Abstract
Heterochromatin is critical for proper centromere and telomere function, and it plays a key role in the transcriptional silencing of specific genomic loci. In fission yeast, the Rik1 protein functions with the Clr4 histone methyltransferase at an early step in heterochromatin formation. Here, we use mass spectrometry and tandem affinity purification of a Rik1-TAP fusion protein to identify Rik1-associated proteins. These studies identify two novel proteins, Raf1 and Raf2, which we find are required for H3-K9 methylation and for transcriptional silencing within centromeric heterochromatin. We also find that subunits of a cullin-dependent E3 ubiquitin ligase are associated with Rik1 and Clr4, and Rik1-TAP preparations exhibit robust E3 ubiquitin ligase activity. Furthermore, expression of a dominant-negative allele of the Pcu4 cullin subunit disrupts regulation of K4 methylation within heterochromatin. These studies provide evidence for a novel Rik1-associated E3 ubiquitin ligase that is required for heterochromatin formation.
Keywords: heterochromatin, Rik1, Clr4, cullin 4, histone methylation
Select chromosomal domains of most eukaryotes are organized into specialized regions of heterochromatin that are conserved in composition and function. In the fission yeast Schizosaccharomyces pombe, heterochromatin plays critical roles in the prevention of chromosome missegregation (Bernard et al. 2001), in the control of conjugation and mating-type switching (Grewal and Moazed 2003), and in the structural packaging of telomeres (Nimmo et al. 1994). Recent studies highlight two genetic pathways that initiate formation and maintain stability of heterochromatin. Within heterochromatic regions, an RNAi-dependent event initiates heterochromatin formation (Volpe et al. 2003), while a histone H3-K9 methylation-dependent system maintains heterochromatin by facilitating the recruitment of HP1, a protein that recognizes this modification and structurally nucleates heterochromatin (Grewal and Elgin 2002; Hall et al. 2002).
Fission yeast centromeres are bipartite in structure, with a critical heterochromatic domain that flanks the central kinetichore-binding core element (Pidoux and Allshire 2004). Methylation of histone H3-K9 within heterochromatin by the Clr4 histone methyltransferase (HMT) (Nakayama et al. 2001) produces a requisite recognition mark for the HP1 proteins Swi6 (Nakayama et al. 2000) and Chp2 (Thon and Verhein-Hansen 2000). These proteins promote both structural spreading of heterochromatin and further H3 methylation from the site of initial nucleation (Grewal and Elgin 2002). This heterochromatin structure is essential for centromere function, as clr4, swi6, or chp1 mutants show defects in chromosome segregation and in recruitment of cohesin to heterochromatin (Nonaka et al. 2002; Partridge et al. 2002). Furthermore, as in higher eukaryotes, introduction of reporter genes within these heterochromatic regions results in their transcriptional silencing (Allshire et al. 1994; Nimmo et al. 1994; Grewal and Klar 1996), presumably through physical occlusion of the gene to transcription factors (Grewal and Elgin 2002).
Proper heterochromatin formation requires a number of additional gene products, including Rik1, several histone deacetylases (Bjerling et al. 2002; Nakayama et al. 2003), subunits of the RITS RNAi-guide complex (Verdel et al. 2004), and a second HMT encoded by set1+ that opposes heterochromatic silencing by methylation of histone H3 at K4 (Roguev et al. 2003). The Rik1 protein functions at an early step in heterochromatin formation, as it is required for proper H3-K9 methylation (Partridge et al. 2002) and Swi6 localization (Ekwall et al. 1996). These observations have led to a prevailing model in which Rik1 recruits Clr4 to a target locus (Grewal and Elgin 2002), likely through a direct physical interaction (Sadaie et al. 2004). In this study, we purified Rik1 from fission yeast extracts, and identify several Rik1-associated proteins, including two novel proteins, Raf1 and Raf2, the Clr4 HMT, and components of a cullin-dependent E3 ubiquitin ligase. This putative Rik1 complex displays H2B-directed polyubiquitylation activity in vitro, and disruption of raf1+ or raf2+ leads to defects in heterochromatic gene silencing in vivo. Interestingly, expression of a dominant-negative allele of the pcu4+ gene specifically elevates H3-K4 methylation within heterochromatin, suggesting that the E3 ligase activity may antagonize the Set1-dependent methylation of histone H3 within heterochromatin.
Results
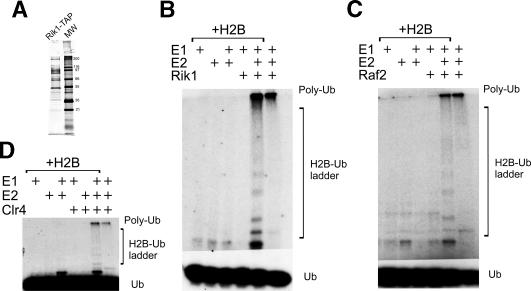
In order to identify proteins that associate with Rik1, we created an S. pombe strain (CYP11) in which sequences encoding a tandem affinity purification (TAP) module were introduced onto the C terminus of the Rik1 gene at its normal chromosomal location. This TAP tag encodes a calmodulin-binding peptide followed by four protein A domains, with an intervening TEV protease site (Tasto et al. 2001). Whole-cell extracts from this Rik1-TAP strain were bound to IgG sepharose resin and cleaved from the resin by TEV protease, and the eluate was then bound to calmodulin resin in the presence of Ca2+. Rik1 and associated proteins were then released from the calmodulin resin by the addition of EGTA (Fig. 1A). Two independent Rik1-TAP preparations were digested with trypsin and analyzed by LC-MS-MS, and peptides were identified by comparing the data to an S. pombe proteome database of predicted tryptic digests. Using this strategy, we identified a number of specific proteins from both characterized and novel ORFs that copurified with Rik1 (Table 1). These include histone H2B, two novel proteins encoded by predicted ORFs, SPCC613.12c and SPCC970.07c, and two previously characterized gene products, Pcu4 and Pip1. In addition, the Clr4 HMT was identified in one of our Rik1-TAP preparations, consistent with a recent study demonstrating an interaction between Clr4 and Rik1 (Sadaie et al. 2004). These proteins are uniquely found within Rik1-TAP preparations. They were not found in several parallel tandem affinity purifications, including TAP isolates of the predicted fission yeast ScSWI/SNF and RSC complexes, or Hrp1-TAP and Hrp3-TAP strains (our unpublished results).
Figure 1.
The Rik1 TAP prep contains a high-molecular-weight E3 ligase activity. (A) SDS PAGE of the Rik1-TAP preparation stained by silver. (B) A sample of the TAP eluate from strain CYP11 (Rik1-TAP) was preincubated on ice with or without human UbcH5b (an E2 enzyme), ATP, radiolabeled ubiquitin, and histone H2B as indicated. Following prebinding, the reactions were initiated with an E1 enzyme, followed by transfer to 30°C. Reaction products were separated by 12% SDS PAGE and conjugates were detected by autoradiography. The CYP11 TAP preparation displayed robust polyubiquitylation activity (as indicated by the ladder of bands ascending from monomeric ubiquitin) that was dependent on the addition of exogenous E1 and E2 enzymes. The reduction of laddering seen in the absence of exogenous H2B suggests that H2B does serve as an in vitro substrate. The components included in each lane are indicated in the panel legend above each lane. (C) A sample of the TAP eluate from strain CYP54 (Raf2-TAP) was assayed for polyubiquitylation activity as described for B. (D) A sample of the TAP eluate from strain CYP52 (Clr4-TAP) was assayed for polyubiquitylation activity as described for B.
Table 1.
Summary of Rik1–TAP preparation LC-MS-MS resultsa
| Number of peptides identified
|
|||
|---|---|---|---|
| ORF (proposed gene name) | TAP prep 1 | TAP prep 2 | Totalb |
| Rik1 | 28 | 16 | 34 |
| pcu4 | 11 | 2 | 13 |
| SPCC613.12c (raf1) | 4 | 4 | 7 |
| SPCC970.07c (raf2) | 6 | 4 | 8 |
| histone H2B | 4 | 4 | 8 |
| Pip1 | 2 | 1 | 2 |
| Clr4 | 3 | 0c | 3 |
Only peptide matches deemed statistically significant following MASCOT database searches are included.
This value represents the sum of the total number of nonidentical peptides isolated from both Rik1-TAP preparations.
Clr4 peptide matches of statistical relevance were identified in only one of the two LC-MS-MS experiments.
The SPCC613.12c ORF encodes an acidic 638 amino acid protein with no apparent Saccharomyces cerevisiae homolog. A BLAST to the NCBI databases reveals strong similarity to other hypothetical ORFs throughout the C-terminal half that encodes several predicted WD repeats. The other novel ORF, SPCC970.07c, encodes a predicted 636-amino acid basic zinc finger protein that bears little similarity to other proteins in the NCBI databases. Recently, both the SPCC613.12c and SPCC970.07c genes were identified in a cytological screen for mutants that disrupt Swi6 localization, and one of these gene products was also found to interact with Rik1 in a two-hybrid assay (R. Martienssen and Z. Cande, pers. comm.). Based on their identification as novel proteins associated with Rik1, we propose the nomenclature raf1+ (Rik1-associated factor) and raf2+ for SPCC613.12c and SPCC970.07c, respectively.
The previously identified pcu4+ (Liu et al. 2003) and pip1+ (Seibert et al. 2002) genes encode components of a cullin-dependent ligase (CDL) that covalently modifies target proteins with ubiquitin. The cullin family of E3 ubiquitin ligases normally function as multisubunit complexes that serve as substrate adaptors for ubiquitin-conjugating (Ubc) enzymes (Petroski and Deshaies 2005). Polyubiquitylation by CDLs through K48 of ubiquitin designates the subsequent degradation of the substrate by the proteasome (Hochstrasser 1996). In SCF, the CDL family prototype, the cullin subunit serves as the primary scaffolding subunit that bridges a substrate-binding subunit (Skowyra et al. 1997) to a ROC1 subunit that recruits the Ubc partner (Zheng et al. 2002). Eukaryotes typically encode multiple orthologous cullin genes that serve specialized roles. These CDLs share a common structural architecture with the cullin subunit serving to anchor a common ROC1 subunit to a variable set of specificity subunits. The pcu4+ gene encodes one of several S. pombe cullin genes (the human cullin-4 homolog) (Liu et al. 2003), while pip1+ encodes a small RING finger protein that is the homolog of ROC1 (Seibert et al. 2002).
The presence of Pcu4 and Pip1 in the Rik1 TAP preparations suggests that Rik1 is associated with a cullin-dependent E3 ubiquitin ligase. Ubiquitylation is a multistep process catalyzed by three enzymes (Hochstrasser 1996). An ubiquitin molecule is first activated by thioesterification at its C terminus by an E1 enzyme, and the ubiquitin is then conjugated by the coordinated action of one of several Ubc (E2) enzymes and an E3 ubiquitin ligase. To test for E3 activity in our Rik1-TAP preparations, we performed ubiquitylation assays using recombinant rabbit E1, recombinant UbcH5b (E2), 32P-labeled ubiquitin, and histone H2B as a substrate. UbcH5b was selected as the E2 based on its previous identification as a partner E2 for a mammalian CUL4A-dependent CDL (Wertz et al. 2004), and H2B was selected as a candidate substrate based on its identification in each of the Rik1-TAP preparations. Addition of the purified Rik1-TAP preparation to these E1/E2 reactions led to the appearance of a ladder of slower migrating 32P-ubiquitin conjugates, including a conjugate that migrated at the separating gel boundary (Fig. 1B). Omission of exogenous H2B eliminated the ladder of ubiquitin conjugates, and the very slowly migrating conjugate was diminished. This residual species is likely to represent self-ligation of polyubiquitin chains, an activity sometimes observed in CDL reactions in the absence of substrate (Jensen et al. 1995; Tan et al. 1999). These results demonstrate that the Rik1-TAP preparations contain a functional E3 ligase, consistent with the identification of Pcu4 and Pip1 as Rik1-interacting proteins.
To clarify whether other Rik1-interacting proteins are also associated with an E3 ligase, we analyzed tandem affinity purified material from Clr4-TAP and Raf2-TAP strains. As shown in Figure 1C, the Raf2-TAP preparation also showed robust polyubiquitylation activity that was partially dependent on exogenously added histone H2B. Similar results were also obtained with a Clr4-TAP preparation (Fig. 1D), although the activity was somewhat less robust. Notably, the copurification of E3 ligase activity is specific for these TAP purifications, as E3 activity is not observed in tandem affinity purifications from multiple TAP preparations of other S. pombe proteins (data not shown).
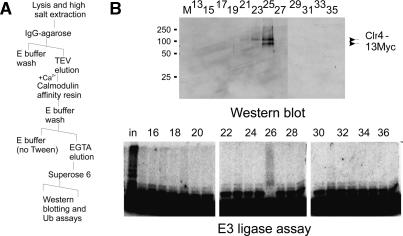
To confirm a direct, stable interaction between Clr4 and the E3 ligase activity, we further fractionated the purified Rik1-TAP preparation on a Superose 6 gel filtration column (see Fig. 2A). In this case, the initial extract was prepared from a Rik1-TAP strain harboring a Myctagged allele of clr4+. As expected from the mass spectrometry results, Clr4–13Myc was detected in the eluate following sequential purification of the Rik1-TAP whole-cell extract on IgG agarose and calmodulin resins. Following application of the calmodulin eluate onto Superose 6, Clr4–13Myc immunoreactivity and E3 ligase activity coeluted at an apparent molecular weight of 700 kDa, verifying that Clr4 and the cullin E3 complex are stably associated in the Rik1-TAP preparations (Fig. 2B). Together, these results suggest that the Rik1-TAP preparation comprises an E3 ligase complex that minimally consists of Rik1, Raf2, Pcu4, Pip1, and Clr4.
Figure 2.
Clr4 is a stable component of the Rik1 E3-containing complex. (A) A schematic of the purification of the material used in B. A Rik1-TAP Clr4–13myc strain (CYP56) was first purified by the TAP strategy and then submitted to additional Superose 6 gel filtration. (B) Concentrated fractions, prepared as in A, were analyzed by Western blot with an anti-9E10 monoclonal antibody (odd fractions only) and by the E3 assay as described above. Both polyubiquitylation activity and Clr4-myc immunoreactivity elute in a high molecular fraction, confirming that the Clr4 protein present in the Rik1-TAP preparation is directly associated with the E3 ligase activity.
We next sought genetic confirmation that the novel Rik1-associated proteins are key for the heterochromatin functions of Rik1. First, we investigated the roles of Raf1 and Raf2 in the transcriptional silencing of transgenes inserted into heterochromatic locations. In S. pombe, silencing of RNA polymerase II transcription occurs at heterochromatic loci that flank centromeric regions (Allshire et al. 1994), within telomeres (Nimmo et al. 1994) and at a specific interval within the mating-type locus (Grewal and Klar 1996). Previous studies have demonstrated that this heterochromatin-based silencing requires the coordinated action of rik1+ (Ekwall and Ruusala 1994), the clr genes (clr1–4,6) (Thon and Klar 1992; Ekwall and Ruusala 1994; Thon et al. 1994; Grewal et al. 1998), and the RNAi machinery (Volpe et al. 2002; Verdel et al. 2004).
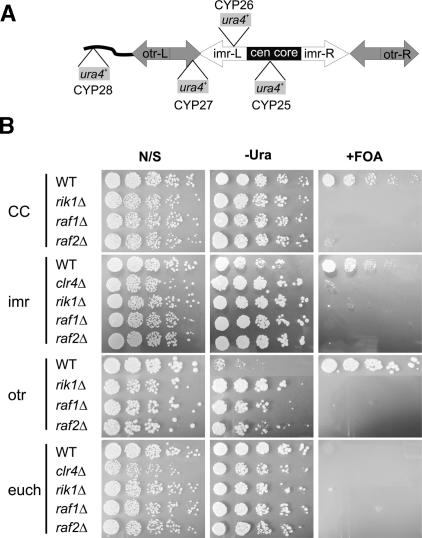
We exploited a set of reporter strains (Allshire et al. 1995) in which ura4+ genes have been inserted into four different genomic loci that are linked to the centromere of chromosome I (cenI). Three of these ura4+ insertions are silenced (central core or “CC”, and the “imr” and “otr” pericentric repeats), while a fourth (“euch”) is located within a euchromatic environment and is expressed (see schematic, Fig. 3A). Derepression of the transgene is readily assessed by growth on media that lacks uracil (–Ura), or by plating on media that contains 5-fluoroorotic acid (FOA), a compound that is toxic to cells that express Ura4. Representative silencing assays for wild-type, rik1Δ, clr4Δ, raf1Δ, and raf2Δ strains are shown in Figure 3B. In the wild-type strain, the ura4+ transgenes were efficiently silenced at all three centric and pericentric loci as evidenced by their poor growth on –Ura medium and the lack of FOA sensitivity (“CC”, “imr”, and “otr” in Fig. 3B). In contrast, all three centromeric loci were strongly derepressed in the clr4Δ, rik1Δ, raf1Δ, and raf2Δ strains. This was most evident on FOA-containing media, where each of these mutants showed strong hypersensitivity. Furthermore, the raf1Δ and raf2Δ mutants displayed enhanced growth on –Ura plates, similar to the phenotype of the rik1Δ and clr4Δ strains. In contrast, deletions of raf1 and raf2 (as well as clr4Δ and rik1Δ) failed to affect growth of the strain harboring the euchromatic reporter locus (“euch” in Fig. 3B), confirming that the effect of raf1+ and raf2+ was specific to heterochromatic loci.
Figure 3.
The raf gene deletions exhibit centromere-specific silencing defects. (A) A schematic diagram of the centromeric and pericentromeric regions of S. pombe chromosome 1. The relative positions of the ura4+ reporter integrants are shown with their corresponding strain designations. The strongest points of repression in cenI correspond to the otr (CYP27), followed by the imr (CYP26) and central core (CYP25) in hierarchical order. Strain CYP28 contains the ura4+ reporter at a euchromatic location. Note that strain CYP27 also contains a swi6 deletion, which partially derepresses this strain and sensitizes this background to the effects of other silencing factor mutations. (B) Overnight cultures of raf1Δ and raf2Δ strains in the indicated reporter backgrounds (illustrated in A) were compared with clr4Δ and rik1Δ control strains. Cells were diluted and spotted onto nonselective supplemented EMMG (N/S sections), EMMG lacking only uracil (–Ura sections), or supplemented EMMG containing 1 g/L 5-fluoroorotic acid (FOA sections). Cells were spotted in serial fourfold dilutions.
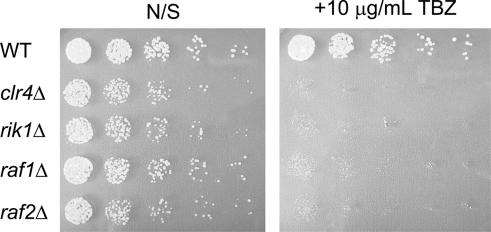
Since heterochromatin disruption also impairs metaphase centromere cohesion and chromosome segregation, strains with disruptions of genes critical for heterochromatin assembly display sensitivity to sublethal concentrations of microtubule depolymerizing agents such as thiabendazole (TBZ). Therefore, we also tested the clr4Δ, rik1Δ, raf1Δ, and raf2Δ strains in the CYP28 background for growth on 10 μg/mL TBZ. As expected, the raf1Δ and raf2Δ strains displayed a strong hypersensitivity to TBZ that was indistinguishable from the rik1Δ and clr4Δ control strains (Fig. 4). Together with the silencing data described above, these results indicate that raf1+ and raf2+ encode novel centromeric silencing components.
Figure 4.
Deletion of the raf genes results in TBZ hypersensitivity. Overnight cultures of raf1Δ and raf2Δ strains in the CYP28 background were compared with clr4Δ, rik1Δ, and wild-type CYP28. Cells were diluted and spotted onto nonselective supplemented EMMG (left panel; “N/S”) or EMMG media containing 10 μg/mL Thiabendazole (right panel; “+TBZ”). Cells were spotted in serial fourfold dilutions.
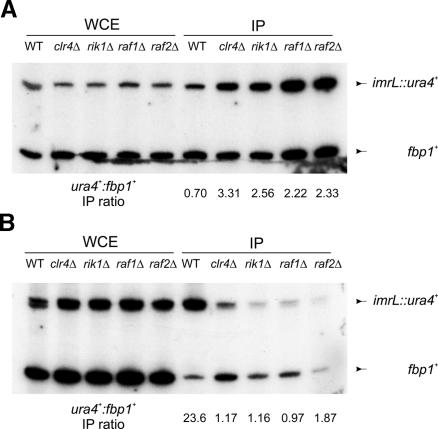
As mentioned above, two hallmarks of heterochromatic loci are an enrichment of histone H3-K9 methylation and a corresponding depletion of histone H3-K4 methylation. Since Rik1 and Clr4 are required for the centromeric pattern of histone H3 methylation (Partridge et al. 2002), we examined H3 methylation in wildtype, raf1Δ, and raf2Δ strains by chromatin immunoprecipitation analysis. Whole-cell extracts from formaldehyde-treated cultures were immunoprecipitated with antibodies directed to either methylated H3-K9 or H3-K4. Immunoprecipitated DNA was quantified by multiplex PCR using primers that recognize the heterochromatic ura4+ cassette or a euchromatic control locus, fbp1+. In wild-type CYP26 cells, the predicted distribution of H3-K4 and H3-K9 methylation was observed—H3-K4 methylation was underrepresented at the ura4+ reporter relative to the fbp1+ locus (Fig. 5A), and H3-K9 methylation was restricted to the silenced ura4+ locus (Fig. 5B). As expected, deletions of clr4 or rik1 led to a loss of H3-K9 methylation and a corresponding increase in H3-K4 methylation at the silenced ura4+ reporter (Fig. 5A,B). Strikingly, the raf1Δ and raf2Δ strains showed similar behavior. In both cases, H3-K9 methylation was reduced to clr4Δ (background) levels and H3-K4 methylation levels within ura4+ mirrored those seen in the rik1Δ and clr4Δ strains (Fig. 5A,B). Together with the silencing assays described above, these results indicate that the newly identified raf1+ and raf2+ genes encode new components of the silencing machinery that contribute to the regulation of histone H3 methylation within heterochromatin.
Figure 5.
The raf1 and raf2 genes contribute to the regulation of histone H3 methylation. (A). Chromatin immunoprecipitation analyses were performed with the indicated strains that each harbor the imrL::ura4+ pericentric reporter (CYP26 background). Immunoprecipitations used antisera against histone H3 dimethyl-Lys 4. Primer sets that amplify the imrL::ura4+ reporter and a reference set that amplifies the fbp1+ euchromatic control locus were multiplexed in PCR reactions. (WCE) Whole-cell extract before immunoprecipitation; (IP) immunoprecipitated DNA. Data were quantified by PhosphorImager and appear below their corresponding lanes expressed as a ratio of ratios using the fbp1+ control for normalization. (B) Chromatin immunoprecipitations were performed with antisera against H3 dimethyl-Lys 9. DNA was multiplexed and quantified as described for A.
Our proteomic and biochemical studies also implicate a Pcu4-dependent CDL in Rik1 function. We next sought confirmation of this prediction using both the previously described heterochromatin silencing assays and chromatin immunoprecipitations. Due to the particularly severe growth defects of pip1Δ and pcu4Δ strains, we were forced to use an allele of pcu4, pcu4-K680R, that impairs post-translational modification of Pcu4 by the ubiquitin-related protein, Nedd8. Neddylation is critical for cullin function in vivo, as substitution of the neddylated residue in either Pcu1 or Pcu4 produces strains with a severe growth defect (Osaka et al. 2000). In the case of pcu1 this allele is dominant-negative for slow growth, whereas the pcu4-K680R allele is recessive for slow growth. However, we were interesting in testing whether the pcu4-K680R allele might be dominant-negative for a subset of Pcu4 function (i.e., in the more specific context of heterochromatin formation and histone methylation). The pcu4-K680R allele was expressed via a low-strength nmt1+ promoter on a ScLEU2 marked plasmid in the CYP26 background. CYP26 cells transformed with pcu4-K680R, a wild-type pcu4+, or an empty vector control were plated onto –Leu and –Leu –Ura media. We observed a minor effect of the pcu4-K680R allele on the growth of CYP26 on –Leu –Ura, consistent with a role for Pcu4 in silencing (data not shown). We hypothesized that the weak phenotype we observed could be due to either poor penetrance of the dominant-negative allele or, alternatively, it could be indicative of a role for Pcu4 that is specific for establishment of heterochromatin rather than its maintenance. Previous studies have shown that two genetically separable pathways govern establishment of heterochromatin at the mating type locus: One requires the RNAi machinery (Hall et al. 2002) and the other requires the stress-response pathway (Jia et al. 2004). While disruption of each single pathway has only a modest effect on the maintenance of heterochromatin, a strong contribution of either pathway is uncovered if heterochromatin at the mating-type locus is first disassembled and then allowed to reassemble in mutant strains.
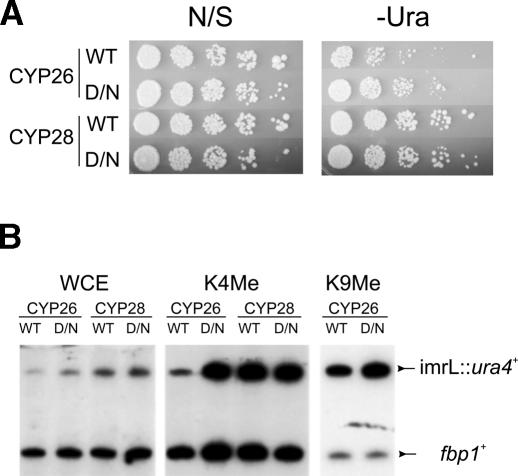
To investigate whether Pcu4 is required for establishment of heterochromatin, CYP26 cells that contained an episome expressing wild-type pcu4+ or the pcu4-K680R allele were selected on media that lacked both leucine and uracil to obtain cells that had spontaneously lost silencing of the ura4+ reporter locus. These Ura+ cultures were then cultured in liquid media that contained uracil for 10 generations prior to replating on –Leu –Ura plates to assess re-establishment of silencing. Strains that contained the wild-type pcu4+ gene were able to efficiently re-silence the ura4+ cassette and thus grew poorly on –Leu –Ura plates. In contrast, the strain that expressed the pcu4-K680R dominant-negative allele failed to efficiently repress the ura4+ cassette and grew well on –Leu –Ura media (Fig. 6A). Interestingly, chromatin immunoprecipitations of identically treated cells showed normal H3-K9 methylation in both cases, but cells expressing pcu4-K680R showed a striking elevation of H3-K4 methylation within heterochromatin, while low-level overexpression of the wild-type Pcu4 resulted in no change in K4 methylation (Fig. 6B). Together, these results indicate that Pcu4 also contributes to silencing within the fission yeast centromere and suggest a specific role for a Pcu4-dependent CDL in the establishment of heterochromatin domains through the prevention of inappropriate H3-K4 methylation.
Figure 6.
A Pcu4 dominant-negative allele impairs heterochromatin formation. (A) Strains CYP26 (containing a ura4+ transgene inserted into imrL of cenI) or CYP28 (a euchromatic insertion) were transformed with an episomal copy of either wild-type pcu4+ (WT lanes) or the pcu4-K680R dominant-negative allele (D/N lanes) expressed from a low-expression nmt1 promoter. Transformants were first plated onto –Leu –Ura solid media to select for derepression of the ura4+ transgene and cultured overnight in –Leu –Ura liquid media, prior to dilution into EMM –Leu. Cells were grown 10 generations in –Leu media to allow for heterochromatin reformation and silencing of the ura4+ cassette and plated in fourfold dilutions onto EMMG –Leu (“nonselective”; N/S section) or EMMG –Leu –Ura (–Ura section) to assess silencing of the transgene. (B) Transformants of CYP26 and CYP28, grown as described in A, were subjected to chromatin immunoprecipitation as described in Materials and Methods. Primer sets that amplify the imrL::ura4+ reporter and a reference set that amplifies the fbp1+ euchromatic control locus were multiplexed in PCR reactions. (WCE) Whole-cell extract; (K4Me) immunoprecipitated with antisera against histone H3 dimethyl-K4; (K9Me) immunoprecipitated with antisera against histone H3 dimethyl-K9.
Discussion
Using a proteomic strategy, we identified a number of proteins that are associated with Rik1 in whole-cell extracts, including the products of two novel genes, raf1+ and raf2+, and three previously identified genes, clr4+, pcu4+, and pip1+. Deletions of either raf gene imparts silencing defects within pericentric heterochromatin and hypersensitivity to a microtubule depolymerizing agent, TBZ. Furthermore, chromatin immunoprecipitations reveal a disruption of histone H3-K9 methylation and a concomitant increase in H3-K4 methylation within pericentromeric regions—all phenotypes that mirror those of a rik1Δ strain. We also demonstrate that Rik1, Raf2, and Clr4 are all associated with an E3 ligase activity, and that disruption of Pcu4 function impairs heterochromatic silencing and aberrantly elevates H3-K4 methylation.
The precise role of the raf gene products in the function of the Rik1-associated CDL remains unclear. Future studies will be needed to address whether the Raf proteins function as substrate adaptors, directly assist in ubiquitylation, or otherwise modulate the activity or assembly of the cullin complex Additionally, the Raf proteins might function coordinately with Rik1 in the recruitment of Clr4 to sites of heterochromatin formation. Interestingly, expression of the dominant-negative pcu4 allele does not disrupt H3-K9 methylation, unlike the raf1 and raf2 deletions. This result suggests that Rik1 might have dual roles in heterochromatin formation through direct recruitment of Clr4, and also through Pcu4-dependent degradation of a factor or factors that promote H3-K4 methylation. If this is the case, the failure of raf deletion strains to properly K9 methylate histone H3 suggests that the primary role of the raf gene products might be Clr4 recruitment. An alternative possibility is that since the dominant-negative pcu4-K680R allele only partially impairs CDL function, any residual activity associated with the wild-type Rik1–Pcu4 complex might be sufficient to prevent full disruption of heterochromatin and subsequent impairment of H3-K9 methylation. It is also formally possible that Pcu4 regulates both K4 and K9 methylation of histone H3, but that neddylation is not essential for its participation in H3-K9 methylation. If either of these possibilities were true, a full deletion of pcu4 might then produce the same impaired H3-K9 methylation seen in rik1 and raf deleted strains. Unfortunately, the severe growth defect associated with a pcu4 deletion prevents us from testing this possibility.
The fission yeast genome encodes a second rik1+ ortholog called ddb1+. Both of these genes share extensive homology over the entirety of their open reading frames (38% similarity and 19% identity over 993 amino acids), and they appear to be members of a larger family of UV-DDB-related proteins. Given this homology with human DDB1, it should not be surprising that rik1+ encodes an E3 ligase subunit. Arabidopsis thaliana DDB1 was identified as a component of an E3 ligase activity in plants (Yanagawa et al. 2004) and in humans, a DDB1 complex targets c-Jun for degradation as part of a CUL4A-dependent CDL (Wertz et al. 2004). Human CUL4A has also been implicated in the degradation of other nuclear factors (Nag et al. 2001, 2004; Zhang et al. 2003), implying a significant role for this complex in nuclear protein turnover, although a requirement for DDB1 in these cases is still unclear. More recently, studies in Caenorhabditis elegans and humans have implicated CUL4A in the maintenance of genome stability in normal cells (Zhong et al. 2003) and after DNA damage (Higa et al. 2003) through the degradation of a DNA replication licensing factor. In this case, DDB1 serves as the specificity subunit for the CUL4A ligase (Hu et al. 2004). This general machinery is conserved in fission yeast, as both Ddb1 (Bondar et al. 2003) and Pcu4 (Liu et al. 2003) have been implicated in DNA damage checkpoint control, and they are critical for the proper maintenance of nucleotide pools (Holmberg et al. 2005).
Previous genetic data from fission yeast also suggest a role for an ubiquitin E3 ligase in heterochromatin formation and silencing. Screens for high-copy suppressors of heterochromatic silencing identified several E2 enzymes, including UbcP3, Ubc15, UbcX, and Rhp6 (Choi et al. 2002; Nielsen et al. 2002). However, in the one case examined, the suppression of silencing due to overexpression of Ubc15 and Rhp6 was still observed in a proteasome mutant (Nielsen et al. 2002), making it unlikely that they function coordinately with a CDL. This is particularly unlikely in the case of Rhp6, the homolog of budding yeast Rad6p, which coordinates H2B monoubiquitylation and H3-K4 methylation (Sun and Allis 2002), as it utilizes an E3 ligase unrelated to cullins (Javerzat et al. 1999; Wood et al. 2003). In contrast, a screen for enhancers of centromeric silencing has identified two proteasome subunits (Javerzat et al. 1999), implying that regulated turnover of one or more proteins is critical for proper control of heterochromatic silencing.
Our results suggest that Rik1 also functions coordinately with Pcu4 in fission yeast to properly maintain the low levels of H3-K4 methylation characteristic of heterochromatin. However, the precise role of Raf1 and Raf2 in CDL function remains unclear, a point accentuated by their requirement for normal levels of both K4 and K9 methylation of histone H3. Whether both these factors are compulsory components of the Rik1 E3 ligase will require a more detailed mapping of the protein–protein interactions between each potential subunit. It is possible that their role is limited to Clr4 recruitment in concert with Rik1. In contrast, given that mammalian DDB1 functions as the substrate adaptor for its CDL (Hu et al. 2004), it seems likely that Rik1 itself directly targets substrate to the E3. While our proteomic and biochemical studies are consistent with the possibility that histone H2B may be a bona fide substrate for the Rik1-associated E3 ligase, it is unclear whether H2B represents a legitimate in vivo substrate of the CDL. Previous studies have shown that monoubiquitylation of H2B is necessary for subsequent K4 methylation of histone H3 (Sun and Allis 2002), establishing precedence for a link between H2B ubiquitylation and H3-K4 methylation. We note, however, that our CDL polyubiquitylates H2B in vitro. Whether this polyubiquitylation occurs in vivo and what its consequences might be remain to be determined. Regardless, we anticipate that the Rik1 CDL will have multiple substrates, and their identification will be an interesting avenue for future studies. In this regard, the elevation of H3-K4 methylation seen in the pcu4K680R dominant-negative mutant suggests that Set1 or a Set1-regulating factor would be intriguing substrate candidates.
Materials and methods
Yeast cultures and strains
For chromatin immunoprecipitations and proteomics, S. pombe liquid cultures were grown on standard YE media supplemented with 100 mg/L each of adenine, histidine, leucine, lysine, and uracil (YES). For spotting assays of ura4+ integrated reporter strains, pombe minimal media (EMM without N2; Q-Biogene/MP Biomedicals) supplemented with 1 g/L glutamate (EMMG) was used as described (Allshire et al. 1994). In selective and nonselective EMMG, auxotrophic supplements were included at 100 mg/L as described for YES, with the exception that +FOA media contained reduced uracil (75 mg/L) and 1 g/L of 5-fluoroorotic acid (Zymo Research). TBZ studies were performed on solid EMMG containing 10 μg/mL thiabendazole. For liquid culture of cells in the pcu4 studies, EMM supplemented with ammonium sulfate (Q-Biogene/MP Biomedicals) was used with nutrients supplemented at the same concentrations described for YES.
Tagged and deleted strains were made by a PCR-based method (Bahler et al. 1998), using either kanamycin or hygromycin resistance cassettes. The TAP tagging construction was performed as described (Tasto et al. 2001). The genotypes and strain designations of all strains used in this study appear in Table 2. In all cases, mutants denoted as deletions (e.g., raf1Δ) refer to replacement of the complete open reading frame. The ura4+ centromere proximal integrants were a kind gift of Dr. Robin Allshire (Wellcome Trust Centre for Cell Biology, Edinburgh, Scotland, UK) and have been previously described (Allshire et al. 1995).
Table 2.
Strains used in this study
| Strain numbera | Genotypeb | Source |
|---|---|---|
| CYP11 | h- leu1-32 ura4D-18 ade6-M210 rik1-TAP::kanr | This study |
| CYP25 (FY336) | h- ade6-M210 leu1-32 ura4-DS/E TM1::ura4+ | R. Allshirea |
| CYP26 (FY496) | h+ ade6-M210 leu1-32 ura4-DS/E imr1L (dg-glu) NcoI::ura4+ oriI | R. Allshirea |
| CYP27 (FY648) | h+ ade6-M210 leu1-32 ura4-DS/E otr1R (dg-glu BamHI-SPeI fragment)SphI::ura4+ swi6Δ | R. Allshirea |
| CYP28 (FY941) | h+ ade6-M210 leu1-32 ura4-DS/E tRNAPhe-otr1L(XhoIBamHI fragment)HpaI::ura4+ | R. Allshirea |
| CYP31 | (CYP25) h- rik1Δ::hygr | This study |
| CYP32 | (CYP25) h- raf1Δ::kanr | This study |
| CYP33 | (CYP25) h- raf2Δ::kanr | This study |
| CYP35 | (CYP27) h+ rik1Δ::hygr | This study |
| CYP36 | (CYP27) h+ raf1Δ::kanr | This study |
| CYP37 | (CYP27) h+ raf2Δ::kanr | This study |
| CYP40 | (CYP26) h+ clr4Δ::kanr | This study |
| CYP42 | (CYP26) h+ rik1Δ::hygr | This study |
| CYP43 | (CYP26) h+ raf1Δ::kanr | This study |
| CYP44 | (CYP26) h+ raf2Δ::kanr | This study |
| CYP45 | (CYP28) h+ clr4Δ::kanr | This study |
| CYP46 | (CYP28) h+ rik1Δ::hygr | This study |
| CYP47 | (CYP28) h+ raf1Δ::kanr | This study |
| CYP48 | (CYP28) h+ raf2Δ::kanr | This study |
| CYP54 | h- leu-32 ura4-D18 ade6-M210 raf2-TAP::kanr | This study |
| CYP56 | (CYP11) h- clr4-13myc::hygr | This study |
The strain number appearing in parentheses refers to the designation from the original strain collection (see Allshire et al. 1995).
The strain number in parentheses refers to the parental background.
Tandem affinity purification and mass spectrometry
Eight to 12 L of a logarithmic culture of CYP11, CYP52, CYP54, or CYP56 were harvested by centrifugation, washed in E buffer (20 mM HEPES at pH 7.4, 350 mM NaCl, 10% glycerol, 0.1% Tween-20), and recentrifuged. The cell pellet was resuspended in a minimal volume of E buffer plus 1× Complete protease inhibitors (Roche) that was sufficient to create a thick cell slurry. The slurry was transferred to a 60-cc tuberculin syringe (without a needle) and ejected in a stream into liquid nitrogen. Frozen cells were lysed by hand grinding with a mortar and pestle in a bed of dry ice. Temperature was maintained by the periodic addition of liquid nitrogen. All subsequent steps were performed at 4°C. The lysate was dissolved in E plus protease inhibitors (20 mL per liter of culture) and cell debris was removed by centrifugation at 2000 × g for 15 min, and then cleared by ultracentrifugation at 235,000 × g for 1 h. The cleared lysate was bound for 2 h at 4°C to 0.6 mL of rabbit IgG-agarose (Sigma) per 4 L of original culture. Unbound material was discarded and the beads were washed with 15 mL of E buffer, followed by 10 mL of E buffer plus 1 mM DTT. Protein was eluted from the beads by rocking overnight in 4 mL (per 0.6 mL of beads) of E buffer plus 1 mM DTT containing 300 units of TEV protease (Invitrogen). The TEV eluate was then supplemented with 2 mM CaCl2 and incubated with rocking for 2 h at 4°C with 0.6 mL of calmodulin resin (Stratagene) per 4 L of original culture. Unbound material was discarded and the beads were washed twice with 10 mL of E buffer containing 2 mM CaCl2. Protein was then eluted from the beads with 2 mL (per 0.6 mL of beads) of E buffer plus 10 mM EGTA.
For mass spectrometry, calmodulin eluates were dialyzed into ammonium bicarbonate, concentrated by lyophilization, digested with trypsin, and analyzed by LC-MS-MS. Minor modifications were made in the TAP purification for samples used for ubiquitylation assays. Following the binding of samples to the calmodulin resin, two washes were performed. The column was first washed with E buffer plus 2 mM CaCl2, followed by a second wash in E buffer plus 2 mM CaCl2 lacking Tween 20. Bound material was then eluted with E buffer plus EGTA lacking Tween 20, dialyzed against Ubc buffer (50 mM HEPES at pH 7.3, 150 mM NaCl, 10% glycerol, 1 mM DTT), and then concentrated to ∼150 μL.
Ubiquitylation assays
Ubiquitin ligation assays were performed at 30°C in a volume of 20 μL of Ub reaction buffer (50 mM HEPES at pH 7.3, 20 mM NaCl, 5 mM MgCl2, 2 mM ATP, 0.2 mM DTT) containing ∼500,000 dpm of ubiquitin radiolabeled with the catalytic subunit of PKA as described previously (Lorick et al. 1999). Reactions contained 10 μL of purified TAP proteins or Ubc buffer (10 μL). The E2 enzyme (800 nM recombinant human UbcH5b; Boston Biochem) and TAP preparations were preincubated at 4°C for 15 min prior to the addition of 90 nM rabbit E1 (Boston Biochem) and transfer of the reaction to 30°C to initiate the reaction. As indicated, H2B was included at 2 μg per reaction. Reactions continued at 30°C for 10 h. Samples were resolved by SDS PAGE and conjugates were detected by autoradiography.
Chromatin immunoprecipitations
Chromatin immunoprecipitations were performed using a protocol described by Pidoux et al. (2004), with modifications as described below: Cells (∼4 × 108) were fixed in 3% formaldehyde for 30 min, washed in Tris-buffered saline (20 mM Tris-Cl at pH 7.4, 150 mM NaCl), and resuspended in 0.4 mL of FA-lysis buffer (50 mM HEPES at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate) plus protease inhibitors. Cells were lysed by hand vortexing with an equal volume of glass beads 8 × 30 sec. This crude lysate was sonicated to shear chromatin and clarified in a microcentrifuge. The remainder of the procedure was unchanged from the original protocol. Quantitative PCR reactions performed with α-32P-dCTP reactions were visualized by autoradiography and quantified by PhosphorImager. Immunoprecipitations were performed with antibodies raised against histone H3 dimethyl-K9 and histone H3 dimethyl-K4 (Upstate Biologicals).
Acknowledgments
We thank Nick Rhind, Dan McCollum, and Robin Allshire for strains and advice, and Rob Martienssen and Zach Cande for sharing data prior to publication. We also thank Allan Weissman for the gift of the GST-ubiquitin and UbcH5b plasmids used for the ubiquitin studies and Tomoki Chiba for the Pcu4 episomal vectors. This work was supported by NIH grant GM54096 to C.L.P. and a Charles A. King Trust Award (Fleet National Bank, Edward Dane and Richard Lovell, Co-Trustees) to P.J.H.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1328005.
References
- Allshire R.C., Javerzat, J.P., Redhead, N.J., and Cranston, G. 1994. Position effect variegation at fission yeast centromeres. Cell 76: 157–169. [DOI] [PubMed] [Google Scholar]
- Allshire R.C., Nimmo, E.R., Ekwall, K., Javerzat, J.P., and Cranston, G. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes & Dev. 9: 218–233. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie III, A., Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. [DOI] [PubMed] [Google Scholar]
- Bernard P., Maure, J.F., Partridge, J.F., Genier, S., Javerzat, J.P., and Allshire, R.C. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542. [DOI] [PubMed] [Google Scholar]
- Bjerling P., Silverstein, R.A., Thon, G., Caudy, A., Grewal, S., and Ekwall, K. 2002. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell. Biol. 22: 2170–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar T., Mirkin, E.V., Ucker, D.S., Walden, W.E., Mirkin, S.M., and Raychaudhuri, P. 2003. Schizosaccharomyces pombe Ddb1 is functionally linked to the replication checkpoint pathway. J. Biol. Chem. 278: 37006–37014. [DOI] [PubMed] [Google Scholar]
- Choi E.S., Kim, H.S., Jang, Y.K., Hong, S.H., and Park, S.D. 2002. Two ubiquitin-conjugating enzymes, Rhp6 and UbcX, regulate heterochromatin silencing in Schizosaccharomyces pombe. Mol. Cell. Biol. 22: 8366–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K. and Ruusala, T. 1994. Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent matingtype loci in fission yeast. Genetics 136: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K., Nimmo, E.R., Javerzat, J.P., Borgstrom, B., Egel, R., Cranston, G., and Allshire, R. 1996. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 109(Pt 11): 2637–2648. [DOI] [PubMed] [Google Scholar]
- Grewal S.I. and Elgin, S.C. 2002. Heterochromatin: New possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12: 178–187. [DOI] [PubMed] [Google Scholar]
- Grewal S.I. and Klar, A.J. 1996. Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell 86: 95–101. [DOI] [PubMed] [Google Scholar]
- Grewal S.I. and Moazed, D. 2003. Heterochromatin and epigenetic control of gene expression. Science 301: 798–802. [DOI] [PubMed] [Google Scholar]
- Grewal S.I., Bonaduce, M.J., and Klar, A.J. 1998. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150: 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I.M., Shankaranarayana, G.D., Noma, K., Ayoub, N., Cohen, A., and Grewal, S.I. 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237. [DOI] [PubMed] [Google Scholar]
- Higa L.A., Mihaylov, I.S., Banks, D.P., Zheng, J., and Zhang, H. 2003. Radiation-mediated proteolysis of CDT1 by CUL4–ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5: 1008–1015. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30: 405–439. [DOI] [PubMed] [Google Scholar]
- Holmberg C., Fleck, O., Hansen, H.A., Liu, C., Slaaby, R., Carr, A.M., and Nielsen, O. 2005. Ddb1 controls genome stability and meiosis in fission yeast. Genes & Dev. 19: 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., McCall, C.M., Ohta, T., and Xiong, Y. 2004. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6: 1003–1009. [DOI] [PubMed] [Google Scholar]
- Javerzat J.P., McGurk, G., Cranston, G., Barreau, C., Bernard, P., Gordon, C., and Allshire, R. 1999. Defects in components of the proteasome enhance transcriptional silencing at fission yeast centromeres and impair chromosome segregation. Mol. Cell. Biol. 19: 5155–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J.P., Bates, P.W., Yang, M., Vierstra, R.D., and Weissman, A.M. 1995. Identification of a family of closely related human ubiquitin conjugating enzymes. J. Biol. Chem. 270: 30408–30414. [DOI] [PubMed] [Google Scholar]
- Jia S., Noma, K., and Grewal, S.I. 2004. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976. [DOI] [PubMed] [Google Scholar]
- Liu C., Powell, K.A., Mundt, K., Wu, L., Carr, A.M., and Caspari, T. 2003. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes & Dev. 17: 1130–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick K.L., Jensen, J.P., Fang, S., Ong, A.M., Hatakeyama, S., and Weissman, A.M. 1999. RING fingers mediate ubiquitin-conjuating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. 95: 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., Bondar, T., Shiv, S., and Raychaudhuri, P. 2001. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 21: 6738–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., Bagchi, S., and Raychaudhuri, P. 2004. Cul4A physically associates with MDM2 and participates in the proteolysis of p53. Cancer Res. 64: 8152–8155. [DOI] [PubMed] [Google Scholar]
- Nakayama J., Klar, A.J., and Grewal, S.I. 2000. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell 101: 307–317. [DOI] [PubMed] [Google Scholar]
- Nakayama J., Rice, J.C., Strahl, B.D., Allis, C.D., and Grewal, S.I. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113. [DOI] [PubMed] [Google Scholar]
- Nakayama J., Xiao, G., Noma, K., Malikzay, A., Bjerling, P., Ekwall, K., Kobayashi, R., and Grewal, S.I. 2003. Alp13, an MRG family protein, is a component of fission yeast Clr6 histone deacetylase required for genomic integrity. EMBO J. 22: 2776–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen I.S., Nielsen, O., Murray, J.M., and Thon, G. 2002. The fission yeast ubiquitin-conjugating enzymes UbcP3, Ubc15, and Rhp6 affect transcriptional silencing of the mating-type region. Eukaryot. Cell 1: 613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo E.R., Cranston, G., and Allshire, R.C. 1994. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J. 13: 3801–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N., Kitajima, T., Yokobayashi, S., Xiao, G., Yamamoto, M., Grewal, S.I., and Watanabe, Y. 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4: 89–93. [DOI] [PubMed] [Google Scholar]
- Osaka F., Saeki, M., Katayama, S., Aida, N., Toh, E.A., Kominami, K., Toda, T., Suzuki, T., Chiba, T., Tanaka, K., et al. 2000. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 19: 3475–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge J.F., Scott, K.S., Bannister, A.J., Kouzarides, T., and Allshire, R.C. 2002. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr. Biol. 12: 1652–1660. [DOI] [PubMed] [Google Scholar]
- Petroski M.D. and Deshaies, R.J. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6: 9–20. [DOI] [PubMed] [Google Scholar]
- Pidoux A.L. and Allshire, R.C. 2004. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res. 12: 521–534. [DOI] [PubMed] [Google Scholar]
- Pidoux A., Mellone, B., and Allshire, R. 2004. Analysis of chromatin in fission yeast. Methods 33: 252–259. [DOI] [PubMed] [Google Scholar]
- Roguev A., Schaft, D., Shevchenko, A., Aasland, R., Shevchenko, A., and Stewart, A.F. 2003. High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J. Biol. Chem. 278: 8487–8493. [DOI] [PubMed] [Google Scholar]
- Sadaie M., Iida, T., Urano, T., and Nakayama, J. 2004. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 23: 3825–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibert V., Prohl, C., Schoultz, I., Rhee, E., Lopez, R., Abderazzaq, K., Zhou, C., and Wolf, D.A. 2002. Combinatorial diversity of fission yeast SCF ubiquitin ligases by homo- and heterooligomeric assemblies of the F-box proteins Pop1p and Pop2p. BMC Biochem. 3: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D., Craig, K.L., Tyers, M., Elledge, S.J., and Harper, J.W. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91: 209–219. [DOI] [PubMed] [Google Scholar]
- Sun Z.W. and Allis, C.D. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108. [DOI] [PubMed] [Google Scholar]
- Tan P., Fuchs, S.Y., Chen, A., Wu, K., Gomez, C., Ronai, Z., and Pan, Z.Q. 1999. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I κ B α. Mol. Cell 3: 527–533. [DOI] [PubMed] [Google Scholar]
- Tasto J.J., Carnahan, R.H., McDonald, W.H., and Gould, K.L. 2001. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast 18: 657–662. [DOI] [PubMed] [Google Scholar]
- Thon G. and Klar, A.J. 1992. The clr1 locus regulates the expression of the cryptic mating-type loci of fission yeast. Genetics 131: 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G. and Verhein-Hansen, J. 2000. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155: 551–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G., Cohen, A., and Klar, A.J. 1994. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics 138: 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A., Jia, S., Gerber, S., Sugiyama, T., Gygi, S., Grewal, S.I., and Moazed, D. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Volpe T., Schramke, V., Hamilton, G.L., White, S.A., Teng, G., Martienssen, R.A., and Allshire, R.C. 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11: 137–146. [DOI] [PubMed] [Google Scholar]
- Wertz I.E., O'Rourke, K.M., Zhang, Z., Dornan, D., Arnott, D., Deshaies, R.J., and Dixit, V.M. 2004. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303: 1371–1374. [DOI] [PubMed] [Google Scholar]
- Wood A., Krogan, N.J., Dover, J., Schneider, J., Heidt, J., Boateng, M.A., Dean, K., Golshani, A., Zhang, Y., Greenblatt, J.F., et al. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11: 267–274. [DOI] [PubMed] [Google Scholar]
- Yanagawa Y., Sullivan, J.A., Komatsu, S., Gusmaroli, G., Suzuki, G., Yin, J., Ishibashi, T., Saijo, Y., Rubio, V., Kimura, S., et al. 2004. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes & Dev. 18: 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Morrone, G., Zhang, J., Chen, X., Lu, X., Ma, L., Moore, M., and Zhou, P. 2003. CUL-4A stimulates ubiquitylation and degradation of the HOXA9 homeodomain protein. EMBO J. 22: 6057–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Schulman, B.A., Song, L., Miller, J.J., Jeffrey, P.D., Wang, P., Chu, C., Koepp, D.M., Elledge, S.J., Pagano, M., et al. 2002. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 416: 703–709. [DOI] [PubMed] [Google Scholar]
- Zhong W., Feng, H., Santiago, F.E., and Kipreos, E.T. 2003. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423: 885–889. [DOI] [PubMed] [Google Scholar]