Short abstract
The B7 family consists of structurally related, cell-surface protein ligands, which bind to the CD28 family of receptors on lymphocytes and regulate immune responses via 'costimulatory' or 'coinhibitory' signals.
Summary
The B7 family consists of structurally related, cell-surface protein ligands, which bind to receptors on lymphocytes that regulate immune responses. Activation of T and B lymphocytes is initiated by engagement of cell-surface, antigen-specific T-cell receptors or B-cell receptors, but additional signals delivered simultaneously by B7 ligands determine the ultimate immune response. These 'costimulatory' or 'coinhibitory' signals are delivered by B7 ligands through the CD28 family of receptors on lymphocytes. Interaction of B7-family members with costimulatory receptors augments immune responses, and interaction with coinhibitory receptors attenuates immune responses. There are currently seven known members of the family: B7.1 (CD80), B7.2 (CD86), inducible costimulator ligand (ICOS-L), programmed death-1 ligand (PD-L1), programmed death-2 ligand (PD-L2), B7-H3, and B7-H4. Members of the family have been characterized predominantly in humans and mice, but some members are also found in birds. They share 20-40% amino-acid identity and are structurally related, with the extracellular domain containing tandem domains related to variable and constant immunoglobulin domains. B7 ligands are expressed in lymphoid and non-lymphoid tissues. The importance of the family in regulating immune responses is shown by the development of immunodeficiency and autoimmune diseases in mice with mutations in B7-family genes. Manipulation of the signals delivered by B7 ligands has shown potential in the treatment of autoimmunity, inflammatory diseases and cancer.
Gene organization and evolutionary history
The seven known members of the B7 family - B7.1, B7.2, ICOS-L, PD-L1, PD-L.2, B7-H3, and B7-H4 - are all transmembrane or glycosylphosphatidylinositol (GPI)-linked proteins characterized by extracellular IgV and IgC domains related to the variable and constant domains of immunoglobulins (Table 1; see [1-3] for detailed reviews on the family). The IgV and IgC domains of B7-family members are each encoded by single exons, with additional exons encoding leader sequences, transmembrane and cytoplasmic domains. B7-H3 is unique in that the major human form contains two extracellular tandem IgV-IgC domains (Table 1) [4]- Differentially spliced forms have been identified for B7.1, B7.2, ICOS-L, B7-H3, and PD-L2 [4-9], although the functional significance of these splice forms is unclear. The cytoplasmic domains are short, ranging in length from 19 to 62 amino-acid residues (see Additional data file 1), and can be encoded by multiple exons. A function for the intracellu-lar domains has not been established, although they all contain serine and threonine residues that could potentially serve as phosphorylation sites in signaling pathways. Recent evidence suggests that B7-family members can signal into the cell on which they are expressed (see below). One member, B7-H4, is GPI-linked to the cell surface [10,11], whereas all others have transmembrane domains.
Table 1.
Structures, expression patterns and receptors of B7 ligands
| Ligand name (alternative names) | Structure | Protein expression pattern in lymphoid cells* | mRNA expression in non-lymphoid tissues or cells | Receptor | Function of ligand-receptor interaction |
| B7.1 (CD80) | Induced in T, B, DCs, and monocytes | Not detected | CD28 CTLA-4 (CD152) |
Costimulation Coinhibition |
|
| B7.2 (CD86) | Constitutive and upregulated upon activation in B, DCs, and monocytes; induced in T | Not detected | CD28 CTLA-4 (CD152) |
Costimulation Coinhibition |
|
| ICOS-L (GL-50, B7 h, B7RP-1, B7H-2) | Constitutive in B, DCs, macrophages, and a subset of T | Lung, liver, kidney, and testes | ICOS | Costimulation | |
| PD-L1 (B7-H1) | Constitutive and upregulated upon activation in B, DCs, and monocytes; induced in T | Placenta, heart, pancreas, lung, liver, and tumor cells (carcinomas and melanomas) | PD-1 Unknown? |
Coinhibition Costimulation |
|
| PD-L2 (B7-DC) | Induced in DCs and monocytes | Heart, placenta, lung, liver keratinocytes, and epithelial cells | PD-1 Unknown? |
Coinhibition Costimulation |
|
| B7-H4 (B7x, B7S1) | Induced in T, B, NK, DCs, and monocytes | Placenta, uterus, testis, kidney, lung, heart, and brain | Unknown | Coinhibition | |
| B7-H3 |  |
Induced in T, B, DCs, and monocytes | Heart, kidney, testes, lung, liver, pancreas, prostate, colon, and osteoblasts | Unknown Unknown |
Costimulation Coinhibition |
*Abbreviations: B, B cells; DCs, dendritic cells; NK, natural killer cells; T, T cells; Unknown?, postulated receptor.
The genes for B7.1 and B7.2 are closely linked, as are the genes for PD-L1 and PD-L2, whereas the genes for ICOS-L, B7-H3 and B7-H4 are unlinked (Table 2). Orthologs of B7.1 and B7.2 have been identified in diverse mammalian species, but genes for other family members remain to be defined in most species beyond primates and rodents (Table 3). Interestingly, genes annotated as B7.1, B7.2, ICOS-L and B7-H4 orthologs have been described in the chicken, Gallus gallus, indicating that the family is conserved in birds (Figure 1; see also Table 3). A BLAST search of the NCBI Fugu rubripes (pufferfish) sequence database suggests that there may also be B7-like proteins in bony fish (data not shown), and CD28-like and CTLA-4 like molecules have been identified in trout (GenBank accession numbers AAW78853 and AAW78854). The CD28-receptor family is also conserved across mammalian species, and chicken orthologs have been described [12], suggesting that these receptor-ligand interactions are critical in the regulation of immune responses in mammals, birds and probably also fish.
Table 2.
Chromosomal locations of B7-family genes
| Chromosomal location | ||
| Gene | Human | Mouse |
| B7.1 | 3q21 | 16 |
| B7.2 | 3q21 | 16 |
| ICOS-L | 21q22.3 | 10 |
| PD-L1 | 9p24 | 19 |
| PD-L2 | 9p24.2 | 19 |
| B7-H3 | 15q23-q24 | 9 |
| B7-H4 | 1p13.1-p12 | 3 |
See the database entries for the accession numbers given in Table 3 for links to genetic mapping data.
Table 3.
Accession numbers for the amino-acid sequences of B7-family proteins from selected species
| Protein | Human | Mouse | Dog | Cat | Cow | Rabbit | Sheep | Pig | Chicken |
| B7.1 | NP_005182 | NP_033985 | NP_001003147 | BAB11687 | CAA71081 | I46690 | AAR27296 | NP_999252 | NP_990415 |
| B7.2 | NP_787058 | NP_062261 | NP_001003146 | BAB11688 | CAC13140 | I46691 | AAR89183 | NP_999387 | XP_425516 |
| ICOS-L | NP_056074 | NP_056605 | N/A | N/A | N/A | N/A | N/A | N/A | XP_413702 |
| PD-L1 | NP_054862 | NP_068693 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| PD-L2 | NP_079515 | NP_067371 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| B7-H3 | NP_079516VC, CAE47548VCVC | NP_598744 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| B7-H4 | NP_078902 | NP_848709 | N/A | N/A | N/A | N/A | N/A | N/A | XP_416546, XP_416760 |
Data obtained from the NCBI Protein Refseq Database [48]. N/A: not available. See the database entries under these accession numbers for well-annotated sequences and references to many of the original publications defining the genes.
Figure 1.
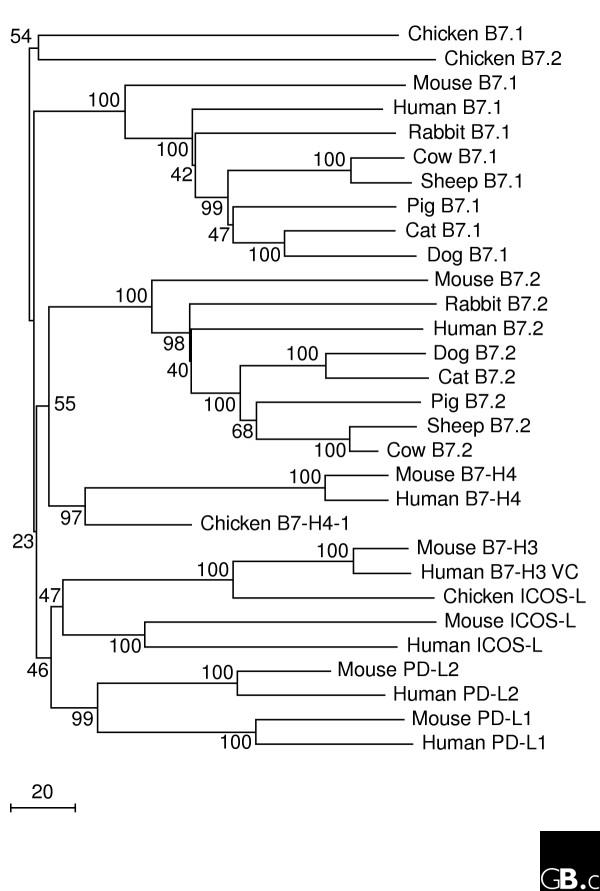
A phylogenetic tree of the B7 family. Sequence alignments were performed using ClustalW implemented in the AlignX module of Vector NTI version 9.1; alignments were imported into Mega 2.1 [47] and a phylogenetic tree was created using the neighbor-joining method (the setting for gaps and missing data was pairwise deletion; the distance method used was number of differences). Numbers at selected nodes indicate the percentage frequencies of branch association on the basis of 1,000 bootstrap repetitions. The scale bar indicates number of residue changes.
Characteristic structural features
Members of the B7 family (Table 1) are predicted to form homodimers at the cell surface, as has been shown for B7.1, although recent data indicate that B7.2 is likely to be a monomer [13]. B7.1 and B7.2 share only about 25% amino-acid identity, yet each interacts with both CD28 and CTLA-4 receptors. Crystal structures have been solved for human B7.1 [14] and human B7.2 [13], both alone and complexed with the CTLA-4 receptor [15,16]. The structures of human B7.1 and B7.2 extracellular domains reveal typical IgV-like and IgC-like domains. Immunoglobulin-superfamily domains are typically about 100 amino acids in length and form a structure characterized by two anti-parallel β sheets, with each sheet composed of three to five anti-parallel strands [17-19]. The distinction between IgV and IgC domains is based on their structures, with IgV domains containing nine anti-parallel strands and IgC domains containing seven anti-parallel strands. Immunoglobulin-superfamily domains are found in many proteins and often serve as protein-protein interaction domains, as is frequently seen for cell-surface proteins in the immune system. For B7.1 and B7.2, the amino-terminal IgV domain is the receptor-binding domain. Interestingly, the amino-acid residues within B7.1 and B7.2 that bind to CTLA-4 are not highly conserved. A comparison of the two structures reveals, however, that the front faces of B7.1 and B7.2 each have shallow cavities that allow the highly conserved MYPPPY motif (in the single letter amino-acid code) in the CD28 and CTLA-4 receptors to fit and bind [13] (see Figure 2).
Figure 2.

Details of the interaction between human CTLA-4 and B7.1. (a) Overview of the interaction. The structure of soluble CTLA-4 monomer (left) interacting with soluble B7.1 monomer (right) is shown as a ribbon diagram, with the molecular surface of the soluble B7.1 monomer overlaid in white. (b) Close-up view of the interaction. The MYPPPY sequence from CTLA-4 (in ball and stick representation) interacts with residues forming a shallow depression in the IgV domain of B7.1. Three out of the five hydrogen bonds formed across the β sheets of the interacting domains are depicted by dashed lines. Reproduced with permission from [15].
B7.1 and B7.2 have non-equivalent roles in immune modulation, in part because of their different interactions with CD28 and CTLA-4. The B7.1-CTLA-4 interaction is of higher affinity than the B7.2-CTLA-4 interaction, whereas CD28 is predicted to bind B7.2 more effectively than B7.1 [20]. Co-crystals of B7.1 with CTLA-4 reveal that B7.1 homodimers have CTLA-4-binding sites that are distal from the B7 dimer interface, allowing formation of a cross-linked B7-CTLA-4 lattice [15] (see Figure 3). In contrast, the data support a monovalent interaction between B7.2 and CD28 [20]. These models predict that the B7.1-CTLA-4 and B7.2-CD28 interactions will be of the greatest functional significance (see [21,22] for further discussion). A crystal structure of the CD28 extracellular domain has recently been reported and shows similarities with that of CTLA-4 [23]. Crystal structures for other B7-family members have not yet been reported. Binding affinities of ICOS-L to its receptor have been reported to be similar to those of CD28-B7.1 interactions [24]. PD-L1 and PD-L2 both have IgV-IgC extracellular domains, and both bind the same PD-1 receptor, although PD-L2 has been reported to bind PD-1 with an affinity 2-6 times higher than the affinity with which PD-L1 binds PD-1 [25]. The crystal structure of the extracellular domain of PD-1 has recently been reported [26].
Figure 3.

Proposed periodic arrangement of CTLA-4 molecules on the surface of a T cell bound to B7-1 molecules on the surface of an antigen-presenting cell. This model is based on the molecular association of CTLA-4 and B7.1 observed in the crystal lattice structure. The proposed lattice structure would enhance the stability of CTLA-4 and B7.1 interactions on the cell surface. C, carboxyl terminus; N, amino terminus. Reproduced with permission from [15].
Localization and function
Expression of B7 ligands
Expression of B7.1, B7.2 and PD-L2 is restricted to lym-phoid cells, whereas ICOS-L, PD-L1, B7-H3 and B7-H4 are also expressed on non-lymphoid cells, suggesting that they have distinct roles in immune function. PD-L1, B7-H3 and B7-H4 expression has also been reported in tumor-cell lines (Table 1) [27]. All B7 ligands are expressed by antigen-presenting cells, including dendritic cells, macrophages and B cells, all of which function in the initiation and amplification of immune responses (Table 1). Low levels of B7.2 protein expression are detected on resting B cells, dendritic cells and macrophages; activation results in enhanced B7.2 expression and de novo expression of B7.1. ICOS-L is constitutively expressed in human monocytes and dendritic cells, and its expression is upregulated in monocytes by interferon-γ [3,28].
Expression of PD-L1 and PD-L2, the independent ligands for the PD-1 receptor, is differentially regulated by cytokines: interleukins IL-4 and IL-13 upregulate PD-L2 [29,30], whereas inflammatory cytokines, primarily interferon-γ, modulate PD-L1 expression [3,31]. PD-L2 expression is restricted to dendritic cells and macrophages, whereas PD-L1 is also expressed by B and T cells and non-lymphoid cells. The distinct tissue expression and cytokine modulation of PD-L1 and PD-L2 suggest non-overlapping roles during an immune response, with PD-L1 predominating in the peripheral tissues and PD-L2 in lymphoid organs.
B7-H3 and B7-H4 are constitutively expressed in murine, but not human, antigen-presenting cells [4,10,11,32,33]. B7-H3 transcripts or protein are not detected in resting human peripheral blood lymphocytes [10,11,34,35], but in murine dendritic cells B7-H3 expression is upregulated by interferon-γ and downregulated by IL-4 [36], suggesting that B7-Hs may regulate immune responses mediated by T helper type 1 inflammatory T cells. The low level of expression of B7-H4 protein in tissues suggests that it may be regulated post-transcriptionally [10,11,34,35]. Inflammatory stimuli and activation downregulate B7-H4 cell-surface expression in murine B cells [10]; the mechanism by which this occurs has yet to be clarified. Receptors for B7-H3 and B7-H4 have not been identified, although expression of these ligands modulates immune responses [4,10,11,32,33,35].
Multiple interactions and multiple functional outcomes
B7 ligands participate in immune responses by providing costimulatory or coinhibitory signals upon binding their receptor. Costimulatory signals are defined as those that act in conjunction with antigen-receptor signals, leading to productive cell activation, growth-factor production, cell expansion and survival (Figure 4a). In contrast, coinhibitory signals attenuate antigen-receptor signals, resulting in decreased cell activation, inhibition of growth-factor production, inhibition of progression of the cell cycle and, in some cases, increased cell death (Figure 4b). The By ligands B7.1 and B7.2 and their receptors CD28 and CTLA-4 exemplify a costimulatory-coinhibitory system that acts to regulate immune responses. Mice deficient in B7.1 and B7.2 have profound deficits in both humoral immune responses (antibody production) and cellular immune responses [37]. Similarly, mice deficient in the CD28 receptor are unable to mount effective immune responses to foreign antigens, infectious pathogens or foreign tissue grafts (allografts) [38]. Thus, the interaction of B7.1 and B7.2 with the CD28 receptor results in costimulatory signals leading to the productive activation, expansion, differentiation and survival of T cells, with resulting effective antibody-mediated and cellular immune responses (Table 1). This productive immune response is kept in balance by the attenuation signal delivered through B7-ligand-CTLA-4 receptor interactions. Thus, in CTLA-4-deficient mice, a severe autoimmune phenotype develops, with death occurring 3-4 weeks after birth as a result of multiorgan destruction [39,40]. Interestingly, the absence of B7.1 and B7.2 signals ablates the autoimmune phenotype in triple-mutant mice deficient in B7.1, B7.2 and CTLA-4 [41]. These findings demonstrate that B7-CTLA-4 interactionscounteract B7-CD28 interactions, resulting in precise regulation of T-cell receptor and CD28 signals, interleukin-2 production and cell-cycle progression.
Figure 4.

Costimulation and coinhibition. The binding of CD28 or CTLA-4 receptors on T cells by B7.1 and B7.2 ligands on antigen-presenting cells (APCs) can lead to either costimulation or coinhibition depending upon the precise expression patterns of the receptors and ligands and on the state of activation of the two cells. (a) CD28 is expressed on resting T cells and can be engaged by either B7.1 or B7.2 on APCs. Current models favor preferential engagement by B7.2, leading to activation of resting T cells. This costimulation leads in the T cell to increased production of growth factors, such as interleukin-2 (IL-2) and increased cell-survival signals, such as Bcl-X. Reverse signaling by CD28 engagement of B7.1 (not shown) or B7.2 ligands resulting in production of interleukin-6 (IL-6) by the APC has also been described. (b) CD28 and CTLA-4 are both expressed on activated T cells, and both receptors on T cells can be engaged by B7.1 or B7.2 on APCs. Current models favor preferential engagement of CTLA-4 by B7.1, resulting in attenuation of T-cell activation. Reverse signaling by CTLA-4 engagement of B7.1 or B7.2 (not shown) ligands resulting in production of indoleamine 2, 3-dioxygenase (IDO) and a reduction in tryptophan levels in the APC has also been described.
Recently, it has been reported that engagement of B7.1 and B7.2 leads to 'reverse signaling' through B7.1 and B7.2 [42,43]. In dendritic cells, engagement of B7.1 or B7.2 with CTLA4.Ig, a soluble CTLA-4-immunoglobulin chimeric protein, leads to production of indoleamine 2,3-dioxygenase (IDO) and decreased tryptophan levels, resulting in inhibition of T-cell proliferation and cell death [42] (Figure 4b). In contrast, engagement of B7.1 and B7.2 with CD28.Ig results in induction of IL-6 production [43] (Figure 4a). Interestingly, this bidirectional signaling could explain the increased survival of cardiac allografts in CD28-/-CTLA-/- mice treated with CTLA4.Ig [44]. Further elucidation of reverse B7 signaling is necessary if we are to understand how this contributes to the overall outcome of receptor-ligand interactions.
In summary, B7.1 and B7.2 can either costimulate or coinhibit immune responses through interaction with their receptors, providing temporal and spatial control of immune responses. Similar costimulatory and coinhibitory paradigms apply to other members of the B7 family, as outlined in Table 1, although details of these pathways are still emerging. Conservation of the B7 family of ligands in species with cellular immune systems underscores the functional importance of B7 ligands in regulating signals delivered by antigen-specific receptors on immune cells.
Frontiers
The B7 family has a critical role in controlling immune responses, as was initially shown for B7.1 and B7.2 and extended by the recent identification and characterization of new B7-family members. Inflammatory stimuli modulate expression of B7 ligands, resulting in the delivery of activating and attenuating signals that determine the nature and extent of subsequent immune responses. Crystal structures, affinity measurements and biological assays have demonstrated how B7.1 and B7.2 deliver specific signals to immune cells resulting in precise regulation of immune function. This detailed level of knowledge is still to be obtained for additional members of the family. The receptors for B7-H3 and B7-H4 remain to be identified, and the available data suggest that there may be additional receptors for PD-L1 and PD-L2. Further clarification is needed before we understand how signals delivered by B7-family ligands are integrated spatially and temporally with those derived from antigen-specific receptors. Therapeutic manipulation of B7 signaling has shown clinical promise [45,46], underscoring the key role that B7-family ligands play in regulating immune responses.
Additional data files
A sequence alignment for the proteins that were used to make the phylogenetic tree in Figure 1 is available in Additional data file 1. This alignment was made using Pileup and the Blosum62 scoring matrix.
Supplementary Material
A sequence alignment for the proteins that were used to make the phylogenetic tree in Figure 1
References
- Sharpe AH, Freeman GJ. The B7-CD28 superfamily. Nat Rev Immunol. 2002;2:116–126. doi: 10.1038/nri727. This article and [3] are comprehensive reviews of the B7 and CD28 families. [DOI] [PubMed] [Google Scholar]
- Schwartz J-CD, Zhang X, Nathenson SG, Almo SC. Structural mechanisms of costimulation. Nat Immunol. 2002;3:427–434. doi: 10.1038/ni0502-427. This article and [21] are reviews of structural interactions among members of the B7 and CD28 families. [DOI] [PubMed] [Google Scholar]
- Carreno BM, Collins M. The B7 family of ligands and its receptors: new pathways for costimulation and inhibition of immune responses. Annu Rev Immunol. 2002;20:29–53. doi: 10.1146/annurev.immunol.20.091101.091806. See [1]. [DOI] [PubMed] [Google Scholar]
- Ling V, Wu PW, Spaulding V, Kieleczawa J, Luxenberg D, Carreno BM, Collins M. Duplication of primate and rodent B7-H3 immunoglobulin V- and C-like domains: divergent history of functional redundancy and exon loss. Genomics. 2003;82:365–377. doi: 10.1016/S0888-7543(03)00126-5. This article and [32] identify a novel isoform of human B7-H3; this article and [33] describe B7-H3 as a negative regulator of T-cell activation. [DOI] [PubMed] [Google Scholar]
- Borriello F, Freeman GJ, Edelhoff S, Disteche CM, Nadler LM, Sharpe AH. Characterization of the murine B7-1 genomic locus reveals an additional exon encoding an alternative cytoplasmic domain and a chromosomal location of chromosome 16, band B5. J Immunol. 1994;153:5038–5048. Characterization of the murine B7.1 gene and identification of differentially spliced forms. [PubMed] [Google Scholar]
- Borriello F, Oliveros J, Freeman GJ, Nadler LM, Sharpe AH. Differential expression of alternate mB7-2 transcripts. J Immunol. 1995;155:5490–5497. Characterization of the murine B7.2 gene and identification of differentially spliced forms. [PubMed] [Google Scholar]
- Guo Y, Wu Y, Zhao M, Kong XP, Liu Y. Mutational analysis and an alternatively spliced product of B7 defines its CD28/CTLA4-binding site on immunoglobulin C-like domain. J Exp Med. 1995;181:1345–1355. doi: 10.1084/jem.181.4.1345. This article defines a novel splice variant of B7.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling V, Wu PW, Miyashiro JS, Marusic S, Finnerty HF, Collins M. Differential expression of inducible costimulator-ligand splice variants: lymphoid regulation of mouse GL50-b and human GL50 molecules. J Immunol. 2001;166:7300–7308. doi: 10.4049/jimmunol.166.12.7300. A description of splice variants of ICOS-L. [DOI] [PubMed] [Google Scholar]
- He XH, Liu Y, Xu LH, Zeng YY. Cloning and identification of two novel splice variants of human PD-L2. Acta Biochim Biophys Sin. 2004;36:284–289. doi: 10.1093/abbs/36.4.284. Identification of splice variants of human PD-L2. [DOI] [PubMed] [Google Scholar]
- Prasad DVR, Richards S, Mai XM, Dong C. B7S1, a novel B7 family member that negatively regulates T cell activation. Immunity. 2003;18:863–873. doi: 10.1016/S1074-7613(03)00147-X. This paper and [11, 35] identify B7-H4 as a negative regulator of T-cell activation. [DOI] [PubMed] [Google Scholar]
- Sica GL, Choi IH, Zhu G, Tamada K, Wang SD, Tamura H, Chapoval AI, Flies DB, Bajorath J, Chen L. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity. 2003;18:849–861. doi: 10.1016/S1074-7613(03)00152-3. See [10]. [DOI] [PubMed] [Google Scholar]
- Koskela K, Arstila TP, Lassila O. Costimulatory function of CD28 in avian gammadelta T cells is evolutionarily conserved. Scand J Immunol. 1998;48:635–641. doi: 10.1046/j.1365-3083.1998.00441.x. Shows that the CD28 costimulatory pathway is functional in chickens. The original papers on the cloning of CD28 are cited in this article. [DOI] [PubMed] [Google Scholar]
- Zhang X, Schwartz JC, Almo SC, Nathenson SG. Crystal structure of the receptor-binding domain of human B7-2: insights into organization and signaling. Proc Natl Acad Sci USA. 2003;100:2586–2591. doi: 10.1073/pnas.252771499. This paper, together with [14-16], describes structural information on soluble and CTLA-4-complexed human B7 ligands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemizu S, Gilbert RJ, Fennelly JA, Collins AV, Harlos K, Jones EY, Stuart DI, Davis SJ. Structure and dimerization of a soluble form of B7-1. Immunity. 2000;12:51–60. doi: 10.1016/S1074-7613(00)80158-2. See [13]. [DOI] [PubMed] [Google Scholar]
- Stamper CC, Zhang Y, Tobin JF, Erbe DV, Ikemizu S, Davis SJ, Stahl ML, Seehra J, Somers WS, Mosyak L. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature. 2001;410:608–611. doi: 10.1038/35069118. See [13]. [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Zhang X, Fedorov AA, Nathenson SG, Almo SC. Structural basis for co-stimulation by the human CTLA-4/B7-2 complex. Nature. 2001;410:604–608. doi: 10.1038/35069112. See [13]. [DOI] [PubMed] [Google Scholar]
- Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. This paper and [18, 19] define and review immunoglobulin domains in proteins. [DOI] [PubMed] [Google Scholar]
- Barclay AN. Membrane proteins with immunoglobulin-like domains - a master superfamily of interaction molecules. Semin Immunol. 2003;15:215–223. doi: 10.1016/S1044-5323(03)00047-2. See [17]. [DOI] [PubMed] [Google Scholar]
- Williams AF, Barclay AN. The immunoglobulin superfamily - domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. See [17]. [DOI] [PubMed] [Google Scholar]
- Collins A, Brodie D, Gilbert R, Iaboni A, Manso-Sancho R, Walse B, Stuart D, van der Merwe P, Davis SJ. The interaction properties of costimulatory molecules revisited. Immunity. 2002;17:201–210. doi: 10.1016/S1074-7613(02)00362-X. Biochemical characterization of the interactions of human CD28 and CTLA-4 with human B7.1 and B7.2. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Ikemizu S, Evans EJ, Fugger L, Bakker TR, van der Merwe PA. The nature of molecular recognition by T cells. Nat Immunol. 2003;4:217–224. doi: 10.1038/ni0303-217. See [2]. [DOI] [PubMed] [Google Scholar]
- Pentcheva-Hoang T, Egen JG, Wojnoonski K, Allison JP. B7-1 and B7-2 selectively recruit CTLA-4 and CD28 to the immunological synapse. Immunity. 2004;21:401–413. doi: 10.1016/j.immuni.2004.06.017. This paper describes how differences in affinity and avidity between B7.1 and B7.2 modulate their interactions with CD28 and CTLA4 at the immune synapse. [DOI] [PubMed] [Google Scholar]
- Evans EJ, Esnouf RM, Manso-Sancho R, Gilbert RJ, James JR, Yu C, Fennelly JA, Vowles C, Hanke T, Walse B, et al. Crystal structure of a soluble CD28-Fab complex. Nat Immunol. 2005;6:271–279. doi: 10.1038/ni1170. The crystal structure of CD28 in complex with an antibody fragment. [DOI] [PubMed] [Google Scholar]
- Brodie D, Collins AV, Iaboni A, Fennelly JA, Sparks LM, Xu XN, van der Merwe PA, Davis SJ. LICOS, a primordial costimulatory ligand? Curr Biol. 2000;10:333–336. doi: 10.1016/S0960-9822(00)00383-3. Characterization of ICOS-L binding affinities for the receptor ICOS. [DOI] [PubMed] [Google Scholar]
- Youngnak P, Kozono Y, Kozono H, Iwai H, Otsuki N, Jin H, Omura K, Yagita H, Pardoll DM, Chen L, et al. Differential binding properties of B7-H1 and B7-DC to programmed death-1. Biochem Biophys Res Comm. 2003;307:672–677. doi: 10.1016/S0006-291X(03)01257-9. Comparison of the binding affinities of the human receptor PD-1 for human PD-L1 and PD-L2. [DOI] [PubMed] [Google Scholar]
- Zhang X, Schwartz JC, Guo X, Bhatia S, Cao E, Lorenz M, Cammer M, Chen L, Zhang ZY, Edidin MA, et al. Structural and functional analysis of the costimulatory receptor programmed death-1. Immunity. 2004;20:337–347. doi: 10.1016/S1074-7613(04)00051-2. Crystal structure of murine PD-1 extracellular domain and comparison with that of CTLA-4. [DOI] [PubMed] [Google Scholar]
- Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol. 2004;4:336–347. doi: 10.1038/nri1349. A comprehensive review of the coinhibitory B7 ligands. [DOI] [PubMed] [Google Scholar]
- Aicher A, Hayden-Ledbetter M, Brady WA, Pezzutto A, Richter G, Magaletti D, Buckwalter S, Ledbetter JA, Clark EA. Characterization of human inducible costimulator ligand expression and function. J Immunol. 2000;164:4689–4696. doi: 10.4049/jimmunol.164.9.4689. Description of the expression and regulation of human ICOS-L on antigen presenting cells. [DOI] [PubMed] [Google Scholar]
- Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, Iwai Y, Long AJ, Brown JA, Nunes R, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2:261–268. doi: 10.1038/85330. This paper and [30] provide the first description of PD-L2. [DOI] [PubMed] [Google Scholar]
- Tseng SY, Otsuji M, Gorski K, Huang X, Slansky JE, Pai SI, Shalabi A, Shin T, Pardoll DM, Tsuchiya H. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J Exp Med. 2001;193:839–846. doi: 10.1084/jem.193.7.839. See [29]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. One of the first reports of the identification of PD-L1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Richards S, Prasad DV, Mai XM, Rudensky A, Dong C. Characterization of mouse and human B7-H3 genes. J Immunol. 2002;168:6294–6297. doi: 10.4049/jimmunol.168.12.6294. See [4]. [DOI] [PubMed] [Google Scholar]
- Prasad DV, Nguyen T, Li Z, Yang Y, Duong J, Wang Y, Dong C. Murine B7-H3 is a negative regulator of T cells. J Immunol. 2004;173:2500–2506. doi: 10.4049/jimmunol.173.4.2500. See [4]. [DOI] [PubMed] [Google Scholar]
- Chapoval AI, Ni J, Lau JS, Wilcox RA, Flies DB, Liu D, Dong H, Sica GL, Zhu G, Tamada K, et al. B7-H3: a co-stimulatory molecule for T cell activation and IFN-gamma production. Nat Immunol. 2001;2:269–274. doi: 10.1038/85339. The first identification of B7-H3 and its description as a costimulatory molecule. [DOI] [PubMed] [Google Scholar]
- Zang X, Loke P, Kim J, Murphy K, Waitz R, Allison JP. B7x: a widely expressed B7 family member that inhibits T cell activation. Proc Natl Acad Sci USA. 2003;100:10388–10392. doi: 10.1073/pnas.1434299100. See [10]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh WK, Gajewska BU, Okada H, Gronski MA, Bertram EM, Dawicki W, Duncan GS, Bukczynski J, Plyte S, Elia A, et al. The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat Immunol. 2003;4:899–906. doi: 10.1038/ni967. This paper describes the regulation of immune responses by B7-H3 and the regulation of B7-H3 expression by cytokines. [DOI] [PubMed] [Google Scholar]
- Borriello F, Sethna MP, Boyd SD, Schweitzer AN, Tivol EA, Jacoby D, Strom TB, Simpson EM, Freeman GJ, Sharpe AH. B7-1 and B7-2 have overlapping, critical roles in immunoglobulin class switching and germinal center formation. Immunity. 1997;6:303–313. doi: 10.1016/S1074-7613(00)80333-7. Describes the function of B7 ligands in immunoglobulin production. [DOI] [PubMed] [Google Scholar]
- Lenschow DJ, Walunas TL, Bluestone JA. CD28/B7 system of T cell costimulation. Annu Rev Immunol. 1996;14:233–258. doi: 10.1146/annurev.immunol.14.1.233. A comprehensive review of B7, CD28 and CTLA4 functions. [DOI] [PubMed] [Google Scholar]
- Tivol EA, Borriello F, Schweitzer AN, Lynch WP, Bluestone JA, Sharpe AH. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity. 1995;3:541–547. doi: 10.1016/1074-7613(95)90125-6. This paper and [40] describe the phenotype of CTLA-4-deficient mice. [DOI] [PubMed] [Google Scholar]
- Waterhouse P, Penninger JM, Timms E, Wakeham A, Shahinian A, Lee KP, Thompson CB, Griesser H, Mak TW. Lymphoproliferative disorders with early lethality in mice deficient in CTLA-4. Science. 1995;270:985–988. doi: 10.1126/science.270.5238.985. See [39]. [DOI] [PubMed] [Google Scholar]
- Mandelbrot DA, McAdam AJ, Sharpe AH. B7-1 or B7-2 is required to produce the lymphoproliferative phenotype in mice lacking cytotoxic T lymphocyte-associated antigen 4 (CTLA-4). J Exp Med. 1999;189:435–440. doi: 10.1084/jem.189.2.435. Demonstration of the requirement for B7 signaling for development of a lymphoproliferative disorder in CTLA-4-deficient mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann U, Orabona C, Fallarino F, Vacca C, Calcinaro F, Falorni A, Candeloro P, Belladonna ML, Bianchi R, Fioretti MC, et al. CTLA-4-Ig regulates tryptophan catabolism in vivo. Nat Immunol. 2002;3:1097–1101. doi: 10.1038/ni846. This paper and [43] describe bidirectional signaling between B7 ligands and their receptors. [DOI] [PubMed] [Google Scholar]
- Orabona C, Grohmann U, Belladonna ML, Fallarino F, Vacca C, Bianchi R, Bozza S, Volpi C, Salomon BL, Fioretti MC, et al. CD28 induces immunostimulatory signals in dendritic cells via CD80 and CD86. Nat Immunol. 2004;5:1134–1142. doi: 10.1038/ni1124. See [42]. [DOI] [PubMed] [Google Scholar]
- Mandelbrot DA, Oosterwegel MA, Shimizu K, Yamada A, Freeman GJ, Mitchell RN, Sayegh MH, Sharpe AH. B7-dependent T-cell costimulation in mice lacking CD28 and CTLA4. J Clin Invest. 2001;107:881–887. doi: 10.1172/JCI11710. Describes the protective effect of CTLA4.Ig, a soluble CTLA-4-immunoglobulin chimeric protein, against allograft rejection in CD28-and CTLA4-deficient mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JM, Westhovens R, Leon M, Di Giorgio E, Alten R, Steinfeld S, Russell A, Dougados M, Emery P, Nuamah IF, et al. Treatment of rheumatoid arthritis by selective inhibition of T-cell activation with fusion protein CTLA4Ig. N Engl J Med. 2003;349:1907–1915. doi: 10.1056/NEJMoa035075. This paper describes the beneficial effect of blocking B7 ligands using CTLA4.Ig in combination with methotrexate in patients with rheumatoid arthritis. [DOI] [PubMed] [Google Scholar]
- Phan GQ, Yang JC, Sherry RM, Hwu P, Topalian SL, Schwartzentruber DJ, Restifo NP, Haworth LR, Seipp CA, Freezer LJ, et al. Cancer regression and autoimmunity induced by cytotoxic T lymphocyte-associated antigen 4 blockade in patients with metastatic melanoma. Proc Natl Acad Sci USA. 2003;100:8372–8377. doi: 10.1073/pnas.1533209100. This paper describes effect of enhancing CTLA-4 signals using anti-CTLA-4 antibody treatment in patients with melanoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEGA: Molecular Evolutionary Genetics Analysis http://www.megasoftware.net/ Software for sequence alignment and phylogenetic analysis developed by Sudhir Kumar and colleagues at Arizona State University, USA.
- NCBI Reference Sequence (RefSeq) http://www.ncbi.nlm.nih.gov/RefSeq/ A non-redundant set of sequences for major research organisms.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A sequence alignment for the proteins that were used to make the phylogenetic tree in Figure 1


