Abstract
Disruptions in the use of skeletal muscle lead to muscle atrophy. After short periods of disuse, muscle atrophy is reversible, and even after prolonged periods of inactivity, myofiber degeneration is uncommon. The pathways that regulate atrophy, initiated either by peripheral nerve damage, immobilization, aging, catabolic steroids, or cancer cachexia, however, are poorly understood. Previously, we found that Runx1 (AML1), a DNA-binding protein that is homologous to Drosophila Runt and has critical roles in hematopoiesis and leukemogenesis, is poorly expressed in innervated muscle, but strongly induced in muscle shortly after denervation. To determine the function of Runx1 in skeletal muscle, we generated mice in which Runx1 was selectively inactivated in muscle. Here, we show that Runx1 is required to sustain muscle by preventing denervated myofibers from undergoing myofibrillar disorganization and autophagy, structural defects found in a variety of congenital myopathies. We find that only 29 genes, encoding ion channels, signaling molecules, and muscle structural proteins, depend upon Runx1 expression, suggesting that their misregulation causes the dramatic muscle wasting. These findings demonstrate an unexpected role for electrical activity in regulating muscle wasting, and indicate that muscle disuse induces compensatory mechanisms that limit myofiber atrophy. Moreover, these results suggest that reduced muscle activity could cause or contribute to congenital myopathies if Runx1 or its target genes were compromised.
Keywords: Atrophy, myopathy, acute myeloid leukemia, hematopoiesis, denervation
The pathways that couple changes in the pattern of myofiber electrical activity to alterations in skeletal muscle gene expression are poorly understood. Disruptions in electrical activity resulting from trauma, immobilization, or aging, ultimately lead to changes in muscle structure and function, including a decrease in myofiber size, termed muscle atrophy (Jagoe and Goldberg 2001; Glass 2003). Although members of the forkhead (FoxO) class of transcription factors have recently been shown to have an important role in regulating atrophy (Sandri et al. 2004; Stitt et al. 2004), it seems likely that additional regulatory programs are activated following muscle disuse and govern muscle wasting.
Atrophic myofibers, due to their smaller cross-sectional area, have a reduced capacity to generate force, but they neither degenerate nor undergo apoptosis. Indeed, atrophic myofibers retain most of the structural features that are characteristic of normal muscle. In contrast, in a variety of congenital myopathies, muscle wasting is far more dramatic. After short periods of inactivity, muscle atrophy is reversible. Even after prolonged periods of disuse, myofiber degeneration remains uncommon, and atrophy can be partially reversed. Taken together, these results raise the possibility that muscle disuse induces compensatory mechanisms to maintain skeletal myofibers and limit atrophy in the absence of innervation-dependent trophic support (Murgia et al. 2000).
Previously, we used a subtractive hybridization and cloning strategy to identify genes that are expressed in skeletal muscle and regulated by innervation (Zhu et al. 1994). We found that Runx1 (AML1), a DNA-binding protein that is homologous to Drosophila Runt and has critical roles in hematopoiesis and leukemogenesis, is poorly expressed in innervated muscle, but strongly induced in muscle shortly after denervation (Zhu et al. 1994). To study the role of Runx1 in skeletal myofibers, we inactivated the runx1 gene selectively in skeletal muscle. We found that induction of Runx1 is required to sustain muscle by preventing denervated myofibers from undergoing autophagy and severe muscle wasting. We used a microarray screen to identify targets of Runx1 and found that only 29 genes are misregulated (greater than or equal to threefold) in runx1 mutant muscle, indicating that misexpression of a small number of genes is responsible for the defects in myofibrillar organization and severe muscle wasting of denervated runx1 mutant muscle. These findings demonstrate that electrical activity regulates autophagy and muscle wasting by controlling expression of a transcription factor that activates and represses muscle gene expression, thereby compensating, in part, for a loss of innervation and limiting the extent of muscle wasting.
Results
Inactivation of Runx1 selectively in skeletal muscle
Mice lacking Runx1 die during mid-embryogenesis (E12), due to hemorrhaging in the central nervous system (Okuda et al. 1996; Wang et al. 1996). To study the role of Runx1 in skeletal myofibers, we circumvented this embryonic lethality by generating mice that carry a loxP-flanked allele of runx1 and a muscle creatine kinase (MCK)::cre transgene (Fig. 1). The runx1 allele contains loxP sites that flank exon 4 (Taniuchi et al. 2002), which encodes the major portion of the DNA-binding domain as well as sequences that are essential for interaction with CBFβ, a Runx-interacting protein that is required for Runx1 activity in vivo (de Bruijn and Speck 2004). Splicing from exon 3, the first coding exon in skeletal muscle (data not shown), to exon 5 or exon 6 alters the reading frame, resulting in a truncated protein. Thus, deletion of exon 4 is likely to result in a runx1-null mutation (see below).
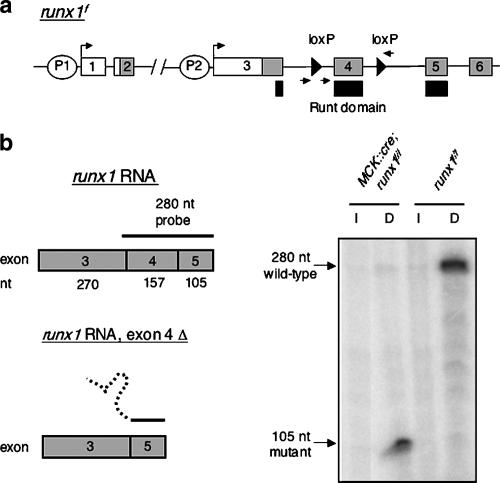
Figure 1.
Runx1 is efficiently inactivated in skeletal muscle. (a) Mice carrying a MCK::cre transgene (Bruning et al. 1998) were crossed with runx1f/f mice (Taniuchi et al. 2002) to generate MCK::cre; runx1f/f mice, as well as runx1f/f control mice. The positions of the distal (P1) and proximal (P2) promoters, the loxP sites (triangles), and the primers used to genotype the mice (arrows) are indicated. (b) RNase protection analysis demonstrates that exon 4 is deleted in MCK::cre; runx1f/f mice. RNA isolated from denervated (4-d) muscles of runx1f/f control mice fully protects the 280-nucleotide (nt) probe from digestion; a low level of protection is observed in innervated muscle, confirming a 50- to 100-fold induction of runx1 following denervation (Zhu et al. 1994). RNA isolated from denervated muscle of MCK::cre; runx1f/f mice protects 105 nt, corresponding to the sequence encoded by exon 5 from digestion; a low level of protection is observed in innervated muscle, demonstrating that the mutant allele, like wild-type runx1, is induced by denervation. Quantitation of these results demonstrates that 96% ± 0.9% (mean ± SEM, n = 3) of the runx1 RNA induced by denervation in MCK::cre; runx1f/f mice lacks the DNA-binding Runt domain.
Runx1 is expressed in developing muscle. but down-regulated by innervation (Zhu et al. 1994), which begins at E12.5. Because the MCK regulatory elements confer only a low level of Cre recombinase expression in embryonic skeletal muscle and substantially greater expression after birth (Bruning et al. 1998; Nguyen et al. 2003), maximal Cre expression is attained at a time when skeletal muscle runx1 expression has already declined. Since severe defects in myogenesis lead to neonatal lethality, this experimental design minimizes the potential for neonatal lethality if Runx1 were to have a key role during myogenesis.
Indeed, MCK::cre; runx1f/f mice were healthy and fertile and born in expected numbers. We assessed the effectiveness of Cre-mediated recombination by measuring runx1 RNA expression in innervated and denervated muscles from MCK::cre; runx1f/f mice and runx1f/f control adult mice. We used an RNase protection assay, which detects both wild-type and mutant runx1 RNAs, and found that 96% of skeletal muscle runx1 is inactivated in MCK::cre; runx1f/f mice (Fig. 1). Moreover, these data demonstrate that runx1 expression is not dependent upon autoregulation, as suggested for runx2 (Drissi et al. 2000), as runx1 is induced to the same extent in runx1 mutant and control muscle.
Denervated runx1 mutant myofibers are severely atrophied
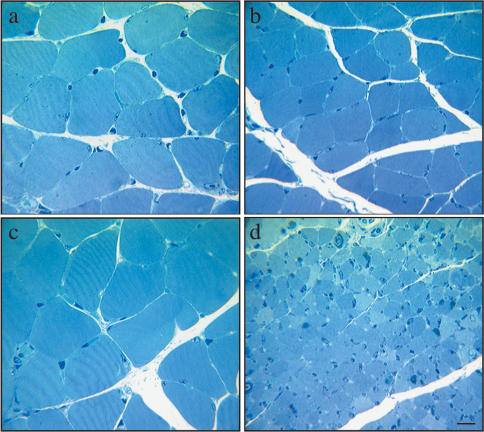
In wild-type mice, peripheral nerve damage (Zhu et al. 1994), as well as limb immobilization (Bodine et al. 2001), lead to an increase in Runx1 expression and muscle atrophy. To determine whether Runx1 regulates muscle atrophy, we denervated hind-limb muscles in mutant and control adult mice and examined innervated and denervated muscles by light microscopy 2 wk later. Figure 2 shows that denervated, runx1 mutant myofibers are severely and unusually atrophic. In the soleus muscles of control runx1f/f mice and runx1f/– mice, which are heterozygous for the mutant allele, a 2-wk denervation period leads to a ∼35% decrease in the crosssectional area of the myofibers (innervated runx1f/f, 690 ± 236 μm2, mean ± SD, n = 218; denervated runx1f/f, 460 ± 150 μm2, n = 226; innervated runx1f/–, 684 ± 259 μm2, n = 489; denervated runx1f/–, 401 ± 142 μm2, n = 434) and a corresponding decrease in muscle wet weight. In the soleus muscles of mice lacking Runx1 (MCK::cre; runx1f/f), denervation leads to a 10-fold decrease in myofiber size (innervated, 702 ± 252 μm2, n = 877; denervated, 82 ± 29 μm2, n = 524) and a similar decrease in muscle wet weight. In addition, the interstitial space between the denervated runx1 mutant myofibers is unusually compacted, indicating that the muscle extracellular matrix is also defective. We noted no signs of apoptosis, as myofibers were neither TUNEL-positive nor stained with antibodies to activated caspase 3 (data not shown). Other muscles, including the tibialis anterior and gastrocnemius muscles, are also affected, and similar changes in myofiber cell size were evident in denervated muscle from myf5cre; runx1f/f mice (data not shown). In contrast, innervated control and runx1 mutant muscles appear similar, indicating that the low level of Runx1 expression in innervated muscle does not regulate myofiber cell size (Fig. 2). These results indicate that Runx1 is required to sustain denervated muscle and to minimize atrophy.
Figure 2.
Runx1mutant denervated myofibers are severely and unusually atrophic. (a,b) In control runx1f/f mice, denervated (14-d) myofibers (b) are ∼30% smaller than innervated myofibers (a). (c,d) In MCK::cre; runx1f/f mice, denervated (14-d) myofibers (d) are severely atrophic and 10-fold smaller than innervated myofibers (c). The size of innervated myofibers is similar in runx1 mutant (c) and control (a) mice. Bar, 10 μm.
The severe wasting of denervated runx1 myofibers is not accompanied by excessive activation of the FoxO-mediated atrophy pathway
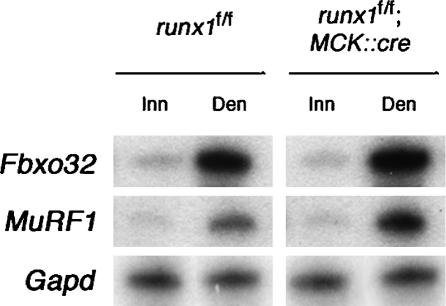
The pathways that regulate atrophy are poorly understood, but muscle disuse leads to a reduction in phosphatidylinositol 3-kinase (PI3K)/Akt activities and a decrease in FoxO phosphorylation, triggering nuclear import of FoxO and activation of FoxO target genes (Sandri et al. 2004; Stitt et al. 2004). To determine whether the severe wasting of denervated runx1 mutant myofibers is due to excessive activation of the FoxO-mediated atrophy pathway, we measured expression of a direct target of FoxO1, atrogin-1 (MAFbx; Fbxo32) (Bodine et al. 2001; Sandri et al. 2004), an E3-ubiquitin ligase, in innervated and denervated muscles from control and runx1 mutant mice. We found that denervation causes a 5.3 ± 0.4-fold (mean ± SEM, n = 3) increase in atrogin-1 expression in control mice and a 8.6 ± 0.4-fold (mean ± SEM, n = 3) increase in atrogin-1 expression in runx1 mutant muscle (Fig. 3). Thus, the severe wasting of denervated runx1 mutant muscle is unlikely to be due to excessive activation of the FoxO pathway.
Figure 3.
The severe atrophy of denervated runx1 mutant myofibers is not accompanied by excessive activation of the FoxO or NFκB pathways. Following denervation, the extent of Fbxo32 (a.k.a. atrogin-1, MAFbx) and MuRF1 induction is similar in MCK::cre; runx1f/f and control runx1f/f mice. As Fbxo32 is a target of FoxO and MuRF1 is induced by NFκB, these results indicate that FoxO and NFκB are similarly activated in denervated muscles from MCK::cre; runx1f/f and control runx1f/f mice. The level of Fbxo32, MuRF1, and Gapd expression in innervated (Inn) and denervated (Den) (3-d) muscles was measured by an RNase protection assay (Zhu et al. 1994).
Atrophy can be induced by NFκB without inducing expression of atrogin-1 (Cai et al. 2004). Instead, NFκB induces expression of MuRF1, an E3-ubiquitin ligase, which, like atrogin-1, is induced by denervation (Bodine et al. 2001; Cai et al. 2004). To determine whether the severe wasting of denervated runx1 mutant myofibers is due to excessive activation of this NFκB pathway, we measured MuRF1 (Trim63; Rnf28) expression in innervated and denervated muscles from control and runx1 mutant mice. Figure 3 shows that denervation leads to a 4.7 ± 0.7-fold (mean ± SEM, n = 2) increase in MuRF1 expression in control mice and a 9.5 ± 0.1-fold (mean ± SEM, n = 2) increase in MuRF1 expression in runx1 mutant muscle (Fig. 3). Thus, the severe wasting of denervated, runx1 mutant muscle is unlikely to be caused by excessive activation of this NFκB pathway. Although Atrogin-1 and MuRF1 may contribute to wasting of runx1 mutant muscle, other pathways are likely to be responsible for the striking decrease in myofiber size in denervated runx1 mutant muscle.
Runx1 induction is required to prevent disused myofibers from undergoing autophagy, myofibrillar disorganization, and severe muscle wasting
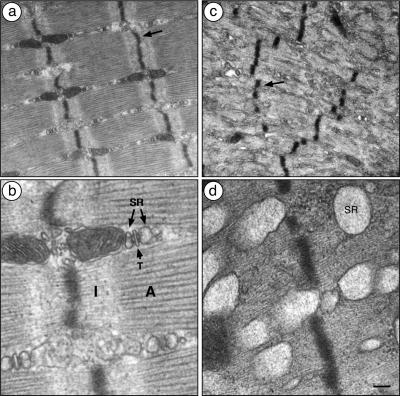
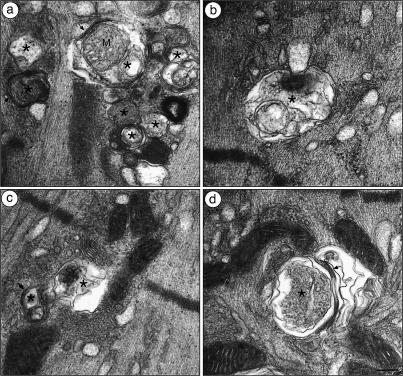
In wild-type mice, disused myofibers have a reduced capacity to generate force, due to their smaller cross-sectional area, but these atrophic myofibers retain most structural features that are characteristic of innervated muscle. We examined runx1 mutant and control soleus muscles by electron microscopy to determine whether denervated runx1 mutant muscles display additional signs of muscle wasting. We found that denervated runx1 mutant myofibers have structural aberrations that are not evident in atrophic denervated control myofibers, but are reminiscent of structural abnormalities found in a variety of congenital myopathies (Fig. 4; Engel 1999; Nishino 2003; Selcen et al. 2004). First, in denervated runx1 mutant muscles, the Z-discs are misaligned, fragmented, and irregularly spaced. Second, the distinctive A- and I-bands, readily evident in innervated and denervated muscles of control mice, are not apparent in denervated runx1 mutant muscles. Instead, the myofibrils appear to contain only thin filaments, presumably composed of actin, and to lack thick filaments composed of myosin. Third, the sarcoplasmic reticulum in denervated runx1 mutant muscle is severely dilated and extends from the Z-disc into the space normally occupied by myofibrils. Fourth, scattered throughout denervated runx1 mutant myofibers are double- or multimembrane vacuoles, which enclose heterogeneous contents including mitochondria and additional membrane-enclosed structures (Fig. 5). These features define these organelles as autophagic vacuoles (Gozuacik and Kimchi 2004), demonstrating that Runx1 is required to prevent denervated myofibers from undergoing autophagy, and indicating that excessive autophagy is responsible for the severe wasting of denervated runx1 mutant myofibers.
Figure 4.
Z-discs are disorganized, thick filaments are absent, and the sarcoplasmic reticulum is dilated in runx1 mutant denervated myofibers. (a,b) In denervated (14-d) muscle of control runx1f/f mice, the Z-discs (arrow) are aligned, the actin-only I-bands (I) and actin + myosin A-bands (A) are readily apparent, and the sarcoplasmic reticulum (SR) and transverse-tubules (T) are situated between the myofibrils. (c,d) In denervated (14-d) MCK::cre; runx1f/f myofibers, the Z-discs are misaligned and fragmented (arrow in c), distinct I- and A-bands are not discernible, and the SR is severely dilated and extends from the Z-disc into the myofibril. Myofibers that had been denervated for 1 wk were similarly, though less severely affected. Bars: a,c, 0.5 μm; b,d, 0.12 μm.
Figure 5.
Autophagic vacuoles are prominent in runx1 mutant denervated myofibers. The vacuoles in runx1 mutant denervated (14-d) muscle (MCK::cre; runx1f/f) are enclosed by two bilayers (arrows) that enclose heterogeneous contents, including mitochondria (a), myofibrils (b), and other membrane-enclosed structures (c,d), hallmark features of autophagic vacuoles (Gozuacik and Kimchi 2004). (*) Autophagic vacuoles; (M) mitochondrion. Bar, 0.12 μm.
Twenty-nine genes, encoding channels, signaling molecules, structural proteins, but not transcription factors, are misexpressed in denervated runx1 mutant muscle
A loss of muscle activity leads to a reduction in signaling that promotes muscle growth (Murgia et al. 2000), but disused muscle does not undergo autophagy. Taken together, these findings suggest four potential mechanisms by which Runx1 prevents autophagy. First, Runx1 may partially counter-balance a loss of muscle activity by inducing the same genes that are induced by muscle activity. Second, Runx1 may induce a different set of genes that compensate for the loss of muscle activity and likewise promote muscle growth/maintenance, thereby preventing autophagy. Third, Runx1 may be required to repress genes that promote autophagy. Fourth, Runx1 may induce genes that encode for structural components of the myofiber, and incomplete or partially assembled structures may lead to a stress response that provokes autophagy. To identify genes that are dependent upon Runx1 expression and regulate muscle structure, we screened oligonucleotide microarrays with RNA from innervated and denervated muscles from wild-type and runx1 mutant mice (Fig. 6). To increase the probability of identifying direct targets of Runx1, we probed microarrays with RNA from skeletal muscle that had been denervated for 3 d, 1 d after runx1 expression is maximal in wild-type mice, but prior to overt structural changes in denervated runx1 mutant muscle. We validated the microarray data by measuring expression of a subset of genes, identified as misregulated in the microarray screen, by RNase protection (Fig. 6).
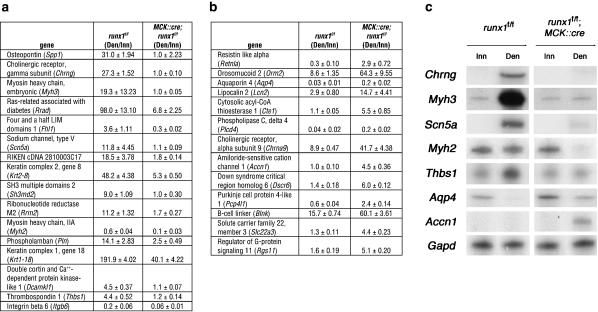
Figure 6.
Twenty-nine genes are misregulated greater than or equal to threefold in runx1 mutant muscle. We calculated the ratio (mean ± SEM, n = 3) of expression, measured on Affymetrix 430 2.0 microarrays (see Materials and Methods), in denervated/innervated (Den/Inn) muscles from runx1f/f control mice and from runx1f/f; MCK::cre mice. (a) Sixteen genes are induced or maintained by Runx1, as the expression ratio is greater in control than runx1 mutant muscle. (b) Thirteen genes are repressed by Runx1, as the expression ratio is less in control than runx1 mutant muscle. (c) We validated the microarray data by measuring expression of a subset of genes, identified as misregulated in the microarray screen, by RNase protection (Zhu et al. 1994). These data confirm that expression of these genes is misregulated in runx1 mutant muscle.
Only 29 genes are misregulated (greater than or equal to threefold) in denervated muscle lacking Runx1, suggesting that only a few genes are responsible for the dramatic muscle wasting observed in runx1 mutant mice. To assess the reliability of the microarray data, we measured expression of six misregulated genes in innervated and denervated muscle from wild-type and runx1 mutant muscle by an RNase protection assay. In each case, the RNase protection data corroborated the results from the microarray analysis, confirming the reliability of the microarray data (Fig. 6).
As most genes are unaffected by Runx1 expression, including genes that are known to be dependent upon innervation including myogenin, MuSK, and the acetylcholine receptor α and δ subunit genes, it is unlikely that Runx1 simply compensates for a loss of innervation. The 29 misregulated genes encode ion channels (five genes), signaling molecules (14 genes), and structural proteins (four genes), but not transcription factors, indicating that the identified genes are good candidates for direct targets of Runx1 (Fig. 6). Sixteen genes are not appropriately up-regulated or maintained in runx1 mutant denervated muscle, suggesting that Runx1 activates their expression. Thirteen genes are expressed at unusually high levels in denervated muscle lacking Runx1, suggesting that Runx1 represses their expression. These findings are consistent with other studies showing that Runx family members can activate or repress target genes (Kramer et al. 1999). Further studies of these Runx1 target genes should lead to a better understanding of their roles in muscle wasting.
Discussion
Disruptions in myofiber electrical activity, including denervation, lead to muscle atrophy (Jagoe and Goldberg 2001; Glass 2003). Although innervation provides trophic signals that are critical for muscle differentiation and growth, atrophic myofibers neither degenerate nor undergo apoptosis, but retain most of the structural features that are characteristic of normal muscle. These results suggest that myofiber size and differentiation are only partially dependent upon innervation or that denervation induces compensatory mechanisms that limit the extent of myofiber atrophy and wasting. Our results demonstrate that Runx1 induction is required to prevent myofibrillar disorganization and severe muscle wasting, indicating that muscle disuse indeed induces pathways that compensate for a loss innervation, thereby preventing myofibrillar disorganization and limiting the extent of muscle atrophy and wasting (Fig. 7). Moreover, our experiments demonstrate that these compensatory pathways depend upon Runx1 induction.
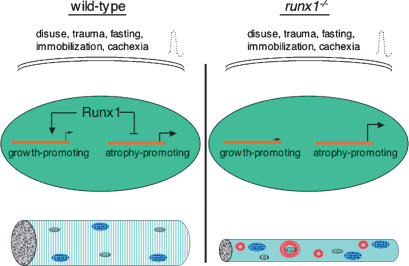
Figure 7.
Model for control of gene expression and muscle wasting by Runx1 in denervated muscle. Muscle disuse, caused by damage to peripheral nerves or by immobilization, as well as nutritional starvation and cancer cachexia, lead to muscle atrophy. In wild-type mice, induction of Runx1 limits the extent of muscle wasting and preserves the structural integrity of myofibrils. In the absence of Runx1 induction, muscle disuse leads to the disorganization of myofibrils, excessive autophagy, and severely atrophic myofibers. Runx1 activates and represses genes in denervated myofibers, and misregulation of these genes in runx1 mutant muscles is responsible for the severe muscle wasting.
Despite the importance of Runx1 in leukemogenesis, only CD4 has been identified as a bona-fide target of Runx1 in hematopoietic cells (Taniuchi et al. 2002). Among the Runx1-dependent genes identified in our microarray screen, we identified several genes, including acetylcholine receptor α9 subunit, osteopontin, orosomucoid 2, lipocalin 2, and amiloride-sensitive cation channel 1, that were not known to be expressed in skeletal muscle. In addition, the microarray screen identified multiple genes, including aquaporin 4, keratin 2–8, and keratin 1–18, that were known to be expressed in muscle, but not known to be regulated by innervation. Moreover, the screen identified multiple genes, including embryonic myosin heavy chain, sodium channel type V, and acetylcholine receptor γ subunit genes, that were known to be regulated by electrical activity, but by unknown transcriptional mechanisms.
Similarly, very few target genes for Runx2 and Runx3 have been identified. Nonetheless, Runx2 activates osteopontin expression in bone and cartilage (Ducy et al. 1997; Komori et al. 1997). Because we find that osteopontin is a target of Runx1 in skeletal muscle, these results demonstrate that the same gene can be a target for different Runx family members in different cell types. Thus, other Runx1-target genes, identified in our screen of skeletal muscle, may be targets for Runx family members in other cell types.
The genes that are misregulated in runx1 mutant muscle provide clues to the causes for the profound structural changes. Notably, expression of myosin heavy-chain IIA (myh2) is not maintained (sixfold decrease) and embryonic myosin heavy chain (myh3) fails to be induced (19-fold decrease) following denervation of runx1 mutant muscle. This reduction in myosin expression may therefore explain the absence of thick filaments and distinct A- and I-bands in runx1 mutant denervated muscle. Two keratin genes (Krt1–18, Krt2–8), which encode subunits of a heterodimer, fail to be appropriately induced (five- and ninefold decrease, respectively) in denervated runx1 mutant muscle. As these keratins are thought to link Z-discs and M-lines with costameres (Ursitti et al. 2004), plasma membrane structures proposed to anchor myofibrils, a reduction in Krt1–18 and Krt2–8 expression may account for the fragmentation and misalignment of Z-discs. Moreover, these findings raise the possibility that a failure to induce myosin and keratin expression not only leads to myofibrillar disorganization, but by leading to structural perturbations, may also trigger a stress response that stimulates autophagy. Alternatively, muscle wasting may occur independently from the myofibrillar defects. As myofiber size can be regulated by signaling proteins, such as IGF-1 (Florini et al. 1991; Goldspink 1999; Musaro et al. 1999; Rommel et al. 2001), the inappropriate expression of genes encoding secreted signaling molecules may contribute to wasting of runx1 mutant denervated muscle. The failure to induce osteopontin and thrombospondin I (31-fold and fourfold decrease, respectively) raises the possibility that their induction may be required to counter-balance the loss of innervation-dependent growth signals and promote muscle growth/maintenance. In addition, overexpression of orosomucoid 2 and lipocalin 2 (seven- and fivefold increase, respectively) in runx1 mutant denervated muscle is consistent with the possibility that their anomalous expression promotes muscle wasting. Further, increased expression of genes that regulate metabolism, such as resistin-like α or cytosolic acyl-CoA-thioesterase (10-fold and fivefold increase, respectively) may stimulate autophagy and contribute to muscle wasting. Given the small number of Runx1 target genes, further studies of these genes should lead to a detailed understanding of the mechanisms that regulate skeletal muscle wasting and may allow for a rational strategy to control autophagy in diseased muscle.
Mammalian Runx proteins contain motifs that allow Runx to associate with coactivators and corepressors, and Drosophila Runx proteins can indeed activate and repress transcription in vivo (Kramer et al. 1999). Although several studies have shown that mammalian Runx proteins can repress transcription in vivo, the evidence for transcriptional activation in vivo is less compelling (Durst and Hiebert 2004; Taniuchi and Littman 2004). We show that Runx1 is required to activate and repress gene expression in mammalian skeletal muscle, and our data indicate that these Runx1-dependent genes are direct targets for Runx1. Thus, these data indicate that mammalian Runx proteins, like their Drosophila counterparts, can activate and repress target genes in vivo.
Autophagy is responsible for the normal bulk degradation of long-lived proteins and organelles, but the program can be overactivated by a variety of stress stimuli, presumably to facilitate cell survival during periods of acute stress (Klionsky and Emr 2000). The pathway for constructing autophagic vacuoles and delivering vacuoles to lysosomes requires the sequential action of a set of genes, identified in yeast and conserved in mammalian cells. The products of these “autophagy genes”, mTOR, class III PI3K, two ubiquitin-like protein conjugation systems, and a cysteine protease, act in a concerted series of post-translational modification steps to form double-membrane autophagosomes and to transfer autophagic vacuoles and their contents to lysosomes (Klionsky and Emr 2000; Gozuacik and Kimchi 2004). The transcriptional changes that initiate and attenuate this autophagy program are poorly understood. Our findings indicate that Runx1 has a role in restraining pathways leading to autophagy, as a failure to up-regulate Runx1 in denervated muscle results in severe muscle wasting accompanied by hallmarks of autophagy.
Although it is currently unclear whether excessive autophagy promotes or prevents cell damage and leads to improvement or worsening of disease outcome, the presence of autophagic vacuoles is a prominent and characteristic structural feature in a variety of congenital myopathies (Engel 1999; Nishino 2003; Selcen et al. 2004), neurodegenerative disorders (Shintani and Klionsky 2004), and cancer (Gozuacik and Kimchi 2004; Shintani and Klionsky 2004). Myofibrillar myopathies, characterized by pathological defects in myofibrillar organization and accumulation of autophagic vacuoles, can be caused by mutations in Z-disc proteins, desmin (Goldfarb et al. 1998; Munoz-Marmol et al. 1998), αB-crystallin (Vicart et al. 1998; Selcen and Engel 2003), or myotilin (Selcen and Engel 2004). Moreover, Danon's disease, which is also typified by the presence of autophagic vacuoles, is caused by mutations in LAMP-2, a lysosomal membrane protein (Nishino et al. 2000; Tanaka et al. 2000). The genes responsible for most congenital myopathies, however, are not yet known. Our findings demonstrate an unexpected role for electrical activity in regulating autophagy and raise the possibility that reductions in muscle activity could cause or contribute to these static muscle-wasting diseases if expression of Runx1 or Runx1-target genes were compromised. Moreover, these findings raise the intriguing possibility that congenital myopathies, which do not follow simple Mendelian inheritance or become evident only late in life, may require two initiating events: a decrease in muscle activity due to trauma, aging, or immobilization, as well as a mutation in Runx1 or its target genes.
Materials and methods
Hind-limb muscles of adult mice were denervated by cutting the sciatic nerve. Mice were perfused with fixative (1% glutaraldehyde, 4% formaldehyde in 0.1M sodium phosphate at pH 7.3), and dissected muscles were immersion-fixed for a further 1 h. The fixed muscles were treated with 1% osmium for 1 h, stained en bloc with saturated aqueous uranyl acetate for 1 h, dehydrated, and embedded in Epon. For light microscopy, crosssections of 2 wk-denervated muscles were stained with toluidine blue; images were captured on a Sony DKC-500 camera, and the cross-sectional area of individual myofibers was measured using NIH ImageJ. We measured muscle wet weight and myofiber size from three mutant and control mice. For electron microscopy, longitudinal sections of 2 wk-denervated muscle were stained with uranyl acetate and lead citrate.
Runx1 RNA expression in innervated and denervated gastrocnemius muscles was measured by RNase protection (Zhu et al. 1994); a uniformly labeled RNA probe, complimentary to the sequences encoded by exons 4, 5, and 18 bp from exon 3, was hybridized to RNA isolated from innervated or 4-d denervated muscle of MCK::cre; runx1f/f mice, or runx1f/f control mice. Runx1+/– mice were generated by crossing runx1f/+ mice with CMV::cre mice. Expression of MuRF1 (AK052911, nucleotides 172–471), Fbxo32 (NM_026346, nucleotides 256–444) and Gapd were measured by RNase protection (Blagden et al. 2004).
Affymetrix mouse genome 430 2.0 microarrays were probed with cDNA copied from RNA isolated from innervated and 3-d denervated tibialis anterior muscles from runxf/f control and MCK::cre; runxf/f male mice. Experiments were performed on three separate microarrays for each of the four experimental conditions. The chips were scanned with an Affymetrix GeneChip Scanner 3000, and the raw data (http://arrayconsortium.tgen.org/np2/viewProject.do?action=viewProject&projectId=64764) were processed with Affymetrix GCOS software. The signals were normalized and analyzed by dChip (Parmigiani 2003). We calculated the ratio of expression in denervated/innervated muscle for runxf/f control and MCK::cre; runxf/f mice.
Results from the microarray screen were validated by measuring expression of selected target genes using an RNase protection assay: Myh2 (NM_144961, nucleotides 1650–1910), Myh3 (XM_354614, nucleotides 2104–2372), Aqp4 (NM_009700, nucleotides 445–895), Accn1 (NM_007384, nucleotides 1406–1672), Thbs1 (NM_011580, nucleotides 298–548), Chrng (NM_009604, nucleotides 696–1244), and Scn5a (NM_021544, nucleotides 76–611).
Acknowledgments
We thank Jack McMahan, Jack Rosenbluth, and Clara Franzini-Armstrong for advice and discussions, Ron Kahn for the MCK::cre mice, David Glass for the sequence of MuRF1 primers, Takeshi Egawa for his assistance, and Ruth Lehmann and lab members for comments on the manuscript. This work was supported by grants from the NIH to S.J.B. and from HHMI to D.R.L. The microarray work was completed through the NINDS NIMH Microarray Consortium (http://arrayconsortium.tgen.org) with the assistance of Zugen Chen.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1318305.
References
- Blagden C.S., Fromm, L., and Burden, S.J. 2004. Accelerated response of the myogenin gene to denervation in mutant mice lacking phosphorylation of myogenin at threonine 87. Mol. Cell. Biol. 24: 1983–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine S.C., Latres, E., Baumhueter, S., Lai, V.K., Nunez, L., Clarke, B.A., Poueymirou, W.T., Panaro, F.J., Na, E., Dharmarajan, K., et al. 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294: 1704–1708. [DOI] [PubMed] [Google Scholar]
- Bruning J.C., Michael, M.D., Winnay, J.N., Hayashi, T., Horsch, D., Accili, D., Goodyear, L.J., and Kahn, C.R. 1998. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol. Cell 2: 559–569. [DOI] [PubMed] [Google Scholar]
- Cai D., Frantz, J.D., Tawa Jr., N.E., Melendez, P.A., Oh, B.C., Lidov, H.G., Hasselgren, P.O., Frontera, W.R., Lee, J., Glass, D.J., et al. 2004. IKKβ/NF-κB activation causes severe muscle wasting in mice. Cell 119: 285–298. [DOI] [PubMed] [Google Scholar]
- de Bruijn M.F. and Speck, N.A. 2004. Core-binding factors in hematopoiesis and immune function. Oncogene 23: 4238–4248. [DOI] [PubMed] [Google Scholar]
- Drissi H., Luc, Q., Shakoori, R., Chuva De Sousa Lopes, S., Choi, J.Y., Terry, A., Hu, M., Jones, S., Neil, J.C., Lian, J.B., et al. 2000. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J. Cell. Physiol. 184: 341–350. [DOI] [PubMed] [Google Scholar]
- Ducy P., Zhang, R., Geoffroy, V., Ridall, A.L., and Karsenty, G. 1997. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 89: 747–754. [DOI] [PubMed] [Google Scholar]
- Durst K.L. and Hiebert, S.W. 2004. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene 23: 4220–4224. [DOI] [PubMed] [Google Scholar]
- Engel A.G. 1999. Myofibrillar myopathy. Ann. Neurol. 46: 681–683. [DOI] [PubMed] [Google Scholar]
- Florini J.R., Ewton, D.Z., and Magri, K.A. 1991. Hormones, growth factors, and myogenic differentiation. Annu. Rev. Physiol. 53: 201–216. [DOI] [PubMed] [Google Scholar]
- Glass D.J. 2003. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat. Cell. Biol. 5: 87–90. [DOI] [PubMed] [Google Scholar]
- Goldfarb L.G., Park, K.Y., Cervenakova, L., Gorokhova, S., Lee, H.S., Vasconcelos, O., Nagle, J.W., Semino-Mora, C., Sivakumar, K., and Dalakas, M.C. 1998. Missense mutations in desmin associated with familial cardiac and skeletal myopathy. Nat. Genet. 19: 402–403. [DOI] [PubMed] [Google Scholar]
- Goldspink G. 1999. Changes in muscle mass and phenotype and the expression of autocrine and systemic growth factors by muscle in response to stretch and overload. J. Anat. 194: 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozuacik D. and Kimchi, A. 2004. Autophagy as a cell death and tumor suppressor mechanism. Oncogene 23: 2891–2906. [DOI] [PubMed] [Google Scholar]
- Jagoe R.T. and Goldberg, A.L. 2001. What do we really know about the ubiquitin–proteasome pathway in muscle atrophy? Curr. Opin. Clin. Nutr. Metab. Care 4: 183–190. [DOI] [PubMed] [Google Scholar]
- Klionsky D.J. and Emr, S.D. 2000. Autophagy as a regulated pathway of cellular degradation. Science 290: 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T., Yagi, H., Nomura, S., Yamaguchi, A., Sasaki, K., Deguchi, K., Shimizu, Y., Bronson, R.T., Gao, Y.H., Inada, M., et al. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89: 755–764. [DOI] [PubMed] [Google Scholar]
- Kramer S.G., Jinks, T.M., Schedl, P., and Gergen, J.P. 1999. Direct activation of Sex-lethal transcription by the Drosophila runt protein. Development 126: 191–200. [DOI] [PubMed] [Google Scholar]
- Munoz-Marmol A.M., Strasser, G., Isamat, M., Coulombe, P.A., Yang, Y., Roca, X., Vela, E., Mate, J.L., Coll, J., Fernandez-Figueras, M.T., et al. 1998. A dysfunctional desmin mutation in a patient with severe generalized myopathy. Proc. Natl. Acad. Sci. 95: 11312–11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia M., Serrano, A.L., Calabria, E., Pallafacchina, G., Lomo, T., and Schiaffino, S. 2000. Ras is involved in nerve-activity-dependent regulation of muscle genes. Nat. Cell. Biol. 2: 142–147. [DOI] [PubMed] [Google Scholar]
- Musaro A., McCullagh, K.J., Naya, F.J., Olson, E.N., and Rosenthal, N. 1999. IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature 400: 581–585. [DOI] [PubMed] [Google Scholar]
- Nguyen Q.G., Buskin, J.N., Himeda, C.L., Shield, M.A., and Hauschka, S.D. 2003. Differences in the function of three conserved E-boxes of the muscle creatine kinase gene in cultured myocytes and in transgenic mouse skeletal and cardiac muscle. J. Biol. Chem. 278: 46494–46505. [DOI] [PubMed] [Google Scholar]
- Nishino I. 2003. Autophagic vacuolar myopathies. Curr. Neurol. Neurosci. Rep. 3: 64–69. [DOI] [PubMed] [Google Scholar]
- Nishino I., Fu, J., Tanji, K., Yamada, T., Shimojo, S., Koori, T., Mora, M., Riggs, J.E., Oh, S.J., Koga, Y., et al. 2000. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 406: 906–910. [DOI] [PubMed] [Google Scholar]
- Okuda T., van Deursen, J., Hiebert, S.W., Grosveld, G., and Downing, J.R. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84: 321–330. [DOI] [PubMed] [Google Scholar]
- Parmigiani G. 2003. The analysis of gene expression data: Methods and software. Springer, New York.
- Rommel C., Bodine, S.C., Clarke, B.A., Rossman, R., Nunez, L., Stitt, T.N., Yancopoulos, G.D., and Glass, D.J. 2001. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell. Biol. 3: 1009–1013. [DOI] [PubMed] [Google Scholar]
- Sandri M., Sandri, C., Gilbert, A., Skurk, C., Calabria, E., Picard, A., Walsh, K., Schiaffino, S., Lecker, S.H., and Goldberg, A.L. 2004. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117: 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcen D. and Engel, A.G. 2003. Myofibrillar myopathy caused by novel dominant negative α B-crystallin mutations. Ann. Neurol. 54: 804–810. [DOI] [PubMed] [Google Scholar]
- ____. 2004. Mutations in myotilin cause myofibrillar myopathy. Neurology 62: 1363–1371. [DOI] [PubMed] [Google Scholar]
- Selcen D., Ohno, K., and Engel, A.G. 2004. Myofibrillar myopathy: Clinical, morphological and genetic studies in 63 patients. Brain 127: 439–451. [DOI] [PubMed] [Google Scholar]
- Shintani T. and Klionsky, D.J. 2004. Autophagy in health and disease: A double-edged sword. Science 306: 990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt T.N., Drujan, D., Clarke, B.A., Panaro, F., Timofeyva, Y., Kline, W.O., Gonzalez, M., Yancopoulos, G.D., and Glass, D.J. 2004. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 14: 395–403. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Guhde, G., Suter, A., Eskelinen, E.L., Hartmann, D., Lullmann-Rauch, R., Janssen, P.M., Blanz, J., von Figura, K., and Saftig, P. 2000. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406: 902–906. [DOI] [PubMed] [Google Scholar]
- Taniuchi I. and Littman, D.R. 2004. Epigenetic gene silencing by Runx proteins. Oncogene 23: 4341–4345. [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Osato, M., Egawa, T., Sunshine, M.J., Bae, S.C., Komori, T., Ito, Y., and Littman, D.R. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111: 621–633. [DOI] [PubMed] [Google Scholar]
- Ursitti J.A., Lee, P.C., Resneck, W.G., McNally, M.M., Bowman, A.L., O'Neill, A., Stone, M.R., and Bloch, R.J. 2004. Cloning and characterization of cytokeratins 8 and 19 in adult rat striated muscle. Interaction with the dystrophin glycoprotein complex. J. Biol. Chem. 279: 41830–41838. [DOI] [PubMed] [Google Scholar]
- Vicart P., Caron, A., Guicheney, P., Li, Z., Prevost, M.C., Faure, A., Chateau, D., Chapon, F., Tome, F., Dupret, J.M., et al. 1998. A missense mutation in the αB-crystallin chaperone gene causes a desmin-related myopathy. Nat. Genet. 20: 92–95. [DOI] [PubMed] [Google Scholar]
- Wang Q., Stacy, T., Binder, M., Marin-Padilla, M., Sharpe, A.H., and Speck, N.A. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. 93: 3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Yeadon, J.E., and Burden, S.J. 1994. AML1 is expressed in skeletal muscle and is regulated by innervation. Mol. Cell. Biol. 14: 8051–8057. [DOI] [PMC free article] [PubMed] [Google Scholar]