Abstract
The intestinal epithelium comprises differentiated cells of four lineages maintained by precursor cells. As the Notch pathway controls the fate of proliferating cells in many systems, we investigated the effect of conditional expression of an activated Notch mutant in intestinal epithelium. An increase in the number of goblet cells occurs within 8 h of induction, due to an effect of Notch on post-mitotic cells, not on precursors. This observation broadens the role of Notch into controlling postmitotic differentiation and indicates that the composition of the epithelium is not solely determined by progenitor cells.
Keywords: Goblet cell, Hes5, Hes1
The epithelium of the small intestine comprises postmitotic differentiated cells arrayed on villi and maintained by proliferative stem cells in the crypts (Radtke and Clevers 2005). The majority of the epithelium is composed of enterocytes, which have an absorptive function. Goblet cells are found in the crypts and the villi. The rarer endocrine cells are dispersed over the villi, while Paneth cells are confined to the base of the crypts (Fig. 1A). Cells constantly migrate from the crypts onto the villi, and then to the villus tip, where they are shed into the gut lumen. The turnover time of the villus epithelium is 2–3 d.
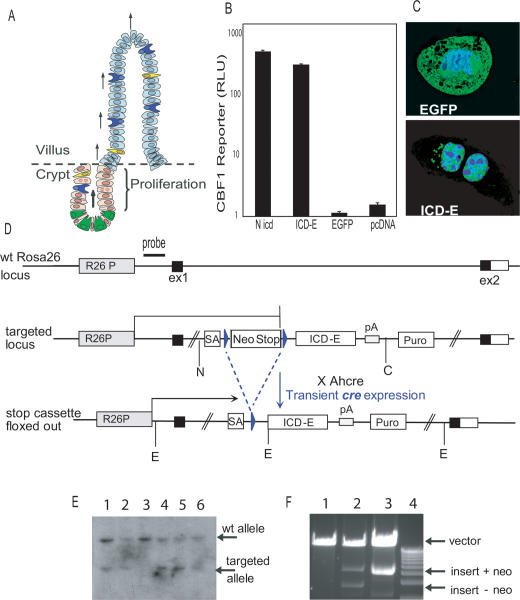
Figure 1.
Construct design, validation, and targeting. (A) Structure of the intestinal epithelium; the cell types shown are enterocytes (pale blue), goblet cells (dark blue), endocrine cells (yellow), Paneth cells (green), and progenitors (pink); arrows indicate the migration of cells from the crypts to the villi. (B) In vitro validation of construct. cDNA encoding mouse Notch 1 cytoplasmic domain (Nicd), ICD-E, EGFP, or empty vector was transfected into CHO cells with a CBF1 DNA-binding motif firefly luciferase reporter vector, and a Renilla luciferase control (Strobl et al. 1997). Error bars indicate SEM. (C) Confocal microscopy showing localization of EGFP and ICD-E (green) in transfected 3T3 cells also stained with DAPI for DNA (blue). (D) Conditional targeting of ICD-E to the Rosa 26 locus. Upstream of the ICD-E fusion a PGK neo pA sequence served as a Stop cassette. This was flanked by LoxP sequences (blue triangles). A splice acceptor (SA) was introduced in front of the floxed Stop cassette to allow normal processing of ICD-E RNA following excision of the Stop cassette by transiently expressed cre from the Ahcre line. The construct was cloned into pRos26-MCS-13 for targeting to the ROSA26 locus. (E) Southern blot analysis of ES cell clones confirmed the targeting of the construct to the ROSA26 locus. The location of the probe used in the Southern blot analysis is shown in D. The upper band corresponds to the EcoRI (E) fragment of the wild-type allele and the lower band to the smaller EcoRI fragment of the targeted allele shown in D. Lanes 1–6 represent ES cell lines CA2, CA3, CB3, CC3, CD1, and CH1, respectively. Mice derived from lines CA2 and CD1 were selected and used in this study; the described phenotype was seen in both lines. (F) In vitro cre-mediated recombination of the targeting construct. The plasmids shown were incubated with or without cre recombinase in vitro, and analyzed after a restriction digest to release the insert from pRosa26-MCS-13. (Lane 1) Linearized pRosa26-MCS-13, no cre incubation. (Lane 2) pRosa26-MCS-13 + insert incubated with cre and subsequently digested with NheI and ClaI (sites N and C, respectively, in D). (Lane 3) pRosa26-MCS-13 + insert digested with NheI and ClaI, no cre incubation. (Lane 4) One-kilobase ladder.
The composition of the villus epithelium is thought to be determined by regulation of crypt progenitor cells. Enterocytes derive from an enterocyte precursor cell, while goblet cells, endocrine cells, and Paneth cells are thought to derive from a common secretory precursor cell. The specification of precursors into enterocyte fate is determined by the transcription factor Hes1, while the differentiation of secretory precursors into goblet or endocrine fate is regulated by Math1 and neurogenin 3 (ngn3) (Jensen et al. 2000; Yang et al. 2001; Jenny et al. 2002; Schonhoff et al. 2004).
Hes1 is a target of the Notch signaling pathway, highlighting Notch as a candidate regulator of intestinal progenitor cell fate. Notch regulates the cell fate decisions of proliferating cells in development and in adult life (Kadesch 2004). Signaling occurs when the transmembrane Notch receptor is bound by ligands expressed on adjacent cells. The intracellular cytoplasmic domain of the receptor (ICD) is cleaved from the transmembrane domain by γ-secretase and translocates to the nucleus, where it binds the transcription factor CBF1 (RBPJ-κ), leading to the recruitment of transcriptional coactivators and expression of Notch-responsive genes such as Hes1 and Hes5 (Jarriault et al. 1995; Furriols and Bray 2001).
In the intestinal epithelium, mRNA encoding Notch ligands, receptors, and the Notch target genes Hes1 and Hes5 is expressed in proliferating crypt cells. Hes5 transcript is also detected in post-mitotic cells in the lower villi (Schroder and Gossler 2002). Zebrafish mutants in which Notch signaling is believed to be disrupted show an increase in the number of goblet cells in the gut epithelium, suggesting that Notch may regulate the fate of cryptal progenitor cells (Crosnier et al. 2005). Here we investigated the effects of conditional expression of an activated Notch mutant on the crypt–villus axis.
Results and Discussion
A mutant of mouse Notch 1 consisting of the cytoplasmic domain fused to EGFP (ICD-E) was constructed and shown to have equivalent activity to the untagged Notch cytoplasmic domain in vitro, in CHO cells (shown), and also in NIH3T3, Cos7, C2C12, and 293T cells (Fig. 1B; data not shown). Fluorescence microscopy showed that in all cell lines tested, ICD-E was localized to the nucleus, although in some cells cytoplasmic staining was also seen (Fig. 1C; data not shown). Western blotting of cells transfected with ICD-E using an anti-Notch-1 antibody confirmed the presence of ICD-E protein of the predicted size (data not shown). ICD-E was then conditionally targeted to the ubiquitously expressed Rosa 26 locus to generate heterozygous R26ICD-E/wt mice (Fig. 1D–F). These animals were crossed with the Ahcre transgenic mouse strain in which the cre recombinase under the control of the Cyp1A promoter is transiently expressed in the intestinal epithelium following intraperitoneal injection of the xenobiotic β-napthoflavone (βNF) (Ireland et al. 2004). The resulting heterozygous Ahcre/R26ICD-E/wt mice express ICD-E in the intestinal epithelium following cre-mediated recombination.
Ahcre/R26ICD-E/wt mice and Ahcre/R26wt/wt control littermates were treated with three doses of βNF at 8-h intervals. Quantitative RT–PCR of intestinal ICD-E mRNA indicated that ICD-E was transcribed within 6 h of the first dose of βNF, rising to a maximum at 16 h, and falling to control levels by 96 h (Fig. 2A). To assess the level of ICD-E mRNA compared with endogenous Notch1 mRNA, we carried out quantitative RT–PCR with two further primer sets (Fig. 2A). Primer set 2, which amplifies a region of native Notch1 mRNA 5′ to the ICD, showed no significant difference in the level of endogenous Notch between control and experimental mice over the time course. In contrast, primer set 3, which amplifies both ICD-E and endogenous RNA, showed a 5.4 ± 1.3 (SEM)-fold increase in total ICD mRNA between experimental and control animals at 24 h (data not shown), suggesting that the level of ICD-E mRNA is about fivefold higher than that of endogenous Notch1.
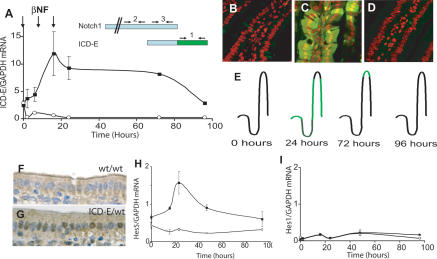
Figure 2.
Inducible expression of ICD-E in adult mouse intestinal epithelium. (A) Ahcre/R26ICDEfl/wt mice and Ahcre/R26wt/wt control mice were injected with βNF (arrows) and sacrificed at the time points shown. Real-time RT–PCR was used to detect ICD-E mRNA in intestinal mRNA. (Solid squares) Ahcre/R26ICD-E/wt mice; (open circles) Ahcre/R26wt/wt controls. Error bars indicate standard error of the mean in two experiments performed in triplicate. (Inset) Location of PCR primers used to amplify ICD-E alone (1), endogenous Notch alone (2), or both RNAs (3). (B–E) Confocal microscopy of the jejunal villi of Ahcre/R26ICD-E/wt mice for ICD-E (green); nuclei were stained with Topro III (red). (B) Uninduced. (C) Twenty-four hour. (D) Seventy-two hour. (E) Summary of location of recombinant epithelium (green) following recombination. (F,G) Immunohistochemical staining for Notch1 ICD at 24 h post-induction. (F) Ahcre/R26wt/wt. (G) Ahcre/R26ICD-E/wt. (H,I) Quantitative PCR analysis of Hes5 (H) and Hes1 (I) mRNA normalized to GAPDH. (Solid squares) Ahcre/R26ICD-E/wt mice; (open circles) Ahcre/R26wt/wt controls. Error bars indicate standard error of the mean in two experiments performed in triplicate.
ICD-E protein expression was confirmed using confocal microscopy (Fig. 2B–E). By 24 h post-induction, green fluorescence was visible in the villi, where nuclear and cytoplasmic localization of ICD-E is seen, consistent with the distribution in cells transfected with ICD-E in vitro (Fig. 2C; data not shown). In contrast, no fluorescence was detected in uninduced Ahcre/R26ICD-E/wt mice (Fig. 2B). Consistent with the loss of ICD-E transcript, fluorescence was confined to the upper villi at 72 h and was absent by 96 h (Fig. 2D,E; data not shown). Faint fluorescence, just above the limit of detection, was seen in the crypts at 24 h, and was lost at later time points (data not shown). Finally, we carried out immunohistochemistry with an antibody that recognizes the N terminus of the Notch 1 cytoplasmic domain. At 24 h, numerous cells with nuclear Notch staining are seen on the villi of the Ahcre/R26ICD-E/wt animals, but these are not seen in the induced Ahcre/R26wt/wt controls (Fig. 2F,G). Thus, ICD-E protein is expressed in the villus post-recombination.
To confirm that ICD-E was functional, we analyzed expression of the Notch target genes Hes1 and Hes5, which require Notch activation for transcription. Hes5 mRNA rose in parallel with ICD-E expression in Ahcre/R26ICD-E/wt mice; Hes1 mRNA was not significantly induced at any time point (Fig. 2H,I). The induction of Hes5 but not Hes1 presumably reflects the fact that these genes are regulated by additional transcription factors as well as by Notch and consequently have overlapping but distinct patterns of expression in the gut and other tissues (Schroder and Gossler 2002; Hatakeyama et al. 2004). At 24 h, the highest expression of ICD-E is in the villi, which normally express Hes5 but not Hes1 (Fig. 2C; Schroder and Gossler 2002).
We then analyzed the phenotype of cells in the recombinant epithelium. Goblet cell numbers were assessed by immunohistochemistry for the goblet-cell-specific marker Muc2 and also by alcian blue and PAS staining, both of which detect mucins. Increased numbers of goblet cells were found on the villi at 8 h post-induction in experimental Ahcre/R26ICD-E/wt mice compared with induced control Ahcre/R26wt/wt animals at the same time point (Fig. 3A–C; data not shown). The maximum increase in goblet cell numbers occurred at 24 h, but by 48 h villus goblet cell numbers had fallen to close to control levels, consistent with the loss of recombinant epithelium. There was only a small change in goblet cell numbers in the crypts at any time point (Fig. 3A). Quantitative PCR for muc2 mRNA showed that transcription rose to a peak at 16 h and decreased to control levels by 72 h, in Ahcre/R26ICD-E/wt mice but not in Ahcre/R26wt/wt control animals.
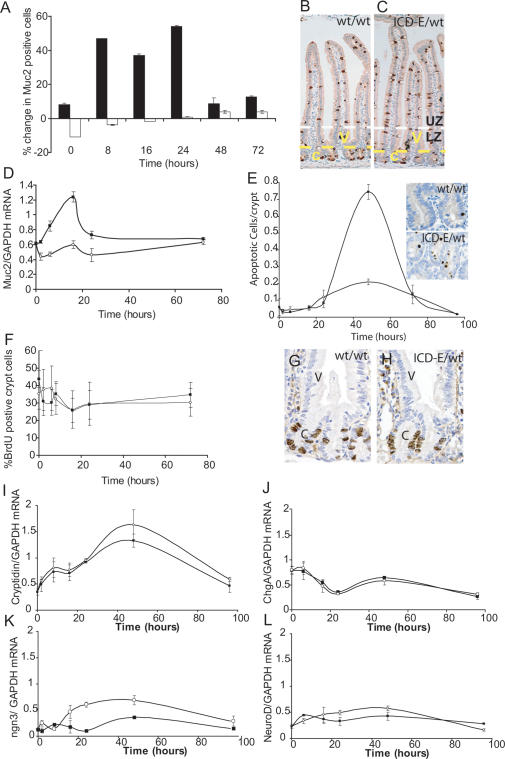
Figure 3.
Phenotype of recombinant epithelium. (A) Immunohistochemical analysis of goblet cell numbers. Values shown are the number of Muc2-positive cells in the crypt (open bars) and the villi (solid bars) in Ahcre/R26ICD-E/wt mice, expressed as a percentage of the corresponding value in Ahcre/R26wt/wt control animals at the same time point. Cell counts of at least 500 cells (crypts) and 3000 cells (villi) were performed on duplicate mice at each time point. (B,C) Appearance of intestine stained for Muc2 (brown) 24 h post-induction. (B) Ahcre/R26wt/wt; (C) Ahcre/R26ICD-E/wt. (c) Crypts; (V) villi. The yellow dotted line is the crypt/villus boundary, and the white dotted line indicates the boundary between the labeled zone (LZ) and unlabeled zone (UZ) (see text and cf. Fig, 4C). (D) Quantitative RT–PCR for Muc2 mRNA following induction. (Solid squares) Ahcre/R26ICD-E/wt mice; (open circles) Ahcre/R26wt/wt controls. (E) Number of Caspase-3-positive apoptotic cells per crypt following recombination. (Solid squares) Ahcre/R26ICD-E/wt mice; (open circles) Ahcre/R26wt/wt controls. Insets show typical crypts in induced Ahcre/R26wt/wt and Ahcre/R26ICD-E/wt animals at 48 h. (F) Percentage of S-phase cells in intestinal crypts following induction assessed by 1 h of pulse BrdU labeling. (Solid squares) Ahcre/R26ICD-E/wt mice; (open circles) Ahcre/R26wt/wt controls. (G,H) Location of S-phase cells 24 h post-induction. (c) Crypts; (V) villi. (I–L) Quantitative RT–PCR for Cryptidin (I), Chromogranin A (J), ngn3 (K), and NeuroD (L).
The increase in goblet cell numbers in the villus compartment following Notch activation is surprisingly rapid. There are five possible explanations for this phenotype:
-
Notch activation induces selective apoptosis of enterocytes resulting in an apparent increase in the number of goblet cells. However, there was no change in the frequency of crypt apoptosis before 24 h, and no apoptosis was detected in the villi at any time. Thus, apoptosis cannot explain the increased frequency of villus goblet cells.
In contrast, at 48 h, a marked increase in apoptosis was observed in the crypts (Fig. 3E). This is consistent with the progressive loss of fluorescent epithelium described above. Following death of recombinant crypt precursors, the fluorescent epithelium is no longer replaced, but migrates to the villus tip, where it is shed; by 72 h fluorescence was confined to the tips of the villi, and by 96 h it was lost completely (Fig. 2B–E). This is not due to Ahcre-induced recombination being restricted to short-lived cryptal progenitors, as in Ahcre/R26EYFP/wt animals, in which EYFP, rather than ICD-E, is expressed following induction, the recombinant epithelium persists for at least 2 mo (Ireland et al. 2004, 2005; data not shown). Furthermore, the persistence of EYFP recombinant epithelium suggests that the apoptosis in ICD-E mice is not due to toxicity following the expression of the EGFP tag, as EYFP and EGFP are highly similar, but is a consequence of Notch activation. The small increase in apoptosis in control animals at 48 h may be due to toxicity from cre recombinase (Loonstra et al. 2001).
Notch activation respecifies the location of the crypt/villus boundary and alters the normal distribution of proliferating goblet cell precursors. To investigate this, S-phase cells were labeled with a single BrdU injection 1 h before mice were sacrificed. No difference was found in the frequency of proliferating cells between ICD-E and control animals (Fig. 3F). More significantly, there was no alteration in the spatial distribution of proliferating cells, indicating that ICD-E has no effect on the upper boundary of the proliferative compartment (Fig. 3G,H; data not shown).
Notch activation respecifies the differentiation of secretory precursor cells from endocrine and Paneth cell fate into goblet cell fate. Quantitative RT–PCR for mRNA of the Paneth cell marker cryptidin (Defcr-rs1) revealed no difference between experimental and control animals (Fig. 3I), and immunohistochemistry revealed no decrease in the number of crypt cells expressing the Paneth cell marker lysozyme at 8, 24, 72, or 96 h post-induction (data not shown). Expression of the endocrine cell marker Chromogranin A did not decrease following induction in experimental animals compared with controls as assayed by quantitative RT–PCR (Fig. 3J), and there was no decrease in the number of synaptophysin-positive cells assessed by immunohistochemistry at 8, 24, 72, or 96 h (data not shown). Finally, we examined expression of neurogenin3 and NeuroD, which regulate the differentiation of endocrine precursor cells. There was a decrease in the level of ngn3 following induction in experimental animals compared with controls, but this does not translate into a change in the number of endocrine cells (Fig. 3K). NeuroD expression was unchanged by Notch activation (Fig. 3L).
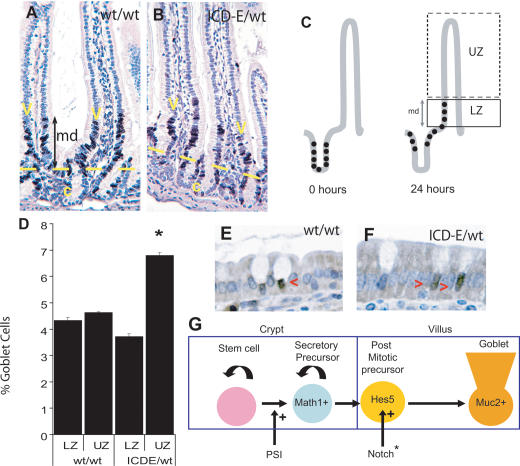
Notch activation may promote rapid migration of goblet cells and/or their precursors from the crypt. We investigated this by treating animals with BrdU at the time of the first βNF injection (0 h) and analyzing the distribution of BrdU-labeled cells 24 h later. BrdU labels the proliferating cells of the crypt, which subsequently migrate onto the villi as they differentiate; the leading edge of BrdU labeling marks the furthest extent of migration of progeny of precursor cells that have divided since ICD-E expression was induced. The position of the leading edge did not vary between control and experimental mice at 24 h (Fig. 4A,B), with the highest point on the villi reached by BrdU-labeled cells (the migration distance [md] being 12.6 ± 3.5 cell positions [mean ± standard deviation]) above the crypts in Ahcre/R26ICD-E/wt mice and 11.0 ± 4.0 in Ahcre/R26wt/wt animals. Thus, differential migration cannot explain the increase in goblet cell numbers at or before 24 h.
A final possibility is that Notch signaling promotes the differentiation of post-mitotic precursor cells. We analyzed goblet cell frequency in the BrdU-labeled zone (LZ) (Fig. 4C), which contains the progeny of proliferative precursors that have differentiated and migrated on to the villus during the 24 h following induction. No increase in goblet cells was found in the LZ (Fig. 4D). In contrast, cells in the upper, unlabeled zone (UZ) (Fig. 4C), which had exited the cell cycle before expression of ICD-E, show an increased frequency of goblet cells that accounts for all the increase seen in Ahcre/R26ICD-E/wt mice (Fig. 4D). Thus, Notch activation increases goblet cell numbers by driving differentiation of post-mitotic cells into mature goblet cells.
Figure 4.
Notch acts on post-mitotic epithelium. (A,B) BrdU cell migration assays; dark-brown stain indicates BrdU labeling. (A) Ahcre/R26wt/wt. (B) Ahcre/R26ICD-E/wt. (md) Migration distance. (C) Migration assays. BrdU is injected at 0 h, labeling proliferating crypt precursor cells (solid dots); after 24 h these cells and their progeny migrate onto the lower villus, and are detected by immunostaining. (LZ) Labeled zone; (UZ) unlabeled zone. (D) The percentage of Muc2-positive goblet cells in LZ and UZ in Ahcre/R26wt/wt and Ahcre/R26ICD-E/wt mice at 24 h post-induction, based on counts of at least 4000 villus cells in duplicate mice. (*) p = 0.01 by two-tailed t-test compared with LZ in Ahcre/R26ICD-E/wt mice. A typical experiment is shown; error bars indicate standard error of the mean. (E–G) Hes5 immunostaining at 8 h post-induction. (E) Ahcre/R26wt/wt. (F) Ahcre/R26ICD-E/wt. Red arrowheads indicate Hes5-positive cells. (G) Summary of the roles of Notch in the goblet cell lineage: (PSI) presenilin inhibitor; (Notch*) activated Notch.
We investigated whether the phenotype seen could be linked with Notch activation. A key regulator of recruitment of crypt precursor cells to the goblet cell lineage is Math1, loss of which results in depletion of goblet cells (Yang et al. 2001). Math1 mRNA levels measured by quantitative RT–PCR at 2, 6, 16, 24, 48, and 96 h following recombination in induced Ahcre/R26ICD-E/wt and Ahcre/R26wt/wt mice are not significantly different in experimental and control mice at any time point (data not shown). Math1 transcription is inhibited by Hes1, so this result is consistent with the lack of induction of Hes1 by ICD-E, and indicates that ICD-E acts independently of Math1 to increase goblet cell numbers. The absence of induction of Hes1 may reflect the low levels of ICD-E expressed in the crypt cells within 24 h of induction (data not shown).
ICD-E was found to induce Hes5 mRNA (Fig. 2H), prompting us to investigate Hes5 expression in experimental and control mice by immunostaining. Nuclear staining of occasional crypt and villus goblet cells was seen in induced control Ahcre/R26wt/wt mice. At 8 h increased numbers of cells exhibiting nuclear staining were seen on the villi of Ahcre/R26ICD-E/wt animals (Fig. 4E,F). There was no significant difference in the percentage of Hes5-positive cells in the lowest 20 cell positions in the villi, corresponding to the LZ in Figure 4C at 8 h (8.4% in both Ahcre/R26ICD-E/wt and Ahcre/R26wt/wt), but there was a substantial increase in the UZ (3.8% of UZ cells in Ahcre/R26ICD-E/wt compared with 0.4% in Ahcre/R26wt/wt; p < 0.0001). Thus the size of the increase and distribution of Hes5-positive cells in the UZ is consistent with ICD-E-induced Notch activation promoting differentiation of post-mitotic Hes5-positive cells on the villi into the goblet cell lineage (Fig. 4G). In contrast, there was no difference between experimental and control animals in their frequency or distribution of Hes1-positive cells detected by immunostaining at 24 h post-induction (data not shown).
These observations also explain the apparent contradiction between ICD-E driving goblet cell differentiation and the results of treatment of rats with presenilin inhibitors (PSI), which block Notch signaling by preventing the cleavage of Notch to release the ICD (Milano et al. 2004; Wong et al. 2004). Two benzodiazepine PSIs cause crypt cell apoptosis within 24 h, followed by an increase in goblet cell numbers in both crypts and villi. Both the agents that affected goblet cell numbers increased levels of Rat atonal 1 (Rath1) mRNA two- to fivefold within 24 h; this and the increase in cryptal goblet cells indicate that PSIs act on cryptal precursor cells (Fig. 4G; Milano et al. 2004). Furthermore, mice that lack Hes1 exhibit both increased numbers of intestinal goblet and endocrine cells, indicating that Hes1 promotes enterocyte differentiation (Jensen et al. 2000). Significantly, the levels of Hes5 mRNA are also increased in Hes1-null mice, consistent with the expression of Figure 4E and F (Jensen et al. 2000). Taken together, the available data suggest that Notch acts in opposing ways at two points in goblet cell development, consistent with other lineages in which Notch plays opposing roles at different stages of differentiation (Morrison et al. 2000; Tanigaki et al. 2001; Robey and Bluestone 2004).
Notch signaling is commonly associated with maintaining cells in a proliferative state, although in proliferating murine keratinocytes, Notch activation induces p21Cip1, leading to cell cycle exit (Rangarajan et al. 2001). Notch signaling regulates dendrite branching in post-mitotic neurons, but has not been reported to regulate the differentiation of post-mitotic cells (Sestan et al. 1999). These results demonstrate that Notch signaling regulates the differentiation of post-mitotic cells and indicate that the composition of the villus epithelium is not solely determined by crypt progenitors.
Materials and methods
Construct design and embryonic stem cell targeting
The EGFP coding sequence was fused to the C-terminal end of the mouse Notch1 intracellular domain to generate ICD-E. A LoxP-sequence-flanked stop cassette was then introduced upstream of the ICD-E Kozak/ATG sequence. The whole transgene was cloned into the pRos26-MCS-13 vector (gift from A. Berns, NKI, Amsterdam) (Vooijs et al. 2001) for targeting to the ROSA26 locus in 129/SV embryonic stem cells. Targeted cell clones were determined by Southern blot.
Generation of Ahcre/R26ICD-E/wt mice
Homozygous mice from the transgenic line Ahcre (Ireland et al. 2004) were crossed to heterozygous R26ICD-E/wt mice to generate heterozygous Ahcre/R26ICD-E/wt mice. Expression of cre recombinase was induced at 2 mo of age by 3× intraperitoneal injections at 0, 8, and 16 h of βNF (80 mg/kg) dissolved in corn oil at 8 mg/mL.
Plasmids and transient transfections
pEGFP, ICD, and ICD-E cDNAs were cloned into pCS2 and NIH 3T3, CHO, Cos7, C2C12, and 293T cells transiently transfected using Fugene (Roche). The Notch-responsive pGa981-6 reporter plasmid, containing 12 RBP-Jκ-binding sites upstream of a minimal β-globin promoter was a gift from Ursula Juste (GSF, Munich) (Strobl et al. 1997). pRL-TK was used as a transfection efficiency control. Firefly and Renilla luciferase activity was measured with a Dual luciferase kit (Promega). All results were normalized to Renilla luciferase activity.
Tissue sample preparation for immunohistochemistry and fluorescence
For immunohistochemistry, the small intestine was dissected and flushed with cold PBS and then fixed in formalin for 24 h and ethanol for 48 h at room temperature. The tissues were paraffin-embedded and sectioned (3 μm). Following dewaxing and rehydration, the sections were pretreated with peroxidase (3% H2O2 in 1× PBS) for 10 min at room temperature. Antigen retrieval was performed by pressure cooking in a sodium citrate buffer (10 mM at pH 6.0) for 3 min. Incubation of antibodies was performed in 3% BSA in 1× PBS for 1 h at room temperature for BrdU (Dako) and overnight at 4°C for Ki-67 (AbCam), Notch1 (Abcam), Hes1 (gift from T. Sudo, Toray Industries, Tokyo, Japan), Hes5 (Chemicon), Synaptophysin (Dako), Lysozyme (Dako), Muc-2 (a gift from Gunnar Hansson, Göteborg University, Gotenberg, Sweden), and Caspase-3 (R&D Systems). Incubation of the biotinylated secondary antibodies was performed at room temperature for 1 h. Strep ABC or EnVision kits (DAKO) were used to amplify the signal; staining was developed using DAB and counterstained with hematoxylin. Goblet cells were also stained histochemically with alcian blue and PAS. All goblet cell, lysozyme, and muc-2-positive cell counts were as a proportion of the total number of nucleated epithelial cells; only crypts or villi that were sagittally sectioned along their entire length were scored. Synaptophysin staining was scored by counting the number of cells/crypt/villus unit in at least 60 sagittally sectioned villi.
For fluorescence, the intestine was dissected and flushed with cold PBS, then 4% paraformaldehyde (PFA) containing a cocktail of protease inhibitors (Roche). The tissues were fixed in PFA for 24 h at 4°C followed by 48 h in 20% sucrose. The tissues were then frozen in liquid nitrogen and sectioned (8–12 μm). The sections were stained with TOPRO-3 (Molecular Probes) and viewed on a confocal microscope.
RT–PCR assays
For RT–PCR analysis, total mRNA was isolated from snap-frozen small intestine using TRIzol according to the manufacturer's instructions. cDNA was synthesized using Invitrogen reagents according to the manufacturer's instructions. Quantitative RT–PCR using Taqman probes (Applied BioSystems) or Sybr Green (for Cryptidin, Notch 1 ICD, Chromogranin A, Neurogenin, and NeuroD) was performed on a Corbett Research RotorGene PCR machine. Primer sequences are available on request.
Acknowledgments
We are very grateful for the assistance of Heather Ireland, Carol Houghton, and Louise Howard in this project. The work was funded by the Association for International Cancer Research, project grant 03-075, Cancer Research UK, and the Medical Research Council. P.J. holds a Cancer Research UK Senior Clinical Fellowship, C609/A3796.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.341705.
References
- Crosnier C., Vargesson, N., Gschmeissner, S., Ariza-McNaughton, L., Morrison, A., and Lewis, J. 2005. Delta–Notch signalling controls commitment to a secretory fate in the zebrafish intestine. Development 132: 1093–1104. [DOI] [PubMed] [Google Scholar]
- Furriols M. and Bray, S. 2001. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol. 11: 60–64. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J., Bessho, Y., Katoh, K., Ookawara, S., Fujioka, M., Guillemot, F., and Kageyama, R. 2004. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 131: 5539–5550. [DOI] [PubMed] [Google Scholar]
- Ireland H., Kemp, R., Houghton, C., Howard, L., Clarke, A.R., Sansom, O.J., and Winton, D.J. 2004. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: Effect of loss of β-catenin. Gastroenterology 126: 1236–1246. [DOI] [PubMed] [Google Scholar]
- Ireland H., Houghton, C., Howard, L., and Winton, D.J. 2005. Cellular inheritance of a Cre-activated reporter gene to determine Paneth Cell longevity in the murine small intestine. Dev. Dyn. (in press). [DOI] [PubMed]
- Jarriault S., Brou, C., Logeat, F., Schroeter, E.H., Kopan, R., and Israel, A. 1995. Signalling downstream of activated mammalian Notch. Nature 377: 355–358. [DOI] [PubMed] [Google Scholar]
- Jenny M., Uhl, C., Roche, C., Duluc, I., Guillermin, V., Guillemot, F., Jensen, J., Kedinger, M., and Gradwohl, G. 2002. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 21: 6338–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J., Pedersen, E.E., Galante, P., Hald, J., Heller, R.S., Ishibashi, M., Kageyama, R., Guillemot, F., Serup, P., and Madsen, O.D. 2000. Control of endodermal endocrine development by Hes-1. Nat. Genet. 24: 36–44. [DOI] [PubMed] [Google Scholar]
- Kadesch T. 2004. Notch signaling: The demise of elegant simplicity. Curr. Opin. Genet. Dev. 14: 506–512. [DOI] [PubMed] [Google Scholar]
- Loonstra A., Vooijs, M., Beverloo, H.B., Allak, B.A., van Drunen, E., Kanaar, R., Berns, A., and Jonkers, J. 2001. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. 98: 9209–9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano J., McKay, J., Dagenais, C., Foster-Brown, L., Pognan, F., Gadient, R., Jacobs, R.T., Zacco, A., Greenberg, B., and Ciaccio, P.J. 2004. Modulation of notch processing by γ-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol. Sci. 82: 341–358. [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Perez, S.E., Qiao, Z., Verdi, J.M., Hicks, C., Weinmaster, G., and Anderson, D.J. 2000. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell 101: 499–510. [DOI] [PubMed] [Google Scholar]
- Radtke F. and Clevers, H. 2005. Self-renewal and cancer of the gut: Two sides of a coin. Science 307: 1904–1909. [DOI] [PubMed] [Google Scholar]
- Rangarajan A., Talora, C., Okuyama, R., Nicolas, M., Mammucari, C., Oh, H., Aster, J.C., Krishna, S., Metzger, D., Chambon, P., et al. 2001. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20: 3427–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey E.A. and Bluestone, J.A. 2004. Notch signaling in lymphocyte development and function. Curr. Opin. Immunol. 16: 360–366. [DOI] [PubMed] [Google Scholar]
- Schonhoff S.E., Giel-Moloney, M., and Leiter, A.B. 2004. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev. Biol. 270: 443–454. [DOI] [PubMed] [Google Scholar]
- Schroder N. and Gossler, A. 2002. Expression of Notch pathway components in fetal and adult mouse small intestine. Gene Expr. Patterns 2: 247–250. [DOI] [PubMed] [Google Scholar]
- Sestan N., Artavanis-Tsakonas, S., and Rakic, P. 1999. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science 286: 741–746. [DOI] [PubMed] [Google Scholar]
- Strobl L.J., Hofelmayr, H., Stein, C., Marschall, G., Brielmeier, M., Laux, G., Bornkamm, G.W., and Zimber-Strobl, U. 1997. Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J κ. Immunobiology 198: 299–306. [DOI] [PubMed] [Google Scholar]
- Tanigaki K., Nogaki, F., Takahashi, J., Tashiro, K., Kurooka, H., and Honjo, T. 2001. Notch1 and Notch3 instructively restrict bFGF-responsive multipotent neural progenitor cells to an astroglial fate. Neuron 29: 45–55. [DOI] [PubMed] [Google Scholar]
- Vooijs M., Jonkers, J., and Berns, A. 2001. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2: 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G.T., Manfra, D., Poulet, F.M., Zhang, Q., Josien, H., Bara, T., Engstrom, L., Pinzon-Ortiz, M., Fine, J.S., Lee, H.J., et al. 2004. Chronic treatment with the γ-secretase inhibitor LY-411,575 inhibits β-amyloid peptide production and alters lymphopoiesis and intestinal cell differentiation. J. Biol. Chem. 279: 12876–12882. [DOI] [PubMed] [Google Scholar]
- Yang Q., Bermingham, N.A., Finegold, M.J., and Zoghbi, H.Y. 2001. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 294: 2155–2158. [DOI] [PubMed] [Google Scholar]