Abstract
CHO1 is a mammalian kinesin-like motor protein of the MKLP1 subfamily. It associates with the spindle midzone during anaphase and concentrates to a midbody matrix during cytokinesis. CHO1 was originally implicated in karyokinesis, but the invertebrate homologues of CHO1 were shown to function in the midzone formation and cytokinesis. To analyze the role of the protein in mammalian cells, we mutated the ATP-binding site of CHO1 and expressed it in CHO cells. Mutant protein (CHO1F′) was able to interact with microtubules via ATP-independent microtubule-binding site(s) but failed to accumulate at the midline of the central spindle and affected the localization of endogenous CHO1. Although the segregation of chromosomes, the bundling of midzone microtubules, and the initiation of cytokinesis proceeded normally in CHO1F′-expressing cells, the completion of cytokinesis was inhibited. Daughter cells were frequently entering interphase while connected by a microtubule-containing cytoplasmic bridge from which the dense midbody matrix was missing. Depletion of endogenous CHO1 via RNA-mediated interference also affected the formation of midbody matrix in dividing cells, caused the disorganization of midzone microtubules, and resulted in abortive cytokinesis. Thus, CHO1 may not be required for karyokinesis, but it is essential for the proper midzone/midbody formation and cytokinesis in mammalian cells.
INTRODUCTION
Mitosis and cytokinesis are unique events in the life cycle of eukaryotic cells in which two basic features of living systems, self-reproduction and cell motility, merge together in the process of equal segregation of genetic material into daughter cells. To separate the duplicated chromosomes, the cell forms a microtubule-containing machine called the spindle, whereas cytokinesis is accomplished by the actin-myosin–based contractile ring that is assembled around the cell equator and constricts inwards at the end of mitosis.
To ensure high-fidelity DNA transmission during cell division, both karyokinesis and cytokinesis must be tightly coordinated. A number of studies have suggested that the temporal and spatial organization of the cytokinetic machine is under the control of the mitotic spindle. Classic experiments done on marine invertebrate eggs established the importance of spindle asters in determining when and where cytokinesis will occur (Rappaport, 1961). Recent studies have also indicated that a spindle midzone composed of highly bundled microtubules, originating from the opposite poles, plays an essential role in cytokinesis. Thus, the creation of a physical barrier between the central spindle and the cell cortex (Cao and Wang, 1996), or the disorganization of central spindles by pharmacological treatment and/or genetic manipulation (Wheatley and Wang, 1996; Adams et al., 1998; Giansanti et al., 1998; Raich et al., 1998; Jantsh-Plunger et al., 2000) caused a failure of cytokinesis in different organisms. A variety of molecules have been shown to localize at the central spindle, including chromosomal passenger proteins (INCENP, TD-60), kinesin-like motor proteins (CENP-E, KLP-3A, MKLP1), protein kinases from the Polo and Aurora families, γ-tubulin (see Field et al., 1999 for a review), and GAP and nucleotide exchange factor for Rho GTPases (Tatsumoto et al., 1999; Jantsch-Plunger et al., 2000). Although many of those proteins were found to be essential for cytokinesis, the exact mechanisms of their function remain to be determined.
Using monoclonal, antimitotic spindle antibodies as probes, we identified a novel antigen located at the spindle midzone of Chinese hamster ovary (CHO) cells (Sellitto and Kuriyama, 1988). The antigen, named CHO1, was shown to associate with the dense midbody matrix during the late stage of cytokinesis. This antigen was found to be a plus-end-directed kinesin-like motor protein present in a wide range of species including human (MKLP1 [Nislow et al., 1992]), Caenorhabditis elegans (Zen-4 [Raich et al., 1998]; ceMKLP1 [Powers et al., 1998]), Drosophila (PAV-KLP [Adams et al., 1998]), sea urchin (KRP110 [Chui et al., 2000]), zebrafish (Chen and Detrich, 1996), Xenopus (Yonetani et al., 1996), and chicken (GgCHO1 and GgMKLP1 [Kuriyama et al., 2002]). Among the members of the MKLP1/CHO1 subfamily, CHO1 is unique in the sense that it contains an actin-interacting domain in the C-terminal tail (Kuriyama et al., 2002).
There has been a contradiction regarding the function of the motor protein in different organisms. Mammalian protein MKLP1 was shown to cross-link and slide antiparallel microtubules in vitro, and, therefore, it was originally thought to function in spindle elongation during anaphase B. Microinjection of monoclonal anti-CHO1 antibody caused mitotic arrest in mammalian cells (Nislow et al., 1990) and sea urchin eggs (Wright et al., 1993). In contrast, genetic analysis of MKLP1/CHO1 homologues in Drosophila and C. elegans did not reveal any inhibitory effects on karyokinesis. Instead, mutations in those genes caused severe disorganization of the central spindle and the inhibition of cytokinesis, suggesting a major role of the motor protein in formation/stabilization of the midzone microtubule bundles (Adams et al., 1998; Raich et al., 1998).
To study the function of CHO1 in mammalian cells, we analyzed the effects caused by the overexpression of an ATP-binding mutant of CHO1 and by the depletion of endogenous CHO1 via RNA-mediated interference. Here we report that CHO1 is required for the formation of the central spindle and midbody matrix, both of which are necessary for completion of cytokinesis in mammalian cells.
MATERIALS AND METHODS
Preparation of cDNA Constructs
The full coding sequence of CHO1 in pBluescript was obtained by immunoscreening of a CHO expression library as previously described (Kuriyama et al., 1994). The following CHO1 constructs (Figure 1) were prepared according to standard molecular cloning techniques (Ausubel et al., 1994): CHO1F (full coding sequence), CHO1F′ (full coding sequence with a mutation in the ATP-binding site), CHO1M (motor domain), CHO1M′ (motor domain with a mutation in the ATP-binding site), CHO1F′ΔT (mutated CHO1 sequence, lacking the tail domain), CHO1F′ΔE20 (mutated CHO1 sequence lacking 110 amino acids, encoded by the exon 20 in the tail domain [Kuriyama et al., 2002]). The cDNA fragments were subcloned into the multicloning site of the eukaryotic expression vector pCMV-HA (Matuliene et al., 1999) and/or pEGFP-C1 (Clontech, Palo Alto, CA), fusing the HA epitope tag and/or GFP to the N terminus of CHO1 constructs.
Figure 1.
A map of CHO1 constructs used for the current study. The total number of amino acids, encoded by each construct, is identified beneath the bold lines, representing the constructs. All constructs are tagged with the HA epitope at the N terminus. X indicates the mutation in the ATP-binding site of CHO1.
Three amino acids in the ATP-binding motif of CHO1 were mutated from GKT to AAA (amino acid positions 117–119) using a Gene Edition in vitro Site-Directed Mutagenesis System (Promega, Madison, WI) according to the manufacturer's protocol. Mutations in the ATP-binding motif have been shown to inhibit the motility of motor proteins in vivo and in vitro (Meluh and Rose, 1990; Nakata and Hirokawa, 1995).
Cell Culture and Synchronization
CHO cells were cultured as monolayers in Ham's F-10 medium containing 10% fetal bovine serum (FBS), as previously described (Matuliene et al., 1999). To obtain cells synchronized at M phase, CHO cells were grown on coverslips to 40–70% confluency and treated with 2.5–5 mM thymidine for 12–16 h to arrest the cell cycle at the S and G1/S stages. After washing out the thymidine, cells were cultured for additional 5 h and then exposed to 0.05 μg/ml nocodazole for 5–6 h to arrest cells at prometaphase. After the drug was carefully washed out from the culture, the cells were incubated further in fresh medium at 37°C for 20–80 min before fixation with −20°C methanol. Fixation at different time points within a 20- 80-min period allowed us to observe cells at the different stages of mitotic progression.
Gene Transfection
Transfection of CHO cells with different CHO1 constructs was performed using either FuGENE 6 (Roche Diagnostics, Indianapolis, IN) or LipofectAMINE (Life Technologies, Gaithersburg, MD) transfection reagents, according to manufacturers' instructions. Purified 0.6–2 μg plasmid DNA was mixed with one of the transfection reagents and applied to the cells grown on coverslips in 3.5-cm culture dishes. After 12–48 h of protein induction, cells were washed with phosphate-buffered saline (PBS) and fixed with −20°C methanol.
Immunofluorescence Microscopy
Immunofluorescence staining was performed as previously described (Kuriyama et al., 1994). The following primary antibodies were used: rat monoclonal anti-HA (dilution 1:50; Roche Diagnostics, Indianapolis, IN), rabbit polyclonal anti-HA (1:50; Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal anti-chicken β-tubulin (1:500; Amersham, Arlington Heights, IL), rabbit polyclonal anti-CHO1 (1:100; Kuriyama et al., 1994), mouse monoclonal anti-CHO1 (1:300; Sellitto and Kuriyama, 1988), rabbit polyclonal anti-CHO1 E20 (1:100; Matuliene and Kuriyama, unpublished data). Primary antibodies were detected with secondary antibodies at dilution 1:200, purchased from Hyclone (Logan, UT; fluorescein-conjugated anti-rat IgG, anti-mouse IgG + IgM, and anti-rabbit IgG), and from Jackson ImmunoResearch (West Grove, PA; Texas red-conjugated anti-rabbit IgG and anti-mouse IgG). To visualize DNA, cells were treated with 4′,6-diamidino-2-phenylindole (DAPI) at 1 μg/ml for 2–5 min. Microscopic observations were made on either an Olympus BH-2 (Lake Success, NY) or a Nikon Eclipse TE300 (Garden City, NY) inverted microscope equipped with epifluorescence optics.
Electron Microscopy
Samples for electron microscopy were prepared according to previously described procedure (Ryu et al., 2000). Briefly, CHO cells, cultured in a plastic culture dish were transfected with the GFP-tagged CHO1F′ and synchronized at the stage of cytokinesis. Cells were fixed with 2% glutaraldehyde in 100PEM (100 mM Pipes at pH 6.9, 1 mM EGTA, 1 mM MgCl2) for 30 min at room temperature. Fixation reaction was quenched with 1 mg/ml NaBH4 in distilled water. Cells expressing GFP-CHO1F′ construct were identified by fluorescence microscopy and their positions were marked by a diamond scribe. After staining with hematoxylin for 5–10 min, cells were postfixed with 1% OsO4 for 30 min, dehydrated through an ethanol series, infiltrated, and embedded using an EMBED 812 kit (Electron Microscopy Sciences, Ft. Washington, PA) according to the manufacturer's protocol. Thin sections were cut with a Reichert Ultracut microtome (Leica, Wien, Austria). After staining with uranyl acetate and lead citrate, specimens were examined with a 100CX transmission electron microscope (JEOL USA, Inc., Peabody, MA).
Small Inhibitory RNA Preparation and Transfection
Twenty-one–nucleotide RNAs (sense: 5′-GGUCAGUAAUACAACGGUGUU and antisense: 5′-CACCGUUGUAUUACUGACCUU) corresponding to the nucleotide positions 138–156 of CHO1 (relative to the start codon) were chemically synthesized and purified by HPLC (Integrated DNA Technologies, Inc., Coralville, IA). To prepare double-stranded RNAs with overhanging 3′ends (small inhibitory RNAs [siRNAs]), 20 μM single strands were incubated in annealing buffer (100 mM potassium acetate, 30 mM HEPES-KOH at pH 7.4, 2 mM magnesium acetate) for 1 min at 90°C and cooled down slowly to 37°C. For RNA-mediated interference (RNAi) assay, 0.5–1 μg of siRNA were mixed with 6 μl of LipofectAMINE reagent (Life Technologies, Gaithersburg, MD) according to the manufacturer's protocol and applied to the CHO cells grown on a coverslip in six-well plates. After 3 h, the transfection medium was replaced with fresh FBS-containing Ham's F-10 medium, and cells were further incubated at 37°C for 24–30 h before fixation. Mock transfections were performed in an identical manner to siRNA transfections, except that siRNA was omitted.
Quantitation of Fluorescence Intensity
To determine the effect of RNAi on the level of CHO1 expression, both mock- and siRNA-transfected cells were synchronized and fixed at M phase 30 h after transfection. Immunofluorescence staining was performed using polyclonal anti-CHO1 antibody and Texas red–conjugated secondary antibody. The fluorescence intensity corresponding to CHO1 was quantitated in 30 of mock- and 100 of siRNA-transfected anaphase cells using MetaMorph digital image analysis software package (version 2.0; Universal Image Co., West Chester, PA) as previously described (Matuliene et al., 1999). To calculate the percentage of CHO1 depletion, the average fluorescence intensity derived from mock-transfected cells was used as 100% of CHO1 expression. The lowest fluorescence intensity detected in siRNAi-transfected cells in which no endogenous CHO1 expression was detected by visual inspection was used as 0% of CHO1 expression.
RESULTS
CHO1 Moves Toward the Spindle Midzone during Anaphase
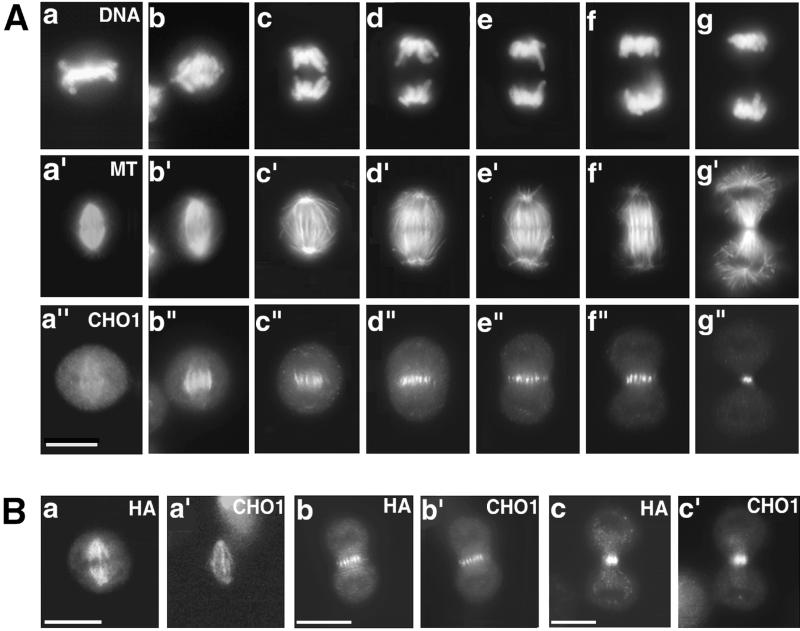
The plus-end directed kinesin-like motor protein CHO1 shows dynamic changes in its subcellular distribution during the cell cycle. The motor protein localizes at the nucleus in interphase cells (Sellitto and Kuriyama, 1988), which is due to the presence of the nuclear localization signal (NLS) at the C terminus of the tail domain (Kuriyama and Matuliene, unpublished). As cells enter M phase, the motor protein becomes associated with the spindle microtubules. In metaphase cells, it is diffusely distributed along the length of spindle fibers (a–a" in Figure 2A). During early anaphase, fluorescence becomes intense at the central region of the spindle where the antiparallel microtubules from the opposite sides of the spindle overlap (Figure 2A, b–b"). As chromosomes move toward the poles, the length of the fluorescent lines shortens (Figure 2A, c–c" to f–f"), and the antigen eventually concentrates to a bright midbody dot at the end of cytokinesis (Figure 2A, g–g").
Figure 2.
(A) Immunolocalization of CHO1 in CHO cells at different stages of mitosis. After synchronization, cells were fixed at the different stages of mitotic progression and stained with DAPI (a–g), antitubulin (a′–g′) and polyclonal anti-CHO1 (a"–g") antibodies. CHO1 changes its distribution from the entire spindle (a") to the midzone region during anaphase (b"–f") and concentrates to a midbody at the end of cytokinesis (g"). (B) Distribution of exogenous CHO1F in transfected cells is identical to the distribution of endogenous CHO1. Cells fixed at metaphase (a, a′), anaphase (b, b′), and telophase (c, c′) were stained with anti-HA (a, b, and c) and polyclonal anti-CHO1 (a′, b′, and c′) antibodies. Bar, 10 μm.
Figure 2B shows the immunolocalization of exogenous, HA-tagged, full-length CHO1 (CHO1F; Figure 1) expressed in CHO cells by transient transfection. Similarly to endogenous CHO1, CHO1F is detected along the spindle fibers during metaphase (Figure 2Ba), then shifts to the midzone region (Figure 2Bb), and eventually concentrates to the midbody at the end of mitosis (Figure 2Bc). The identical immunolocalization pattern was obtained by staining with polyclonal anti-CHO1 antibody that recognizes the tail domain of the motor protein (Figure 2B, a′, b′, and c′).
Motor Activity Is Essential for the Localization of CHO1 at the Center of Spindles and Midbodies
To analyze the role of CHO1 motor activity in mitotic cells, we created a mutant protein in which the amino acid sequence in the ATP-binding consensus motif was changed from GKT to AAA (amino acid positions 117–119). Although nonmutated motor domain alone (CHO1M, Figure 1) was detected in association with the microtubule network (Figure 3A), the mutated motor domain (CHO1M′, Figure 1) showed a dramatically reduced microtubule-binding activity in vivo (compare Figure 3, B and B′). However, the full-length mutated protein (CHO1F′; Figure 1) was still capable of efficient microtubule binding in both mitotic (Figure 3, C–E) and interphase (Figure 3, F–F") cells. These observations suggest the presence of additional ATP-independent microtubule binding site(s) in the CHO1 sequence. This is consistent with our previous findings that the N-terminal half of the CHO1 sequence can interact with microtubules in both ATP-dependent and ATP-independent manners in vitro (Kuriyama et al., 1994). To eliminate the chance that CHO1F′ distributes along the microtubules simply via dimerization with endogenous CHO1, we expressed the truncated polypeptide in which the NLS-containing tail domain was deleted from the CHO1F′ sequence (CHO1F′ΔT, Figure 1). Figure 3, G–G", demonstrates that although endogenous CHO1 is confined inside the nucleus (Figure 3G′), CHO1F′ΔT still binds and even bundles microtubules (Figure 3G), suggesting that CHO1 is able to interact with microtubules in the ATP-independent manner in vivo.
Figure 3.
ATP-binding mutant of CHO1 (CHO1F′) interacts with microtubules via ATP-independent microtubule binding site(s) but does not concentrate at the midline of the central spindle during anaphase. (A) CHO1M associates with microtubules when expressed in CHO cells. (B) CHO1 M′ generally does not display microtubule binding (compare HA staining in B and tubulin staining in B′). CHO1F′ interacts with microtubules in both mitotic (C, metaphase; D, anaphase; E, telophase) and interphase cells (F). Although CHO1F′ contains the NLS and is generally seen within the nucleus, in some interphase cells with the high level of expression, CHO1F′ can also be detected in the cytoplasm (F). CHO1F′ΔT strongly binds and bundles microtubules (G), whereas endogenous CHO1 is confined within the nucleus (G′). This indicates that microtubule binding by mutated CHO1F′ and CHO1F′ΔT constructs is not due to the dimerization with endogenous CHO1 but is due to the ATP-independent microtubule binding site(s) present in the motor protein. CHO1F′ fails to concentrate at the midline of the central spindle during anaphase (D) and distributes along the entire intercellular bridge at the end of cytokinesis (E). Staining was performed using monoclonal anti-HA (A and C–E), polyclonal anti-HA (B, F, and G), monoclonal antitubulin (B′ and F′), monoclonal anti-CHO1 antibodies (G′) and DAPI (F" and G"). Bars, 10 μm.
Although CHO1F′ associates with the spindle fibers in mitotic cells (Figure 3, C–E), it can no longer shift toward the equator of the cell. Instead of concentrating at the midline of the central spindle, as does wild-type CHO1F (Figure 2Bb), CHO1F′ remains in a wide region of the spindle midzone during anaphase B (Figure 3D) and distributes along the entire intercellular bridge at the end of cytokinesis (Figure 3E). These results suggest that mechanochemical motor activity of CHO1 is required for the proper localization of the protein to the midline of central spindles and midbodies.
Overexpression of CHO1F′ Inhibits Completion of Cytokinesis in Mammalian Cells
To assess the role of CHO1 in mammalian cells, we examined the mitotic profiles in cells expressing high levels of CHO1F′. Figure 4 shows synchronized transfected cells fixed at 50–80 min after release from nocodazole treatment. In these cells, CHO1F′ failed to translocate to the cell equator and remained distributed along the entire midzone fibers and astral microtubules throughout mitosis. As shown in Figure 4, A–A", the excess of mutated protein did not cause any inhibitory effects on bipolar spindle formation and chromosome segregation. Statistical analysis did not reveal significant differences between the length of the mitotic spindles in both transfected and control cells. The average pole to pole distance at the end of anaphase B was found to be 11.9 ± 1.7 and 11.4 ± 2.4 μm (p = 0.05; n = 2 × 15) in control and CHO1F′-expressing cells, respectively. The bundling of midzone microtubules and the initiation of cytokinesis also occurred normally in the cells expressing mutant protein (Figure 4, A′ and A"), suggesting that ATPase activity of CHO1 is not essential for the formation of microtubule bundles at the midzone region. However, the completion of cytokinesis was strongly affected. Figure 4, B–F, shows that the daughter cells flattened out and entered interphase, while still connected by the cytoplasmic bridges of various sizes and lengths. Although some bridges were long and thread-like (Figure 4, B and B′), the majority of such linkers were thick, forming an isthmus or neck between the two parts of the cytoplasm (Figure 4, C–F). The cytoplasmic bridges included microtubule bundles along which CHO1F′ was distributed. Although cells generally showed normal chromosome segregation, lagging chromosomes were occasionally detected in the middle of the cytoplasmic bridges (Figure 4, E–E").
Figure 4.
High level of CHO1F′ expression inhibits completion of cytokinesis in CHO cells. Although the midzone bundles are forming in cells expressing CHO1F′ (A′ and A"), the daughter cells remain connected by the cytoplasmic bridges after mitosis (B–F). Cells expressing mutant protein are stained by anti-HA (A′, B, C, D′, E′, and F) and either anti-CHO1 polyclonal (A", B′, D", and F′) or antitubulin (C′ and E") antibodies. DNA is visualized by DAPI staining (A and E). Bars, 10 μm
In contrast to CHO1F′, the cells overexpressing wild-type CHO1F seemed to undergo normal cell division (Figures 2B, c and c′, and see Figure 7, C–C"), suggesting that cytokinesis defects were caused specifically by the expression of mutated protein. To confirm the inhibitory effect of CHO1F′, we counted the number of nuclei in cells expressing either CHO1F or CHO1F′ (Table 1). At 48 h after transfection, nearly 45% of CHO1F′-expressing cells were found to contain more than one nucleus, whereas only ∼8% multinucleation was detected in the cells expressing wild-type CHO1F. The majority (80–90%) of multinucleated cells had two nuclei of similar size (Figure 5), suggesting that transfected cells underwent abortive cytokinesis after proper chromosome segregation. Because daughter cells connected by the cytoplasmic bridge (Figure 4) were seen only for the few hours after division, it is likely that the cleavage furrows initiated in CHO1F′-expressing cells (Figure 4) eventually regress, causing the formation of binucleate cells (Figure 5). It is noteworthy that the expression of CHO1F also induced multinucleation above the control level (∼8% vs. ∼1.5%), suggesting that the proper level and/or timing of CHO1 expression is important for normal progression of cytokinesis.
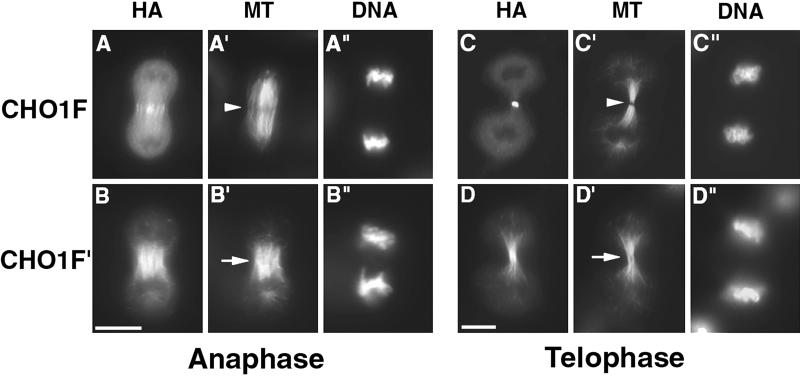
Figure 7.
Formation of stem body and midbody matrices is inhibited in dividing cells expressing mutated CHO1. CHO cells, transfected with CHO1F (A and C) and CHO1F′ (B and D) constructs were fixed at anaphase (A and B) and telophase (C and D) stages and stained using anti-HA (A–D), antitubulin (A′–D′) antibodies, and DAPI (A"–D"). The stem body and midbody matrix is easily detected in cells expressing wild-type CHO1F (arrowheads in A′ and C′). However, in cells expressing mutant protein, the dense matrix material is missing, based on the absence of dark spots in tubulin staining (arrows in B′ and D′). Bars, 10 μm.
Table 1.
Expression of CHO1F′ causes multinucleation in CHO cells
| Expressed DNA constructs | Cells with more than one nucleus (%) | p value | Number of counted cells |
|---|---|---|---|
| CHO1F′ | 44.6 ± 11.2 | 0.05 | 3 × 500 |
| CHO1F | 7.8 ± 6.7 | 0.05 | 3 × 500 |
| No expression (control) | 1.5 ± 0.6 | 0.01 | 6 × 500 |
Cells were transfected with CHO1F and CHO1F′ constructs and cultured for 48 h before fixation. Fixed cells were immunostained with anti-HA antibodies to visualize cells expressing exogenous proteins. The number of nuclei was counted in 500 cells expressing CHO1F and CHO1F′ constructs and in 500 nonexpressing (control) cells, present on the same coverslips. Experiment was repeated three times.
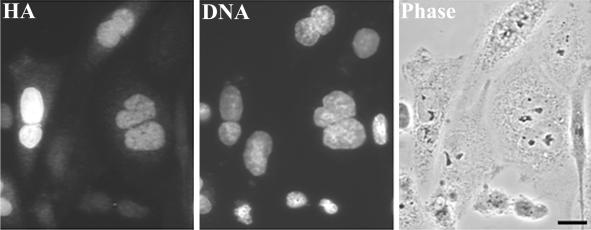
Figure 5.
Expression of CHO1F′ induces binucleation in CHO cells (also see Table 1). Cells were fixed at 48 h after transfection with CHO1F′ construct and stained with anti-HA antibodies and DAPI to visualize the nuclei. Cells were observed by the fluorescence and phase contrast microscopy. Bar, 10 μm.
Endogenous CHO1 Is Dislocated from the Midzone/midbody in Cells Expressing Mutant CHO1
To study the mechanism by which CHO1F′ inhibits the completion of cytokinesis, we examined the localization of endogenous CHO1 in cells expressing mutant protein. Because CHO1 appears to exist as a dimer (Kuriyama et al., 1994), we hypothesized that CHO1F′ can dimerize with the endogenous CHO1 via α-helical, coiled-coil stalk domains and sequester the endogenous protein from the central spindles and the midbodies.
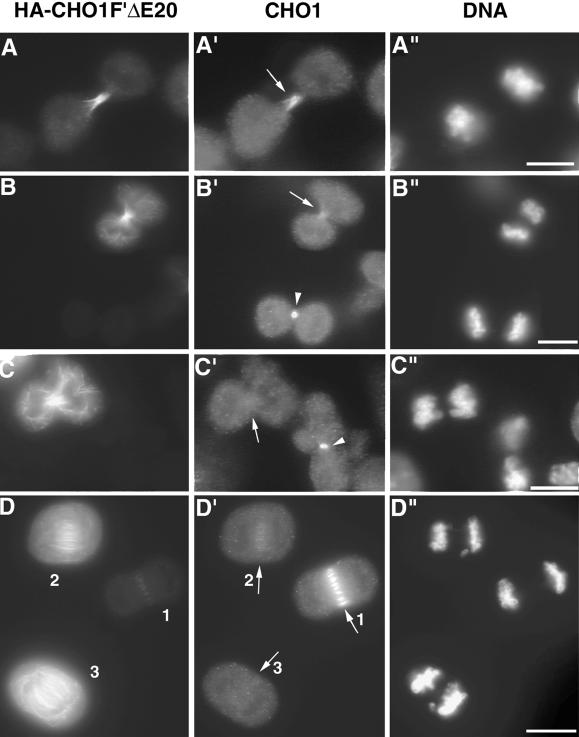
To test the effect of the mutated protein on the localization of endogenous CHO1, CHO cells were transfected with CHO1F′ΔE20 construct (Figure 1), which expresses the HA-tagged CHO1F′ lacking 110 amino acid residues (E20) in the middle of the C-terminal tail. Although CHO1F′ΔE20 polypeptide has the distribution pattern identical to that of CHO1F′, it cannot be recognized by the antibody raised against the E20 sequence (Matuliene and Kuriyama, unpublished results). This difference allowed us to distinguish between the distribution of endogenous CHO1 and exogenous CHO1F′ΔE20 by probing with E20 and HA antibodies, respectively. Figure 6, A–A", shows a dividing cell expressing relatively low level of the mutated protein. In this cell, CHO1F′ΔE20 is detected along the entire intercellular bridge formed between two daughter cells (Figure 6A), similar to CHO1F′ at a low level of expression (Figure 3E). Significantly, the endogenous CHO1 also shows an abnormal distribution along with the HA-tagged mutant molecule (arrow in Figure 6A′). When the expression level of mutated protein increases and the protein starts to distribute along the length of midzonal and astral microtubules (Figure 6B), the labeling of midzone/midbody area by E20 antibody becomes very weak (arrow in Figure 6B′). At the highest levels of mutant protein expression (Figure 6C), the E20 staining at region of central spindle is almost undetectable (arrow in Figure 6C′), indicating that endogenous CHO1 is displaced from the midzone region and dispersed throughout the cell. Because the E20 antibody detects the normal localization of endogenous CHO1 in control cells (arrowheads in Figure 6, B′ and C′), a failure to detect endogenous protein in transfected cells is not due to insufficient immunostaining.
Figure 6.
Mutant CHO1 inhibits the accumulation of endogenous CHO1 at the midzone/midbody region. (A–D) The effect of CHO1F′ΔE20 expression on the localization of endogenous CHO1. When the expression of CHO1F′ΔE20 increases from low (A) to medium (B) and high levels (C), the endogenous CHO1 becomes depleted from midzone/midbody area of the dividing cells (arrows in B′ and C′). (D–D") Three anaphase cells expressing different levels of mutated protein in the same field. The higher the level of CHO1F′ΔE20 expression (D), the weaker the accumulation of endogenous protein at the midzone region (arrows 1 to 3 in D′). Staining was performed using anti-HA (A–D) and anti-CHO1 E20 (A′–D′) antibodies and DAPI (A"–D"). Bars, 10 μm.
Displacement of endogenous CHO1 from the midzone region was also seen in anaphase cells. In one field shown in Figure 6, D–D", there are three anaphase cells expressing different levels of CHO1F′Δ20. The cell with a very low level of exogenous protein (1 in Figure 6D) has a proper localization of endogenous CHO1 (arrow 1 in Figure 6D′). However, the medium level of CHO1F′ΔE20 expression (2 in Figure 6D) causes a significant reduction of endogenous protein at the central spindle (arrow 2 in Figure 6D′), and the high level of expression (3 in Figure 6D) completely inhibits proper localization of CHO1 (arrow 3 in Figure 6D′). Correlation between the decrease in the fluorescence level of CHO1 at the central spindle and the increase in the fluorescence level of CHO1F′ΔE20 was further confirmed by a quantitation of fluorescence intensity using MetaMorph image analysis software package (unpublished data). These results suggest that at a high level of expression the mutant protein could be acting in a dominant negative manner by inhibiting the accumulation of endogenous CHO1 at the midline of the central spindle and midbody.
Overexpression of CHO1F′ Inhibits Organization of the Electron Dense Midbody Matrix
To identify morphological defects caused by the mislocalization of CHO1, we analyzed the midzone structure in cells overexpressing mutant protein. Figure 7 shows anaphase (panels A and B) and telophase (panels C and D) cells expressing either CHO1F (panels A and C) or CHO1F′ (panels B and D) constructs. Double-staining with anti-HA and antitubulin antibodies revealed the presence of the microtubule bundles in anaphase cells expressing both CHO1F and CHO1F′. However, in cells expressing mutant protein, the dark areas corresponding to stem bodies were hardly detectable (arrow in B′). The stem bodies form during anaphase at the regions of the antiparallel polar microtubule overlap (McIntosh and Landis, 1971). These structures are associated with dense amorphous material (matrix), which cannot be penetrated by antitubulin antibodies and, therefore, appear as dark spots in the tubulin staining (arrowhead in Figure 7A′). By the end of cytokinesis, the stem body matrices are compacted to form a single midbody matrix (arrowhead in Figure 7C′). However, in cells expressing mutated CHO1, the dense midbody matrix does not form, based on the even tubulin staining at the region of the intercellular bridge (arrow in Figure 7D′). This finding suggests that mislocalization of CHO1 protein inhibits formation of the dense midbody matrix in dividing cells.
To confirm the absence of the midbody matrix in CHO1F′-expressing cells, we analyzed the midbody structure by electron microscopy. CHO cells were transfected with the GFP-tagged CHO1F′. Conjugation with GFP did not cause any changes in the intracellular distribution of the mutant protein throughout the cell cycle (unpublished data). Figure 8 illustrates a control (panels A and B) and a transfected cell (panels C and D) seen at both low (panels A and C) and high (panels B and D) magnifications. Cells are at a late stage of cell division, and nuclei have already reformed in each daughter cell. Although control cells are round and connected by an intercellular bridge with a typical dense midbody matrix in the center (arrows in Figure 8, A and B), the transfected daughter cells are connected by a thick cytoplasmic bridge, indicating incomplete cytokinesis (Figure 8, C and D). The cells expressing mutant protein are quite irregular in shape and contain abnormal cytoplasmic structures as indicated by the arrowheads in Figure 8D. Microtubules arranged in parallel are seen in the middle of the bridge connecting two transfected daughter cells (Figure 8D). Although few areas of the electron dense material are sporadically dispersed along the microtubules (small arrows in Figure 8D), the midbody matrix is not organized in a regular pattern as in the control cell (Figure 8B), where the electron dense material is flanked by low-density regions on either side. Similar results were obtained after analyzing four other cells expressing mutant protein. The electron dense matrix in all those cells was either absent or very weak and disorganized. From these results, we conclude that mutant CHO1 may inhibit the completion of cytokinesis, at least in part by interfering with the organization of electron dense midbody matrix in the center of the intercellular bridge.
Figure 8.
Expression of CHO1F′ inhibits the organization of electron dense midbody matrix. Thin section electron micrographs show a control cell (A and B) and a cell expressing CHO1F′ (C and D) at the late stage of cell division. Both cells are seen at low (A and C) and high (B and D) magnifications. Although the electron dense midbody matrix can be easily detected in the intercellular bridge connecting two control cells (arrows in A and B), the electron-dense structure is completely disorganized in the cell expressing CHO1F′. Bars: A and C, 5 μm; B and D, 0.5 μm.
RNAi Directed against Endogenous CHO1 Affects Cytokinesis in CHO Cells
To confirm the role of CHO1 in cytokinesis, we performed the RNAi assay recently developed for mammalian cultured cells (Elbashir et al., 2001). Base-paired 21-nucleotide siRNAs corresponding to the nucleotide positions 138–156 of CHO1 were synthesized and introduced into CHO cells via liposome-mediated transfection. Immunofluorescence staining with anti-CHO1 antibody showed a dramatic decrease in the fluorescence intensity in siRNA-treated cells 30 h after transfection. In comparison with mock-transfected cells, the fluorescence intensity derived from endogenous CHO1 was reduced by 75–100% in 43% of siRNA-transfected cells, by 50–75% in 39% of siRNA-transfected cells, by 25–50% in 13% of siRNA-transfected cells, and by 0–25% in 5% of siRNA-transfected cells, as determined by the quantitation of fluorescence intensity using MetaMorph image analysis software package.
To analyze the effect of CHO1 depletion on mitosis, siRNA-transfected cells were synchronized and fixed at the different stages of mitotic progression. Figure 9 shows both mock- (panels A, C, and F) and siRNA-transfected (panels B, D, G, E, and H) mitotic cells double-stained with antitubulin (Figure 9, A–H) and anti-CHO1 (Figure 9, A′–H′) antibodies. We could not detect any abnormalities in prometaphase or metaphase cells expressing almost undetectable levels of endogenous CHO1 (Figure 9, B–B", compared with control, A–A"). However, the proper formation of central spindle in RNAi-affected cells was inhibited. As shown in Figure 9, D-D" and G-G", the bundles of midzone microtubules were still forming, but the stem body/midbody matrix was hardly detectable in cells with the significantly reduced level of CHO1 (arrowheads in panels D and G, compared with arrows in panels C and F), confirming the role of CHO1 in matrix formation. When CHO1 was almost entirely depleted from CHO cells (Figure 9, E′, H′), the midzone microtubule bundles became severely disorganized (Figure 9, E and H), suggesting that CHO1 is essential for maintaining the organization of central spindle in mammalian cells. Although initiation of cytokinesis occurred normally in CHO1-depleted cells (Figure 9, D′, E′, G′, and H′), the completion of cytokinesis was inhibited, resulting in the formation of binucleate/multinucleate cells. It was found, that 30 h after transfection ∼50% of cells transfected with siRNA had more than one nucleus (49.6 ± 3.3%, p = 0.05, n = 3 × 500), whereas the level of multinucleation in mock-transfected cells never exceeded 1.7%.
Figure 9.
RNAi directed against endogenous CHO1 interferes with the organization of central spindle and the formation of midbody matrix in CHO cells. Mock- and siRNA-transfected cells were synchronized and fixed at the different stages of mitosis 30 h after transfection. Cells were visualized by staining with antitubulin (A–H), anti-CHO1 polyclonal (A′-H′) antibodies, and DAPI (A"-H"). To correctly represent the amount of endogenous CHO1 in all cells, both mock- and siRNA-transfected cells were fixed and immunostained in an identical manner, and all images were captured and developed using identical exposure and conditions. RNAi directed against endogenous CHO1 affects neither the formation of bipolar spindle (B–B") nor chromosome segregation (D", E", G", and H") in CHO cells. However, the reduced level of CHO1 expression (D′ and G′) diminishes the matrix formation in anaphase (arrowhead in D) and telophase (arrowhead in G) cells in comparison with control (arrows in C and F). Almost complete depletion of CHO1 (E′ and H′) causes the severe disorganization of central spindles (E and H).
DISCUSSION
Microtubule-binding Sites of CHO1
The kinesin-like protein CHO1 is a midbody matrix component, which plays an essential role in organization of the midbody matrix and the completion of cytokinesis in mammalian cells. The protein shows dynamic changes in its subcellular distribution at the different stages of cell cycle. During interphase, it is seen inside the nucleus, which is due to the presence of the NLS at the C terminus of the tail domain. The NLS has already been identified in the tail domain of human MKLP1 at the amino acid positions 710–961 (Deavours and Walker, 1999). In CHO1 we were able to identify a region of 27 amino acids (positions 899–925) capable of recruiting the motor protein to the nucleus (unpublished results). At the onset of M phase, the protein becomes associated with spindle microtubules. In vitro cosedimentation experiments provided evidence that the N-terminal half of the protein, which covers the motor domain and the one third of central stalk, can interact with microtubules in both ATP-dependent and ATP-independent manners (Kuriyama et al., 1994). This is consistent with our current results of domain analysis in vivo. When the motor domain alone (CHO1M) is expressed in CHO cells, it can always be detected in association with the microtubule network (Figure 3A). Mutation in the ATP-binding site of CHO1M dramatically reduced the microtubule binding (Figure 3B), indicating that the motor domain alone interacts with microtubules, primarily in the ATP-dependent manner. Nonetheless, in some cells, we were able to detect a weak microtubule-binding activity of CHO1M′ (unpublished data), which suggested that the motor domain of CHO1 includes the ATP-independent microtubule-binding site as well. CHO1F′ΔT retained much more intense microtubule-binding capacity than CHO1M′ (Figure 3G), but the stalk domain alone never showed any affinity to microtubules (unpublished data). These observations allowed us to predict that the ATP-independent microtubule-binding site resides in the C terminus of the motor domain, close to the stalk. Indeed, the deletion of 147 amino acids (positions 276–422) from the C-terminal side of the motor domain completely abolished microtubule binding of the CHO1F′ΔT construct (Matuliene and Kuriyama, unpublished results). Consistent with our results, a similar region of the motor domain has already been implicated in the ATP-independent microtubule binding in both kinesin heavy chain (Yang et al., 1989) and kinesin-like protein Ncd (Moore et al., 1996).
Interaction between CHO1 Molecules
Kinesin heavy chain and the majority of kinesin-like motor proteins are believed to form dimers/oligomers via α-helical, coiled-coil stalk domains (de Cuevas et al., 1992; Miki et al., 2001). We have previously shown that CHO1 forms a dimer complex when expressed in Sf9 cells (Kuriyama et al., 1994). In this study, we demonstrated the effect of the mutated protein on the localization of endogenous CHO1 (Figure 6). One possible mechanism for the displacement of endogenous protein from the midzone/midbody region could be a stalk-mediated dimerization between endogenous CHO1 and mutated protein subunits. In agreement with that, the expression of both CHO1F′ and CHO1F constructs with the deleted stalk domains and also motor domains alone (CHO1M and CHO1M′) had no effect on the localization of the endogenous protein and the completion of cytokinesis (our unpublished results). We speculate that immotile CHO1F′ interferes with the motility of endogenous CHO1 and reduces the local accumulation of the motor protein and its associated factors at the midline of the central spindle, thus affecting both the density of midbody matrix and cytokinesis. However, the exact mechanism by which CHO1F′ affects cytoplasmic division and the localization of endogenous CHO1 remains to be determined.
In cells with a low level of CHO1F′ΔE20 expression, the mutant protein and endogenous CHO1 were not diffusely distributed along the entire spindle fibers but were concentrated toward the central region of the spindle and the intercellular bridge (Figure 6A and cell 1 in 6D). This observation suggests that the mutant and the endogenous CHO1 could be forming a dimer complex, which is partially functional and capable of translocation toward the equator of the cell. In contrast, at a high level of mutant protein expression, no accumulation of exogenous, as well as endogenous, protein could be observed at the midline of the central spindle (Figure 6, C and C′, cell 3 in 6, D and D′), implying that the CHO1-CHO1F′ heterodimer might be completely immotile when a large excess of the mutant protein is present. This difference could be due to the amount of mutant homodimers interacting with microtubules during mitosis. As the abundance of mutant protein bound to the polar microtubules increases, more of the endogenous CHO1-containing dimers could be forced to stay in the cytoplasm and, therefore, their accumulation at the midzone region may not be easily detectable. Another possibility is that CHO1 is not a dimer but a tetramer, as has been suggested by the studies of the CHO1 homologue KRP110 in sea urchin eggs (Chui et al., 2000). If so, one or two mutated molecules in the tetramer may not interfere with the motility of CHO1 as severely as three or four mutated monomers. Alternatively, CHO1 may function as a dimer, which has to be associated with other proteins in order to localize properly at the central spindle. In favor of this notion are the recent findings that the Aurora-related kinase Air-2 and the GAP for Rho family GTPases Cyk-4 are both required for the localization of Zen-4 at the central spindle of C. elegans embryos (Jantsch-Plunger et al., 2000; Severson et al., 2000). It is reasonable to speculate that the excess of nonmotile mutant homodimers may deplete the pool of those binding partners in dividing cells and prevent the localization of endogenous CHO1-containing dimers at the midzone region.
The Function of CHO1 in Cell Division
Because MKLP1 has been shown to cross-link and slide the antiparallel microtubules in vitro, the motor protein has been implicated in the spindle elongation during anaphase B (Nislow et al., 1992). Microinjection of the CHO1-specific monoclonal antibodies into mammalian cells and sea urchin eggs caused prophase or metaphase arrest depending on the time of injection (Nislow et al., 1990; Wright et al., 1993). In contrast, neither mutation in the ATP-binding site of CHO1 nor the depletion of CHO1 by RNAi caused a detectable effect on karyokinesis, suggesting that CHO1 is not essential for chromosome segregation in mammalian cells. We cannot rule out the possibility that the inhibition or depletion of CHO1 may have had transient effects on spindle function and morphology, which could have been missed in our fixed time point analysis of mitotic progression. Real time video microscopy will be required to determine if CHO1 plays any role in karyokinesis. Nevertheless, the role of CHO1 in cytokinesis appears to be essential, consistent with the function of CHO1 homologues in Drosophila and C. elegans (Adams et al., 1998; Powers et al., 1998; Raich et al., 1998; Severson et al., 2000). Similarly to the null mutations of pavarotti and zen-4 (Adams et al., 1998; Raich et al., 1998), almost complete depletion of endogenous CHO1 from mitotic cells caused the disorganization of central spindles (Figure 9, E and H). This observation suggests that, in a concert with other midzone components, CHO1 is functioning in the formation of midzone microtubule bundles in mammalian cells. In contrast, the ATP-binding mutant of CHO1 did not inhibit the bundling of midzone microtubules. This difference could be due to the presence of the wild-type protein in CHO1F′-expressing cells and the fact, that mutation in the ATP-binding site did not inactivate the function of CHO1 completely. As shown in Figure 3G, the mutated protein retained the capability to bind and bundle microtubules, suggesting that the ATPase activity is not essential for microtubule bundling. We speculate that multiple ATP-independent microtubule binding sites, created in the CHO1 molecule via stalk-mediated dimerization, are sufficient for microtubule cross-linking activity.
Although the bundling of midzone microtubules could be a primary function of CHO1, its role in the midbody matrix formation seems to be equally important for the completion of cytoplasmic division. As shown in Figure 4, cytokinesis of CHO1F′-expressing cells remained incomplete, although midzone microtubule bundles were present. We also observed the regression of cleavage furrows in endogenous CHO1-depleted cells in which microtubule bundles were present but the midbody matrix was weak or undetectable by fluorescence microscopy. These observations suggest that the presence of midzone bundles cannot support the completion of cytokinesis, unless a sufficient amount of CHO1 concentrates to the narrow central region of these bundles and participates in the midbody matrix formation. Therefore, the motor activity of CHO1, which facilitates the accumulation of CHO1 at the midline of the central spindle, plays a crucial role in the formation of the midbody matrix and the completion of cytokinesis.
It seems that the midbody, which is discarded and eventually deteriorates after cytokinesis, is not simply a remnant of the mitotic apparatus. Rather, it could be a key structure in controlling the division of the cytoplasm. In agreement with this, a large number of proteins have been shown to localize at the midbody during cytokinesis (Rattner, 1992; Field et al., 1999; Straight and Field, 2000 for reviews). Although some of these proteins may be important for carrying certain signals from the different parts of the spindle/cell, others could serve as the strictly structural components necessary to build the different regions of the intercellular bridge. Because CHO1 is a motor protein, it is plausible that it not only plays a structural role by cross-linking microtubules together but also carries certain cargoes as it moves toward the plus ends of midzone microtubules. Possible candidate cargo molecules would be CYK-4 (Jantsch-Plunger et al., 2000) and small G-protein Arf (Boman et al., 1999; Skop et al., 2001), which have been shown to interact with CHO1 and to be important for cytokinesis.
The Function of the Midbody Matrix
Although an electron dense midbody structure was identified by electron microscopy over four decades ago, little is known about the molecular composition of this structure. SDS-PAGE analysis of isolated midbodies revealed α and β tubulins as major constituents along with other ∼35 minor components (Mullins and McIntosh, 1982). Extraction of the isolated midbodies with an ionic detergent (Sarkosyl NL-30) was shown to solubilize the midbody microtubules leaving the central, dense matrix zone of the midbody intact (Mullins and McIntosh, 1982). CHO1 has been determined to be a component in this dense Sarkolyl-insoluble midbody matrix (Sellitto and Kuriyama, 1988). Although immunofluorescence microscopy revealed the presence of many different proteins at the same/similar region as CHO1 during cell division (Rattner, 1992; Jantsch-Plunger et al., 2000), CHO1 appears to be the only protein characterized as a component of midbody matrix at this date.
Although several studies have indicated the correlation between the disorganization of the midbody matrix and the regression of the cleavage furrows (King and Akai, 1971; Mullins and Biesele, 1977), the exact role of the midbody matrix in the process of cytokinesis remains uncertain. Because microtubules are required for the completion of cytokinesis in animal cells (Larkin and Danilchik, 1999; Straight and Field, 2000), the dense matrix material may function in stabilizing microtubules within the intercellular bridge by gluing them together. Indeed, midbody microtubules are much more resistant to disruption by physical or chemical agents than are other parts of the mitotic spindle (Salmon et al., 1976). In addition, the midbody matrix may directly link microtubules to the cell cortex. Several studies have suggested that the midbody matrix could be a structure holding microtubules and the cortex together after the cessation of the furrowing and the disassembly of the contractile ring (Mullins and Biesele, 1973, 1977). This could be an important function, because the actual separation of the daughter cells has been shown to occur up to several hours after the midbody formation (Sanger et al., 1985). Consistent with these observations are the experiments involving conditional loss-of-function alleles of zen-4, which established the requirement of ZEN-4 for the late cytokinesis and the maintenance of cell separation through much of the subsequent interphase (Severson et al., 2000).
It is widely believed that the insertion of membrane vesicles is important for the progression and completion of the cleavage furrows (Straight and Field, 2000). Several evidences suggest that the component(s) of the midbody matrix may facilitate the membrane fusion events. It has been previously demonstrated that CHO1 can directly interact with the small G-protein Arf, known for its function in the membrane trafficking (Boman et al., 1999). The injection of the antibody raised against the Arf-binding domain of CHO1 into dividing Ptk1 cells, caused a very late regression of the cleavage furrows, even although the formation of the midbody matrix was not affected (Matuliene and Kuriyama, manuscript in preparation). Intriguingly, Skop et al., (2001) recently reported that brefeldin A, which inhibits vesicle secretion by targeting the G-protein Arf, also specifically inhibits the terminal stage of cytokinesis in C. elegans. Thus, it is possible that in addition to its structural function, the midbody matrix may have an active role in the completion of cytoplasmic division by facilitating membrane fusion events.
ACNOWLEDGMENTS
The authors thank Dr. Richard Linck (University of Minnesota) for his critical reading of the manuscript. This work was supported by National Institutes of Health grant GM73510 to R.K.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–10–0504. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–10–0504.
REFERENCES
- Adams RR, Tavares AAM, Salzberg A, Bellen HJ, Glover DM. pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 1998;12:1483–1494. doi: 10.1101/gad.12.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: John Wiley & Sons, Inc; 1994. [Google Scholar]
- Boman AL, Kuai J, Zhu X, Chen J, Kuriyama R, Kahn RA. Arf proteins bind to mitotic kinesin-like protein 1 (MKLP1) in a GTP-dependent fashion. Cell Motil Cytoskeleton. 1999;44:119–132. doi: 10.1002/(SICI)1097-0169(199910)44:2<119::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Cao LG, Wang YL. Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol Biol Cell. 1996;7:225–232. doi: 10.1091/mbc.7.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Detrich HW., III The kinesin superfamily of zebrafish embryos. Mol Biol Cell. 1996;7:396a. [Google Scholar]
- Chui KK, Rogers GC, Kashina AM, Wedaman KP, Sharp DJ, Nguyen DT, Wilt F, Scholey JM. Roles of two homotetrameric kinesins in sea urchin embryonic cell division. J Biol Chem. 2000;275:38005–38011. doi: 10.1074/jbc.M005948200. [DOI] [PubMed] [Google Scholar]
- Deavours BE, Walker RA. Nuclear localization of C-terminal domains of the kinesin-like protein MKLP-1. Biochem Biophys Res Commun. 1999;260:605–608. doi: 10.1006/bbrc.1999.0952. [DOI] [PubMed] [Google Scholar]
- de Cuevas M, Tao T, Goldstein LS. Evidence that the stalk of Drosophila kinesin heavy chain is an alpha-helical coiled coil. J Cell Biol. 1992;116:957–965. doi: 10.1083/jcb.116.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschi T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Field C, Li R, Oegema K. Cytokinesis in eukaryotes: a mechanistic comparison. Curr Opin Cell Biol. 1999;11:68–80. doi: 10.1016/s0955-0674(99)80009-x. [DOI] [PubMed] [Google Scholar]
- Giansanti MG, Bonaccorsi S, Williams B, Williams EV, Santolamazza C, Goldberg ML, Gatti M. Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes Dev. 1998;12:396–410. doi: 10.1101/gad.12.3.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger V, Gonczy P, Romano A, Schnabel H, Hamill D, Schnabel R, Hyman AA, Glotzer M. CYK-4: A Rho family GTPase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol. 2000;149:1391–1404. doi: 10.1083/jcb.149.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RC, Akai H. Spermatogenesis in Bombyx mori. I. The canal system joining sister spermatocytes. J Morphol. 1971;134:47–56. doi: 10.1002/jmor.1051340104. [DOI] [PubMed] [Google Scholar]
- Kuriyama R, Dragas-Granoic S, Maekawa T, Vassilev A, Khodjakov A, Kobayashi H. Heterogeneity and microtubule interaction of the CHO1 antigen, a mitosis-specific kinesin-like protein. Analysis of subdomains expressed in insect Sf9 cells. J Cell Sci. 1994;107:3485–3499. doi: 10.1242/jcs.107.12.3485. [DOI] [PubMed] [Google Scholar]
- Kuriyama R, Gustus C, Uetake Y, Terada Y, Matuliene J. CHO1, a mammalian kinesin-like protein, interacts with F-actin and is involved in the terminal phase of cytokinesis. J Cell Biol. 2002;156:783–790. doi: 10.1083/jcb.200109090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin K, Danilchik MV. Microtubules are required for completion of cytokinesis in sea urchin eggs. Dev Biol. 1999;214:215–226. doi: 10.1006/dbio.1999.9395. [DOI] [PubMed] [Google Scholar]
- Matuliene J, Essner R, Ryu J-H, Hamaguchi Y, Baas PW, Haraguchi T, Hiraoka Y, Kuriyama R. Function of a minus-end-directed kinesin-like motor protein in mammalian cells. J Cell Sci. 1999;112:4041–4050. doi: 10.1242/jcs.112.22.4041. [DOI] [PubMed] [Google Scholar]
- McIntosh JR, Landis SC. The distribution of spindle microtubules during mitosis in cultured mammalian cells. J Cell Biol. 1971;49:468–497. doi: 10.1083/jcb.49.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KLF, genes in mouse and human. Proc Natl Acad Sci USA. 2001;98:7004–7011. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD, Song H, Endow SA. A point mutation in the microtubule binding region of the Ncd motor protein reduces motor velocity. EMBO J. 1996;13:3306–3314. [PMC free article] [PubMed] [Google Scholar]
- Mullins JM, Biesele JJ. Cytokinetic activities in a human cell line: the midbody and intercellular bridge. Tissue Cell. 1973;5:47–61. doi: 10.1016/s0040-8166(73)80005-9. [DOI] [PubMed] [Google Scholar]
- Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-98s cells. J Cell Biol. 1977;73:672–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins JM, McIntosh JR. Isolation and initial characterization of the mammalian midbody. J Cell Biol. 1982;94(3):654–661. doi: 10.1083/jcb.94.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T, Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- Nislow C, Sellitto C, Kuriyama R, McIntosh JR. A monoclonal antibody to a mitotic MAP blocks mitotic progression. J Cell Biol. 1990;111:511–522. doi: 10.1083/jcb.111.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J, Bossinger O, Rose D, Strome S, Saxton W. A nematode kinesin required for cleavage furrow advancement. Curr Biol. 1998;8:1133–1136. doi: 10.1016/s0960-9822(98)70470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich WB, Moran AN, Rothman JH, Hardin J. Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol Biol Cell. 1998;9:2037–2049. doi: 10.1091/mbc.9.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport R. Experiments concerning the cleavage stimulus in sand dollar eggs. J Exp Zool. 1961;148:81–89. doi: 10.1002/jez.1401480107. [DOI] [PubMed] [Google Scholar]
- Rattner JB. Mapping the mammalian intercellular bridge. Cell Motil Cytoskeleton. 1992;23:231–235. doi: 10.1002/cm.970230402. [DOI] [PubMed] [Google Scholar]
- Ryu J-H, Essner R, Ohta T, Kuriyama R. Filamentous polymers induced by overexpression of a novel centrosomal protein, Cep135. Micros Res Technol. 2000;49:478–486. doi: 10.1002/(SICI)1097-0029(20000601)49:5<478::AID-JEMT10>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Salmon ED, Goode D, Maugel TK, Bonar DB. Pressure-induced depolymerization of spindle microtubules. III. Differential stability in HeLa cells. J Cell Biol. 1976;69:443–454. doi: 10.1083/jcb.69.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger JM, Pochapin MB, Sanger JW. Midbody sealing after cytokinesis in embryos of the sea urchin Arbacia punctulata. Cell Tissue Res. 1985;240:287–292. doi: 10.1007/BF00222337. [DOI] [PubMed] [Google Scholar]
- Sellitto C, Kuriyama R. Distribution of a matrix component of the midbody during the cell cycle in Chinese hamster ovary cells. J Cell Biol. 1988;106:431–439. doi: 10.1083/jcb.106.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, Hamill DR, Carter JC, Schumacher J, Bowerman B. The Aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr Biol. 2000;10:1162–1171. doi: 10.1016/s0960-9822(00)00715-6. [DOI] [PubMed] [Google Scholar]
- Skop AR, Bergmann D, Mohler WA, White JG. Completion of cytokinesis in C. elegans requires a brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr Biol. 2001;11:735–746. doi: 10.1016/s0960-9822(01)00231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Field CM. Microtubules, membranes and cytokinesis. Curr Biol. 2000;10:R760–R770. doi: 10.1016/s0960-9822(00)00746-6. [DOI] [PubMed] [Google Scholar]
- Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phase, and involved in cytokinesis. J Cell Biol. 1999;147:921–927. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley SP, Wang Y. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J Cell Biol. 1996;135:981–989. doi: 10.1083/jcb.135.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BD, Terasaki M, Scholey JM. Roles of kinesin and kinesin-like proteins in sea urchin embryonic cell division: evaluation using antibody microinjection. J Cell Biol. 1993;123:681–689. doi: 10.1083/jcb.123.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JT, Laymon RA, Goldstein LS. A three-domain structure of kinesin heavy chain revealed by DNA sequence and microtubule binding analysis. Cell. 1989;56:879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]
- Yonetani A, Walczak C, Verma S, Mitchison TJ. XCHO1, a Xenopus kinesin-related protein involved in bipolar spindle assembly. Mol Biol Cell. 1996;7:211a. [Google Scholar]