Abstract
Initially, it was believed that glycolysis and DNA damage repair (DDR) were two distinct biological processes that independently regulate tumor progression. The former metabolic reprogramming rapidly generates energy and generous intermediate metabolites, supporting the synthetic metabolism and proliferation of tumor cells. While the DDR plays a pivotal role in preserving genomic stability, thus resisting cellular senescence and cell death under both physiological and radio-chemotherapy conditions. Recently, an increasing number of studies have shown closely correlation between these two biological processes, and then promoting tumor progression. For instance, lactic acid, the product of glycolysis, maintains an acidic tumor microenvironment that not only fosters cell proliferation and invasion but also facilitates DDR by enhancing AKT activity. Here, we provide a comprehensive overview of the enzymes and metabolites involved in glycolysis, along with the primary methods for DDR. Meanwhile, this review explores existing knowledge of glycolysis enzymes and metabolites in regulating DDR. Moreover, considering the significant roles of glycolysis and DDR in tumor development and radio-chemotherapy resistance, the present review discusses effective direct or indirect therapeutic strategies targeted to glycolysis and DDR.
Keywords: Glycolysis, DNA damage repair, Genomic stability, Tumor, Resistance
Background
Distinct from normal tissue cells, solid tumor cells have a tendency to utilize glycolysis to produce ATPs (adenosine triphosphates) rather than aerobic respiration, even in the presence of oxygen-rich conditions, which is widely known as the “Warburg effect” [1, 2]. Multiple factors are believed to responsible for this phenomenon. Primarily, solid tumor cells generally initiate glycolysis to fulfill the hypoxic and energy shortage tumor microenvironment (TME), and the alteration of signaling pathways and glycolysis related enzymes making tumor cells tend to utilize glycolysis to metabolize glucose. In addition, the ability of tumor cells to uptake glucose is more than ten times that of normally differentiated cells, and glycolysis possess a shorter while faster metabolic pathways than that of oxidative phosphorylation [3]. Besides providing ATPs for tumor cells, glycolysis can also generate various intermediate metabolites for nucleotide, protein as well as fatty acid metabolisms [3, 4]. As the product of glycolysis lactic acid gradually acidifies the TME, which in turn disrupting the immune environment and extracellular matrix, and therefore advancing and exacerbating the migration and invasion ability of tumors [5–7]. Deprivation of glycolysis by inhibiting glucose transporter proteins (Gluts) and glycolysis related proteins induce apoptotic cell death in several kinds of cancer cells [8–10]. Although there are still conflicting aspects, the alteration of glycolysis can modulate autophagy, which is generally considered as a pro-survival mechanism. Moreover, increasing investigations indicate glycolysis play a role in modulating DNA damage and DNA damage repair (DDR). For instance, several glycolysis enzymes promote the DDR upon both chemotherapy agents and radiation [8, 9, 11, 12], and the glycolysis metabolites function in reducing the reactive oxygen species (ROS) thereby reducing the probability of DNA damage [13, 14]. In addition, cellular senescence, characterized by metabolic deregulation and macromolecular damage causing genomic instability, arises as a stress response to diverse stimuli. Gorgoulis et al. emphasized the necessity of elucidating the mechanisms underlying cellular senescence, revealing the crucial involvement of DNA damage responses and protein stress in initiating senescence, frequently accompanied by metabolic shifts towards glycolysis [15, 16]. Therefore, the metabolic reprogramming of glycolysis in tumor cells not only promotes the progression of tumors, but also resists the efficacy of radio-chemotherapy. In the current review, we will discuss the impact of glycolytic pathways on DDR, and their roles in regulating tumor progression and therapy.
The primary process of glycolysis
Glycolysis represents the initial enzymatic degradation process of glucose within eukaryotic cells, during which a single glucose molecule degrades into two pyruvate molecules and releases two ATP molecules. Glycolysis is recognized as a primitive and fundamental means of energy acquisition, serving as the common metabolic pathway for glucose catabolism. Subsequently, pyruvate will be metabolized by the oxidative phosphorylation process to generate carbon dioxide and water, and release a large amount of ATP under aerobic conditions. However, under hypoxia and mitochondrial dysfunction conditions, pyruvate is enzymatically converted to lactic acid by lactate dehydrogenase (LDH). This transformation relies on NADH (nicotinamide adenine dinucleotide reduced form) as a hydrogen donor, and then regenerates NAD+ (nicotinamide adenine dinucleotide) during the process. Therefore, lactic acid serves as a valuable indicator for monitoring glycolytic activity.
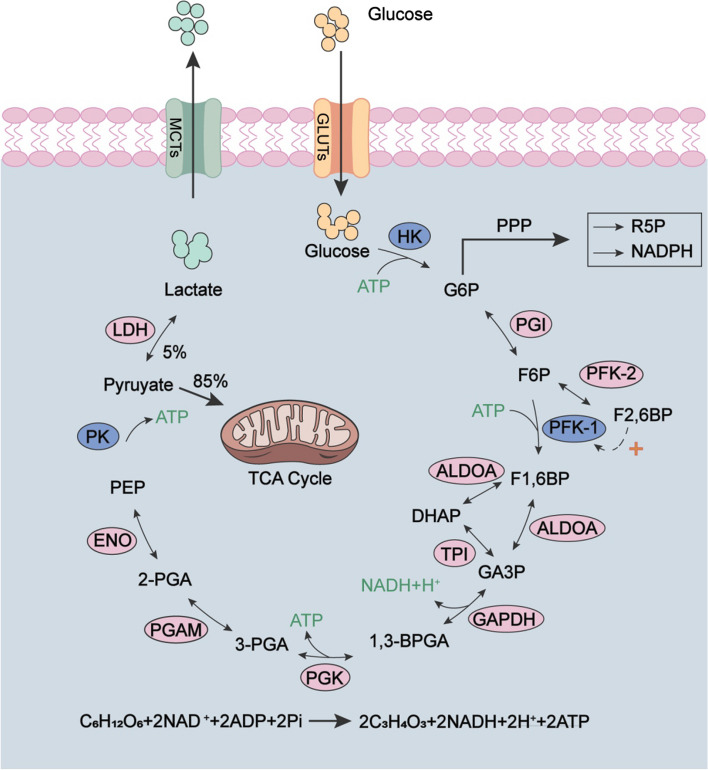
As shown in Fig. 1, glycolysis process comprises ten reactions that can be segregated into two distinct stages: the preparatory phase for fermentation and the subsequent energy-releasing stage [17, 18]. The preparatory phase of glycolysis comprises five key reactions. 1) Glucose is converted to glucose-6-phosphate (G6P) by hexokinase (HK), an irreversible process consuming an ATP molecule. This reaction activates glucose molecules and ensures the intracellular glucose is effectively captured, and therefore prevents its efflux of the cell. 2) G6P is isomerized to fructose-6-phosphate (F6P) by phosphoglucose isomerase (PGI), while this reaction is reversible. 3) F6P is converted to fructose-1,6-bisphosphate (F1,6BP) by 6-phosphofructo-1-kinase (PFK-1), an irreversible step requiring an ATP. Meanwhile, F6P can be reversible catalyzed to fructose-2,6-bisphosphate (F2,6BP) by 6-phosphofructo-2-kinase (PFK-2), which plays an important role in regulating glycolysis by modulating the activity of PFK-1. 4) F1,6BP is reversible transformed into glyceraldehyde-3-phosphate (GA3P) and dihydroxyacetone phosphate (DHAP) by aldolase. 5) DHAP is reversible isomerized to GA3P by triosephosphate isomerase (TPI), as it can’t be utilized by glycolysis. During this preparatory phase, one glucose molecule is converted into two GA3P and consumes two ATP molecules.
Fig. 1.
Schematic diagram of glycolysis process. Enzymes: GLUTs (glucose transporter proteins), HK (hexokinase), PGI (phosphoglucose isomerase), PFK-1 (6-phosphofructo-1-kinase), PFK-2 (6-phosphofructo-2-kinase), ALDOA (aldolase A), TPI (triosephosphate isomerase), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), PGK (phosphoglycerate kinase), PGAM (phosphoglycerate mutase), ENO (enolase), PK (pyruvate kinase), LDH (lactate dehydrogenase), MCTs (monocarboxylate transporters). Glycolytic intermediates: G6P (glucose-6-phosphate), F6P (fructose-6-phosphate), F2,6BP (fructose-2,6-bisphosphate), F1,6BP (fructose-1,6-bisphosphate), GA3P (glyceraldehyde-3-phosphate), DHAP (dihydroxyacetone phosphate), 1,3-BPGA (1,3-bisphosphoglycerate), 3-PGA (3-phosphoglycerate), 2-PGA (2-phosphoglycerate), PEP (phosphoenolpyruvate), NADH (nicotinamide adenine dinucleotide reduced form), NAD+ (nicotinamide adenine dinucleotide). Abbreviation: PPP (pentose phosphate pathway), TCA cycle (Krebs cycle), NADPH (nicotinamide adenine dinucleotide phosphate, reduced form), R5P (ribulose-5-phosphate). The overall reaction equation for the glycolysis process was shown
The following five reactions constitute the energy-releasing stage. 6) GA3P is dehydrogenated by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in the presence of inorganic phosphate (Pi) to produce 1,3-diphosphoglycerate (1,3-BPGA), and simultaneously reduce 1 molecule of NAD+ to NADH. 7) Under the catalysis of phosphoglycerate kinase (PGK), 1,3-BPGA is converted to 3-phosphoglycerate (3-PGA) and generates the first ATP molecule. 8) 3-PGA is converted to 2-PGA under the action of phosphoglycerate mutase (PGAM). 9) 2-PGA undergoes dehydration to generate phosphoenolpyruvate (PEP) by enolase. These four reactions are all reversible. 10) With the help of pyruvate kinase (PK), PEP transfers phosphate groups to ADP, and then generates pyruvate and ATP. This is the second ATP generated during glycolysis. As one glucose molecule is converted into two GA3P, so totally four ATP molecules and two NADH molecules are generated during the energy-releasing stage, and the total reaction of glycolysis is shown in Fig. 1. For detailed information on the agonists and antagonists of glycolytic-related enzymes, please refer to the following literatures [2, 17, 18], and we will not delve into them further here.
Among all the catalytic enzymes participated in glycolysis, HK, PFK-1 and PK function as the rate-limiting enzymes [2, 17]. As PFK-1 characters the lowest catalyze activity, thus serving as the gatekeeper for glycolysis. Its activity is effectively allosteric inhibit by citrate, ATP, as well as its catalytic product F1,6BP, while ructose-2,6-bisphosphate (F2,6BP) potently aggregates its activity [3]. Cellular F2,6BP is synthetized by 6-phosphofructo-2-kinase (PFK-2) from F6P. Notably, PFK-2 simultaneously catalyze the inverse reaction, the hydrolysis of F2,6BP to F6P, thus exhibiting phosphatase activity and being referred to as fructose-2,6-bisphosphatase (FBPase-2). Although not directly involved in the glycolysis process, PFK-2 is widely approved to be a crucial modulator of the glycolysis process. In addition, there are also other proteins closely related to the glycolytic pathway, such as Glut, MCTs (monocarboxylate transporters) and LDH. Prior to the commencement of the glycolytic pathway, glucose is transported from the extracellular to the intracellular by trans-membrane Glut, which is widely expressed in nearly all cells. LDH converts pyruvate and NADH to lactic acid and NAD+, therefore leading to the acidic tumor microenvironment (TME), which can promote tumor invasion and angiogenesis [19]. Interestingly, lactic acid can participate in the occurrence and development of tumors as a carcinogenic factor [5, 20]. Moreover, it negatively regulates the function and metabolic growth of immune cells, ultimately achieving "immune escape" and avoiding surveillance by immune checkpoints [21].
Primary mechanisms of DDR in eukaryotic cells
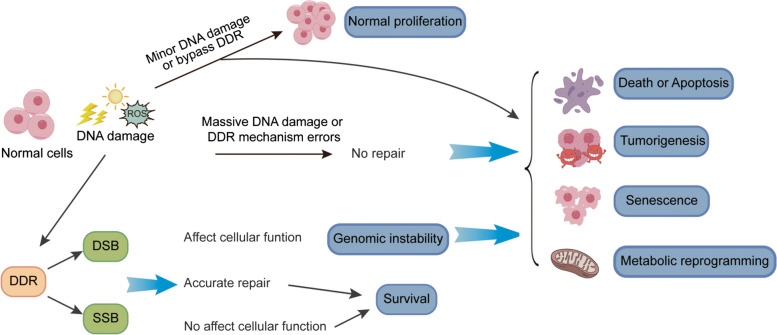
Upon stimulation by external and internal environment factors, such as exogenous chemical carcinogens, ultraviolet, ionizing radiation, endogenous chemical carcinogens, and oxidative stress, normal cells generally undergo DNA base mutations/damage, DNA inter-strand crosslinks, DNA strand breaks and/or chromosomal variations [22]. Among all the DNA damage mechanisms, DNA double-strand break (DSB) is the relatively serious mechanism, which need timely and appropriate repair. Otherwise, cells will emerge cell cycle arrest or even develop into cell death, or introduce mutations and chromosomal translocations, making the cell prone to either senescence or tumorigenesis [23–25]. For cancer cells, DNA damage and its repair are important therapeutic targets, such as radiotherapy, alkylating agents and platinum based drugs, etc. As shown in Fig. 2, cell fate is determined by the balance of DNA damage and its repair. Massive DNA damage, when left unrepaired or subject to incorrect repair processes that substantially affect cellular function, can give rise to genomic instability, ultimately culminating in phenomena such as cell death, tumorigenesis, senescence, or metabolic reprogramming. Apart from causing damage to the DNA within chromosomes, mtDNA (mitochondrial DNA) is susceptible to attacks by endogenous damaging factors, such as ROS, resulting in dysfunction of the mitochondrial respiratory chain [26, 27]. In eukaryotic cells, homologous recombination repair (HRR) and non-homologous end joining (NHEJ) are frequently discussed and investigated for DSB repair. Several mechanisms are utilized for mismatch and single-strand break (SSB) repair, including DNA direct repair (DR), mismatch repair (MMR), base excision repair (BER), nucleotide excision repair (NER).
Fig. 2.
Cell fate is determined by the balance of DNA damage and its repair. External and internal environmental factors can elicit diverse forms of DNA damage. While some injuries exert minimal impact on cells, others, coupled with inadequate/unsuitable DNA repair, may result in cell cycle arrest, cell death, mutations, and chromosomal translocations, ultimately fostering senescence or tumorigenesis. Under optimal or nearly optimal repair conditions, cells have the capacity to recuperate to relatively normal states and successfully conclude their predetermined life cycle, maintaining genomic stability and ensuring proper cellular function. The delicate balance between DNA damage and its repair thus plays a pivotal role in determining the ultimate fate of cells
HRR and NHEJ
HRR possess a high fidelity repair mechanism, which predominantly occurs in the S and G2 stages of the cell cycle, mainly because the sister chromatids function as template is spatially close during this period. Therefore, although this type of HRR can be error-free, the actual occurrence rate of it in cells is not high. In some cases, non-sisters chromatids can also be used as templates for HRR, which may lead to point mutations or gene conversions at injury sites [28]. NHEJ does not require homology and repairs damages at any phase of the cell cycle, however, this repair method inevitably leads to the deletion of DNA bases in the vast majority cases. HRR and NHEJ share both competitive and complementary relationships, as plasmid DNA with DSB could be repaired by either method in cells, and either defect would lead to the enrichment of another method [28, 29]. Although the NHEJ proteins are recruited to DSB more rapidly than HRR proteins, both sets of factors are present at the DSB during significant period [30]. In fact, several proteins are involved in both HRR and NHEJ processes, such as BRCA1/2 (breast cancer susceptibility proteins 1/2), DNA-PK (DNA-dependent protein kinase), ATM (Ataxia telangiectasia mutated kinase), Rad50, Rad18, H2AX, PARP-1, etc. [28, 31, 32]. The significant enhancement in γ-H2AX (phosphorylation of H2AX at Ser139) is one of the typical biomarker for DSB, which is widely used for DSB detection in scientific investigation. When DSB are aroused, H2AX is rapidly phosphorylated by PI3K-related kinases, including ATM, DNA-PK and ATR (ataxia telangiectasia and Rad3-related kinase), which then amplify DNA damage signal and recruit functional proteins for DDR, such as MDC1 [33, 34]. However, former studies indicated that γ-H2AX is not indispensable for HRR and NHEJ, as its absence failed to block the HRR and NHEJ [33, 35, 36]. The role of γ-H2AX in DDR can be referenced here [33].
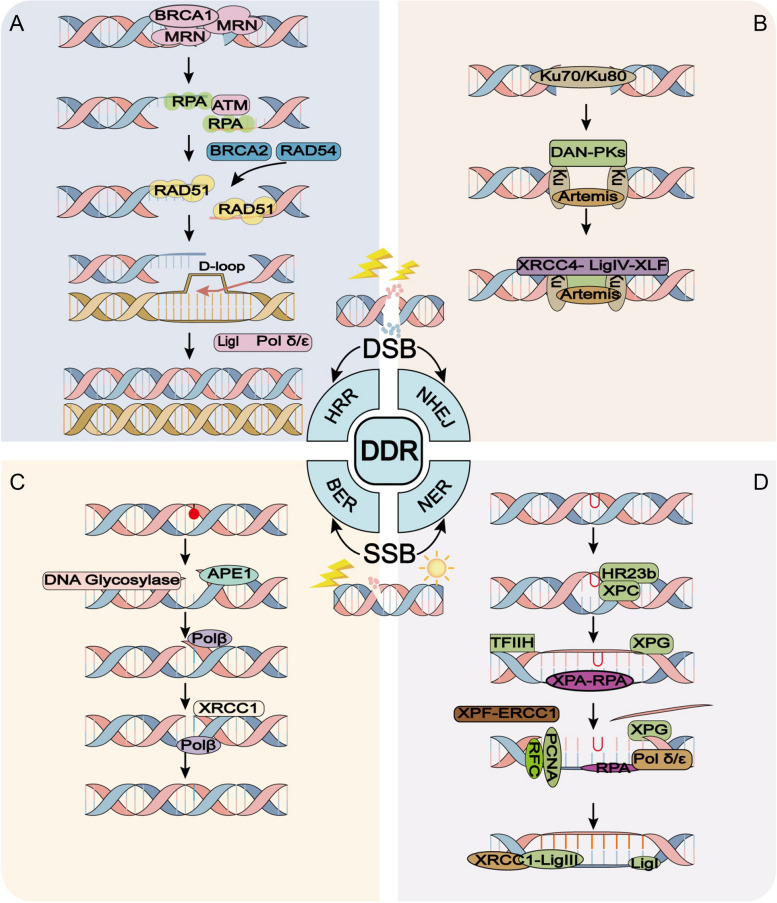
During the HRR (Fig. 3A), the MRN complex (MRE11-Rad50-NBS1) acts as a DSB sensor and participates in the initial phase upon DNA damage. This complex rapidly recruits to DSB ends via Rad50, and then resects the end of broken DNA via MRE11 with the help of BRCA1 to generate 3’-end ssDNA (single-strand DNA), RPA covers and stabilizes these single chain regions. Generally, ATM is stabilized by DNA-PK and existed in an inactivated dimer form. NBS1 recruits and disrupts the dimer form of ATM, which then transformed to activate form by auto-phosphorylation at S1981, S367 and S1893 sites [37]. Subsequently, activated ATM anchors at the DSB and catalyzes the phosphorylation of downstream effectors, such as H2AX. With the help of Rad54 and BRCA2, the recombinase Rad51 is recruited to the DSB site and replaced RPA [38]. After disconnecting from BRCA2, Rad51 polymerized and forms a filament nearby the ssDNA, and then the ssDNA-Rad51 filament enters the template and forms D-loop intermediates to complete the HRR [39].
Fig. 3.
Brief schematic models of common DNA damage repair pathways. DNA double-strand break (DSB) and single-strand break (SSB) represent two prevalent and critical forms of DNA damage, and numerous repair pathways have been studied extensively. A and B In the case of DSB, homologous recombination repair (HRR) and non-homologous end joining (NHEJ) are commonly employed repair methods. HRR boasts a high-fidelity repair mechanism, whereas NHEJ frequently results in the deletion of DNA bases in the majority of instances. C and D For SSB, base excision repair (BER) and nucleotide excision repair (NER) are frequently utilized repair methods. BER is relatively straightforward, whereas NER is more intricate
When it comes to NHEJ (Fig. 3B), the Ku70-Ku80 heterodimer is the first factor binding to DSB in NHEJ pathway in most cases. On the one hand, the abundant Ku70-Ku80 heterodimer have a high affinity for binding to DSB ends and therefore preventing its excessive excision. On the other hand, the Ku70-Ku80 heterodimer recruit DNA-PK and Artemis nuclease to the DSB ends, both of them play a role in DNA end processing. The end processing of DNA includes nucleotide addition, modification and excision, which is necessary for the following DNA repair and the reason for NHEJ prones to cause genome rearrangement. Once the DNA-PK is recruited, it is activated by auto-phosphorylation, and phosphorylates other targets including H2AX [40–42]. Subsequently, XRCC4, XLF (XRCC4-like factor) and DNA ligase IV anchor to the DSB ends and perform break sealing. Besides the canonical NHEJ pathway above, there is an alternative NHEJ (alt-NHEJ) pathway functions independent of Ku70-Ku80. During this process, the MRN complex assists in processing the DSB ends [28, 43], and then PARP-1, XRCC1 and DNA ligase III are utilized to rejoining the break ends. This pathway requires 5–25 bp micro-homology and characters slow efficiency [44]. For those looking forward to further understanding HRR and NHEJ proteins and detailed processes, please refer to the following literatures [28, 31].
In addition, another mechanism based on DSB is SSA (Single Strand Annealing), which belongs to the homologous directed repair mechanism. It uses homologous repeat sequences on the DSB flank for repair, and the process of ligation is often prone to errors due to the removal of DNA repeat sequences. Unlike HRR, SSA does not require chain invasion and donor sequence; therefore it does not require RAD51 support [45]. In addition, the RAD52-RPA complex participates in the core response of SSA, making RAD52 a highly valuable therapeutic target in homologous recombination deficient BRCA1 or BRCA2 deficient cells [46].
For DNA SSB
This intricate mechanism for DNA SSB repair involves several steps and a myriad of proteins, ensuring the accurate restoration of damaged DNA. Initially, DNA damage is detected by a network of sensor proteins, such as poly(ADP-ribose) polymerase (PARP), which recognizes SSB [47]. Upon detecting an SSB, PARP-1 swiftly localizes to the damage site and activates its catalytic function, using NAD⁺ as a substrate to synthesize poly(ADP-ribose) (PAR) polymers [48, 49]. This process, known as PARylation, serves as a vital signaling mechanism that orchestrates the recruitment of various DNA repair factors to the site of damage [48, 49]. Besides, PARP-1's PARylation activity can influence chromatin structure, promoting a more relaxed state that facilitates access of repair enzymes to damaged DNA. This chromatin remodeling function is crucial for efficient DNA repair, as it ensures that repair factors can reach and repair damage within the tightly packed chromatin.
One of the primary repair mechanisms is base excision repair (BER), which addresses damaged bases (Fig. 3C) [50]. This process commences with the recognition and excision of the damaged base by DNA glycosylases, like uracil-DNA glycosylase (UDG), forming an apyrimidinic (AP) site. Subsequently, AP endonucleases, like APE1, cleave the DNA backbone at the AP site, creating a single-nucleotide gap. This gap is then filled in by DNA polymerase β, which utilizes the intact DNA strand as a template. Finally, DNA ligase III, aided by XRCC1, seals the nick, completing the BER process. In cases where the damage is more extensive or the BER pathway is overwhelmed, the cell may resort to the more comprehensive nucleotide excision repair (NER) mechanism (Fig. 3D) [51]. NER recognizes a wider spectrum of lesions, including bulky adducts and UV-induced damage. The NER process can be divided into two sub pathways: Global Genome NER (GG-NER) and Transcription Coupled NER (TC-NER) (only GG-NER will be discussed below). The process involves excision of the damaged DNA segment (typically 22–30 nucleotides) by endonucleases, followed by gap-filling by polymerase δ or ε and ligation by DNA ligase I or III. After XPC complex recognizes damage, XPA-RPA confirms and stabilizes the damage, followed by endonucleases cutting DNA at the 5 'and 3' ends of the damage. DNA polymerase δ or ε and ligase I or III are responsible for filling and connecting, while XPG, XPF, TFIIH, etc. promote the process of NER [52, 53]. Moreover, when DNA replication encounters SSB, the cell employs a specialized mechanism called post-replication repair (PRR), often mediated by error-prone trans-lesion synthesis (TLS) polymerases like polymerase η [54]. TLS allows DNA replication to bypass the damage site, albeit with a risk of introducing mutations.
Glycolytic enzymes in regulating DNA damage
Former studies show that DNA damage and repair processes exert profound effects on cellular metabolism. DNA lesions can disrupt metabolic pathways, while efficient DNA repair mechanisms are crucial for restoring metabolic homeostasis and ensuring cell survival. The intricate interplay between DNA integrity and metabolism underscores the importance of maintaining genomic stability for overall metabolic health. During the tumorigenesis process, extensive DNA damage and repair may be important factors leading to metabolic reprogramming [55, 56]. Conversely, glycolysis, particularly the activity of glycolytic enzymes, plays a role in modulating DDR mechanisms. These enzymes can influence the availability of metabolites and energy necessary for DNA repair processes, impacting the cell's ability to respond to and recover from DNA damage. Therefore, while DNA damage and repair can affect glycolysis, glycolytic enzymes also play a crucial role in influencing DDR mechanisms.
HK2
As the first rate-limiting enzyme in glycolysis, HK, particularly isoform 2 (HK2), emerges as a multifaceted player in cellular responses to DNA damage, albeit with indirect mechanisms. It has been established that HK2 localizes to the DNA in acute myeloid leukemia and normal hematopoietic stem and progenitor cells, where it contributes to maintaining stemness [12]. This unusual subcellular localization suggests that HK2 may participate in nuclear processes beyond its canonical role in glycolysis. By modulating gene expression or interacting with chromatin-associated proteins 53BP1 and Rad51, HK2 could influence the cellular response to DNA damage, potentially enhancing survival and proliferation under stressful conditions [12]. In embryonal rhabdomyosarcoma, hyperactive AKT1 signaling promotes tumor progression and DNA repair [57]. By increasing glucose uptake and flux through the glycolytic pathway, hyperactive AKT1 enhances the cell's metabolic flexibility, allowing it to withstand DNA damage and continue proliferating. Therefore, HK2 inhibitors could represent a promising therapeutic strategy to sensitize cells to DNA-damaging agents [57]. Similarly, inhibiting HK2 kinase can reverse DNA damage repair in NSCLC cells regulated by CAFs and increase radiation sensitivity [58]. Loss of BRCA1 in ovarian cancer cells initiates a metabolic shift towards glycolysis [59]. This metabolic reprogramming, driven by HK2 and other glycolytic enzymes, provides ovarian cancer cells with the energy and biosynthetic intermediates necessary to withstand chemotherapy and progress towards malignancy. Therefore, targeting glycolytic pathways, particularly HK2, may present a promising opportunity for chemoprevention [59]. 3-Bromopyruvate, a glycolytic inhibitor, regulates the status of glycolysis and modulates the sensitivity of human hepatocellular carcinoma cells to BCNU, a chemotherapeutic agent [60]. This finding underscores the importance of glycolysis in modulating cellular responses to DNA-damaging chemotherapy. By inhibiting HK2 and disrupting glycolysis, 3-Bromopyruvate enhances the efficacy of BCNU, suggesting a potential therapeutic synergy [60]. Meanwhile, 2-DG (2-Deoxy-D-glucose), another inhibitor of HK2, has been shown to modulate cellular sensitivity to DNA-damaging agents, such as radiation and chemotherapy, underscoring the intricate connection between glycolysis and DNA damage responses [61]. Beyond cancer, HK2 and glycolysis have also been implicated in neurodegenerative diseases such as Alzheimer's disease [62]. Here, biological sex and DNA repair deficiencies drive systemic metabolic remodeling and brain mitochondrial dysfunction, which are closely tied to glycolysis and HK2 activity. Although the direct link between HK2 and DNA damage in Alzheimer's disease remains to be fully elucidated, this research highlights the broader implications of glycolytic dysregulation in disease pathogenesis [62].
PFK-2/PFKFB3
Although serving as the gatekeeper for glycolysis, the role of PFK-1 in DNA damage and DDR is still uncovered. However, robust researches revealed that PFK-2 plays essential roles in regulating DNA damage and DDR. Among all four isoforms of PFK-2/FBPase-2, PFKFB3 (6-phosphofructo-2-kinase/ fructose-2,6-bisphosphatase isoform 3) characters the highest ratio (> 700) of kinase/phosphatase activity. Generally, it’s highly expressed in rapidly proliferation cells, and its activity and expression are induced by various cellular signals, such as hypoxia, growth factor and hormone. Moreover, an increasing number of studies indicate its involvement in regulating nuclear damage and repair processes, although its roles remain controversial.
Da et al. emphasized PFKFB3's potential as a prognostic and tumor microenvironment biomarker in human cancers, and its expression is correlated with several signaling pathways, including cell cycle and DNA damage response [63]. Moreover, Xiao et al. reported that PFKFB3 inhibition induced cell death and synergistically enhanced chemosensitivity of both carboplatin and cisplatin in endometrial cancer cells by enhancing DNA damage as well as reducing RAD51 expression [8]. Our former study reported that blocking the necroptosis pathway attenuated PFKFB3 inhibitor-induced cell viability loss and genome instability in colorectal cancer cells, suggesting that PFKFB3 inhibition may enhance the efficacy of chemotherapy by exacerbating DNA damage [9]. Inhibiting either PFKFB3 or TIGAR individually led to an elevation in DNA damage in HeLa cells, and the phenotypic effect was notably more pronounced in cells deficient with both proteins [64]. Interestingly, PFKFB3 knockdown increased the AKT-mTOR activity in this research, which is distinct from most studies [64].
Gustafsson et al. found that targeting PFKFB3 radio-sensitized cancer cells and suppressed HRR, a crucial DNA repair pathway [65]. Shi et al. reported that inhibiting PFKFB3 hinders the growth of hepatocellular carcinoma by disrupting DNA repair mechanisms via AKT signaling, implicating AKT as a mediator of PFKFB3's effects on DNA repair and cancer progression. This study highlights the complexity of signaling networks involved in PFKFB3's function and underscores the potential for targeting multiple pathways to achieve therapeutic benefit [66]. Ninou et al. showed that PFKFB3 inhibition sensitizes cancer cells to DNA crosslinking chemotherapies by suppressing Fanconi anemia repair, further emphasizing PFKFB3's role in regulating specific DNA repair pathways [67]. Recently, Sun et al. reported that PFKFB3 translocate to sites of DNA damage induced by oxidative stress, facilitating DNA repair processes through its interaction with the MRN-ATM signaling pathway [68]. Meanwhile, they also point out that PFKFB3 is involved in regulating the process of cellular senescence [68].
Importantly, PFKFB3's effects on DNA damage and repair are not uniform across all cell types and contexts. While inhibition of PFKFB3 often enhances DNA damage and sensitizes cancer cells to therapeutic interventions, its role in other type cells can be more nuanced. In Fanconi anemia hematopoietic stem cell, p53-TIGAR axis suppresses glycolysis and therefore reduced DNA damage, while ectopic expression of PFKFB3 resisted p53-TIGAR mediated metabolic reprogramming [69]. Although they failed to monitor the role of PFKFB3 in modulating DNA damage, it is highly likely that PFKFB3 promotes DNA damage under these conditions, highlighting the need for a careful assessment of PFKFB3's function in different cellular environments. Another study unveiled that p53 orchestrates DNA repair and nucleotide synthesis by repressing PFKFB3 expression, thereby fostering the pentose phosphate pathway and leading to a more efficacious DDR mechanism [70]. In these studies, PFKFB3 may play a negative role in regulating DDR.
PKM2
Among the primarily four isoforms of PK mammalian cells, PKM2 (pyruvate kinase muscle 2 isoform) is highly expressed in cancer cells and functions as a master regulator of cancer reprogramming. Beyond, despite potential contradictions, previously studies have indicated that PKM2 is implicated in regulating cellular senescence, potentially due to variations in cell type and PKM2 forms [71, 72]. The upregulation of PKM2, along with its altered subcellular localization, has been correlated with multiple hallmarks of cancer, such as perpetual proliferative signaling, evasion from apoptosis, the activation of invasion and metastasis pathways, and the deregulation of energetic metabolism. PKM2 exists in two forms in tumors: a tetramer that promotes glycolysis, fueling tumor energy metabolism, and a dimer that enters the DNA to regulate gene expression, activating Wnt signaling pathways and influencing DDR. This morphological switch is vital for tumor survival, growth, and adaptation. Additionally, although its involvement in regulating nuclear damage and the subsequent repair processes is recognized, the precise mechanisms by which it functions in these pathways continue to be a topic of ongoing discussion and research.
PKM2 directly interacts with p53, and disrupts p53’s phosphorylation by ATM. As a result, the expression of PKM2 confers a proliferative advantage to tumor cells when confronted with DNA damaging stimuli [73]. In non-small cell lung cancer (NSCLC) cells, inhibition of PKM2 triggers a cascade of events, including the upregulation of p53, ROS production, DNA damage, and downregulation of PARP-1, while its overexpression attenuates the DNA damage, mitochondrial fission, and cell viability loss aroused by agent [74, 75]. Knockdown of PKM2 enhances the radiosensitivity of cervical cancer cells, and enhanced the p-ATM and γ-H2AX, suggesting that PKM2 may confer resistance to radiation-induced DNA damage [76]. Upon DNA damage induced by radiation, PKM2 interact with FACT complex (SPT16 and SSRP1), and the pyruvate produced by PKM2 directly bind to SSRP1, therefore enhancing FACT-mediated chromatin loading of γ-H2AX. This promotion of γ-H2AX’s chromatin loading facilitates DNA repair and tumor cell survival [77]. Downregulation of PKM2 by siRNA or small molecular inhibitor enhances the effects of olaparib, a PARP-1 inhibitor, by amplifying the p-ATM and γ-H2AX levels, and PKM2 inhibition disrupts the nuclear accumulation of BRCA1, thereby compromising the cell's ability to effectively repair DNA damage by HRR [78]. Another study also uncovered that ATM phosphorylates PKM2 at T328 site, and pT328-PKM2 promotes HRR-mediated DNA DSB repair by phosphorylating CtIP at T126 site, enhancing CtIP recruitment to DSB and facilitating DNA end resection [79].
The abovementioned researches indicate that PKM2 inhibits DNA damage and promotes DDR, however, there are also some studies suggest that PKM2 plays a completely opposite role. For instance, Xia et al. demonstrated nuclear PKM2 interacts with H2AX and then phosphorylates it at S139 site, and depletion of PKM2 reduces the level of γ-H2AX under DNA damage conditions [80]. Another study indicated PKM2 binds to DDB2 (DNA Damage-Binding Protein 2), a critical component of the NER pathway that specifically recognizes UV-induced DNA damage, and this interaction impairs the efficient assembly of the NER complex and consequently compromise the cell's ability to repair UV-induced DNA damage. However, this study claimed PKM2 negatively correlates with cell survival upon UV irradiation, which is inconsistent with the general perspective [81].
Other glycolytic enzymes
As for several other glycolytic enzymes, such as PGI, TPI, PGK, and enolase may indirectly impact DNA damage through metabolic alterations, but their direct regulating of DNA damage has not been well established yet. While PGAM has been suggested to modulate the DNA damage response, there is a paucity of conclusive experimental data to definitively support this assertion [82].
Aldolase, particularly Aldolase A and B, exhibits complex roles in regulating DNA damage. Aldolase A has been shown to promote radioresistance by upregulating glycolysis post-irradiation, providing metabolic intermediates for DNA repair pathways [83, 84]. Specifically, it facilitates the activity of DNA-PKcs and ATM kinases, which are critical for DSB repair [85]. In contrast, by impairing DNA MMR mechanisms, Aldolase B contributes to the accumulation of DNA damage, ultimately triggering apoptosis in colorectal cancer cells [86].
GAPDH, usually used as the internal reference, are unveiled to involve in the cellular response to DNA damage. In DNA damaged Saccharomyces cerevisiae cells, the peptide derived from GAPDH secreted into the microenvironment can enhance cell survival [87]. Its phosphorylation mediated by Src kinase regulates its nuclear translocation, enhancing resistance to DNA damage in mammalian cells [88]. Furthermore, GAPDH expression is modulated by natural flavonoids like naringin, which aids in DNA repair by facilitating NER machinery, thereby mitigating UVB-induced DNA damage [89]. Moreover, GAPDH interacts with chromatin-associated proteins and other enzymes, to sustain nucleotide salvage pathways critical for DNA repair under stress conditions [90, 91]. Interestingly, GAPDH's overexpression protects against neurovascular degeneration following retinal injury, highlighting its potential therapeutic implications in mitigating DNA damage-related disorders [92]. Conversely, its inappropriate nuclear accumulation can lead to non-apoptotic cell death, a phenomenon mitigated by CIB1 through AKT and ERK signaling [93]. Studies also indicate that GAPDH can physically interact with DNA repair enzymes, such as apurinic/apyrimidinic endonuclease I (APE1), facilitating the protection of cells against oxidant-induced DNA damage [94, 95]. Ferreira E et al. further underscores the necessity of GAPDH for efficient repair of cytotoxic DNA lesions in bacteria, suggesting a conserved function across species [96]. Conversely, pyruvate, a metabolic intermediate closely tied to GAPDH function, can mitigate cell stress and genotoxicity, underscoring the therapeutic potential of modulating GAPDH-related pathways in response to DNA damage [97].
Beyond these enzymes directly involved in the glycolysis process, there are also some other proteins intimately related to glycolysis, such as hypoxia inducible factor (HIF). As a transcription factor, HIF activates genes encoding key enzymes involved in the glycolytic pathway, including HK2, PFKFB3, PKM2 and LDH [98, 99]. By upregulating these glycolytic proteins, HIF promotes glucose uptake, accelerates glucose breakdown, and enhances lactate production, thereby providing energy for cells in oxygen-deprived environments. This adaptation to hypoxia is particularly evident in cancer cells, where HIF-mediated glycolysis supports tumor growth, invasion, and metastasis [100]. Furthermore, HIF can also directly regulate DDR, with evidence suggesting its involvement in modulating the expression of genes critical for DNA repair mechanisms [101–103]. Here, we will not further summary the roles of these proteins in regulating DNA damage.
Glycolytic intermediate metabolites in regulating DNA damage
Intermediate metabolites generated during glycolysis are also involved in the pentose phosphate pathway (PPP), thereby they can indirectly influence DNA damage through this mechanisms. In the PPP, G6P is converted into 6-phosphogluconolactone, which further leads to the generation of NADPH (nicotinamide adenine dinucleotide phosphate, reduced form) and ribose-5-phosphate (R5P). NADPH functions as an important antioxidant within cells, maintaining the reduced state of glutathione (GSH) and protecting cells against oxidative stress damage [104], which is one of the triggering factors of DNA damage. GSH functions as a fundamental antioxidant in preserving intracellular redox equilibrium. By eliminating ROS, it safeguards cells against oxidative stress. Moreover, GSH holds a vital position in regulating various modes of cell death during cancer therapy by altering ROS concentrations, underscoring its importance in cellular redox regulation and disease mechanisms. Beyond this mechanism, several studies have also clarified some other mechanisms. Li et al. reported that nuclear F1,6BP directly interacts with HMGB1 (High Mobility Group Box 1), disrupting its association with chromatin and thereby inhibiting HMGB1's ability to support DNA repair processes [105]. Therefore, F1,6BP sensitizes cancer cells to chemotherapy-induced DNA damage, thereby inhibiting tumor growth and enhancing the efficacy of chemotherapeutic agents. The relationship between pyruvate and DNA damage is an area of active research that has uncovered several intriguing connections, and it has been shown to play a role in modulating cellular responses to DNA damage in various ways. In glioblastoma cells, pyruvate facilitates the γ-H2AX loading to chromatin mediated by the FACT complex, therefore promotes the efficient initiation of DNA repair and resulting in the radiation resistance of these cells [77]. Another study showed that pyruvate can act as a precursor for GSH biosynthesis through oxidative metabolism in hepatocellular carcinoma cells, which is an important antioxidant that protects against oxidative stress-induced DNA damage [106]. Meanwhile, pyruvate dehydrogenase 1α (PDHE1α) orchestrates the rapid conversion of pyruvate into acetyl-CoA (acetyl-coenzyme A), a vital step in supporting local chromatin acetylation during the critical phase of DNA damage repair. This enzymatic process fosters the development of relaxed chromatin configurations, thereby enhancing the accessibility of essential repair factors and ultimately facilitating the efficient restoration of DNA integrity [107].
During glycolysis, the key substrate and product directly involved in energy generation are NAD+ and NADH, respectively. NADH is a critical cofactor in cellular respiration, which functions as an electron carrier, transferring electrons through the electron transport chain, ultimately leading to the production of ATP, the cell's energy currency. Moreover, NADH exists in a reduced state and can reduce intracellular ROS levels, thereby reducing the occurrence of DNA damage. However, there are indications from certain studies that NADH might contribute to the process of DNA damage by mediating oxidative stress [13, 14]. Numerous research endeavors have underscored the pivotal role of NAD+ in the intricate mechanisms of DNA damage repair, notably by serving as an essential cofactor for PARP enzymatic activity. PARP enzymes employ NAD+ as their substrate to mediate the attachment of ADP-ribose moieties from NAD+ onto acceptor proteins, notably histones and chromatin-bound proteins in proximity to DNA lesions. The PARylation process serves as a crucial signaling mechanism, expediting the recruitment of DNA repair proteins to the site of damage and fine-tuning their functional properties, thereby significantly enhancing the overall efficiency and effectiveness of the DNA repair process [108]. Moreover, NAD+ plays a pivotal role in orchestrating protein–protein interactions vital for DNA repair. Age-related declines in NAD+ levels have been implicated in the accumulation of DNA damage and the progression of aging [109]. Further researches have shown that restoring NAD+ levels through supplementation effectively enhances the repair mechanisms of DNA damage incurred by radiation, oxidative stress, as well as the natural aging process [110, 111]. In summary, NAD+ plays a direct role in DNA repair processes, particularly through PARP-1 activation, while NADH's primary contribution is indirect, through maintaining cellular energy levels essential for DNA repair and other cellular functions.
Tumor therapeutic agents targeting glycolysis cells. Chemotherapeutics such as cisplatin and doxorubicin, and radiation induce and DNA damage
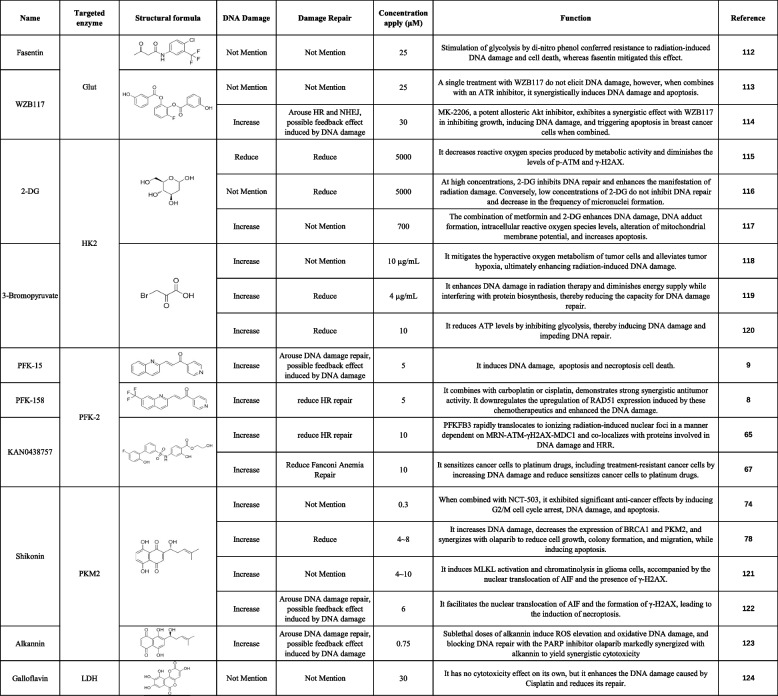
DNA damage is a well-established mechanism to trigger cell death in cancer DNA lesions, activating cellular stress responses that can culminate in apoptosis. Targeting DNA repair pathways, such as PARP inhibitors in BRCA-mutated tumors, further exacerbates DNA damage and enhances therapeutic efficacy. Recently, the exploration of therapeutic agents targeting glycolysis and DNA damage has garnered significant attention in the scientific community, particularly within the realm of oncology research. This approach leverages the unique metabolic features of cancer cells, which often rely heavily on aerobic glycolysis for energy production, alongside targeting DNA integrity, a fundamental aspect of cell survival. Multiple inhibitors of glycolytic enzymes have been investigated, including Glut [112–114], HK2 [115–120], PFK-2 [8, 9, 65, 67], PKM2 [74, 78, 121–123] and LDH [124] (Table 1). These inhibitors have shown promising antitumor effects in preclinical models, demonstrating reduced proliferation and induction of apoptosis in cancer cells.
Table 1.
The combination of glycolysis inhibitors and DNA damaging agents presents a compelling therapeutic strategy, and their synergistic effects have sparked a surge of interest in exploring the underlying mechanisms and optimizing these combinations for clinical application (Fig. 4). One such advancement lies in the understanding of how glycolysis inhibition modulates the tumor's redox status. Cancer cells often exhibit elevated levels of ROS due to their altered metabolism. Glycolysis inhibitors, by disrupting ATP production and the associated antioxidant defenses, can exacerbate oxidative stress, making cancer cells more susceptible to DNA damage. For instance, studies have shown that PKM2 inhibitor treatment increases intracellular ROS levels, induces DNA damage and ROS-dependent apoptosis [74].
Fig. 4.
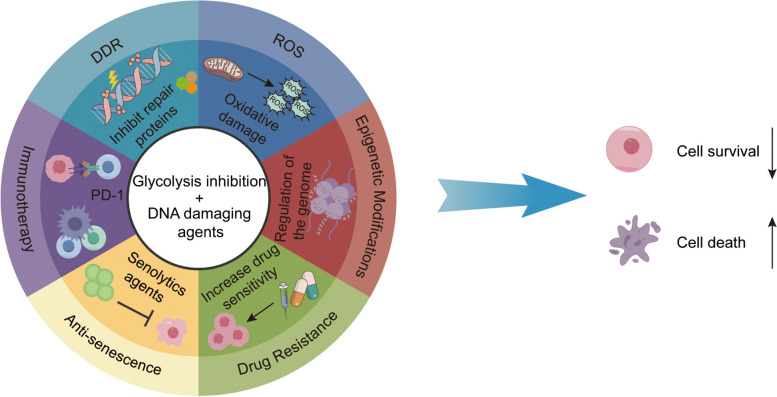
Mechanism of glycolysis inhibition & DNA damage in tumor therapy. DDR: DNA damage repair; ROS: reactive oxygen species. The combination of glycolysis inhibitors and DNA damaging agents constitutes a highly persuasive therapeutic approach, with their synergistic effects potentially attributed to the following mechanisms: 1) diminishing the efficacy of DDR; 2) elevating intracellular ROS levels; 3) modifying epigenetic patterns; 4) overcoming drug resistance by disrupting survival pathways; 5) resisting cellular senescence mechanisms that promote survival; and 6) enhancing the immune system's capabilities
Another emerging area of research focuses on the interaction between glycolysis inhibition and epigenetic modifications. Epigenetic regulators, such as histone deacetylases (HDACs), play crucial roles in maintaining chromatin structure and gene expression. Recent evidence suggests that glycolytic metabolism influences chromatin structure by modulating HDAC activity, either directly through metabolic intermediates or indirectly via changes in HDAC expression and regulation. This crosstalk between metabolism and epigenetics represents a crucial layer of regulation that fine-tunes cellular responses to environmental cues and maintains homeostasis. Inhibiting glycolysis leads to chromatin condensation, hindering DNA repair and enhancing cancer cells' sensitivity to DNA-damaging drugs, suggesting a novel therapeutic strategy for cancer treatment [125].
Moreover, the use of glycolysis inhibitors in combination therapies may help to overcome drug resistance. Cancer cells that develop resistance to DDR inhibitors or other targeted therapies often do so by activating alternative survival pathways, including those related to metabolism. By blocking glycolysis, these alternative pathways may be compromised, restoring sensitivity to the primary therapeutic agent.
Wang et al. reported that exosomal PKM2 may serve as a promising biomarker and therapeutic target for cisplatin resistance in NSCLC cells, and the PKM2 inhibitor significantly reversed the resistance of cisplatin [11]. Chen et al. conclusively demonstrated that the utilization of LDH inhibitors effectively circumvents chemotherapy resistance by diminishing the efficacy of DNA repair mechanisms. Furthermore, they pinpointed the suppression of lactate production as an encouraging and potentially groundbreaking therapeutic approach for cancer management [124, 126]. Gonzalez et al. demonstrated that mannose inhibits tumor growth and potentiates chemotherapy by competing with glucose for uptake, accumulating as mannose-6-phosphate, and disrupting glucose metabolism in glycolysis, TCA cycle, pentose phosphate pathway, and glycan synthesis. Consequently, it alters Bcl-2 protein levels, rendering cells more susceptible to death [127].
In addition, resisting cellular senescence mechanisms that promote survival represents a novel approach in combined therapies targeting glycolysis and DNA damage. During the process of anti-tumor therapy, cells may activate senescence-related pathways to evade death, resulting in surviving senescent cells that can subsequently lead to treatment failure and tumor recurrence [128, 129]. By modulating glycolysis, a metabolic pathway often deregulated in senescent cells, therapies aim to disrupt the senescent metabolic phenotype, which includes reduced glycolysis and increased autophagy [130]. Simultaneously, DNA damage triggers senescence through the activation of p53 and other senescence-associated signaling pathways, leading to cell cycle arrest and the secretion of senescence-associated secretory phenotype (SASP) factors [131]. Therefore, targeting the DNA damage response, which is activated in senescent cells, can synergize with glycolysis inhibition to mitigate the senescent phenotype. The interplay between glycolysis and DNA damage responses in senescence is bidirectional. Senescent cells exhibit altered glycolysis, which can disrupt the energy supply necessary for DNA repair, further exacerbating DNA damage and senescence [132]. Conversely, DNA damage can also disrupt glycolysis, leading to metabolic stress and the induction of senescence. By targeting both glycolysis and DNA damage responses, combined therapies aim to reprogram cellular metabolism and DNA repair mechanisms to counteract senescence and promote a more favorable therapeutic outcome in cancer [133, 134].
Finally, the integration of immunotherapy into combined therapies targeting glycolysis and DNA damage is a rapidly evolving field. Immune checkpoint inhibitors, such as anti-PD-1 and anti-CTLA-4 antibodies, have revolutionized cancer treatment by unleashing the immune system's ability to recognize and eliminate cancer cells. Recent studies suggest that glycolysis inhibition can enhance the efficacy of immunotherapy by promoting the infiltration and activation of immune cells within the tumor microenvironment [135, 136]. When coupled with immunotherapy, this synergistic effect can result in more potent antitumor immune responses and ultimately yield better patient outcomes. To overcome challenges such as poor bio-availability and nonspecific toxicity, nanoparticle-based delivery systems have emerged as promising carriers for glycolytic inhibitors and DNA damaging agents. These systems can selectively target tumor tissues, enhancing drug accumulation and reducing off-target effects. For example, polymeric nanoparticles encapsulating 2-DG and doxorubicin have demonstrated enhanced antitumor efficacy in breast cancer [137].
The clinical translation of these combinatorial strategies is actively ongoing, with several promising trials underway. Researchers are exploring various combinations, including glycolysis inhibitors with PARP inhibitors, ATR inhibitors, as well as with standard-of-care chemotherapeutics and radiation. The goal is to identify the most effective and tolerable combinations that can improve patient outcomes and prolong survival. Combination therapies targeting glycolysis and DNA damage pathways offer promising potential to improve treatment outcomes for cancer patients, especially those with resistant or advanced disease. However, further research is needed to optimize dosing schedules, identify predictive biomarkers, and rigorously assess long-term safety and efficacy in clinical trials. The success of these strategies depends on factors such as agent selectivity and potency, timing and sequence of administration, and the specific genetic and metabolic profiles of the cancer. Consequently, a personalized approach to therapy, guided by biomarkers and patient-specific data, is crucial for optimizing these combinatorial strategies and maximizing their clinical benefit.
Prospection
Looking forward, the exploration of the intricate interplay between glycolysis enzymes and metabolites in regulating DNA damage repair represents a promising frontier in tumor therapy. Not only does this relationship govern the cell's ability to repair DNA damage, but DNA damage itself can significantly impact glycolysis, altering the metabolic pathway within cells. By unraveling the intricate molecular mechanisms that orchestrate this bidirectional crosstalk, we can harness this knowledge to develop novel therapeutic strategies that selectively target cancer cells. Future research endeavors should be towards the identification and development of drugs or therapeutic strategies that concurrently target both glycolysis and DNA damage, aiming to achieve a dual-pronged attack on cancer cells. By strategically inhibiting or modulating the activity of these regulators, we aspire to sensitize cancer cells to the effects of DNA-damaging agents, thereby amplifying therapeutic efficacy. This approach could simultaneously disrupt the cancer cell's ability to adapt its glycolytic machinery in response to DNA damage, further compromising its survival. By exploiting this bidirectional relationship, we aspire to amplify therapeutic efficacy while minimizing detrimental impacts on healthy tissues. Moreover, the discovery of predictive biomarkers that correlate with the activity of these glycolytic regulators, as well as their response to DNA damage, could revolutionize personalized medicine. These biomarkers would enable us to tailor therapies not only to the unique characteristics of individual tumors but also to their dynamic responses to DNA damage, ensuring patients receive the most appropriate and effective treatments. As our comprehension of this intricate network deepens, we will gain a more comprehensive understanding of how DNA damage and glycolysis are intertwined, enhancing our ability to harness this knowledge for the benefit of cancer patients. In conclusion, targeting glycolysis and DNA damage in cancer therapy represents a dual-pronged approach with significant therapeutic potential. The combination of these strategies, facilitated by advanced drug delivery systems, holds the key to overcoming treatment resistance and improving clinical outcomes. Ongoing research into the molecular mechanisms underlying these interactions and the development of novel agents will continue to propel this field forward, ushering in a new era of precision oncology.
Conclusion
The interplay between glycolysis and DDR in cancer progression and therapy resistance is summarized. Glycolysis, which is crucial for tumor cell energy production and proliferation, exhibits a significant correlation with DDR, which is essential for maintaining genomic stability. The review highlights how glycolysis enzymes and metabolites regulate DDR, suggesting a synergistic role in tumor growth. It discusses therapeutic approaches targeting glycolysis and DDR to overcome treatment resistance. By presenting current research status and limitations, the study aims to spur further investigation into these processes. Understanding their interactions is crucial for advancing effective cancer treatment strategies, highlighting their pivotal roles in cancer biology and therapy.
Acknowledgements
Not applicable.
Authors' contributions
Conception: Siyuan Yan. Original manuscript drafting: Fengyao Sun and Jianxing Ma. Figure and table construction: Fengyao Sun and Wen Li. Investigation: Wen Li, Ruihang Du, Mingchan Liu, and Yi Cheng. Manuscript amending: Siyuan Yan. All authors reviewed the manuscript and approved the submitted version.
Funding
This work was supported by grants from the Shandong Provincial Natural Science Foundation (ZR2023MH329), and the Project of Shandong Province Higher Educational Youth Innovation Science and Technology Program (2023KJ263).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. [DOI] [PubMed] [Google Scholar]
- 2.Bose S, Zhang C, Le A. Glucose Metabolism in Cancer: The Warburg Effect and Beyond. Adv Exp Med Biol. 2021;1311:3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ros S, Schulze A. Balancing glycolytic flux: the role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab. 2013;1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBerardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ippolito L, Morandi A, Giannoni E, Chiarugi P. Lactate: A Metabolic Driver in the Tumour Landscape. Trends Biochem Sci. 2019;44:153–66. [DOI] [PubMed] [Google Scholar]
- 6.Rabinowitz JD, Enerback S. Lactate: the ugly duckling of energy metabolism. Nat Metab. 2020;2:566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Certo M, Tsai CH, Pucino V, Ho PC, Mauro C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat Rev Immunol. 2021;21:151–61. [DOI] [PubMed] [Google Scholar]
- 8.Xiao Y, Jin L, Deng C, Guan Y, Kalogera E, Ray U, Thirusangu P, Staub J, Sarkar Bhattacharya S, Xu H, et al. Inhibition of PFKFB3 induces cell death and synergistically enhances chemosensitivity in endometrial cancer. Oncogene. 2021;40:1409–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan SY, Li QQ, Zhang DR, Wang XW, Xu Y, Zhang C, Guo DL, Bao YH. Necroptosis pathway blockage attenuates PFKFB3 inhibitor-induced cell viability loss and genome instability in colorectal cancer cells. American Journal Of Cancer Research. 2021;11:2062. [PMC free article] [PubMed] [Google Scholar]
- 10.Gao C, Yan X, Wang B, Yu L, Han J, Li D, Zheng Q. Jolkinolide B induces apoptosis and inhibits tumor growth in mouse melanoma B16F10 cells by altering glycolysis. Sci Rep. 2016;6:36114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, Zhao C, Xu F, Zhang A, Jin M, Zhang K, Liu L, Hua Q, Zhao J, Liu J, et al. Cisplatin-resistant NSCLC cells induced by hypoxia transmit resistance to sensitive cells through exosomal PKM2. Theranostics. 2021;11:2860–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas GE, Egan G, Garcia-Prat L, Botham A, Voisin V, Patel PS, Hoff FW, Chin J, Nachmias B, Kaufmann KB, et al. The metabolic enzyme hexokinase 2 localizes to the nucleus in AML and normal haematopoietic stem and progenitor cells to maintain stemness. Nat Cell Biol. 2022;24:872–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oikawa S, Hirosawa I, Tada-Oikawa S, Furukawa A, Nishiura K, Kawanishi S. Mechanism for manganese enhancement of dopamine-induced oxidative DNA damage and neuronal cell death. Free Radic Biol Med. 2006;41:748–56. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi H, Hirao Y, Kawanishi S, Kato S, Mori Y, Murata M, Oikawa S. Rosmarinic acid, a natural polyphenol, has a potential pro-oxidant risk via NADH-mediated oxidative DNA damage. Genes Environ. 2024;46:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C, Campisi J, Collado M, Evangelou K, Ferbeyre G, et al. Cellular Senescence: Defining a Path Forward. Cell. 2019;179:813–27. [DOI] [PubMed] [Google Scholar]
- 16.Gorgoulis VG, Pefani DE, Pateras IS, Trougakos IP. Integrating the DNA damage and protein stress responses during cancer development and treatment. J Pathol. 2018;246:12–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganapathy-Kanniappan S, Geschwind JFH. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013;12:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papachristodoulou D, Snape A, Elliott WH, Elliott DC. Cellular energy metabolism. In: Biochemistry and Molecular Biology. Oxford University Press; 2018. https://catalog.loc.gov/vwebv/search?searchCode=LCCN&searchArg=2017960067&searchType=1&permalink=y.
- 19.Krstic J, Deutsch A, Fuchs J, Gauster M, Sparovec TG, Hiden U, Krappinger JC, Moser G, Pansy K, Szmyra M, et al. (Dis)similarities between the Decidual and Tumor Microenvironment. Biomedicines. 2022;10:1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574:575–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen DS, Mellman I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity. 2013;39:1–10. [DOI] [PubMed] [Google Scholar]
- 22.Sokolova O, Naumann M. Crosstalk Between DNA Damage and Inflammation in the Multiple Steps of Gastric Carcinogenesis. Curr Top Microbiol Immunol. 2019;421:107–37. [DOI] [PubMed] [Google Scholar]
- 23.Johnston LH, White JH, Johnson AL, Lucchini G, Plevani P. The yeast DNA polymerase I transcript is regulated in both the mitotic cell cycle and in meiosis and is also induced after DNA damage. Nucleic Acids Res. 1987;15:5017–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. [DOI] [PubMed] [Google Scholar]
- 25.Basu AK. DNA Damage, Mutagenesis and Cancer. Int J Mol Sci. 2018;19:970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu L, Hu C, Xu M, Yu J, He H, Lin J, Sha H, Lu B, Engelender S, Guan M, Song Z. ATAD3B is a mitophagy receptor mediating clearance of oxidative stress-induced damaged mitochondrial DNA. EMBO J. 2021;40:e106283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Correia-Melo C, Marques FD, Anderson R, Hewitt G, Hewitt R, Cole J, Carroll BM, Miwa S, Birch J, Merz A, et al. Mitochondria are required for pro-ageing features of the senescent phenotype. EMBO J. 2016;35:724–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–47. [DOI] [PubMed] [Google Scholar]
- 29.Roth DB, Wilson JH. Relative rates of homologous and nonhomologous recombination in transfected DNA. Proc Natl Acad Sci U S A. 1985;82:3355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J Cell Biol. 2005;170:341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan J, Sun X, Zhao H, Guan H, Gao S, Zhou PK. Double-strand DNA break repair: molecular mechanisms and therapeutic targets. MedComm. 2020;2023(4):e388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galita G, Sarnik J, Brzezinska O, Budlewski T, Poplawska M, Sakowski S, Dudek G, Majsterek I, Makowska J, Poplawski T. The Association between Inefficient Repair of DNA Double-Strand Breaks and Common Polymorphisms of the HRR and NHEJ Repair Genes in Patients with Rheumatoid Arthritis. International Journal Of Molecular Sciences. 2024;25:2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan J, Adamski R, Chen J. Focus on histone variant H2AX: to be or not to be. FEBS Lett. 2010;584:3717–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue C, Greene EC. DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing. Trends Genet. 2021;37:639–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci U S A. 2002;99:8173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Czornak K, Chughtai S, Chrzanowska KH. Mystery of DNA repair: the role of the MRN complex and ATM kinase in DNA damage repair. J Appl Genet. 2008;49:383–96. [DOI] [PubMed] [Google Scholar]
- 38.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J Biol Chem. 2003;278:42729–32. [DOI] [PubMed] [Google Scholar]
- 39.Wright WD, Shah SS, Heyer WD. Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem. 2018;293:10524–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stucki M, Jackson SP. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst). 2006;5:534–43. [DOI] [PubMed] [Google Scholar]
- 41.Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol. 2005;25:10842–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2005;24:949–61. [DOI] [PubMed] [Google Scholar]
- 43.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7:2902–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Y, Xu D. Repair pathway choice for double-strand breaks. Essays Biochem. 2020;64:765–77. [DOI] [PubMed] [Google Scholar]
- 45.Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang CC, Greenhough LA, Masino L, Maslen S, Bajrami I, Tuppi M, Skehel M, Taylor IA, West SC. Mechanism of single-stranded DNA annealing by RAD52-RPA complex. Nature. 2024;629:697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peralta-Leal A, Rodriguez-Vargas JM, Aguilar-Quesada R, Rodriguez MI, Linares JL, de Almodovar MR, Oliver FJ. PARP inhibitors: new partners in the therapy of cancer and inflammatory diseases. Free Radic Biol Med. 2009;47:13–26. [DOI] [PubMed] [Google Scholar]
- 48.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. BioEssays. 2004;26:882–93. [DOI] [PubMed] [Google Scholar]
- 49.Virag L. 50Years of poly(ADP-ribosyl)ation. Mol Aspects Med. 2013;34:1043–5. [DOI] [PubMed] [Google Scholar]
- 50.Grundy GJ, Parsons JL. Base excision repair and its implications to cancer therapy. Essays Biochem. 2020;64:831–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Apelt K, Lans H, Scharer OD, Luijsterburg MS. Nucleotide excision repair leaves a mark on chromatin: DNA damage detection in nucleosomes. Cell Mol Life Sci. 2021;78:7925–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuss JO, Cooper PK. DNA repair: dynamic defenders against cancer and aging. PLoS Biol. 2006;4:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oksenych V, Coin F. The long unwinding road: XPB and XPD helicases in damaged DNA opening. Cell Cycle. 2010;9:90–6. [DOI] [PubMed] [Google Scholar]
- 54.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li FL, Liu JP, Bao RX, Yan G, Feng X, Xu YP, Sun YP, Yan W, Ling ZQ, Xiong Y, et al. Acetylation accumulates PFKFB3 in cytoplasm to promote glycolysis and protects cells from cisplatin-induced apoptosis. Nat Commun. 2018;9:508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dylgjeri E, Kothari V, Shafi AA, Semenova G, Gallagher PT, Guan YF, Pang A, Goodwin JF, Irani S, McCann JJ, et al. A Novel Role for DNA-PK in Metabolism by Regulating Glycolysis in Castration-Resistant Prostate Cancer. Clin Cancer Res. 2022;28:1446–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Codenotti S, Zizioli D, Mignani L, Rezzola S, Tabellini G, Parolini S, Giacomini A, Asperti M, Poli M, Mandracchia D, et al. Hyperactive Akt1 Signaling Increases Tumor Progression and DNA Repair in Embryonal Rhabdomyosarcoma RD Line and Confers Susceptibility to Glycolysis and Mevalonate Pathway Inhibitors. Cells. 2022;11:2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang H, Zhang K, Qiu L, Yue J, Jiang H, Deng Q, Zhou R, Yin Z, Ma S, Ke Y. Cancer-associated fibroblasts facilitate DNA damage repair by promoting the glycolysis in non-small cell lung cancer. Biochim Biophys Acta Mol Basis Dis. 2023;1869:166670. [DOI] [PubMed] [Google Scholar]
- 59.Chiyoda T, Hart PC, Eckert MA, McGregor SM, Lastra RR, Hamamoto R, Nakamura Y, Yamada SD, Olopade OI, Lengyel E, Romero IL. Loss of BRCA1 in the Cells of Origin of Ovarian Cancer Induces Glycolysis: A Window of Opportunity for Ovarian Cancer Chemoprevention. Cancer Prev Res (Phila). 2017;10:255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun X, Sun G, Huang Y, Hao Y, Tang X, Zhang N, Zhao L, Zhong R, Peng Y. 3-Bromopyruvate regulates the status of glycolysis and BCNU sensitivity in human hepatocellular carcinoma cells. Biochem Pharmacol. 2020;177:113988. [DOI] [PubMed] [Google Scholar]
- 61.Kalia VK, Prabhakara S, Narayanan V. Modulation of cellular radiation responses by 2-deoxy-D-glucose and other glycolytic inhibitors: implications for cancer therapy. J Cancer Res Ther. 2009;5(Suppl 1):S57-60. [DOI] [PubMed] [Google Scholar]
- 62.Demarest TG, Varma VR, Estrada D, Babbar M, Basu S, Mahajan UV, Moaddel R, Croteau DL, Thambisetty M, Mattson MP, Bohr VA. Biological sex and DNA repair deficiency drive Alzheimer’s disease via systemic metabolic remodeling and brain mitochondrial dysfunction. Acta Neuropathol. 2020;140:25–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Da Q, Huang L, Huang C, Chen Z, Jiang Z, Huang F, Shen T, Sun L, Yan Z, Ye X, et al. Glycolytic regulatory enzyme PFKFB3 as a prognostic and tumor microenvironment biomarker in human cancers. Aging (Albany NY). 2023;15:4533–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simon-Molas H, Calvo-Vidal MN, Castano E, Rodriguez-Garcia A, Navarro-Sabate A, Bartrons R, Manzano A. Akt mediates TIGAR induction in HeLa cells following PFKFB3 inhibition. FEBS Lett. 2016;590:2915–26. [DOI] [PubMed] [Google Scholar]
- 65.Gustafsson NMS, Farnegardh K, Bonagas N, Ninou AH, Groth P, Wiita E, Jonsson M, Hallberg K, Lehto J, Pennisi R, et al. Targeting PFKFB3 radiosensitizes cancer cells and suppresses homologous recombination. Nat Commun. 2018;9:3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi WK, Zhu XD, Wang CH, Zhang YY, Cai H, Li XL, Cao MQ, Zhang SZ, Li KS, Sun HC. PFKFB3 blockade inhibits hepatocellular carcinoma growth by impairing DNA repair through AKT. Cell Death Dis. 2018;9:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ninou AH, Lehto J, Chioureas D, Stigsdotter H, Schelzig K, Akerlund E, Gudoityte G, Joneborg U, Carlson J, Jonkers J, et al. PFKFB3 Inhibition Sensitizes DNA Crosslinking Chemotherapies by Suppressing Fanconi Anemia Repair. Cancers. 2021;13:3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun D, Chen S, Li S, Wang N, Zhang S, Xu L, Zhu S, Li H, Gu Q, Xu X, Wei F. Enhancement of glycolysis-dependent DNA repair regulated by FOXO1 knockdown via PFKFB3 attenuates hyperglycemia-induced endothelial oxidative stress injury. Redox Biol. 2023;59:102589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X, Wu L, Zopp M, Kopelov S, Du W. p53-TP53-Induced Glycolysis Regulator Mediated Glycolytic Suppression Attenuates DNA Damage and Genomic Instability in Fanconi Anemia Hematopoietic Stem Cells. Stem Cells. 2019;37:937–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Franklin DA, He Y, Leslie PL, Tikunov AP, Fenger N, Macdonald JM, Zhang Y. p53 coordinates DNA repair with nucleotide synthesis by suppressing PFKFB3 expression and promoting the pentose phosphate pathway. Sci Rep. 2016;6:38067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bie J, Li R, Li Y, Song C, Chen Z, Zhang T, Tang Z, Su L, Zhu L, Wang J, et al. PKM2 aggregation drives metabolism reprograming during aging process. Nat Commun. 2024;15:5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu Y, Tang L, Huang H, Yu Q, Hu B, Wang G, Ge F, Yin T, Li S, Yu X. Phosphoglycerate dehydrogenase activates PKM2 to phosphorylate histone H3T11 and attenuate cellular senescence. Nat Commun. 2023;14:1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xia L, Wang XR, Wang XL, Liu SH, Ding XW, Chen GQ, Lu Y. A Novel Role for Pyruvate Kinase M2 as a Corepressor for P53 during the DNA Damage Response in Human Tumor Cells. J Biol Chem. 2016;291:26138–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang KX, Lu H, Wang XM, Liu QX, Hu JX, Liu Y, Jin MH, Kong DX. Simultaneous suppression of PKM2 and PHGDH elicits synergistic anti-cancer effect in NSCLC. Front Pharmacol. 2023;14:1200538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ding Z, Xi J, Zhong M, Chen F, Zhao H, Zhang B, Fang J. Cynaropicrin Induces Cell Cycle Arrest and Apoptosis by Inhibiting PKM2 to Cause DNA Damage and Mitochondrial Fission in A549 Cells. J Agric Food Chem. 2021;69:13557–67. [DOI] [PubMed] [Google Scholar]
- 76.Lin Y, Zhai H, Ouyang Y, Lu Z, Chu C, He Q, Cao X. Knockdown of PKM2 enhances radiosensitivity of cervical cancer cells. Cancer Cell Int. 2019;19:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu S, Cao R, Tao B, Wu P, Peng C, Gao H, Liang J, Yang W. Pyruvate Facilitates FACT-Mediated gammaH2AX Loading to Chromatin and Promotes the Radiation Resistance of Glioblastoma. Adv Sci (Weinh). 2022;9:e2104055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou S, Li D, Xiao D, Wu T, Hu X, Zhang Y, Deng J, Long J, Xu S, Wu J, et al. Inhibition of PKM2 Enhances Sensitivity of Olaparib to Ovarian Cancer Cells and Induces DNA Damage. Int J Biol Sci. 2022;18:1555–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sizemore ST, Zhang M, Cho JH, Sizemore GM, Hurwitz B, Kaur B, Lehman NL, Ostrowski MC, Robe PA, Miao W, et al. Pyruvate kinase M2 regulates homologous recombination-mediated DNA double-strand break repair. Cell Res. 2018;28:1090–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xia L, Qin K, Wang XR, Wang XL, Zhou AW, Chen GQ, Lu Y. Pyruvate kinase M2 phosphorylates H2AX and promotes genomic instability in human tumor cells. Oncotarget. 2017;8:109120–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xie X, Wang M, Mei J, Hu F, Ding F, Lv L. Pyruvate kinase M2 interacts with DNA damage-binding protein 2 and reduces cell survival upon UV irradiation. Biochem Biophys Res Commun. 2015;467:427–33. [DOI] [PubMed] [Google Scholar]
- 82.Johannessen TA, Mukherjee J. Phosphoglycerate mutase 1 (PGAM1) overexpression promotes radio- and chemoresistance in gliomas by activating the DNA damage response. Mol Cell Oncol. 2021;8:1875804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou J, Lei N, Qin B, Chen M, Gong S, Sun H, Qiu L, Wu F, Guo R, Ma Q, et al. Aldolase A promotes cervical cancer cell radioresistance by regulating the glycolysis and DNA damage after irradiation. Cancer Biol Ther. 2023;24:2287128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Q, Qin T, Bi Z, Hong H, Ding L, Chen J, Wu W, Lin X, Fu W, Zheng F, et al. Rac1 activates non-oxidative pentose phosphate pathway to induce chemoresistance of breast cancer. Nat Commun. 2020;11:1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sobanski T, Suraweera A, Burgess JT, Richard I, Cheong CM, Dave K, Rose M, Adams MN, O’Byrne KJ, Richard DJ, Bolderson E. The fructose-bisphosphate, Aldolase A (ALDOA), facilitates DNA-PKcs and ATM kinase activity to regulate DNA double-strand break repair. Sci Rep. 2023;13:15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lian J, Xia L, Chen Y, Zheng J, Ma K, Luo L, Ye F. Aldolase B impairs DNA mismatch repair and induces apoptosis in colon adenocarcinoma. Pathol Res Pract. 2019;215:152597. [DOI] [PubMed] [Google Scholar]
- 87.Zhao X, Lian X, Liu Y, Zhou L, Wu B, Fu YV. A Peptide Derived from GAPDH Enhances Resistance to DNA Damage in Saccharomyces cerevisiae Cells. Appl Environ Microbiol. 2022;88:e0219421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ci S, Xia W, Liang W, Qin L, Zhang Y, Dianov GL, Wang M, Zhao X, Wu C, Alagamuthu KK, et al. Src-mediated phosphorylation of GAPDH regulates its nuclear localization and cellular response to DNA damage. FASEB J. 2020;34:10443–61. [DOI] [PubMed] [Google Scholar]
- 89.Das RN, Balupillai A, David E, Santhoshkumar M, Muruhan S. Naringin, a Natural Flavonoid, Modulates UVB Radiation-Induced DNA Damage and Photoaging by Modulating NER Repair and MMPS Expression in Mouse Embryonic Fibroblast Cells. J Environ Pathol Toxicol Oncol. 2020;39:191–9. [DOI] [PubMed] [Google Scholar]
- 90.Krynetskaia NF, Phadke MS, Jadhav SH, Krynetskiy EY. Chromatin-associated proteins HMGB1/2 and PDIA3 trigger cellular response to chemotherapy-induced DNA damage. Mol Cancer Ther. 2009;8:864–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grolla AA, Miggiano R, Di Marino D, Bianchi M, Gori A, Orsomando G, Gaudino F, Galli U, Del Grosso E, Mazzola F, et al. A nicotinamide phosphoribosyltransferase-GAPDH interaction sustains the stress-induced NMN/NAD(+) salvage pathway in the nucleus. J Biol Chem. 2020;295:3635–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cai R, Xue W, Liu S, Petersen RB, Huang K, Zheng L. Overexpression of glyceraldehyde 3-phosphate dehydrogenase prevents neurovascular degeneration after retinal injury. FASEB J. 2015;29:2749–58. [DOI] [PubMed] [Google Scholar]
- 93.Leisner TM, Moran C, Holly SP, Parise LV. CIB1 prevents nuclear GAPDH accumulation and non-apoptotic tumor cell death via AKT and ERK signaling. Oncogene. 2013;32:4017–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hou X, Snarski P, Higashi Y, Yoshida T, Jurkevich A, Delafontaine P, Sukhanov S. Nuclear complex of glyceraldehyde-3-phosphate dehydrogenase and DNA repair enzyme apurinic/apyrimidinic endonuclease I protect smooth muscle cells against oxidant-induced cell death. FASEB J. 2017;31:3179–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Azam S, Jouvet N, Jilani A, Vongsamphanh R, Yang X, Yang S, Ramotar D. Human glyceraldehyde-3-phosphate dehydrogenase plays a direct role in reactivating oxidized forms of the DNA repair enzyme APE1. J Biol Chem. 2008;283:30632–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ferreira E, Gimenez R, Canas MA, Aguilera L, Aguilar J, Badia J, Baldoma L. Glyceraldehyde-3-phosphate dehydrogenase is required for efficient repair of cytotoxic DNA lesions in Escherichia coli. Int J Biochem Cell Biol. 2015;60:202–12. [DOI] [PubMed] [Google Scholar]
- 97.Dad A, Jeong CH, Pals JA, Wagner ED, Plewa MJ. Pyruvate remediation of cell stress and genotoxicity induced by haloacetic acid drinking water disinfection by-products. Environ Mol Mutagen. 2013;54:629–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kierans SJ, Taylor CT. Regulation of glycolysis by the hypoxia-inducible factor (HIF): implications for cellular physiology. J Physiol. 2021;599:23–37. [DOI] [PubMed] [Google Scholar]
- 99.Yeung SJ, Pan J, Lee MH. Roles of p53, MYC and HIF-1 in regulating glycolysis - the seventh hallmark of cancer. Cell Mol Life Sci. 2008;65:3981–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Elzakra N, Kim Y. HIF-1alpha Metabolic Pathways in Human Cancer. Adv Exp Med Biol. 2021;1280:243–60. [DOI] [PubMed] [Google Scholar]
- 101.Chen W, Xiao Z, Zhao Y, Huang L, Du G. HIF-1alpha inhibition sensitizes pituitary adenoma cells to temozolomide by regulating MGMT expression. Oncol Rep. 2013;30:2495–501. [DOI] [PubMed] [Google Scholar]
- 102.Sherapura A, Siddesh BM, Malojirao VH, Thirusangu P, Avin BRV, Kumari NS, Ramachandra YL, Prabhakar BT. Steroidal alkaloid solanidine impedes hypoxia-driven ATM phosphorylation to switch on anti-angiogenesis in lung adenocarcinoma. Phytomedicine. 2023;119:154981. [DOI] [PubMed] [Google Scholar]
- 103.Wozny AS, Gauthier A, Alphonse G, Malesys C, Varoclier V, Beuve M, Brichart-Vernos D, Magne N, Vial N, Ardail D, et al. Involvement of HIF-1alpha in the Detection, Signaling, and Repair of DNA Double-Strand Breaks after Photon and Carbon-Ion Irradiation. Cancers (Basel). 2021;13:3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhen X, Zhang M, Hao S, Sun J. Glucose-6-phosphate dehydrogenase and transketolase: Key factors in breast cancer progression and therapy. Biomed Pharmacother. 2024;176:116935. [DOI] [PubMed] [Google Scholar]
- 105.Li Y, Fu Y, Zhang Y, Duan B, Zhao Y, Shang M, Cheng Y, Zhang K, Yu Q, Wang T. Nuclear Fructose-1,6-Bisphosphate Inhibits Tumor Growth and Sensitizes Chemotherapy by Targeting HMGB1. Adv Sci (Weinh). 2023;10:e2203528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roudier E, Bachelet C, Perrin A. Pyruvate reduces DNA damage during hypoxia and after reoxygenation in hepatocellular carcinoma cells. FEBS J. 2007;274:5188–98. [DOI] [PubMed] [Google Scholar]
- 107.Zhang J, Chen F, Tian Y, Xu W, Zhu Q, Li Z, Qiu L, Lu X, Peng B, Liu X, et al. PARylated PDHE1alpha generates acetyl-CoA for local chromatin acetylation and DNA damage repair. Nat Struct Mol Biol. 2023;30:1719–34. [DOI] [PubMed] [Google Scholar]
- 108.Sousa FG, Matuo R, Soares DG, Escargueil AE, Henriques JA, Larsen AK, Saffi J. PARPs and the DNA damage response. Carcinogenesis. 2012;33:1433–40. [DOI] [PubMed] [Google Scholar]
- 109.Canto C, Menzies KJ, Auwerx J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 2015;22:31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Trammell SA, Schmidt MS, Weidemann BJ, Redpath P, Jaksch F, Dellinger RW, Li Z, Abel ED, Migaud ME, Brenner C. Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat Commun. 2016;7:12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yoshino J, Baur JA, Imai SI. NAD(+) Intermediates: The Biology and Therapeutic Potential of NMN and NR. Cell Metab. 2018;27:513–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bhatt AN, Chauhan A, Khanna S, Rai Y, Singh S, Soni R, Kalra N, Dwarakanath BS. Transient elevation of glycolysis confers radio-resistance by facilitating DNA repair in cells. BMC Cancer. 2015;15:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Erber J, Steiner JD, Isensee J, Lobbes LA, Toschka A, Beleggia F, Schmitt A, Kaiser RWJ, Siedek F, Persigehl T, et al. Dual Inhibition of GLUT1 and the ATR/CHK1 Kinase Axis Displays Synergistic Cytotoxicity in KRAS-Mutant Cancer Cells. Cancer Res. 2019;79:4855–68. [DOI] [PubMed] [Google Scholar]
- 114.Li YL, Weng HC, Hsu JL, Lin SW, Guh JH, Hsu LC. The Combination of MK-2206 and WZB117 Exerts a Synergistic Cytotoxic Effect Against Breast Cancer Cells. Front Pharmacol. 2019;10:1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tanaka T, Kurose A, Halicka HD, Traganos F, Darzynkiewicz Z. 2-deoxy-D-glucose reduces the level of constitutive activation of ATM and phosphorylation of histone H2AX. Cell Cycle. 2006;5:878–82. [DOI] [PubMed] [Google Scholar]
- 116.Jain VK, Kalia VK, Sharma R, Maharajan V, Menon M. Effects of 2-deoxy-D-glucose on glycolysis, proliferation kinetics and radiation response of human cancer cells. Int J Radiat Oncol Biol Phys. 1985;11:943–50. [DOI] [PubMed] [Google Scholar]
- 117.Hou XB, Li TH, Ren ZP, Liu Y. Combination of 2-deoxy d-glucose and metformin for synergistic inhibition of non-small cell lung cancer: A reactive oxygen species and P-p38 mediated mechanism. Biomed Pharmacother. 2016;84:1575–84. [DOI] [PubMed] [Google Scholar]
- 118.He Y, Chen H, Li W, Xu L, Yao H, Cao Y, Wang Z, Zhang L, Wang D, Zhou D. 3-Bromopyruvate-loaded bismuth sulfide nanospheres improve cancer treatment by synergizing radiotherapy with modulation of tumor metabolism. J Nanobiotechnology. 2023;21:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fu Z, Liu Z, Wang JX, Deng LF, Wang H, Tang W, Ni DL. Interfering biosynthesis by nanoscale metal-organic frameworks for enhanced radiation therapy. Biomaterials. 2023;295:122035. [DOI] [PubMed] [Google Scholar]
- 120.Rai Y, Yadav P, Kumari N, Kalra N, Bhatt AN. Hexokinase II inhibition by 3-bromopyruvate sensitizes myeloid leukemic cells K-562 to anti-leukemic drug, daunorubicin. Biosci Rep. 2019;39:BSR20190880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ding Y, He C, Lu S, Wang X, Wang C, Wang L, Zhang J, Piao M, Chi G, Luo Y, et al. MLKL contributes to shikonin-induced glioma cell necroptosis via promotion of chromatinolysis. Cancer Lett. 2019;467:58–71. [DOI] [PubMed] [Google Scholar]
- 122.Wang XY, Fan LW, Wang XZ, Luo TF, Liu LL. Cyclophilin A contributes to shikonin-induced glioma cell necroptosis and promotion of chromatinolysis. Sci Rep. 2022;12:14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chang MX, Wang HG, Niu JJ, Song Y, Zou ZH. Alkannin-Induced Oxidative DNA Damage Synergizes With PARP Inhibition to Cause Cancer-Specific Cytotoxicity. Front Pharmacol. 2020;11:610205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Manerba M, Di Ianni L, Fiume L, Roberti M, Recanatini M, Di Stefano G. Lactate dehydrogenase inhibitors sensitize lymphoma cells to cisplatin without enhancing the drug effects on immortalized normal lymphocytes. Eur J Pharm Sci. 2015;74:95–102. [DOI] [PubMed] [Google Scholar]
- 125.Liu XS, Little JB, Yuan ZM. Glycolytic metabolism influences global chromatin structure. Oncotarget. 2015;6:4214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen H, Li Y, Li H, Chen X, Fu H, Mao D, Chen W, Lan L, Wang C, Hu K, et al. NBS1 lactylation is required for efficient DNA repair and chemotherapy resistance. Nature. 2024;631:663–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gonzalez PS, O’Prey J, Cardaci S, Barthet VJA, Sakamaki JI, Beaumatin F, Roseweir A, Gay DM, Mackay G, Malviya G, et al. Mannose impairs tumour growth and enhances chemotherapy. Nature. 2018;563:719–23. [DOI] [PubMed] [Google Scholar]
- 128.Zampetidis CP, Galanos P, Angelopoulou A, Zhu Y, Polyzou A, Karamitros T, Kotsinas A, Lagopati N, Mourkioti I, Mirzazadeh R, et al. A recurrent chromosomal inversion suffices for driving escape from oncogene-induced senescence via subTAD reorganization. Mol Cell. 2021;81(4907–4923):e4908. [DOI] [PubMed] [Google Scholar]
- 129.Evangelou K, Belogiannis K, Papaspyropoulos A, Petty R, Gorgoulis VG. Escape from senescence: molecular basis and therapeutic ramifications. J Pathol. 2023;260:649–65. [DOI] [PubMed] [Google Scholar]
- 130.Zhang F, Guo J, Yu S, Zheng Y, Duan M, Zhao L, Wang Y, Yang Z, Jiang X. Cellular senescence and metabolic reprogramming: Unraveling the intricate crosstalk in the immunosuppressive tumor microenvironment. Cancer Commun (Lond). 2024;44:929–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kang C, Xu Q, Martin TD, Li MZ, Demaria M, Aron L, Lu T, Yankner BA, Campisi J, Elledge SJ. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349:aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang L, Pitcher LE, Yousefzadeh MJ, Niedernhofer LJ, Robbins PD, Zhu Y. Cellular senescence: a key therapeutic target in aging and diseases. J Clin Invest. 2022;132:e158450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang Z, Gao J, Xu C. Targeting metabolism to influence cellular senescence a promising anti-cancer therapeutic strategy. Biomed Pharmacother. 2024;177:116962. [DOI] [PubMed] [Google Scholar]
- 135.Yu T, Liu ZY, Tao QX, Xu X, Li XY, Li Y, Chen MX, Liu RF, Chen DW, Wu M, Yu JM. Targeting tumor-intrinsic SLC16A3 to enhance anti-PD-1 efficacy via tumor immune microenvironment reprogramming. Cancer Lett. 2024;589:216824. [DOI] [PubMed] [Google Scholar]
- 136.Yan J, Li WX, Tian H, Li B, Yu XY, Wang GH, Sang W, Dai YL. Metal-Phenolic Nanomedicines Regulate T-Cell Antitumor Function for Sono-Metabolic Cancer Therapy. ACS Nano. 2023;17:14667–77. [DOI] [PubMed] [Google Scholar]
- 137.Islamian JP, Hatamian M, Aval NA, Rashidi MR, Mesbahi A, Mohammadzadeh M, Jafarabadi MA. Targeted superparamagnetic nanoparticles coated with 2-deoxy-D-gloucose and doxorubicin more sensitize breast cancer cells to ionizing radiation. Breast. 2017;33:97–103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.