Abstract
The product of the gene mutated in Bloom's syndrome, BLM, is a 3′–5′ DNA helicase belonging to the highly conserved RecQ family. In addition to a conventional DNA strand separation activity, BLM catalyzes both the disruption of non-B-form DNA, such as G-quadruplexes, and the branch migration of Holliday junctions. Here, we have characterized a new activity for BLM: the promotion of single-stranded DNA (ssDNA) annealing. This activity does not require Mg2+, is inhibited by ssDNA binding proteins and ATP, and is dependent on DNA length. Through analysis of various truncation mutants of BLM, we show that the C-terminal domain is essential for strand annealing and identify a 60 amino acid stretch of this domain as being important for both ssDNA binding and strand annealing. We present a model in which the ssDNA annealing activity of BLM facilitates its role in the processing of DNA intermediates that arise during repair of damaged replication forks.
INTRODUCTION
Bloom's syndrome (BS) is a rare genetic disorder associated with several abnormalities, including proportional dwarfism, sunlight sensitivity and a predisposition to cancers of most types. The gene mutated in BS, BLM, encodes a protein comprising 1417 amino acids that is a member of the highly conserved RecQ family (1,2). This family includes Sgs1p of Saccharomyces cerevisiae, Rqh1p of Schizosaccharomyces pombe, and the WRN, RECQ1, RECQ4 and RECQ5 proteins in humans. WRN and RECQ4 are defective in Werner's and Rothmund–Thomson syndromes, respectively, which are disorders not only associated with cancer predisposition but also with some features of premature aging (2). Where tested, all RecQ family members are DNA helicases that translocate along DNA strands in the 3′–5′ direction. A number of studies have shown that RecQ helicases unwind a wide variety of different oligonucleotide-based DNA substrates, including forked duplexes, four-way junctions modeling the Holliday junction, and simple 3′-tailed duplexes. Moreover, RecQ helicases can even unwind non-B-form DNA structures, such as G-quadruplexes (3–7).
RecQ helicases are defined by a highly conserved domain that contains seven signature motifs (hereafter referred to as the helicase domain) (2). These sequence motifs are also found, in a related form, in many other RNA or DNA helicases from different families. Outside of the helicase domain there is much less sequence similarity between RecQ family members. Nevertheless, there are two additional identifiable domains that are found in some, but not all, RecQ family proteins. One of these, the RQC (RecQ C-terminal) domain, is apparently unique to the RecQ family and is found in BLM. Recent X-ray crystallographic analysis of Escherichia coli RecQ protein indicates that the RQC domain forms a so-called winged-helix structure that is implicated in the binding of DNA (8). Indeed, a previous study showed that the RQC domain of WRN binds several DNA structures, including a forked duplex and a four-way junction (9). Interestingly, in some RecQ helicases, the RQC domain also seems to direct protein–protein interactions (10–12). A second sequence feature found in certain RecQ helicases, including BLM, is the HRDC (helicase and RNaseD C-terminal) domain (13). This domain is poorly conserved at the primary sequence level, but seemingly has a well-conserved structural fold comprising five α-helices and a number of basic residues that constitute a putative DNA binding surface (14). This domain is not unique to RecQ helicases, being present also in RNaseD homologs and, in a related form, in some DNA polymerases and recombinases. In those RecQ helicases where the RQC and HRDC domains are present, it is possible that interactions with different DNA substrates are influenced by either or both of these putative auxiliary DNA binding regions in the enzyme. Nevertheless, because some RecQ enzymes only contain the helicase domain, and lack the RQC and HRDC domains, it seems highly likely that the helicase domain is both necessary and sufficient for at least some enzymatic activities. The functional role(s) of the RQC and HRDC domains can only be the subject of speculation at this stage, but a possible candidate is to extend the range of alternate DNA structures, such as G-quadruplexes or Holliday junctions, that can be recognized by a particular RecQ helicase.
Our studies of the RecQ helicase family are focused on the BLM protein. We have previously shown that BLM is a DNA structure-specific helicase that unwinds a wide variety of DNA molecules, although it is incapable of unwinding a blunt-ended DNA duplex (3,15). Nevertheless, it can disrupt a four-way junction modeling the Holliday junction recombination intermediate even if that substrate has blunt termini (15). Consistent with this activity being dependent upon branch migration, BLM has been shown to catalyze branch migration of bona fide Holliday junctions generated by the RecA recombinase (15). Here, we report a new activity for the BLM protein: the promotion of single-stranded DNA (ssDNA) annealing. We have characterized the biochemical properties of this strand annealing activity and have identified a short region of the C-terminal domain of BLM that is essential for this function. We provide evidence that strand annealing is influenced by ssDNA binding proteins, by nucleotide co-factors and by DNA length.
MATERIALS AND METHODS
Plasmids
Plasmids pJP71, pJP74 and pJP75, which were used for the expression of the truncated BLM variants, BLM642-1290, BLM642-1417 and BLM642-1350, respectively, were constructed as described previously (16). Briefly, the region of the BLM cDNA encoding residues 642–1290, 642–1350 or 642–1417 was amplified by PCR with primers introducing NcoI and SapI sites. The PCR product was digested with NcoI and SapI, and inserted between the NcoI and SapI sites of pTXB3 (NEB). Constructs for the expression of BLM213-1417 and BLM213-1267 have been described previously (17).
Protein purification
BLM, BLM213-1417 and BLM213-1267 were expressed as C-terminally hexahistidine-tagged proteins in the protease-deficient yeast strain JEL1 (MATα leu2 trp1 ura3-52 prb1-1122 pep4-3 his3::PGAL10-GAL4) and were purified as described previously (18). BLM642-1290, BLM642-1350 and BLM642-1417 were produced as C-terminal fusion proteins with the Mxe-CBD affinity tag in the E.coli BL21-Codonplus-(DE3)-RIL strain (Stratagene) and were purified as described previously (16). Human replication protein A (RPA) was kindly provided by Dr A Vindigni. E.coli ssDNA binding protein (SSB) was purchased from USB. All concentrations of proteins are given in terms of moles of monomer.
DNA substrates
All oligonucleotides used in this study were purchased from Sigma Genosys and were purified by PAGE. X12-1 (5′-CGGGTCAACGTGGGCAAAGATGTCCTAGCAATGTAATCGTCTATGAGACG-3′) and its complement X12-2 (5′-GACGCTGCCGAATTCTGGCTTGCTAGGACATCTTTGCCCACG TTGACCCG-3′) were used for the generation of the 31 bp forked duplex. DF-1 (5′-TGTAATCGTCTAT GAGACGCGGGTCAACGTGGACATCTGCAAAGATGTCCTAGCAATGTAATCGTCTATGAGACG-3′) and its complement DF-2 (5′-GACGCTGCCGAATTCTGGCTTGCTAGGACATC TTTGCAGATGTCCACGTTGACCCGGACGCTGCCGAATTCTGGC-3′) were used for the generation of the twin-forked duplex. DS-50-1 (5′-GGCAAAGATGTCCTAGCAACGGGTCAACGTGGGCAAAGATGTCCTAGCAA-3′) and its complement DS-50-2 (5′-TTGCTAGGACATCTTTGCCCACGTTGACCCGTTGCTAGGACATCTTT GCC-3′) were used for the generation of the 50 bp duplex; DS-31-1 (5′-CGGGTCAACGTGGGCAAAG ATGTCCTAGCAA-3′) and its complement DS-31-2 (5′-TTGCTAGGACATCTTTGCCCACGTTGACCCG-3′) were used for the generation of 31 bp duplex; DS-15-1 (5′-GGCAAAGATGTCCTA-3′) and its complement DS-15-2 (5′-TAGGACATCTTTGCC-3′) were used for the generation of the 15 bp duplex. X12-1, DF-1 DS-50-1, DS-31-1 and DS-15-1 were labeled at the 5′ end using T4 polynucleotide kinase (NEB) and [γ-32P]ATP. The forked duplex used in helicase assays was generated by the annealing of X12-1 and X12-2. X12-1 was 5′-32P-labeled before annealing. All concentrations of DNA substrates are given in terms of moles of oligonucleotides.
Helicase assays
The 31 bp forked duplex was prepared as described previously (18). Reactions (20 μl) were carried out at 37°C in buffer H (50 mM Tris–HCl, pH 7.5, 50 mM NaCl, 1 mM DTT and 0.1 mg/ml BSA) supplemented with 2 mM MgCl2 and 2 mM ATP for 30 min in the presence of the indicated amounts of BLM and 1 nM of 32P-end-labeled forked duplex substrate. Where indicated, SSB and RPA were added at a concentration of 60 and 3 nM, respectively. Reactions were stopped, and the samples were de-proteinized by the addition of stop buffer (50 mM EDTA, 1% SDS and 0.1 mg/ml proteinase K) and incubated at 37°C for 10 min. The reaction products were resolved on a native 10% (w/v) polyacrylamide gel (acrylamide to bis-acrylamide ratios 19:1) run in TBE at 30 mA per gel for 1 h at 4°C. The radiolabeled DNA was visualized using a PhosphorImager and the percentage DNA unwinding was quantified using ImageQuant software.
Strand annealing assays
DNA strand annealing activity was measured using fully or partially complementary oligonucleotides (at a concentration of 1 nM each), one of which was 5′-32P-end-labeled. Reactions (20 μl) were carried out in buffer H supplemented with 2 mM MgCl2 unless otherwise indicated for 30 min at 37°C with the indicated concentrations of BLM. For time-course experiments, an 80 μl reaction was initiated and 10 μl aliquots were removed at defined time points. Reactions were stopped by the addition of stop buffer (50 mM EDTA, 1% SDS and 0.1 mg/ml of proteinase K) and incubated for 10 min at 37°C. Where indicated, RPA was added (0.75–48 nM) to the reaction. The reaction products were analyzed as described for the helicase assay.
Gel retardation assays
Radiolabeled oligonucleotide (1 nM) was incubated with BLM or its truncated derivatives in 50 mM triethanolamine, pH 7.5, 2 mM MgCl2, 2 mM ATPγS, 50 mM NaCl, 1 mM DTT and 0.1 μg/ml BSA for 20 min at 37°C. Glutaraldehyde (0.25%) was then added and the reaction was incubated for a further 10 min at 37°C. The protein–DNA complexes were resolved on a 5% (w/v) native polyacrylamide gel (acrylamide to bis-acrylamide ratios 19:1) run in TBE at 150 V for 70 min at 4°C. The radiolabeled DNA was visualized using a PhosphorImager and the percentage ssDNA bound was quantified using ImageQuant software.
RESULTS
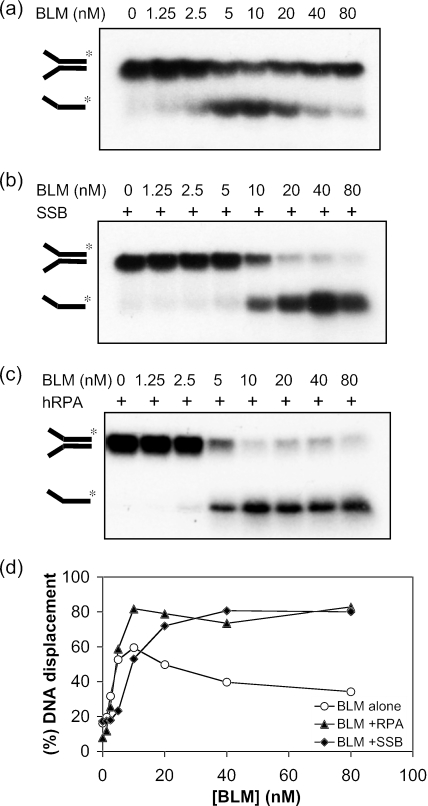
During an analysis of the DNA helicase activity of BLM, we observed that the appearance of the ssDNA product of unwinding followed an unusual pattern. The extent of ssDNA product formation increased with increasing enzyme concentration up to a point, beyond which the level of unwound product declined substantially. An example of such a pattern is shown in Figure 1a and quantified in Figure 1d for a forked DNA duplex substrate. In this particular case, the level of the ssDNA product of unwinding peaked in reactions containing ∼10 nM BLM and then declined significantly until it was only marginally above background in reactions containing 80 nM BLM (Figure 1a and quantified in Figure 1d). One explanation for this phenomenon was that high concentrations of BLM were able to promote the re-annealing of the ssDNA products of the unwinding reaction. Consistent with this was the observation that addition of either E.coli SSB or human RPA, two unrelated ssDNA binding proteins, could overcome the apparent decline in helicase activity seen at high BLM concentrations (Figure 1b–d). Previously, it was shown that BLM helicase activity is stimulated by RPA (19), but not by SSB, making it unlikely that the similar effects of RPA and SSB shown in Figure 1 involved direct stimulation of BLM helicase activity.
Figure 1.
Inhibition of BLM helicase activity occurs at high protein concentrations, which is relieved by ssDNA binding proteins. (a) Unwinding of 1 nM of a 31 bp forked duplex catalyzed by different concentrations of BLM, as indicated above the lanes. (b) Unwinding of 1 nM of a 31 bp forked duplex catalyzed by BLM in the presence of 60 nM SSB or (c) in the presence of 3 nM RPA. In (a–c), the positions of the forked duplex and the ssDNA product of unwinding are indicated on the left. (d) Quantification of the helicase activity of BLM from the data in (a–c). All reactions were incubated for 30 min at 37°C.
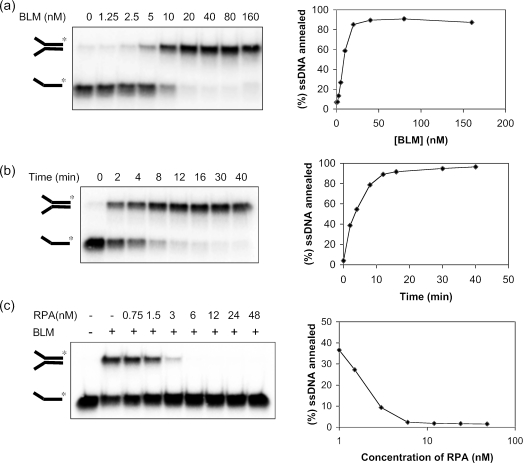
To analyze this putative ssDNA annealing activity of BLM more directly, we incubated different concentrations of BLM with the two single-stranded 50mer oligonucleotides (each at 1 nM) that were utilized to create the forked duplex used in the experiment shown in Figure 1. One of these oligonucleotides was 5′-end-labeled to allow strand annealing to be monitored. We found that BLM promoted efficient annealing of the two oligonucleotides. Using BLM concentrations of ≥20 nM, ∼90% of the labeled ssDNA was annealed to its complement (Figure 2a). Analysis of the kinetics of this reaction using a fixed BLM concentration (20 nM) indicated that 50% of the labeled oligonucleotide was annealed in <4 min (Figure 2b), using the reaction conditions described in Materials and Methods. Consistent with the notion that this reaction reflected genuine ssDNA annealing, addition of increasing concentrations of RPA progressively diminished the level of the annealed forked duplex product (Figure 2c). This confirmed that the ability of RPA to overcome the decline in helicase activity seen at high BLM concentrations (Figure 1c) was due to the inhibition of the ssDNA annealing activity of BLM.
Figure 2.
BLM promotes annealing of ssDNA. (a) Effect of BLM concentration on the annealing of two ssDNA molecules to generate a forked duplex. Reactions were incubated for 30 min. (b) Time course of ssDNA annealing in reactions containing 20 nM BLM. (c) Effect of increasing concentrations of RPA on ssDNA annealing catalyzed by 10 nM BLM. In (a–c), the percentage of ssDNA annealed was quantified and the data are presented graphically on the right of the corresponding dataset.
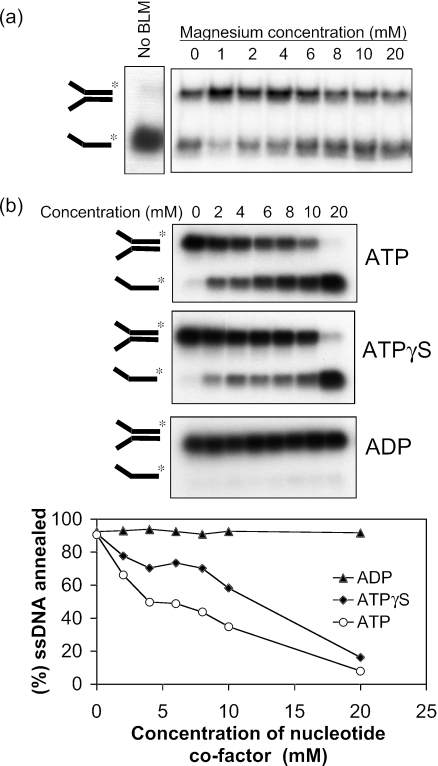
BLM requires Mg2+ and ATP to catalyze DNA unwinding (18). We tested whether there was a similar co-factor requirement for the strand annealing function. As shown in Figure 3a, strand annealing promoted by BLM did not depend on the presence of Mg2+. Indeed, the strand annealing activity was resistant to incubation with 50 mM EDTA (data not shown). Although there was a slight stimulation of strand annealing activity at a Mg2+ concentration of 1 mM, Mg2+ concentrations >4 mM caused a mild inhibition of the reaction. In marked contrast to the absolute requirement for ATP hydrolysis in DNA unwinding, increasing ATP concentrations strongly inhibited the DNA strand annealing reaction (Figure 3b). A similar, if slightly less dramatic, inhibition was seen with the poorly hydrolyzable ATP analog, ATPγS. In contrast, concentrations of ADP up to 20 mM had no detectable inhibitory effect on the reaction (Figure 3b). Hence, the ssDNA annealing reaction promoted by BLM has different Mg2+ and adenine nucleotide co-factor requirements from those needed to support BLM helicase activity.
Figure 3.
Effect of Mg2+ and adenine nucleotide co-factors on ssDNA annealing. (a) Strand annealing by BLM (10 nM) as a function of increasing Mg2+ concentration. The reaction shown in the left panel was performed in the absence of BLM. (b) Strand annealing by BLM (10 nM) as a function of increasing concentration of ATP, ATPγS or ADP, as indicated. Graph below shows quantification of the data.
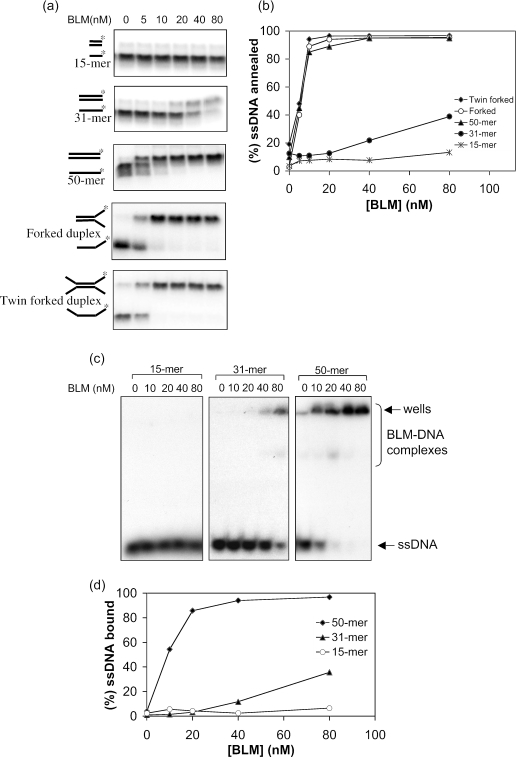
Next, we asked whether oligonucleotide length influenced the efficiency of BLM-mediated strand annealing (Figure 4). We found that complementary 15mer oligonucleotides were not annealed to any detectable extent, while a 31mer pair could be annealed, albeit very inefficiently, requiring a BLM concentration of >40 nM. Increasing the oligonucleotide length to 50 nt substantially increased the extent of annealing, particularly in reactions containing a low BLM concentration (<20 nM). We then asked whether the annealing reaction required that the two oligonucleotides be fully complementary. To analyze this, we used two related oligonucleotide pairs. One pair comprised a 31mer complementary region and a non-complementary 19mer tail, which could form a forked partial duplex, and the other comprised the same 31mer complementary region flanked by two 19mer non-complementary regions that could form a twin-forked duplex (Figure 4a). Both oligonucleotide pairs could be annealed efficiently by BLM (Figure 4a and b), indicating that the annealing reaction does not require the oligonucleotides to be fully complementary, and that the complementary portion of the oligonucleotide need not be located at the ends of the DNA molecule. Despite containing only 31 nt of complementary sequence, these oligonucleotides were annealed as efficiently as the fully complementary 50mers, and far more efficiently than the fully complementary 31mer pair (Figure 4a and b). These data indicate that, for a fixed length of complementary sequence, the overall length of the oligonucleotide strongly influences the efficiency of the annealing reaction.
Figure 4.
Strand annealing of different ssDNA structures by BLM. (a) In each case, different concentrations of BLM (indicated above the lanes) were incubated with the ssDNA species indicated on the left of each autoradiogram. The positions of the unannealed ssDNA and the annealed fully duplex or partial duplex products are indicated on the left. (b) Quantification of the data in (a). (c) Gel retardation assays with increasing quantities of BLM and the ssDNA oligonucleotides indicated above the wells. The positions of the ssDNA and the retarded BLM–DNA complexes are shown on the right. (d) Quantification of the data from (c).
Next, we addressed whether the differences in annealing efficiency noted above for different lengths of ssDNA were reflected in differences in the ability of BLM to form a stable complex with each DNA substrate. To achieve this, we used gel retardation assays with a 15mer, a 31mer and a 50mer single-stranded oligonucleotide. As shown in Figure 4c and d, BLM formed a stable complex with the 50mer and, to a much lesser extent, with the 31mer, but no complex with the 15mer could be detected. These data suggest that the DNA length-dependence of the ssDNA annealing reaction may be a reflection of the relative ability of BLM to bind stably to DNA molecules of different length.
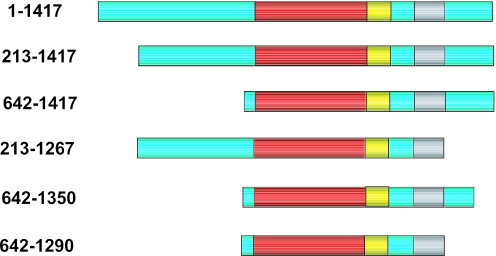
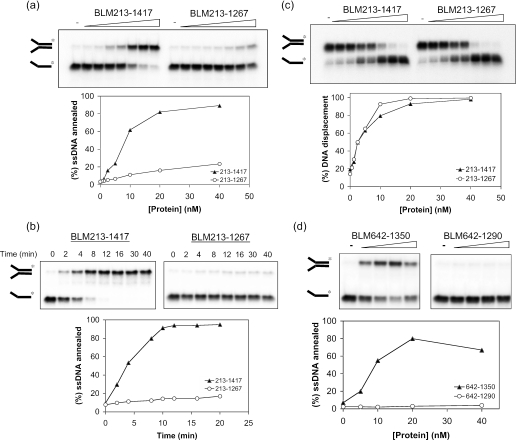
In order to identify functional domains in BLM required for the ssDNA annealing reaction, we purified a series of truncated versions of BLM, which are shown diagrammatically in Figure 5. These variants all contain the central helicase and RQC domains, but lack part or all of the N-terminal domain, and in some cases also part of the C-terminal domain. Initially, we analyzed whether strand annealing required the N- and C-terminal portions of BLM. A BLM variant lacking the N-terminal 212 amino acids (BLM213–1417) was proficient at strand annealing (Figure 6a and b), as indeed was BLM642–1417 that lacked the entire N-terminal domain (data not shown). In contrast, BLM213–1267 was barely able to catalyze strand annealing above the background level seen in the absence of BLM (Figure 6a and b) and showed a >10-fold reduced rate of strand annealing in comparison with BLM213–1417. This difference between these two BLM variants was specific for the strand annealing function of BLM, since the intrinsic helicase activity of each protein was comparable (Figure 6c). Importantly, these helicase assays were performed in the presence of SSB in order to prevent ssDNA annealing.
Figure 5.
Schematic representation of the full-length BLM (1–1417) and truncated BLM variants used in this study. Amino acid residue numbers of each protein are indicated on the left. The positions of the helicase (red), RQC (yellow), HRDC (gray) domains are indicated. Green boxes denote poorly conserved regions.
Figure 6.
The C-terminal domain of BLM, between residues 1290 and 1350, is required for ssDNA annealing. (a) Strand annealing as a function of increasing protein concentration for BLM213–1417 and BLM213–1267. Reactions were incubated for 30 min. The graph below shows quantification of the data. (b) Time course of ssDNA annealing by 20 nM BLM213–1417 or BLM213–1267. The graph below shows quantification of the data. (c) Comparison of the helicase activity of BLM213–1417 and BLM213–1267. Assays were as described in Figure 1. Graph below shows quantification of the data. (d) Strand annealing as a function of increasing protein concentration for BLM642–1290 and BLM642–1350. Graph below shows quantification of the data.
The results presented thus far would indicate that the C-terminal domain of BLM between residues 1267 and 1417 is important for its strand annealing function. Given that the entire N-terminal domain of BLM is dispensable for ssDNA annealing, we used BLM variants commencing at residue 642 to analyze whether the annealing function could be localized to a particular region of the C-terminal domain of BLM. We found that a protein truncated by only 67 C-terminal residues (BLM642–1350) could promote efficient strand annealing (Figure 6d). In contrast, removal of an additional 60 residues (to create BLM642–1290) generated a protein that had no detectable annealing activity (Figure 6d). The removal of residues 1291–1350 appear to specifically affect the strand annealing function of BLM since BLM642–1290 protein is an active helicase (16).
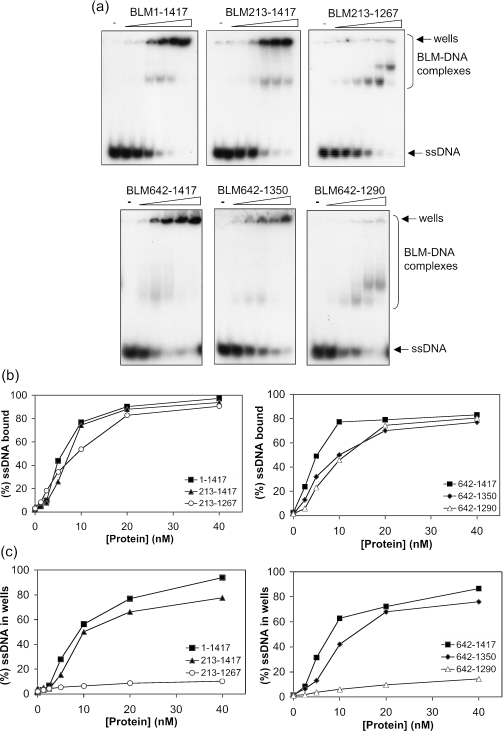
Since we had shown above that a correlation exists between DNA binding efficiency and an ability to promote annealing of different lengths of ssDNA, we next asked if those truncation mutants of BLM that were unable to promote annealing might have an alteration in their intrinsic ssDNA binding properties. Using gel retardation assays we found that, although there was no quantitative difference in the total amount of ssDNA bound by full-length BLM and its truncated derivatives, there was a clear qualitative difference in the nature of the retarded complexes when comparing strand annealing-proficient and -deficient variants. Using BLM, BLM213–1417, BLM642–1417 or BLM642–1350 at concentrations that promote efficient strand annealing, most of the retarded DNAs could not be resolved in the gel, indicative of the formation of large protein–DNA complexes (Figure 7). In contrast, the retarded species seen in reactions with the two strand annealing-defective variants, BLM213–1267 and BLM642–1290, were readily resolved within the gel (Figure 7). Taken together, these data indicate that the first 641 amino acids of BLM are not important for ssDNA binding or annealing, but that the 60 residues between positions 1290 and 1350 are essential for strand annealing and for the formation of higher-order protein–DNA complexes.
Figure 7.
The C-terminal domain of BLM is required for the formation of higher-order protein–DNA complexes. (a) Gel retardation assays of BLM1–1417, BLM213–1417, BLM213–1267 (upper panel), BLM642–1417, BLM642–1350 and BLM642–1290 (lower panel) using a 50mer ssDNA oligonucleotide. The positions of the gel wells and of BLM–DNA complexes that could be resolved in the gel are indicated on the right. (b) Quantification of total ssDNA bound by BLM and its truncated derivatives. (c) Quantification of the ssDNA that was retarded in the gel wells. For clarity, the data are represented on two graphs in each case.
DISCUSSION
We report a new activity for the BLM helicase in showing that BLM promotes efficient annealing of complementary ssDNA molecules. This activity does not require Mg2+ or ATP, the co-factors essential for BLM to perform its helicase function. We have shown that significant DNA length dependence exists for the BLM strand annealing activity, which appears to derive from differences in the ability of BLM to form stable complexes with ssDNA of different lengths. Most interestingly, this length dependence does not require that the annealed strands be fully complementary. This may suggest that the mechanism by which BLM promotes ssDNA annealing is for it to bind non-specifically to the two ssDNA molecules and then to bring them into close proximity via protein–protein interactions between BLM molecules bound to the individual oligonucleotides. The length dependence may, therefore, reflect the number of BLM molecules that can bind simultaneously to each ssDNA molecule. Using different BLM variants, we found a strong correlation exists between ssDNA annealing activity and an ability to form higher-order protein–DNA complexes that were not resolved on polyacrylamide gels. In line with the comments above, we would propose that the formation of these large molecular weight complexes is a function of the binding of multiple BLM molecules to each ssDNA oligonucleotide.
BLM may promote ssDNA annealing in a manner similar to that of the RAD52 recombination protein. RAD52 forms oligomeric rings that bind ssDNA on their outer surface, and the annealing reaction requires that multiple rings engage their bound ssDNA in a large complex (20,21). BLM is oligomeric in solution, and previous studies have identified 4- and 6-fold symmetric ring structures that can form in the absence of DNA (22). However, the oligomeric state of the catalytically active form of BLM remains to be confirmed. Certain truncated forms of BLM are monomeric in solution and yet can still perform DNA unwinding (16), suggesting that the ability to form oligomers is not obligatory for helicase function. An interesting avenue for future research will be to address whether oligomerization of BLM is required for its ssDNA annealing function.
Recent data indicate that BLM is not alone among RecQ helicases in promoting strand annealing. Garcia et al. (23) showed that human RECQ5β also possesses a DNA strand annealing activity that is strongly inhibited by RPA. However, we have data to indicate that such a strand annealing activity is not conserved in E.coli RecQ (C.F. Cheok, L. Wu and I.D. Hickson, unpublished data). This suggests that the annealing function is unlikely to be intrinsic to the highly conserved helicase and RQC domains, but rather is dictated by one of the poorly conserved N- or C-terminal regions that flank these domains in BLM. Consistent with this, the C-terminal domains of BLM and RecQ5β are required for ssDNA annealing activity. Interestingly, the C-terminal domains of these two proteins, however, show little sequence similarity. Notably, the RecQ5β C-terminal domain lacks both the 60 amino acid region found in the C-terminal domain of BLM, which we have shown is essential for ssDNA annealing activity, and an HRDC domain, which in BLM lies adjacent to the critical 60 amino acid region. Moreover, there are obvious differences in the effects of nucleotide analogs on the respective activities of the two proteins. Most notably, the annealing activity of RECQ5β is inhibited by ATPγS, but not by ATP or ADP, whereas BLM is inhibited by both ATPγS and ATP. While this manuscript was in preparation, we became aware of an electronic paper in press containing findings that overlap with those presented here. Machwe et al. (24) reported that RecQ family members, including BLM, promote DNA strand pairing as well as DNA unwinding. Our findings are in general agreement with those of Machwe et al. (24), except we did not find that BLM requires Mg2+ for its strand annealing activity. Moreover, in the present work, we have significantly extended our knowledge of the strand annealing function of BLM. In particular, we have mapped a domain in BLM vital for strand annealing activity and have defined structural features of the DNA substrates that are required for BLM to mediate this function.
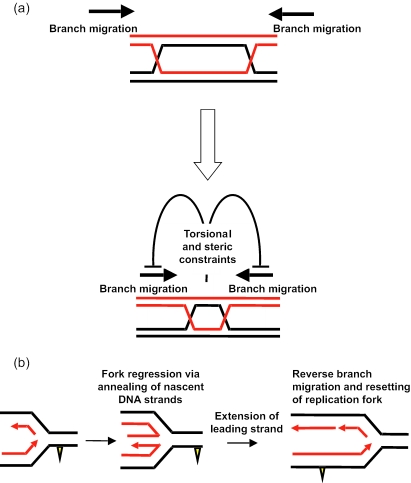
An important issue to address is why should RecQ helicases catalyze apparently antagonistic reactions: DNA unwinding and ssDNA annealing? Although the precise cellular role of any RecQ helicases is not fully elucidated, our previously published evidence indicates that BLM co-operates with topoisomerase IIIα to resolve DNA intermediates that arise during recombinational repair of replication forks (25). The ability of BLM to promote strand annealing of molecules with non-complementary ends indicates that BLM can promote strand annealing at internal sites in DNA and may not require DNA end recognition. One potential role for ssDNA annealing activity may be to promote the processing of Holliday junctions. BLM and topoisomerase IIIα together catalyze double Holliday junction dissolution (25), a process that requires the juxtaposition of two individual Holliday junctions. As two Holliday junctions are brought together in close proximity, the torsional stress generated by the convergence of the two junctions may be relieved by topoisomerases acting ahead of each migrating Holliday junction. The strand annealing activity of BLM acting on the strands behind each junction may function to overcome the torsional constraints imposed by two converging Holliday junctions (Figure 8a). Furthermore, as the two junctions are brought together in close proximity, steric hindrance may prevent either junction from being able to assume the necessary square planar configuration. Rather, each junction may assume the configuration in which the two duplexes are stacked, which is inhibitory to classical branch migration. Under these conditions, the ssDNA annealing activity of BLM may be required to promote an atypical form of Holliday junction branch migration in order to generate a hemicatenane, the presumed substrate for double Holliday junction dissolution. Our recent data indicate that the two strand annealing defective derivatives of BLM described here are also defective in catalyzing Holliday junction dissolution (26), although whether there is a mechanistic link between these two functions is unclear at this stage.
Figure 8.
Models for the possible role of the strand annealing function of BLM in replication fork maintenance. (a) Branch migration model. Schematic representation of how torsional stress and steric hindrance may impede the convergence of two Holliday junctions by branch migration. The strand annealing activity of BLM acting on strands behind each junction may act to overcome these constraints and thereby facilitate the juxtaposition of two Holliday junctions. See text for details. (b) Fork regression model. Template strands are shown in black and nascent strands in red. The yellow triangle depicts a fork-blocking adduct on the leading strand template. See text for details.
A second possible model (Figure 8b) for the role of the ssDNA annealing function specifically links BLM function with repair of damaged replication forks. Lesions on the leading strand template for DNA replication likely block fork progression. One proposal is that lesion bypass can occur via template switching in which the fork regresses via the annealing of the nascent strands to form a so-called chicken-foot structure (15,27). After extension of the leading strand and/or lesion removal, the regressed fork (a four-way junction) can be reversed by branch migration to re-set an active fork. We propose that BLM could catalyze both the fork regression step, by promoting annealing of the nascent strands, and the reversal of the regressed fork, using its helicase/branch migration function. In this way, BLM recruited to the arrested fork could promote repair and hence restoration of productive DNA replication. Clearly, further work is needed to identify whether these models have validity.
Acknowledgments
The authors thank Dr A.Vindigni for human RPA, Dr P. McHugh for helpful comments on the manuscript and members of the Genome Integrity Group for useful discussions and technical help. This work was supported by Cancer Research UK and the Swiss National Science Foundation. Funding to pay the Open Access publication charges for this article was provided by JISC. C.F.C. acknowledges the support of A*STAR (Singapore).
Conflict of interest statement. None declared.
REFERENCES
- 1.German J. Bloom's syndrome. Dermatol. Clin. 1995;13:7–18. [PubMed] [Google Scholar]
- 2.Hickson I.D. RecQ helicases: caretakers of the genome. Nature Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 3.Mohaghegh P., Karow J.K., Brosh R.M., Jr, Bohr V.A., Hickson I.D. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huber M.D., Lee D.C., Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J.L., Harrison R.J., Reszka A.P., Brosh R.M., Jr, Bohr V.A., Neidle S., Hickson I.D. Inhibition of the Bloom's and Werner's syndrome helicases by G-quadruplex interacting ligands. Biochemistry. 2001;40:15194–15202. doi: 10.1021/bi011067h. [DOI] [PubMed] [Google Scholar]
- 6.Sun H., Karow J.K., Hickson I.D., Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 7.Wu X., Maizels N. The substrate-specific inhibition of RecQ helicases. Nucleic Acids Res. 2001;29:1765–1771. doi: 10.1093/nar/29.8.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein D.A., Zittel M.C., Keck J.L. High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Kobbe C., Thoma N.H., Czyzewski B.K., Pavletich N.P., Bohr V.A. Werner syndrome protein contains three structure-specific DNA binding domains. J. Biol. Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 10.von Kobbe C., Harrigan J.A., May A., Opresko P.L., Dawut L., Cheng W.H., Bohr V.A. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol. Cell. Biol. 2003;23:8601–8613. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Opresko P.L., von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 12.Brosh R.M., Jr, von Kobbe C., Sommers J.A., Karmakar P., Opresko P.L., Piotrowski J., Dianova I., Dianov G.L., Bohr V.A. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morozov V., Mushegian A.R., Koonin E.V., Bork P. A putative nucleic acid-binding domain in Bloom's and Werner's syndrome helicases. Trends Biochem. Sci. 1997;22:417–418. doi: 10.1016/s0968-0004(97)01128-6. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z., Macias M.J., Bottomley M.J., Stier G., Linge J.P., Nilges M., Bork P., Sattler M. The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure Fold Des. 1999;7:1557–1566. doi: 10.1016/s0969-2126(00)88346-x. [DOI] [PubMed] [Google Scholar]
- 15.Karow J.K., Constantinou A., Li J.L., West S.C., Hickson I.D. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl Acad. Sci. USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janscak P., Garcia P.L., Hamburger F., Makuta Y., Shiraishi K., Imai Y., Ikeda H., Bickle T.A. Characterization and mutational analysis of the RecQ core of the Bloom's syndrome protein. J. Mol. Biol. 2003;330:29–42. doi: 10.1016/s0022-2836(03)00534-5. [DOI] [PubMed] [Google Scholar]
- 17.Wu L., Davies S.L., North P.S., Goulaouic H., Riou J.F., Turley H., Gatter K.C., Hickson I.D. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 18.Karow J.K., Chakraverty R.K., Hickson I.D. The Bloom's syndrome gene product is a 3′–5′ DNA helicase. J. Biol. Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 19.Brosh R.M., Jr, Li J.L., Kenny M.K., Karow J.K., Cooper M.P., Kureekattil R.P., Hickson I.D., Bohr V.A. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 20.Singleton M.R., Wentzell L.M., Liu Y., West S.C., Wigley D.B. Structure of the single-strand annealing domain of human RAD52 protein. Proc. Natl Acad. Sci. USA. 2002;99:13492–13497. doi: 10.1073/pnas.212449899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd J.A., Forget A.L., Knight K.L. Correlation of biochemical properties with the oligomeric state of human rad52 protein. J. Biol. Chem. 2002;277:46172–46178. doi: 10.1074/jbc.M207262200. [DOI] [PubMed] [Google Scholar]
- 22.Karow J.K., Newman R.H., Freemont P.S., Hickson I.D. Oligomeric ring structure of the Bloom's syndrome helicase. Curr. Biol. 1999;9:597–600. doi: 10.1016/s0960-9822(99)80264-4. [DOI] [PubMed] [Google Scholar]
- 23.Garcia P.L., Liu Y., Jiricny J., West S.C., Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machwe A., Xiao L., Groden J., Matson S.W., Orren D.K. RECQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J. Biol. Chem. 2005;280:23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 25.Wu L., Hickson I.D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 26.Wu L., Chan K.L., Ralf C., Bernstein D.A., Garcia P.L., Bohr V.A., Janscak P., Keck J.L., Hickson I.D. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005 doi: 10.1038/sj.emboj.7600740. doi:10.1083/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGlynn P., Lloyd R.G., Marians K.J. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc. Natl Acad. Sci. USA. 2001;98:8235–8240. doi: 10.1073/pnas.121007798. [DOI] [PMC free article] [PubMed] [Google Scholar]