In this essay, I convey the results of some of our recent studies of reproduction in the model plant, Arabidopsis thaliana. Our work has been influenced by the investigations of a large and interactive community of researchers studying a variety of plant systems, including lily, tobacco, maize, and plants from the mustard family (for recent reviews, see Cheung et al., 2000; Dixit and Nasrallah, 2001; Johnson and Preuss, 2002). This essay does not attempt to review the large body of work from the field; rather, it focuses on the questions of interest to my laboratory.
I have long been fascinated by the mechanisms that regulate cell–cell communication. For a higher plant cell, the modes of interaction are quite distinct from what we commonly see in animals. Higher plant cells typically do not migrate; instead, they spend their entire lives next to the same cells, all connected by rigid extracellular walls. Pollen grains are an exception: These cells are, in fact, small organisms. They consist of a large vegetative cell that carries within its cytoplasm two intact sperm cells, complete with their own plasma membranes and walls. To accomplish fertilization, the sperm are delivered through a pollen tube to eggs buried deep within the flower. Pollen tubes expand rapidly, invade female tissues, navigate across multiple cell layers, and finally rupture, delivering the resident sperm (for recent review, see Johnson and Preuss, 2002).
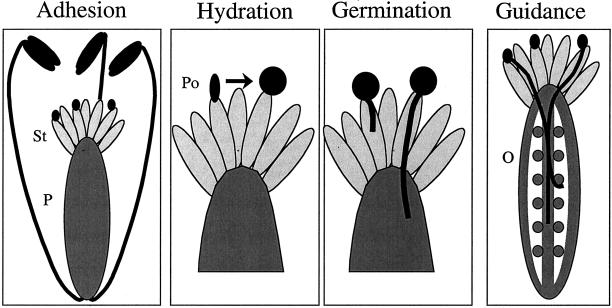
Throughout this journey, female cells continuously check the pollen's identity, inhibiting foreign pollen, while promoting fertilization by pollen from compatible plants. Thus, unlike higher animals, where visual cues, pheromones, and behavior dominate the choice of mating partners, plants choose their mates by relying on cell–cell interactions. Our work has focused on identifying the signaling molecules responsible for this cellular mating dance. Such signals are most likely specific to each plant species; consequently, we anticipate that evolution of these molecules is at least one mechanism that drives speciation. There are several molecular barriers to interspecies pollination, acting at four main checkpoints (Figure 1): 1) receptive stigma cells on the surface of the female reproductive structure (the pistil) regulate adhesion to pollen grains, 2) stigmas cells also control water traffic to the desiccated pollen soon after adhesion has occurred, 3) ovary tissues monitor the growth and invasion of pollen tubes, and 4) unfertilized ovules precisely guide pollen tubes to the eggs. Each of these steps requires the interplay of signaling molecules; pollen grains are recognized by female cells, and in turn, female cells stimulate or inhibit pollen progression.
Figure 1.
Major checkpoints in pollination. Cell–cell signaling interactions dominate the choice of plant mating partners. Plants select the most appropriate pollen (Po) by regulating adhesion to the female stigma cells (St), hydration of desiccated pollen on the stigma surface, germination of a pollen tube, guidance of pollen tubes into the pistil (P), and ultimately targeting of pollen tubes to the ovules (O, gray circles) that contain the eggs.
One of the more surprising discoveries of the past few years was the remarkable nature of the adhesive interactions between pollen and the stigma (Zinkl et al., 1999). Once thought to be a passive event, we now know that plants use this step to recognize compatible pollen grains. Despite a dry cell surface, pollen of an appropriate species binds tightly upon contacting a stigma cell. This adhesion reaction is extremely strong—the binding force is 10−7 N per pollen grain, large enough for the stigma to capture pollen as it passes by on a gentle breeze or an insect's bristles. Thus, from the very first step, cell–cell interactions dominate plant mating.
Much of our work is now focused on identifying the adhesive molecules that mediate pollen–stigma binding. Reconstitution experiments demonstrate that on the male side, the adhesives reside in the pollen wall, the exine. Unlike previously characterized adhesion proteins, exine adhesives are extremely resistant to treatments with proteases, strong acids, and heat, and they also resist extraction with aqueous and organic solutions, suggesting nonproteinaceous molecules (Zinkl et al., 1999). Taken together, the chemical analyses performed to date point toward a lipophilic moiety that mediates adhesion, perhaps a lipopolysaccharide. The female receptor for the pollen identification tag is not yet known, and one of the exciting challenges will be to determine if, on the stigma surface, adhesion is mediated by proteins or by other polymers.
Exine components alone are not sufficient for pollen recognition—the hydrophobic coat embedded in the surface of the exine also plays a vital role. When the dry surfaces of pollen and stigma cells come into contact, the coat becomes highly motile, with its protein and lipid moieties congealing at the cellular interface (Elleman et al., 1992). This massive migration of coat material creates a patch through which water moves. It is a marvelous mechanism for establishing communication in a nonaqueous setting.
The pollen coat is extremely hydrophobic and can be extracted from the exine with cyclohexane or other organic solvents. We used this method to characterize the composition of the Arabidopsis coat, purifying sufficient material to define the resident lipids and proteins. Gas chromatography and mass spectroscopy indicate the lipids often have a chain length of 26 or more carbons, a make-up that suggests a waxy, immotile substance that is inconsistent with the observed pollen coat motility (Preuss et al., 1993). Consequently, other constituents of the coat, most likely the coat proteins, are needed to solubilize and mobilize the lipid moieties. We used Edman degradation sequencing to identify the entire complement of pollen coat proteins in Arabidopsis (Mayfield et al., 2001). Comparison with the complete sequence generated by the Arabidopsis genome project revealed only a few proteins. The most abundant class of coat proteins is the pollen oleosins, glycine-rich proteins (GRPs) with an oil-binding domain. Another abundant class of pollen coat proteins (the extracellular lipases, or EXLs) is highly similar to family II lipases, proteins that are often active in extracellular environments. The combination of coat proteins and lipids somehow interact with the stigma cell, providing the recognition cues and perhaps a mechanical conduit that leads to pollen hydration.
A cluster of six tandem genes encodes the pollen coat GRPs, and another cluster of six genes encodes the EXLs. Although clusters of genes are not uncommon, the Arabidopsis genome contains only 40 gene families with clusters of six or more genes. The genes within each cluster of GRP or EXL genes likely descended from a common ancestor; they have similar promoter sequences, exon and intron boundaries, and highly similar domains. The EXL proteins resemble each other throughout. In contrast, although the GRP proteins all have a highly conserved oleosin domain, the second exon is highly variable, indicating these genes have diverged considerably from each other since their duplication.
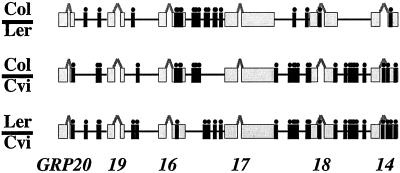
Proteins that mediate gamete interactions often have repetitive structures that allow for progressive changes, making it possible for populations to diverge and generate new species. Such repetitive motifs in mating proteins have been observed in many species, from Chlamydomonas to abalone to primates (Ferris et al., 1997; Vacquier 1998; Wyckoff et al., 2000). Because the GRP proteins have diverged from each other, we investigated whether they are, in fact, evolving faster than an average Arabidopsis protein. We sequenced the same genomic cluster in six different populations, and found an abundance of insertions and deletions (Figure 2) (Mayfield et al., 2001). Despite these changes, the reading frame of all six proteins was maintained, indicating that each likely plays an important role. We also found many single nucleotide polymorphisms, again suggesting that this genomic region is under a selection for a high rate of change, up to 10-fold greater than the average Arabidopsis gene. Taken together, the location of GRP proteins on the pollen surface, the extremely high polymorphism frequency between strains, the high levels of insertions and deletions, and the maintenance of all six GRP genes strongly implicates a role for these proteins in species recognition. Indeed, genetic evidence indicates that the most abundant GRP protein, GRP17, plays a direct role in communication with the stigma (Mayfield and Preuss, 2000). Pollen lacking GRP17 are not recognized quickly and therefore cannot compete effectively with wild type, a disadvantage that, in nature, would be significant enough to eliminate such mutations in just a few generations.
Figure 2.
Location of insertions and deletions in the GRP cluster. Comparisons of DNA sequences from the Columbia (Col), Landsberg (Ler), and Cape Verdi (Cvi) ecotypes. The GRP genes consist of two exons (gray boxes) and a single intron (gray lines); insertion and deletion differences between ecotypes are indicated by filled circles above a line (black).
To examine speciation in a more direct manner, we are extending our analyses to other plants. The Brassica family, which includes Arabidopsis, provides an enormous resource. In the evolutionary space of 20 million years (approximately the time since gorillas, chimps, and humans diverged), over 3000 Brassica species have evolved, providing enormous resources for comparing mating components. The adhesives that bind pollen grains are able to distinguish among pollen grains within this family, but the pollen hydration mechanisms are less selective. Brassica oleracea stigmas do not recognize Arabidopsis pollen, although Arabidopsis stigmas support hydration and growth of B. oleracea pollen tubes.
We sequenced the GRP cluster in B. oleracea, identifying the syntenic region from a bacterial artificial chromosome library (Mayfield et al., 2001). Typical B. oleracea and Arabidopsis genes are 75–85% identical, and the genes flanking the GRP cluster show that level of homology. Yet the genes within the GRP cluster are highly divergent from their Arabidopsis counterparts, so much so that homologous relationships are difficult to detect. Other than the oleosin lipid binding domain, these proteins have little in common, supporting the model that GRPs are rapidly evolving proteins that play a role in species-specific recognition.
The extreme lack of conservation between B. oleracea and Arabidopsis GRPs makes it impossible to postulate models for the mechanisms of evolutionary change. To further dissect this problem, additional genomic resources are required; consequently, we are constructing bacterial artificial chromosome libraries from 10 species, choosing plants both in the same genera, as well as plants just outside the Brassica family. These libraries will be a useful resource, not only for our research, but also for others in the plant community with an interest in comparative genomics.
Over the past years, the power of genetic analysis in Arabidopsis has identified many genes required for reproductive cell–cell interactions. Coupled with robust assays that monitor each stage of pollination, the molecules that mediate sexual communication within this species are becoming clear. With the completion of the Arabidopsis genome project, the door for extending these investigations to the mechanisms of speciation in flowering plants is now open. Identifying the homologues of Arabidopsis reproductive genes in near relatives promises to reveal how pollen adhesion, hydration, tube germination, guidance, and sperm delivery are modified during the ongoing evolutionary process.
ACKNOWLEDGMENTS
I thank the American Society for Cell Biology and the Promega Corporation for this generous award. I also recognize the contributions of the members of my laboratory to this work, including Aretha Fiebig, Mark Johnson, Jacob Mayfield, Shuh-ichi Nishikawa, Ravishankar Palanivelu, Gary Rudgers, Robert Swanson, Laura Wilhelmi-Brass, and Gregory Zinkl. These studies have been supported in part by the National Science Foundation, by the Department of Energy, by the Searle Scholars Foundation, and by the Howard Hughes Medical Institute
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.ES–01–0001. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.ES–01–0001.
REFERENCES
- Cheung A, Wu H, Di Stillo V, Glaven R, Chen C, Wong E, Ogdahl J, Estavillo A. Pollen-pistil interactions in Nicotiana tabacum. Ann Bot. 2000;85:29–37. [Google Scholar]
- Dixit R, Nasrallah JB. Recognizing self in the self-incompatibility response. Plant Physiol. 2001;125:105–108. doi: 10.1104/pp.125.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman CJ, Franklin Tong V, Dickinson HG. Pollination in species with dry stigmas the nature of the early stigmatic response and the pathway taken by pollen tubes. New Phytol. 1992;121:413–424. doi: 10.1111/j.1469-8137.1992.tb02941.x. [DOI] [PubMed] [Google Scholar]
- Ferris PJ, Pavlovic C, Fabry S, Goodenough UW. Rapid evolution of sex-related genes in Chlamydomonas. Proc Natl Acad Sci USA. 1997;94:8634–8639. doi: 10.1073/pnas.94.16.8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Preuss D. Plotting a course. Multiple signals guide pollen tubes to their targets. Dev Cell. 2002;2:273–281. doi: 10.1016/s1534-5807(02)00130-2. [DOI] [PubMed] [Google Scholar]
- Mayfield JA, Fiebig A, Johnstone S, Preuss D. Gene families from the Arabidopsis thaliana pollen coat proteome. Science. 2001;292:2482–2485. doi: 10.1126/science.1060972. [DOI] [PubMed] [Google Scholar]
- Mayfield JA, Preuss D. Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat Cell Biol. 2000;2:128–130. doi: 10.1038/35000084. [DOI] [PubMed] [Google Scholar]
- Preuss D, Lemieux B, Yen G, Davis RW. A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signaling during fertilization. Genes Dev. 1993;7:974–985. doi: 10.1101/gad.7.6.974. [DOI] [PubMed] [Google Scholar]
- Vacquier VD. Evolution of gamete recognition proteins. Science. 1998;281:1995–1998. doi: 10.1126/science.281.5385.1995. [DOI] [PubMed] [Google Scholar]
- Wyckoff GJ, Wang W, Wu CI. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- Zinkl GM, Zwiebel BI, Grier DG, Preuss D. Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development. 1999;126:5431–5440. doi: 10.1242/dev.126.23.5431. [DOI] [PubMed] [Google Scholar]