Abstract
The failure of cells to repair damaged DNA can result in genomic instability and cancer. To efficiently repair chromosomal DNA lesions, the repair machinery must gain access to the damaged DNA in the context of chromatin. Here we report that both the RSC and Swi/Snf ATP-dependent chromatin-remodeling complexes play key roles in double-strand break (DSB) repair, specifically by homologous recombination (HR). RSC and Swi/Snf are each recruited to an in vivo DSB site but with distinct kinetics. We show that Swi/Snf is required earlier, at or preceding the strand invasion step of HR, while RSC is required following synapsis for completion of the recombinational repair event.
Keywords: Chromatin remodeling, DSB repair, HR, RSC, SWI/SNF
The high-fidelity transmission of genetic information is essential for proper cell growth and viability. Chromosomal DNA lesions resulting from replication errors or from a variety of environmental and endogenous insults such as radiation, chemicals, or free radicals must be promptly and accurately repaired in order to maintain genomic integrity. Errors in any of the steps involved in the recognition or repair of damaged DNA may result in the rearrangement or gain or loss of chromosomes or tumorigenesis (Stewart et al. 1999; Pierce et al. 2001).
Double-strand breaks (DSBs), the most serious of eukaryotic DNA lesions, are repaired by two distinct and complementary pathways: nonhomologous end joining (NHEJ) and homologous recombination (HR). In NHEJ, the two ends of a DSB are reunited in the absence of sequences homologous to the break site by the double-stranded DNA (dsDNA) end-binding heterodimer KU (Yku70p/Yku80p in budding yeast) and DNA ligase IV and its cofactor XRCC4 (Dnl4p and Lif1p in budding yeast) (Critchlow and Jackson 1998; Valerie and Povirk 2003). In HR, the ends of the broken chromosome must locate an intact undamaged homologous template whose DNA sequences are then copied to repair the DSB. HR is mediated by the evolutionarily conserved RAD52 epistasis group proteins, which include budding yeast Rad51p, Rad52p, Rad54p, Rad55p, Rad57p, Rad59p, Mre11p, Rad50p, and Xrs2p (Symington 2002). Approximately 90% of the DSBs in Saccharomyces cerevisiae and 50% of those in mammalian mitotic cells are repaired by HR (Paques and Haber 1999; Johnson and Jasin 2000). Recombinational repair in yeast involves the initial resection of the 5′ ends of DNA flanking the break by an exonuclease, followed by binding of the replication protein A (RPA) single-stranded DNA (ssDNA)-binding protein to the exposed 3′ ssDNA. The Rad51p strand exchange protein then displaces RPA to form the presynaptic filament (New et al. 1998), a process mediated by Rad52p (Sung 1997; Shinohara and Ogawa 1998), the Rad55p/Rad54p heterodimer (Sung et al. 2000; Symington 2002), and Rad54p (Wolner et al. 2003). Following its successful search for a homologous sequence, Rad51p facilitates the presynaptic filament's invasion of the homologous template. DNA replication of the homologous template is then followed by ligation to the distal broken DNA end (Paques and Haber 1999). Recently, studies demonstrating the interdependent recruitment of checkpoint and recombinational repair proteins to DSBs in vivo have provided a molecular time-frame of DSB repair events (Sugawara et al. 2003; Wolner et al. 2003; Lisby et al. 2004).
These two highly conserved pathways must repair DSBs in the context of chromatin, implicating enzymes that modulate chromatin structure in DNA repair (see Peterson and Cote 2004). Indeed, histone acetyltransferase and histone deacetylase complexes are implicated in the repair of DSBs by either NHEJ or HR (Bird et al. 2002; Qin and Parthun 2002; Downs et al. 2004). ATP-dependent nucleosome-remodeling factors are also implicated in repair: The Rad54p enzyme functions in multiple steps of DSB repair by HR (Alexiadis and Kadonaga 2002; Alexeev et al. 2003; Wolner and Peterson 2005), INO80 has recently been linked to DSB repair (Morrison et al. 2004; van Attikum et al. 2004), and mutants of SWR1 are sensitive to DNA damaging agents (Mizuguchi et al. 2004).
Despite emerging roles for chromatin-remodeling enzymes in DSB repair, the paucity of mechanistic data demonstrating how and in which steps of the DSB repair pathways ATP-dependent chromatin-remodeling enzymes function prompted us to examine the roles of the yeast RSC and related Swi/Snf complexes in repair of DSBs. RSC is essential for cell viability and has been implicated in transcriptional regulation (Angus-Hill et al. 2001) and chromosome transmission (Tsuchiya et al. 1998; Hsu et al. 2003; Baetz et al. 2004), and more recently in sister chromatid cohesion (Baetz et al. 2004; Huang et al. 2004). Swi/Snf functions in regulating transcription. Although in vitro experiments have linked Swi/Snf to the nucleotide excision repair pathway (Hara and Sancar 2002), its in vivo function in DNA repair, and in particular DSB repair, is unknown.
Here we present evidence that both RSC and Swi/Snf complexes play key roles in DNA DSB repair by HR. RSC and Swi/Snf associate with a DSB site with different kinetics, suggesting functional differences between the remodelers. Indeed, following a DSB, Swi/Snf is recruited to the donor sequence and is essential for repair at a step during or prior to synapsis, whereas RSC function is required for HR repair at a step post-synapsis.
Results and Discussion
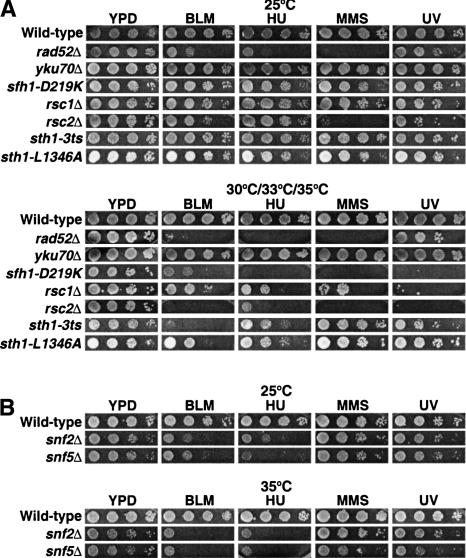
To investigate the role of the RSC ATP-dependent remodeling complex in DNA repair, we first compared the growth of several temperature-sensitive rsc mutants to wild-type cells on rich medium in the presence of the DNA damaging agents bleomycin, hydroxyurea, methyl methane sulfonate, or ultraviolet (UV) radiation, each of which can directly or indirectly induce DSBs. Bleomycin generates free radicals that can cause DSBs. Hydroxyurea lowers the dNTP pool, thereby collapsing replication forks, and activates the intra-S checkpoint. Methylmethane sulfonate (MMS) alkylates purines, which, when left unrepaired, can result in DSBs after replication. UV irradiation primarily causes pyrimidine dimers, which, when left unrepaired, can occasionally create DSBs following replication. rsc mutants were hypersensitive to all three of the DNA damaging agents and to UV exposure when grown at their respective semipermissive temperatures (Fig. 1A). An earlier study also reported enhanced sensitivity of rsc1 and rsc2 mutants to γ irradiation (Bennett et al. 2001), suggesting further that RSC functions directly in DNA DSB repair.
Figure 1.
rsc and swi/snf mutants are hypersensitive to genotoxic agents. (A) Tenfold serial dilutions of wild-type (BLY76), rad52Δ (BLY772), yku70Δ (BLY773), sfh1-D219K (BLY650), rsc1Δ (BLY788), rsc2Δ (BLY662), sth1-3ts (BLY49), and sth1-L1346A (BLY491) strains were spotted onto rich YPD medium in the absence or presence of 3 mU/mL bleomycin, 50 mM hydroxyurea, 0.01% MMS, or 75 J/m2 UV light. Cells were incubated for 3–4 d at permissive (25°C) or at each of their respective semipermissive (33°C for rad52Δ, yku70Δ and sfh1-D219K; 35°C for rsc1Δ, rsc2Δ, sth1-3ts, and sth1-L1346A) temperatures; wild-type cells were grown at 30°C. (B) Tenfold serial dilutions of wild-type (BLY1), snf2Δ (BLY35), and snf5Δ (BLY3) strains were spotted onto YPD medium in the absence or presence of 3 mU/mL bleomycin, 50 mM hydroxyurea, 0.01% MMS, or 75 J/m2 UV radiation and incubated at 25°C and 35°C for 3–4 d.
Interestingly, mutants of the related Swi/Snf chromatin-remodeling complex, snf5Δ and snf2Δ, were also hypersensitive to bleomycin and hydroxyurea and only moderately sensitive to MMS and UV irradiation at 25°C and 35°C (Fig. 1B). As expected, the rad52Δ recombination mutant, but not the yku70Δ end-joining mutant, also showed decreased viability in the presence of bleomycin, hydroxyurea, MMS, or UV light. The variability in sensitivities among mutants could reflect the different types of mutations or the inherent functional differences of subunits (Angus-Hill et al. 2001). Together, these results are consistent with roles for both RSC and Swi/Snf in the repair of DSBs.
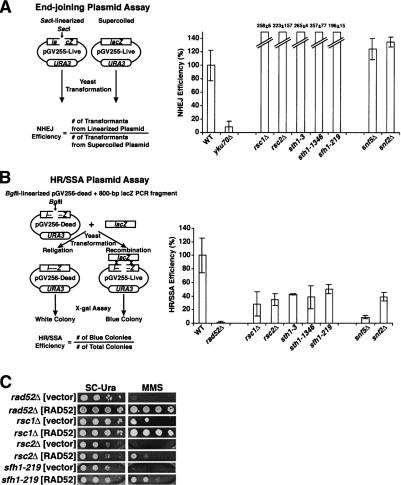
Because eukaryotic DSBs can be repaired by NHEJ or HR, we evaluated the roles of rsc or swi/snf mutants in either repair pathway using epistasis analysis and plasmid-based repair assays. The enhanced hypersensitivity of rsc rad52 and rsc yku70 double mutants to bleomycin, hydroxyurea, and MMS suggests that RSC may function both in NHEJ and HR (Supplementary Fig. 1). We also assayed plasmid end joining in five rsc and two swi/snf mutants using a standard plasmid recircularization assay (Erdemir et al. 2002). The sth1, sfh1, rsc1, and rsc2 mutants of RSC, as well as the snf2 and snf5 mutants of Swi/Snf, were able to religate the linearized plasmid in vivo, inexplicably better than or as well as wild-type cells. As expected, the control yku70Δ cells could not rejoin a linearized plasmid, while rad52Δ cells recircularized linearized plasmids as efficiently as wild-type cells (Fig. 2A; Boulton and Jackson 1996; data not shown). Thus, these rsc and swi/snf mutants were not defective in end joining of linearized plasmids, although it is still possible that RSC and Swi/Snf might be important in the context of chromosomal NHEJ.
Figure 2.
rsc and swi/snf mutants are competent for plasmid end joining but defective in repair by HR/SSA. (A) rsc and swi/snf mutants are capable of repair by plasmid end joining. Wild-type (BLY76), yku70Δ (BLY773), rsc?Δ (BLY788), rsc2Δ (BLY662), sth1-3ts (BLY49), sth1-L1346A (BLY491), sfh1-219 (BLY650), snf5Δ (BLY3), and snf2Δ (BLY16) strains transformed in parallel with the SacI-linearized or supercoiled pGV255-live plasmid were plated onto selective SC-Ura medium at 30°C for 4–5 d. Values represent the ratios of the number of transformants from linearized plasmid to the number of transformants from supercoiled plasmid, normalized to values obtained from wild-type strains. The means and standard deviations of at least three independent experiments are plotted. (B) rsc and swi/snf mutants are defective for HR/SSA. The same strains as in A and rad52Δ (BLY772) were cotransformed with the BglII-linearized pGV256-dead plasmid and an 800-bp lacZ PCR fragment amplified from pGV255-live, plated onto SC-Ura medium at 30°C for 4–5 d, and assayed for β-galactosidase activity. Values represent the ratios of blue colonies to total colonies, normalized to values obtained from wild-type strains. The means and standard deviations of at least three independent experiments are plotted. (C) RAD52 overexpression partially rescues the MMS hypersensitivity of rsc mutants. rad52Δ (BLY772), rsc1Δ (BLY788), rsc2Δ (BLY662), and sfh1-219 (BLY650) strains transformed with either pRS426:RAD52 or pRS426 alone were 10-fold serially diluted and spotted onto SC-Ura medium in the absence or presence of 0.01% MMS at 35°C (or at 33°C for sfh1-219) for 3–4 d.
HR was first examined using a plasmid-based repair assay in which successful recombination or single-strand annealing (SSA) with an 800-bp PCR fragment produces full-length functional β-galactosidase (Erdemir et al. 2002). We found that each of the rsc and swi/snf mutants tested achieved only 10%–50% of the repair of wild-type cells, suggesting that RSC and Swi/Snf play important roles in repair of DSBs by either HR or SSA. As expected, rad52Δ cells were completely unable to carry out repair, while yku70Δ cells were proficient in repair (Fig. 2B; Paques and Haber 1999; data not shown). Thus, analysis of the sth1, sfh1, rsc1, and rsc2 alleles uncovered defects in the repair of DSBs by either HR or SSA.
To gain further evidence for a role for RSC in DSB repair, we examined whether overexpression of a protein involved in HR could rescue the hypersensitivity of rsc mutants to genotoxic agents. Remarkably, we found that rsc1Δ, rsc2Δ, and sfh1-219 cells transformed with high-copy plasmids expressing RAD52 exhibited partial to nearly wild-type resistance to MMS (Fig. 2C).
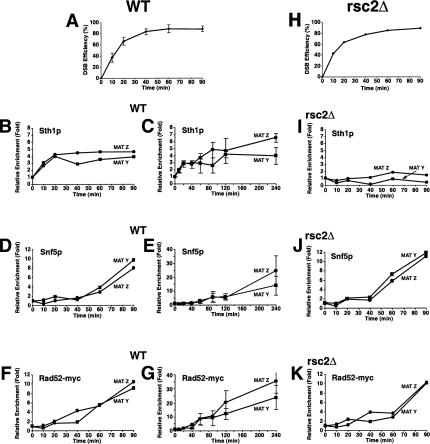
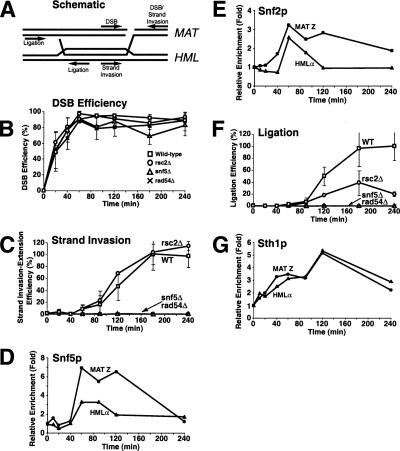
We predicted that if RSC and Swi/Snf function directly in DSB repair by HR, then they should be recruited to DSB sites. To test this, we exploited mating-type switching in yeast in which the HO endonuclease creates a site-specific DSB at the mating-type (MAT) locus to induce HR from the silent HML and HMR mating loci. Initially, we examined the kinetics of Sth1p, Snf5p, and Rad52p recruitment to both sides of the DSB site in strains lacking both the HML and HMR homologous donor sequences (Fig. 3) so that the initial binding of proteins to the DSB site would be more readily observed. Remarkably, Sth1p was rapidly recruited to both sides of the DSB site within 10 min of DSB induction and reached maximal levels by 20 min (Fig. 3B), suggesting that RSC function is required early in DSB repair. Snf5p was also recruited to the same MAT Y and MAT Z sequences, but in contrast to RSC, appeared only after 40 min of DSB induction (Fig. 3D). Recruitment of both Snf5p and Snf2p, the ATPase subunit of the Swi/Snf complex, to the DSB site increased steadily over the next 4 h, whereas Sth1p's presence at the DSB site increased only slightly during the same period (Fig. 3C,E; data not shown), suggesting that Swi/Snf plays a relatively later role in DSB repair. Similar to Swi/Snf, Rad52-myc13p was recruited to the DSB site within 40 min of HO induction and its association with the broken DNA ends increased steadily for at least 4 h (Fig. 3F,G), as shown previously (Wolner et al. 2003; Miyazaki et al. 2004).
Figure 3.
RSC and Swi/Snf are recruited to an in vivo DSB but neither is required for Rad52p's recruitment to a DSB. (A,H) Kinetics of DSB induction. Wild-type (BLY755) or rsc2Δ (BLY780) cells grown to mid-logarithmic phase in 2% raffinose at 30°C were shifted to 2% galactose to induce DSBs and genomic DNA harvested at the indicated times. The averages and standard deviations of five wild-type or three rsc2Δ mutant time-course experiments are plotted. (B,D,F) Aliquots of wild-type cells from A were processed for ChIP using anti-Sth1p, anti-Snf5p, or anti-Rad52p-13myc antibodies at the indicated times. All ChIPs were quantified by real-time PCR analysis and normalized to the time 0 values and to input DNA. Levels of CEN-proximal (MAT Y) or CEN-distal (MAT Z) DNAs immunoprecipitated by anti-Sth1p and anti-Snf5p were additionally normalized to those at the PRE1 locus. (C,E,G) Long time courses of anti-Sth1p, anti-Snf5p, or anti-Rad52-myc13p ChIP analysis show continuous recruitment of Snf5p and Rad52-myc13p to a DSB. (I–K) ChIP assays using anti-Sth1p, anti-Snf5p, or anti-Rad52-myc13p antibodies were performed on rsc2Δ (BLY780) cells as described for wild-type cells. All ChIP data are representative of at least three independent time-course experiments except for anti-Rad52-myc ChIPs, which were done twice. Error bars represent standard deviations.
To investigate repair of the DSB by gene conversion in the wild-type and remodeling mutants, we used strains containing HMLα as the donor template. This would permit us to examine repair of the HO-induced DSB at MATa by recombination with the homologous HMLα locus. The HR repair of chromosomal DNA DSBs is a multiple-step process that includes recruitment of DNA repair enzymes to a DSB site, strand invasion of a homologous template, and ligation. RSC's presence at a DSB site prior to the DNA repair enzymes suggested that it might remodel chromatin to facilitate recruitment of the DNA repair machinery. However, we found that in the absence of RSC2 (which prevented the recruitment of Sth1p to the DSB site), Rad52-myc13p, one of the first HR repair enzymes to be recruited to a DSB site (Sugawara et al. 2003), was still recruited to both sides of the DSB with wild-type kinetics and at wild-type levels, suggesting that Rad52p recruitment is independent of RSC function (Fig. 3I,J). Rad52p's recruitment to the DSB also occurred in the absence of Swi/Snf (Supplementary Fig. 2). Similarly, we found that Swi/Snf's association with the DSB was also independent of RSC (Fig. 3K).
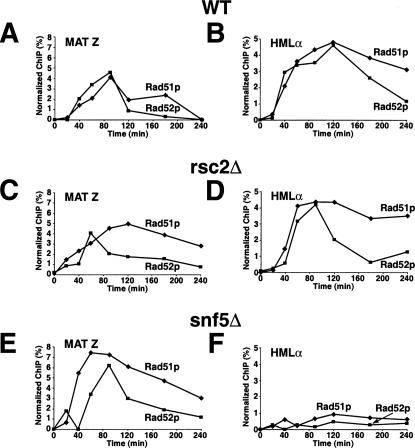
Because recruitment of Rad52p to a site-specific DSB was unimpaired in the chromatin-remodeling mutants, we next examined whether synapsis between MAT and HML occurs in rsc or swi/snf mutants by comparing the association of Rad51p and Rad52p proteins with HMLα in rsc2Δ, snf5Δ, and wild-type cells by chromatin immunoprecipitation (ChIP) (Fig. 4). In rsc2Δ mutants, the levels of Rad51p and Rad52p proteins at both MAT Z and HMLα sites were comparable to those in wild-type cells (Fig. 4C,D). In sharp contrast, in snf5Δ mutants, although wild-type or slightly higher levels of Rad51p and Rad52p were recruited to MAT Z, levels of these proteins at HMLα were reduced at least fivefold (Fig. 4E,F). Importantly, recruitment of Rad51p to HMLα can be restored in snf5Δ mutants carrying a CEN-SNF5 plasmid but not a CEN plasmid alone (Supplementary Fig. 3). These results suggest an important role for Swi/Snf at or just preceding synapsis formation. Interestingly, we found that the snf5Δ cells that survive following 16 h of DSB induction (survival was comparable to wild-type cells) cannot switch mating type (Supplementary Fig. 4), consistent with the lack of synapsis in these cells.
Figure 4.
Synapsis formation between MAT and HML is defective in snf5 mutants but not rsc2 mutants. Wild-type (BLY747), rsc2Δ (BLY784), or snf5Δ (BLY786) strains carrying the HMLα donor sequence were treated with 2% galactose to induce expression of HO endonuclease, and then with 2% dextrose after 1 h to repress HO. Chromatin prepared from wild-type (A,B), rsc2Δ (C,D), or snf5Δ (E,F) cells at the indicated time points was immunoprecipitated with anti-Rad51p or anti-Rad52p antibodies. The immunoprecipitated DNAs were quantified by real-time PCR amplification using primers specific to MAT Z or HMLα regions and normalized to ACT1 promoter sequences.
We next asked whether either remodeling complex is necessary for extension of the invading MAT DNA strand on the homologous HMLα donor site by new DNA polymerization (Fig. 5). Notably, in the rsc2Δ mutant, extension of the invading strand occurred with wild-type kinetics (Fig. 5C). As predicted, snf5Δ mutants failed to extend invading strands, as these mutants failed to establish synapsis and in this regard, were indistinguishable from rad54Δ mutants (Fig. 5C). We also found that both RSC and Swi/Snf were recruited to the homologous donor sequence by 40 min and 1 h, respectively, preceding the appearance of the primer extension product. Binding of both RSC and Swi/Snf remained high for ∼1 h and then decreased to nearly baseline levels once primer extension products appeared (Fig. 5, cf. D,E,G and C). These data are consistent with roles for both remodelers in recombinational repair, and further suggest that Swi/Snf might clear nucleosomes on the HML donor in order to expose nucleosomal DNA to the homology-searching complex.
Figure 5.
rsc and swi/snf are defective at distinct steps of HR repair. (A) Schematic diagram of an HR intermediate during mating-type switching. Primers used to monitor DSB, strand invasion-extension, and completion of repair are indicated. (B,C,F) DSBs were induced in wild-type (BLY747), rsc2Δ (BLY784), snf5Δ (BLY786), and rad54Δ (BLY804) strains as described in Figure 4. Genomic DNA was isolated at the indicated time points and monitored by quantitative real-time PCR analysis for efficiency of DSB formation (B), primer extension (C), and completion of repair (F) (see Supplemental Material for details). Primer extension and completion of repair were arbitrarily set at 100% for the highest wild-type level. All values were normalized to an ACT1 internal control. The values are the averages of three independent experiments except for experiments carried out in snf5Δ and rad54Δ strains, which were each done twice. Error bars indicate one standard deviation. (D,E,G) Chromatin was also isolated from wild-type (BLY747) strains at the indicated time points and immunoprecipitated with anti-Snf5p, anti-Snf2p, or anti-Sth1p antibodies, followed by quantitative real-time PCR analysis as described in Figure 3.
Following strand invasion and resolution of the Holliday intermediates, DNA ligation completes the recombinational repair of the DNA DSB. Importantly, in rsc2Δ mutants, this latter post-synaptic DNA repair step was defective, achieving only 20% of wild-type cells, suggesting that RSC plays a role in HR repair at a step(s) following the initiation of new DNA synthesis by the invading 3′ single-stranded tail. snf5Δ mutants were entirely defective in post-synaptic repair since these cells could not perform strand invasion (Fig. 4F). Thus, RSC and Swi/Snf are both required for HR repair of DSBs, but function at different stages: Swi/Snf is required at or just preceding strand invasion, while RSC is required following extension of the invading strand.
Although we found that rsc2Δ mutants were defective in a post-synaptic step of HR, RSC's rapid association with both sides of a DSB site suggests that RSC also plays an early role during HR. Interestingly, RSC and the Mre11/Rad50/Xrs2 complex, ATM-related Tel1p kinase, and ATR-related Mec1p kinase are recruited to DSB sites with similar kinetics (this study; Lisby et al. 2004), raising the possibility that an early MRX-, Tel1p-, or Mec1p-mediated modification of chromatin is a prerequisite for RSC's association with DSBs; a comparable chromatin modification could be responsible for Swi/Snf's later recruitment. Recent studies implicating RSC in the cohesion of sister chromatids (Baetz et al. 2004; Huang et al. 2004) and those demonstrating a requirement of cohesion for DSB repair (Strom et al. 2004; Unal et al. 2004) suggest that RSC might be necessary for DNA damage-induced cohesion. One model to explain RSC's later role in completing HR repair, which is supported by its association with HMLα donor sites, is that RSC's remodeling activity is required for the post-synapsis dissociation of the invading DNA from donor DNA prior to ligation.
Interestingly, rsc30 and rsc8 were recently identified in a genetic screen for mutants that are defective in NHEJ (Shim et al. 2005). Therefore, RSC appears to play a broad role in the repair of DSBs, facilitating repair by both HR and NHEJ; individual subunits might differentially contribute to HR or NHEJ, or distinct RSC isoforms might be involved in distinct repair pathways. Future experiments will elucidate the mechanism(s) by which RSC is directed to repair DSBs by HR or NHEJ.
Swi/Snf binds to a DSB site later than RSC at about the time that Rad52p and Rad54p HR repair enzymes bind (Sugawara et al. 2003; Wolner et al. 2003; Miyazaki et al. 2004) and is required to achieve synapsis between MAT and HMLα loci. Therefore, the very modest swi/snf mutant sensitivity to MMS and insensitivity to GAL-HO expression (Supplementary Fig. 4) were surprising; one possible explanation is that the relative balance of DSB repair is upset in swi/snf mutants to favor end joining.
Both Swi/Snf and RSC also function in transcription. Although swi/snf and rsc mutants do not affect expression of DNA repair enzymes under the conditions assayed (Sudarsanam et al. 2000; Angus-Hill et al. 2001), Swi/Snf is required for the DNA damage transcriptional response (DUN response) (see Sharma et al. 2003). Thus, while the results presented here in both nonswitching and switching strains strongly suggest that Swi/Snf and RSC participate directly in early and late steps in HR repair, global changes in transcription factor or repair factor levels mediated by these remodeling complexes likely contribute at least partly to the overall efficiency of DSB repair.
The requirement of two distinct ATP-dependent remodelers in HR repair emphasizes the crucial role of chromatin remodelers in repairing DNA in the context of chromatin. In addition, the separate roles of RSC and Swi/Snf demonstrate the specificity with which two ATP-dependent chromatin remodelers can function in the same process, providing new insight into both the mechanism of action of chromatin-remodeling complexes in DNA damage repair and the cellular coordination necessary to maintain genome integrity.
Materials and methods
Strains, media, and plasmids
S. cerevisiae strains used in the study are listed in Supplementary Table 1. Mutant and plasmid construction and additional methods are described in the Supplemental Material.
Plasmid-based DNA end-joining and HR assays
In vivo plasmid-based end-joining assays and in vivo plasmid-based HR/SSA assays were performed as previously described (Erdemir et al. 2002).
Chromatin immunoprecipitation
Asynchronous cultures were grown overnight at 30°C in YEP media containing 2% raffinose. When cultures reached mid-logarithmic phase, expression of the HO endonuclease was induced by the addition of 2% galactose. Chromatin was prepared at the indicated time points as previously described (Geng and Laurent 2004) and immunoprecipitated with anti-Sth1, anti-Snf5, anti-Snf2, anti-myc, affinity-purified anti-Rad51 (generously provided by L. Symington, Columbia University, New York, NY), or anti-Rad52 (Santa Cruz Biotechnology, Inc.) antibodies. The levels of immunoprecipitated DNAs were measured by quantitative real-time PCR as described (Geng and Laurent 2004). The relative enrichment was determined by the fold increase of ChIPed DNA relative to that at time 0. IP efficiencies of MAT Z and HMLα sequences using anti-Rad51 and anti-Rad52 antibodies are shown relative to those of an ACT1 promoter sequence. For each time course, HO-induced DSBs were verified by quantitative real-time PCR of genomic DNA. Primers used for DSB analyses and ChIPs are listed in Supplementary Table 2.
Acknowledgments
We thank Jing-mei Hsu for integrating the sfh1-219 mutation and for critical input throughout this project; Jim Haber, Rodney Rothstein, Stephane Marcand, Lorraine Symington, and Ugur Yavuzer for strains or plasmids; and the Symington laboratory for generously providing affinity-purified anti-Rad51 antibody. Rodney Rothstein is kindly acknowledged for very useful criticism of the manuscript. We also thank Sang Eun Lee for helpful discussion. B.L. dedicates this manuscript to the memory of his father, John Laurent. This work was supported by a grant from the National Institutes of Health (GM56700).
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1273105.
References
- Alexeev A., Mazin, A., and Kowalczykowski, S.C. 2003. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 10: 182–185. [DOI] [PubMed] [Google Scholar]
- Alexiadis V. and Kadonaga, J.T. 2002. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes & Dev. 16: 2767–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus-Hill M.L., Schlichter, A., Roberts, D., Erdjument-Bromage, H., Tempst, P., and Cairns, B.R. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7: 741–751. [DOI] [PubMed] [Google Scholar]
- Baetz K.K., Krogan, N.J., Emili, A., Greenblatt, J., and Hieter, P. 2004. The ctf13-30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol. Cell. Biol. 24: 1232–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.B., Lewis, L.K., Karthikeyan, G., Lobachev, K.S., Jin, Y.H., Sterling, J.F., Snipe, J.R., and Resnick, M.A. 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29: 426–434. [DOI] [PubMed] [Google Scholar]
- Bird A.W., Yu, D.Y., Pray-Grant, M.G., Qiu, Q., Harmon, K.E., Megee, P.C., Grant, P.A., Smith, M.M., and Christman, M.F. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415. [DOI] [PubMed] [Google Scholar]
- Boulton S.J. and Jackson, S.P. 1996. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15: 5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Critchlow S.E. and Jackson, S.P. 1998. DNA end-joining: From yeast to man. Trends Biochem. Sci. 23: 394–398. [DOI] [PubMed] [Google Scholar]
- Downs J.A., Allard, S., Jobin-Robitaille, O., Javaheri, A., Auger, A., Bouchard, N., Kron, S.J., Jackson, S.P., and Cote, J. 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell. 16: 979–990. [DOI] [PubMed] [Google Scholar]
- Erdemir T., Bilican, B., Cagatay, T., Goding, C.R., and Yavuzer, U. 2002. Saccharomyces cerevisiae C1D is implicated in both non-homologous DNA end joining and homologous recombination. Mol. Microbiol. 46: 947–957. [DOI] [PubMed] [Google Scholar]
- Geng F. and Laurent, B.C. 2004. Roles of SWI/SNF and HATs throughout the dynamic transcription of a yeast glucose-repressible gene. EMBO J. 23: 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara R. and Sancar, A. 2002. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol. Cell. Biol. 22: 6779–6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.M., Huang, J., Meluh, P.B., and Laurent, B.C. 2003. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol. Cell. Biol. 23: 3202–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Hsu, J.M., and Laurent, B.C. 2004. The RSC nucleosome-remodeling complex is required for cohesin's association with chromosome arms. Mol. Cell 13: 739–750. [DOI] [PubMed] [Google Scholar]
- Johnson R.D. and Jasin, M. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19: 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Barlow, J.H., Burgess, R.C., and Rothstein, R. 2004. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713. [DOI] [PubMed] [Google Scholar]
- Miyazaki T., Bressan, D.A., Shinohara, M., Haber, J.E., and Shinohara, A. 2004. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 23: 939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Shen, X., Landry, J., Wu, W.H., Sen, S., and Wu, C. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348. [DOI] [PubMed] [Google Scholar]
- Morrison A.J., Highland, J., Krogan, N.J., Arbel-Eden, A., Greenblatt, J.F., Haber, J.E., and Shen, X. 2004. INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119: 767–775. [DOI] [PubMed] [Google Scholar]
- New J.H., Sugiyama, T., Zaitseva, E., and Kowalczykowski, S.C. 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391: 407–410. [DOI] [PubMed] [Google Scholar]
- Paques F. and Haber, J.E. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C.L. and Cote, J. 2004. Cellular machineries for chromosomal DNA repair. Genes & Dev. 18: 602–616. [DOI] [PubMed] [Google Scholar]
- Pierce A.J., Stark, J.M., Araujo, F.D., Moynahan, M.E., Berwick, M., and Jasin, M. 2001. Double-strand breaks and tumorigenesis. Trends Cell Biol. 11: S52–S59. [DOI] [PubMed] [Google Scholar]
- Qin S. and Parthun, M.R. 2002. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 22: 8353–8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V.M., Li, B., and Reese, J.C. 2003. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery. Genes & Dev. 17: 502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim E.Y., Ma, J.-L., Oum, J.H., Yanex, Y., and Lee, S.E. 2005. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks.Mol. Cell. Biol. 25: 3934–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A. and Ogawa, T. 1998. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391: 404–407. [DOI] [PubMed] [Google Scholar]
- Stewart G.S., Maser, R.S., Stankovic, T., Bressan, D.A., Kaplan, M.I., Jaspers, N.G., Raams, A., Byrd, P.J., Petrini, J.H., and Taylor, A.M. 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99: 577–587. [DOI] [PubMed] [Google Scholar]
- Strom L., Lindroos, H.B., Shirahige, K., and Sjogren, C. 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16: 1003–1015. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P., Iyer, V.R., Brown, P.O., and Winston, F. 2000. Whole-genome expresion analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 97: 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Wang, X., and Haber, J.E. 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12: 209–219. [DOI] [PubMed] [Google Scholar]
- Sung P. 1997. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272: 28194–28197. [DOI] [PubMed] [Google Scholar]
- Sung P., Trujillo, K.M., and Van Komen, S. 2000. Recombination factors of Saccharomyces cerevisiae. Mutat. Res. 451: 257–275. [DOI] [PubMed] [Google Scholar]
- Symington L.S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya E., Hosotani, T., and Miyakawa, T. 1998. A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res. 26: 3286–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal E., Arbel-Eden, A., Sattler, U., Shroff, R., Lichten, M., Haber, J.E., and Koshland, D. 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16: 991–1002. [DOI] [PubMed] [Google Scholar]
- Valerie K. and Povirk, L.F. 2003. Regulation and mechanisms of mammalian double-strand break repair. Oncogene 22: 5792–5812. [DOI] [PubMed] [Google Scholar]
- van Attikum H., Fritsch, O., Hohn, B., and Gasser, S.M. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119: 777–788. [DOI] [PubMed] [Google Scholar]
- Wolner B. and Peterson, C.L. 2005. ATP-dependent and ATP-independent roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J. Biol. Chem. 280: 10855–10860. [DOI] [PubMed] [Google Scholar]
- Wolner B., van Komen, S., Sung, P., and Peterson, C.L. 2003. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell 12: 221–232. [DOI] [PubMed] [Google Scholar]