A) or denatured (as described in Materials and Methods) before the chromatography (B). The fractions were treated with the sample buffer without the reducing reagent DTT before being separated by SDS/PAGE. TrJ Pmp22 was visualized by Western blot with anti-c-Myc antibody. In A, both uncrosslinked monomers and crosslinked dimers of TrJ Pmp22 were detected in the dimer peak. However, in B, the uncrosslinked TrJ Pmp22 dimers now appeared in the monomer peak, whereas the crosslinked TrJ Pmp22 dimers still eluted in the dimer peak.
Transfected COS7 cells lysed in RIPA buffer showed different patterns of aggregation. Wt Pmp22 eluted partly as a monomer, because denaturation did not lower its molecular mass (Fig. 6, lanes 1 and 4). Also, wt-Pmp22 eluted at the same position as denatured Tr and TrJ Pmp22 (Fig. 6, lanes 5 and 6). The apparent molecular mass of wt Pmp22 in RIPA buffer was less than in Nonidet P-40 buffer. A second species was observed in the wt Pmp22 analysis (Fig. 6, lane 1, oligomer). Because the dimer comigrates on these columns at the same position as the TrJ Pmp22 dimer (unpublished data), this second component is significantly larger than a dimer and probably contains 6–8 monomers. In the RIPA buffer TrJ Pmp22 eluted principally as a monomer with only a fraction of oligomer. In contrast, Tr Pmp22 again was found almost entirely in higher molecular mass aggregates, as in the Nonidet P-40 buffer.
Figure 6.
Gel filtration chromatography demonstrating aggregation of wt, TrJ, and Tr Pmp22 in RIPA buffer. COS7 cells transiently transfected with Myc-tagged wt, TrJ, or Tr Pmp22 were lysed in RIPA buffer. The lysates were separated on an Amersham Pharmacia Superdex 200 sizing column (rows 1–3) or incubated for 10 min at 95°C in the presence of 2% SDS before filtration chromatography (rows 4–6). Fractions were analyzed by Western blot using anti-c-Myc antibody. Most wt Pmp22 eluted in the monomer peak and oligomer peak (row 1). TrJ Pmp22 is eluted mainly in the monomer peak with a small amount in the oligomer peak (row 2). Most of the Tr Pmp22 eluted from the column as high molecular aggregates with only a small proportion in the monomer peak (row 3). Denatured wt, TrJ, and Tr Pmp22 all eluted only in the position of the Pmp22 monomer peak (rows 4–6).
Coexpression of wt and Mutant Pmp22s Alters the Pattern of Complex Formation of Both.
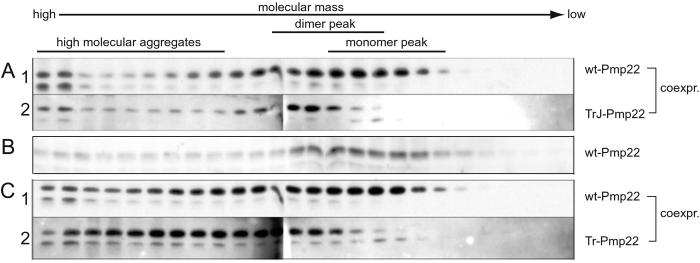
Given that Tr Pmp22 forms heterodimers with wt Pmp22, it seemed likely that the accumulation of Tr Pmp22 in the ER also would be accompanied by the accumulation of wt Pmp22. To test this idea we analyzed lysates containing mixtures of wt and mutant Pmp22s by chromatography in the Nonidet P-40 lysis buffer using two different Pmp22 tags. When HA-tagged wt Pmp22 was coexpressed with Myc-tagged TrJ Pmp22, the elution pattern of the wt protein displayed characteristics of both wt and TrJ Pmp22 (Fig. 7A). In particular, HA-tagged wt Pmp22 not only eluted at the position of the monomer but also was found at the position of the dimer, a complex not detected when wt Pmp22 was expressed alone. This dimer therefore likely contains both wt and TrJ Pmp22. Likewise, the presence of the Tr-wt Pmp22 heterodimer could be deduced from the appearance of HA-tagged wt Pmp22 and Myc-tagged Tr Pmp22 in the dimer fraction of the analysis of these two coexpressed proteins (Fig. 7B). The controls for these experiments came from the coimmunoprecipitation experiments, which showed that no heterodimer or oligomers formed after simple mixing of two proteins. Unique to the coexpressed HA-tagged wt Pmp22 and Myc-tagged Tr Pmp22 were the presence of a series of heterooligomers, larger than the heterodimers, that sequestered a fraction of the wt Pmp22.
Figure 7.
Wt Pmp22 forms high molecular weight complexes with the mutant Pmp22s. COS7 cells expressing HA-tagged wt and Myc-tagged TrJ Pmp22 (A) or HA-tagged wt and Myc-tagged Tr Pmp22 (C) were lysed in Nonidet P-40 buffer and separated on an Amersham Pharmacia Superdex 200 column. An analysis of HA-tagged wt Pmp22 alone is shown in B. Fractions were analyzed by Western blot using antibodies against HA (rows A1, B, and C1) or Myc (rows A2 and C2). A significant amount of wt Pmp22 shifted from the monomer peak (B) into the dimer peak and high molecular aggregates in both mixtures (rows A1 and C1). The pattern of eluted TrJ Pmp22 remained unchanged (A2), whereas a significant amount of Tr Pmp22 shifted toward and into the dimer peak (C2). [Compare with the elution patterns of the homologous mutant Pmp22 proteins (Fig. 4, rows 2 and 3)].
Discussion
Aggregate formation by wt Pmp22 and its mutant forms is of considerable interest for several reasons. The size of Pmp22 complexes in a cell could provide information of possible functions and interactions of the protein. For example, another tetraspan protein found in peripheral myelin, connexin 32, forms hexamers in the process of creating hemichannels (19), and the analysis of wt Pmp22 in RIPA buffer indicates that it too can form oligomers in this size range. Moreover, the recent discovery that Pmp22 is present in tight junctions in epithelial tissues (20) suggests a possible role for these oligomers. In another context protein aggregation is often a key determinant in the disease process, and it is known that wt Pmp22 forms aggresomes (21), unique intracellular aggregates that are toxic to the cell (22, 23). Finally, the propensity of wt Pmp22 to form heterodimers and heterooligomers means that through aggregation the mutant forms also may influence the trafficking of wt Pmp22. How mutations affect Pmp22 aggregation, the aggresome formation, and the trafficking of wt Pmp22 may well provide clues to the mechanisms behind the ensuing demyelination diseases.
The Tr and TrJ Mutations Do Not Affect Two Properties of the Pmp22 Protein.
Similar to wt Pmp22, Tr and TrJ Pmp22 can be crosslinked efficiently to form dimers. Because Tr Pmp22 does not leave the ER, dimerization likely occurs as nascent Pmp22 peptide chains are formed, and this is not significantly affected by misfolding caused by either mutation. Nor do the two mutations affect the ability of Tr and TrJ Pmp22 to coimmunoprecipitate with wt Pmp22 either as heterodimers or heterooligomers, and as a consequence wt and mutant Pmp22 traffic together. The fact that it is the mutant Pmp22 in each instance that determines the trafficking pattern of the heterodimer, e.g., into the IC with TrJ Pmp22 (13), suggests that the mutant protein retains its abnormal conformation in the heterodimer. If the wt Pmp22 imposed its normal conformation on the mutant Pmp22, then the trafficking of the heterodimer would mimic that of the wt Pmp22 homodimer, and it would be directed mainly to the Schwann cell membrane.
Tr and TrJ Pmp22 Differ in Their Aggregation.
Where the two mutants do differ is in the degree of aggregation. Aggregation was assessed by size chromatography of detergent lysates of cells expressing either the Tr or TrJ Pmp22 alone or in combination with wt Pmp22. Although such analyses do not necessarily indicate the state of Pmp22 in the cell, they do allow comparisons of the degree of aggregation of the three proteins under specific conditions. One lysis buffer containing 0.1% Nonidet P-40 was chosen, because wt Pmp22 eluted mainly as a monomer providing a standard for comparison. Substantial differences in the aggregation of the mutant Pmp22s were observed. Under these conditions, TrJ Pmp22 eluted mainly as a dimer with some higher aggregates, whereas Tr Pmp22 was essentially all in the form of higher aggregates. The marked tendency of Tr Pmp22 to form higher aggregates compared with the other two Pmp22s was observed also in a different (RIPA) lysis buffer. The greater capacity of Tr Pmp22 to aggregate compared with TrJ Pmp22 may be one determinant that affects trafficking.
As noted above, Tr Pmp22 fails to exit the ER, whereas TrJ Pmp22 leaves the ER and accumulates in the IC between the ER and the Golgi, a behavior that is between that of wt Pmp22 and Tr Pmp22. The significant tendency of Tr Pmp22 compared with TrJ Pmp22 to aggregate also may be the determining factor in making the Tr mutation dominant. In support of this idea is the finding that the specific ubiquitin-containing aggregates or aggresomes (22) form more readily when the proteosome is inhibited in the Schwann cells expressing Tr Pmp22 than TrJ Pmp22 or wt Pmp22 (M. Ryan, E.M.S., and L. Notterpek, unpublished data). Moreover, these aggregates have the capacity to inhibit the ubiquitin-proteosome degradation pathway (23) leading to further aggregation that provides the toxic gain of function leading to cell death. An alternative possibility to explain the dominant nature of the Tr mutation is that Tr Pmp22 sequesters more wt Pmp22 into heterodimers or oligomers than does TrJ Pmp22, effectively nullifying most or all of the function of the wt Pmp22 allele. Quantitation of the distribution of the various forms of wt Pmp22 in the mixtures with TrJ and Tr Pmp22 does not, however, support this suggestion. In both instances, about the same amount (≈70%) of wt Pmp22 was sequestered into dimers or oligomers with either TrJ or Tr Pmp22. Disruption in the trafficking of wt Pmp22 therefore seems to play less of a role in the pathogenicity of the mutant Pmp22 than does aggregation.
Acknowledgments
We are indebted to Drs. James R. Lupski and Steven S. Scherer for their critical reviews of the manuscript. This work was supported by grants from National Institutes of Health (National Institute of Neurological Disorders and Stroke), the Muscular Dystrophy Association, the Christopher Reeve Paralysis Association, and the McGowan Charitable Trust (to E.M.S.) and the Swiss National Science Foundation and the Novartis Jubilaumsstiftung (to A.R.T.).
Abbreviations
- PMP22
peripheral myelin protein 22
- Tr
Trembler
- TrJ
Trembler J
- wt
wild type
- ER
endoplasmic reticulum
- IC
intermediate compartment
- HA
hemagglutinin
- RIPA
radioimmunoprecipitation assay
- DSP
dithiobis(succinimidyl proprionate)
References
- 1.Snipes G J, Suter U, Welcher A A, Shooter E M. J Cell Biol. 1992;117:225–238. doi: 10.1083/jcb.117.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lupski J R, Garcia C A. In: The Metabolic and Molecular Bases of Inherited Disease. 8th Ed. Scriver C, Beaudet A, Valle D, Sly W, Childs B, Kinzler K, Vogelstein B, editors. New York: McGraw–Hill; 2001. pp. 5759–5788. [Google Scholar]
- 3.Manfioletti G, Ruaro M E, Del Sal G, Philipson L, Schneider C. Mol Cell Biol. 1990;10:2924–2930. doi: 10.1128/mcb.10.6.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zoidl G, D'Urso D, Blass-Kampmann S, Schmalenbach C, Kuhn R, Muller H W. Cell Tissue Res. 1997;287:459–470. doi: 10.1007/s004410050770. [DOI] [PubMed] [Google Scholar]
- 5.Suter U, Welcher A A, Ozcelik T, Snipes G J, Kosaras B, Francke U, Billings-Gagliardi S, Sidman R L, Shooter E M. Nature (London) 1992;356:241–244. doi: 10.1038/356241a0. [DOI] [PubMed] [Google Scholar]
- 6.Suter U, Moskow J J, Welcher A A, Snipes G J, Kosaras B, Sidman R L, Buchberg A M, Shooter E M. Proc Natl Acad Sci USA. 1992;89:4382–4386. doi: 10.1073/pnas.89.10.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valentijn L J, Baas F, Wolterman R A, Hoogendijk J E, van den Bosch N H, Zorn I, Gabreels-Festen A W, de Visser M, Bolhuis P A. Nat Genet. 1992;2:288–291. doi: 10.1038/ng1292-288. [DOI] [PubMed] [Google Scholar]
- 8.Henry E W, Sidman R L. J Neurogenet. 1983;1:39–52. doi: 10.3109/01677068309107071. [DOI] [PubMed] [Google Scholar]
- 9.Henry E W, Cowen J S, Sidman R L. J Neuropathol Exp Neurol. 1983;42:688–706. doi: 10.1097/00005072-198311000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Adlkofer K, Naef R, Suter U. J Neurosci Res. 1997;49:671–680. doi: 10.1002/(SICI)1097-4547(19970915)49:6<671::AID-JNR2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Adlkofer K, Frei R, Neuberg D H, Zielasek J, Toyka K V, Suter U. J Neurosci. 1997;17:4662–4671. doi: 10.1523/JNEUROSCI.17-12-04662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Urso D, Prior R, Greiner-Petter R, Gabreels-Festen A A, Muller H W. J Neurosci. 1998;18:731–740. doi: 10.1523/JNEUROSCI.18-02-00731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tobler A R, Notterpek L, Naef R, Taylor V, Suter U, Shooter E M. J Neurosci. 1999;19:2027–2036. doi: 10.1523/JNEUROSCI.19-06-02027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naef R, Suter U. Neurobiol Dis. 1999;6:1–14. doi: 10.1006/nbdi.1998.0227. [DOI] [PubMed] [Google Scholar]
- 15.Hanemann C O, D'Urso D, Gabreels-Festen A A, Muller H W. Brain. 2000;123:1001–1006. doi: 10.1093/brain/123.5.1001. [DOI] [PubMed] [Google Scholar]
- 16.Sanders C R, Ismail-Beigi F, McEnery M W. Biochemistry. 2001;40:9453–9459. doi: 10.1021/bi010894f. [DOI] [PubMed] [Google Scholar]
- 17.Naef R, Adlkofer K, Lescher B, Suter U. Mol Cell Neurosci. 1997;9:13–25. doi: 10.1006/mcne.1997.0604. [DOI] [PubMed] [Google Scholar]
- 18.D'Urso D, Ehrhardt P, Muller H W. J Neurosci. 1999;19:3396–3403. doi: 10.1523/JNEUROSCI.19-09-03396.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diez J A, Ahmad S, Evans W H. Eur J Biochem. 1999;262:142–148. doi: 10.1046/j.1432-1327.1999.00343.x. [DOI] [PubMed] [Google Scholar]
- 20.Notterpek L, Roux K J, Amici S, Yazdanpour A, Rahner C, Fletcher B S. Proc Natl Acad Sci USA. 2001;98:14404–14409. doi: 10.1073/pnas.251548398. . (First Published November 20, 2001; 10.1073/pnas.251548398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Notterpek L, Ryan M C, Tobler A R, Shooter E M. Neurobiol Dis. 1999;6:450–460. doi: 10.1006/nbdi.1999.0274. [DOI] [PubMed] [Google Scholar]
- 22.Johnston J A, Ward C L, Kopito R R. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bence N F, Sampat R M, Kopito R R. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]