Abstract
Formation of the bipolar mitotic spindle relies on a balance of forces acting on the spindle poles. The primary outward force is generated by the kinesin-related proteins of the BimC family that cross-link antiparallel interpolar microtubules and slide them past each other. Here, we provide evidence that Stu1p is also required for the production of this outward force in the yeast Saccharomyces cerevisiae. In the temperature-sensitive stu1–5 mutant, spindle pole separation is inhibited, and preanaphase spindles collapse, with their previously separated poles being drawn together. The temperature sensitivity of stu1–5 can be suppressed by doubling the dosage of Cin8p, a yeast BimC kinesin–related protein. Stu1p was observed to be a component of the mitotic spindle localizing to the midregion of anaphase spindles. It also binds to microtubules in vitro, and we have examined the nature of this interaction. We show that Stu1p interacts specifically with β-tubulin and identify the domains required for this interaction on both Stu1p and β-tubulin. Taken together, these findings suggest that Stu1p binds to interpolar microtubules of the mitotic spindle and plays an essential role in their ability to provide an outward force on the spindle poles.
INTRODUCTION
The process of chromosome segregation is performed by the mitotic spindle, a complex structure composed of microtubules and associated proteins. All spindle microtubules have their minus ends associated with the spindle pole, but three different classes of spindle microtubules are defined by the position of their microtubule plus ends. Kinetochore microtubules extend from the spindle pole to the chromosomes, where they attach to chromosomes through their kinetochores (McDonald et al., 1992; McEwen et al., 1997). Interpolar microtubules extend from one spindle pole into the central spindle and associate with interpolar microtubules from the opposite pole (Ding et al., 1993; Mastronarde et al., 1993; Winey et al., 1995). Astral microtubules extend toward the cell periphery, where their plus ends interact with the cell cortex and play an important role in positioning the spindle (Stearns, 1997; Busson et al., 1998; Skop and White, 1998; Bloom, 2000).
The formation and maintenance of a bipolar spindle relies on a balance of forces acting on the spindle poles (Wittmann et al., 2001). The primary outward force is generated by the plus end–directed, kinesin-related proteins of the BimC family. These homotetrameric proteins have motor domains at each end and are believed to cross-link antiparallel interpolar microtubules and slide them past each other (Kashina et al., 1996). Support for this model comes from the localization of the Drosophila BimC homolog, KLP61F, to overlapping microtubules within the early embryo mitotic spindle (Sharp et al., 1999a). In addition, perturbation of the function of the BimC motors inhibits spindle pole separation in animal cells (Blangy et al., 1995; Kapoor et al., 2000), Drosophila (Heck et al., 1993; Sharp et al., 1999b), and Saccharomyces cerevisiae (Hoyt et al., 1992; Saunders and Hoyt, 1992).
In this report, we examine the role of Stu1p in the spindle pole separation in the yeast S. cerevisiae. Stu1p belongs to a family of proteins that includes the Drosophila Orbit/Mast protein (Inoue et al., 2000; Lemos et al., 2000), the human CLASP1 and CLASP2 proteins (Akhmanova et al., 2001), and uncharacterized ORFs in Schizosaccharomyces pombe and Caenorhabditis elegans. Orbit/Mast has been shown to be a microtubule-associated protein that localizes to the mitotic spindle and is required for bipolar spindle organization. In contrast, the CLASPs localize to the plus ends of interphase microtubules and stabilize them, but they have not been shown to play a role in spindle assembly. Previously, we identified alleles of STU1 as suppressors of a mutation in TUB2, the gene that encodes β-tubulin (Pasqualone and Huffaker, 1994). Here, we show that Stu1p is necessary for both the assembly and maintenance of the yeast mitotic spindle. In addition, we characterize the microtubule-binding properties of Stu1p and identify the domains required for this interaction on both Stu1p and β-tubulin.
MATERIALS AND METHODS
Yeast Strains and Media
Yeast growth media were prepared as described by Sherman (1991). Cells were treated with 4 μg/ml α-factor or 0.2 mM hydroxyurea to arrest them in G1 or S phase, respectively.
Yeast strains used in this study are listed in Table 1. A PGAL-STU1-GFP strain (CUY1293) was created as follows. Overlapping PCR was used to fuse the C-terminal coding region of STU1 to yGFP (Cormack et al., 1997). The final PCR product was cloned into pDP12 (Pasqualone and Huffaker, 1994) to create a full-length STU1 fused to green fluorescent protein (GFP). A PGAL-STU1-GFP fusion was generated by ligating the ORF of the STU1-GFP fusion fragment to the GAL1/10 promoter. The resultant PGAL-STU1-GFP fusion was cloned into an integrating plasmid with a URA3 marker (pRS306; Sikorski and Hieter, 1989) and integrated into a STU1/stu1-Δ1::HIS3 diploid strain (CUY547) at the URA3 locus. Ura+ transformants were sporulated and Ura+ His+ haploids obtained by tetrad dissection.
Table 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| CUY25 | MATa ade2 his3-Δ200 leu2-3,112 ura3-52 | This laboratory |
| CUY26 | MATα his3-Δ200 leu2-3,112 ura3-52 | This laboratory |
| CUY446 | MATα cin8∷LEU2 his3-Δ200 leu2-3,112 lys2-801 ura3-52 | Hoyt laboratory |
| CUY546 | CUY25 × CUY26 | This laboratory |
| CUY547 | MATa/MATα stu1-Δ1∷HIS3/STU1 ade2/ADE2 his3-Δ200/his3-Δ200 leu2-3,112/leu2-3,112 ura3-52/ura3-52 | This laboratory |
| CUY983 | MATa stu1-Δ1∷HIS3 ade2 ade3-24 his3-Δ200 leu2-3,112 ura3-52 (pDP96: ARS4 CEN6 STU1 ADE3 URA3) | This laboratory |
| CUY998 | MATa stu1-5 ade2-101 his3-Δ200 leu2-3,112 ura3-52 | This study |
| CUY999 | MATα stu1-5 his3-Δ200 leu2-3,112 ura3-52 | This study |
| CUY1000 | MATa stu1-5 his3-Δ200 leu2-3,112 ura3-52 | This study |
| CUY1001 | CUY999 X CUY1000 | This study |
| CUY1139 | MATa spc42Δ1∷LEU2 TRP1∷SPC42-GFP (3x) ade2-1 can1-100 his3-11,15 leu2-3,112 ssd1-Δ2 trp1-1 ura3 | Kilmartin laboratory |
| CUY1150 | MATa mad2Δ stu1-5 his3-Δ200 leu2-3,112 ura3-52 | This study |
| CUY1158 | MATa stu1-5 spc42Δ1∷LEU2 TRP1∷SPC42-GFP (3x) can1-100 his3 leu2-3,112 ssd1-Δ2 trp1-1 ura3 | This study |
| CUY1293 | MATa PGAL-STU1-GFP∷URA3 stu1-Δ1:HIS3 his3-Δ200 leu2-3,112 ura3-52 | This study |
| CUY1310 | MATa stu1-5∷URA3∷stu1Δ ACT1∷HIS3 ade2-101 his3-Δ200 leu2-3,112 ura3-52 | This study |
| CUY1311 | MATa stu1-5 TUB1∷URA3∷tub1Δ ade4 his3-Δ200 leu2-3,112 lys2-801 ura3-52 | This study |
| CUY1312 | MATa stu1-5 TUB3∷URA3∷TUB3 ade2 his3-Δ200 leu2-3,112 ura3-52 trp1-1 | This study |
| CUY1324 | MATα stu1-5 CIN8∷HIS3∷CIN8 his3-Δ200 leu2-3,112 ura3-52 | This study |
| CUY1385 | MATa/MATα STU1/STU1-13myc∷HIS3MX6 ADE2/ade2-101 his3-Δ200/his3-Δ200 leu2-3,112/leu2- 3,112 ura3-52/ura3-52 | This study |
| CUY1386 | MATa STU1-13myc∷HIS3MX6 ade2-101 his3-Δ200 leu2-3,112 ura3-52 | This study |
| Y190 | MATa URA3∷PGAL-lacZ LYS2∷PGAL-HIS3 cyhR ade2-101 gal4 gal80 his3 leu2-3,112 trp1-901 ura3-52 | Elledge laboratory |
CUY1385 and CUY1386 containing STU1–13myc were constructed by use of the one-step PCR-mediated technique for modification of chromosomal genes (Longtine et al., 1998); CUY546 and CUY25 were the parent strains, respectively.
Isolation of Temperature-Sensitive Mutations in STU1
Mutagenic PCR conditions were a modification of those described previously (Cadwell and Joyce, 1992). Each mutagenic reaction contained 30 pmol of each PCR primer, 10 ng of pDP94 template DNA, 50 mM KCl, 10 mM Tris-HCl, pH 8.3, 0.01% gelatin, 7 mM MgCl2, 0.5 mM MnCl2, 5 U of Taq polymerase, 0.2 mM dGTP, 0.2 mM dATP, 1 mM dTTP, and 1 mM dCTP in 100 μl reaction volume. pDP94 was constructed by blunt-end ligation of a 6.4-kb genomic BamHI-SalI fragment containing STU1 into the XbaI and SacI sites of pRS415 (Sikorski and Hieter, 1989).
Three pairs of oligonucleotide primers were used to amplify three separate but overlapping regions of STU1: primer set 1, 5′-GCTGGCAATTATAAACACAAGT-3′ and 5′-GATAACTCATCAGTCAAGTCG-3′; primer set 2, 5′-AGTTCTTCCTTTTCTCCACAG-3′ and 5′-ACACTTACTATTTCGTCATT-3′; and primer set 3, 5′-GAACGATCTCATGCCTAAAAT-3′ and 5′-GGAGTCTTTGGAATACTAAGG-3′. Mutagenic PCR products were incorporated into the full-length STU1 coding sequence by homologous recombination in vivo as follows (Muhlrad et al., 1992). From pDP94, three gapped plasmids were isolated by cutting with restriction enzymes in the following pairwise combinations: SacII and PstI (pDP94SP), PstI and XbaI (pDP94PX), and XbaI and SacI (pDP94XS). Each gapped plasmid was cotransformed with its corresponding PCR product into CUY983. Transformants were plated directly onto selective media containing 6 μg/ml adenine at the nonpermissive temperature, 37°C.
Temperature-sensitive stu1 alleles were identified by a colony color-sectoring screen (Sundberg et al., 1996). Transformants were screened for those that remained solid red at 37°C and could sector white at 26°C. In addition, all temperature-sensitive candidates were screened for viability at 26 and 37°C on 5-FOA (Boeke et al., 1984), which selects against pDP96; cells containing a stu1 temperature-sensitive allele on plasmid pDP94 will grow on 5-FOA at 26°C but not at 37°C. The stu1 temperature-sensitive alleles were integrated at the STU1 locus by the two-step gene replacement method (Boeke et al., 1984).
Fluorescence and Electron Microscopy
Immunofluorescence staining of yeast was performed as previously described (Pasqualone and Huffaker, 1994). Rat anti-yeast α-tubulin polyclonal antibody, YOL1/34, was obtained from Accurate Antibodies (Westbury, NY), rabbit anti-yeast β-tubulin antibody 206 was a gift from F. Solomon (Massachusetts Institute of Technology, Cambridge, MA), rabbit anti-yeast γ-tubulin antibody was a gift from T. Stearns (Stanford University, Stanford, CA), and 9E10 anti-myc antibody was obtained from Covance Research Products (Berkeley, CA). Cy3-conjugated goat anti-mouse secondary antibody and fluorescein-conjugated goat anti-rabbit and anti-rat secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
Spc42-GFP was observed in live cells and in cells that had been fixed in 3.7% formaldehyde for 30 min. The distance between two Spc42-GFP dots in a single cell was determined by 3D reconstruction of a z-series stack of images taken at 0.5-μm intervals.
Cells were prepared for electron microscopy as described by Byers and Goetsch (1991).
Plasmid Constructs for In Vitro Transcription and Translation
pDP106 contains STU1 under the control of the T7 promoter for in vitro transcription. The entire STU1 gene was amplified by PCR using the following oligonucleotide primers: 5′-GAGGTACCTTCT-TCAGAAATAATGTCGTC-3′ (upstream primer; Met1 codon in bold, KpnI site italic) and 5′-GGAGTCTTTGGAATACTAAGG-3′ (downstream primer; hybridizes ∼580 base pairs [bp] downstream of the STU1 stop codon). The PCR product was digested with KpnI, which cleaves within the 5′ extension of the upstream primer, and with SacI, which cleaves 98 bp past the STU1 stop codon. This restriction fragment was then subcloned into the corresponding sites of pBluescript II SK+ to create plasmid pDP106.
A nested series of deletions that removed the C-terminal coding region of STU1 were created by digesting pDP106 with SacI and EcoRI followed by ExoIII nuclease digestion. The end points in the STU1 sequence were estimated from the mobility of restriction fragments on polyacrylamide gels, except for C716, whose end point was determined by sequencing. Because the endogenous STU1 stop codon was destroyed by this procedure, stop codons in all three reading frames were provided by downstream sequence in the pBluescript II SK+ multiple cloning site.
A series of deletions that removed the amino-terminal coding region of STU1 was constructed by various methods. pN308 was created by digesting pDP106 with KpnI and ClaI, treating with ExoIII nuclease to remove ∼320 bp past the ClaI site, and religating. pN670 was created by digesting pDP106 with KpnI and PstI, making blunt ends with T4 DNA polymerase, and religation. In pN308 and pN670, the methionine codons at positions 308 and 670, respectively, are the first in-frame methionine codons in the corresponding mRNA transcripts and therefore become the initiation codons. To create pN461 and pN569, novel methionine initiation codons were introduced at amino acid positions 461 and 569, respectively, by PCR using the following upstream primers: 5′-CCGCTCGAGATGATAAATGAGAAAACCGTAACACC-3′ and 5′-CCGCTCGAGATGAACTATCAAGTTTCCAGGGTGTC-3′. The downstream primer used with both of these primers was the same as that described for the construction of pDP106. The PCR products were digested with XhoI, which cleaves within the 5′ extension of each sense primer (italic), and with SacI, which cleaves 98 bp past the STU1 stop codon. The restriction fragments were then subcloned into the corresponding sites of pBluescript II SK+. For all plasmid constructs generated by PCR, at least two independent plasmid clones were isolated for the in vitro microtubule-binding assay to ensure that the PCR did not introduce mutations that would alter microtubule binding properties.
Microtubule-Cosedimentation Assay
Synthesis of radiolabeled Stu1p peptides by in vitro transcription and translation and in vitro microtubule cosedimentation assays were performed as previously described (Wang and Huffaker, 1997).
Immunoprecipitation
A fusion protein containing the 517 C-terminal amino acids of Stu1p fused to maltose-binding protein was expressed in Escherichia coli and purified on amylose resin. Rabbit antiserum to this polypeptide was produced by the Center for Research Animal Resources at Cornell University (Ithaca, NY).
Immunoprecipitation experiments were done as described previously (Chen et al., 1998), except that PME buffer (0.1 M PIPES, 1 mM magnesium chloride, 2 mM EGTA, pH 6.9) was used in place of PBS.
Two-Hybrid Assays
The pAS2 and pATCII vectors were obtained from S. Elledge (Baylor College of Medicine, Houston, TX).
pLY39 contains PADH1-GAL4BD-TUB2 and PADH1-TUB1 and was created as follows. The promoter of the ADH1 gene was amplified from pAS2 by PCR using primers that introduced an SstI and SstII at the 5′ and 3′ ends, respectively. The 0.7-kb SstI–SstII fragment was cloned into pRS304 (Sikorski and Hieter, 1989) to create pLY30. A genomic fragment containing TUB1 was amplified from yeast chromosomal DNA by PCR using primers that introduced a BamHI and PstI site at each end. The 1.6-kb BamHI–PstI fragment was then cloned into the same sites in pLY30 to create pLY32. pLY35 was created by replacing the 0.3-kb SalI–NaeI fragment of pRS424 with the 3.0-kb SalI–NaeI fragment from pAS2. The SstI–PstI fragment, which contains PADH1-TUB1, was cut from pLY32, blunted with T4 DNA polymerase, and then inserted into the SstII and PstI digested and blunted pLY35 to create pLY37. Finally, a genomic DNA fragment containing TUB2 was amplified from yeast chromosomal DNA by PCR using primers that introduced a NcoI and SmaI site at each end. The resulting 1.7-kb NcoI–SmaI fragment was ligated into the same sites of pLY37 to make pLY39.
Mutant tub2 alleles tub2–409 through tub2–422 were cloned into the two-hybrid vector as follows: PCR was used to amplify a 0.45-kb fragment from the plasmids containing the tub2 mutations (Reijo et al., 1994). These fragments were digested with NcoI and KpnI and ligated into the large NcoI–KpnI fragment of pLY39. These fragments were sequenced to ensure that no errors were introduced by PCR. Mutant alleles tub2–423 through tub2–462 were cloned into the two-hybrid vector as follows. A 1.2-kb KpnI–SunI fragment from pLY39 was replaced by the 1.2-kb KpnI–SunI fragment containing these tub2 mutations (Reijo et al., 1994).
GAL4BD-TUB1 was constructed as follows: TUB1 was amplified from yeast chromosomal DNA by PCR using primers that introduced NcoI and SmaI sites at each end. The NcoI-SmaI digested PCR fragment was then inserted into pACTII to give rise to pLY42.
GAL4AD-STU1(308–718) was created as follows: PCR was used to amplify the part of STU1 that encodes amino acids 308–718 from genomic DNA. The lower primer was designed so that amino acid 718 is followed by a stop codon. The PCR product was cloned into pACTII to create pLY62. β-Galactosidase assays were performed on Y190 yeast containing pLY62 and a plasmid carrying one of the GAL4BD-tub2 mutant alleles.
Screen for Spontaneous Suppressors of stu1–5
Two hundred individual colonies (∼106 cells per colony) of strain CUY999 were each resuspended in 100 μl sterile water, spread onto separate YPD plates, and incubated at 37°C. Colonies that arose were picked and retested for growth at 37°C. Each ts+ strain was mated to CUY1310. The resulting diploids were then sporulated, and tetrads were dissected to determine whether suppression segregated as a single locus, and if so, whether the mutations are intragenic (linked to STU1 locus) or linked to TUB2 (which is adjacent to the marked ACT1 locus of CUY1310). Candidates not linked to STU1 or TUB2 were then tested for linkage to TUB1 or TUB3 by crossing to CUY1311 and CUY1312, respectively. For all genetic crosses, we dissected at least 10 tetrads, and we defined genes as linked if no recombinants (i.e., all parental ditypes) were observed.
Each of the tub2 suppressor alleles was amplified from genomic DNA by PCR. PCR products were purified with QIAGEN PCR purification kit (QIAGEN, Chatsworth, CA) and sequenced by use of three primers that spanned the length of the TUB2 gene. Sequencing was done by BioResource Center at Cornell University (Ithaca, NY).
Screen for Overexpression Suppressors of stu1–5
stu1–5 strain CUY999 was transformed with a YCp genomic library (Wang and Huffaker, 1997). Twenty thousand transformants were replicated, plated, and tested for growth at 35°C. Candidates were then streaked onto YPD plates to allow loss of the plasmid and retested for growth at 35°C. Plasmids were isolated from strains that showed plasmid-dependent temperature sensitivity, transformed back into CUY999, and again assayed for suppression of stu1–5 at 35°C.
Two plasmids, p5–39 and p7–19, were found to suppress stu1–5. Inserts from these plasmids were sequenced by use of T3 and T7 primers. The inserts contain overlapping regions of chromosome V: p5–39, spanning bp 34586–42941, and p7–19, spanning 35260–44831. The 7681-bp overlap includes the full-length genes CIN8, PRB1, and SOM1. A 6.1-kb ClaI fragment from p5–39 that contains only the full-length CIN8 was subcloned to pRS313 (Sikorski and Hieter, 1989) to create pLY4. CUY999 cells carrying pLY4 grow at 35°C.
To create a strain with an extra copy of CIN8, we subcloned the 4.4-kb SalI-XbaI fragment that contains CIN8 from pMA1260 into the integrating plasmid pRS303 (Sikorski and Hieter, 1989). The resulting plasmid was linearized by digestion with SphI and transformed into the stu1–5 strains, CUY998. One transformant was mated to CUY446 (cin8Δ::LEU2) to confirm by linkage that the extra CIN8 copy was integrated at the CIN8 locus. PCR amplification using primers that flank the chromosomal region containing the CIN8 duplication confirmed that only a single extra copy of CIN8 had been integrated.
RESULTS
Isolation of Temperature-Sensitive Mutations in STU1
To investigate the role of Stu1p in vivo, we created temperature-sensitive alleles of STU1 using PCR-mediated mutagenesis and a colony color screen as described in MATERIALS AND METHODS. Three overlapping regions of STU1 were mutagenized independently. The N-terminal region included the coding sequence for amino acids 1–715, the midregion included amino acids 564-1077, and the C-terminal region included amino acids 923-1513. Using this approach, we obtained 120 stu1ts alleles: 4 in the N-terminal region, 28 in the midregion, and 88 in C-terminal region.
These stu1ts strains were examined by immunofluorescence microscopy for defects in microtubule assembly and chromosome segregation after a shift to the restrictive temperature (37°C) for 3 h. All displayed similar phenotypes regardless of whether mutagenesis had occurred in the N-terminal region, midregion, or C-terminal coding region of STU1. For this reason, we chose one allele, stu1–5, for a detailed analysis of the stu1ts phenotype. For these studies, the stu1–5 allele was integrated at the STU1 locus, replacing the endogenous STU1 gene.
stu1–5 Cells Lack Normal Bipolar Spindles
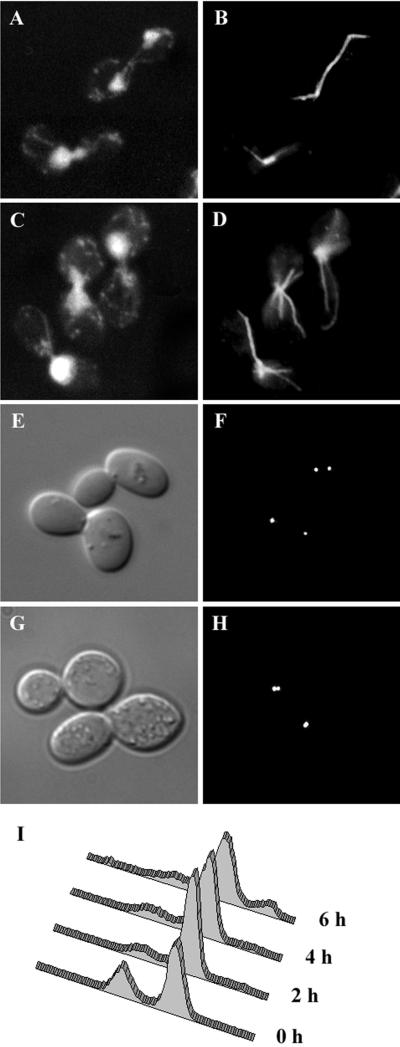
To assess the effect of the stu1–5 mutation on chromosome segregation, stu1–5 cells were incubated at 37°C for 3 h. After this treatment, nearly 80% of cells were large-budded, and 95% of these contained a single mass of chromosomal DNA located at the bud neck (Figure 1C). Flow cytometry demonstrated that most of these cells contained a G2 content of DNA (Figure 1I). These characteristics are typical of a G2/M-phase cell cycle arrest. After 6 h at 37°C, the percentage of large-budded cells decreased to ∼65%, and the percentage of unbudded cells increased correspondingly. Approximately one-third of the unbudded cells contained little or no chromosomal DNA, as determined by DAPI staining. These cells were most likely produced when large-budded cells containing unsegregated DNA proceeded through cell division. In these cases, the amount of DNA inherited by each daughter cell probably depends on the position of the DNA in the bud neck at the time of cytokinesis. Consistent with this interpretation, flow cytometry revealed a small percentage of cells with less than G1 and greater than G2 content of DNA after 6 h at 37°C.
Figure 1.
Phenotype of stu1–5 cells. (A–D) stu1–5 strain (CUY1000) was grown at 22°C (A and B) and shifted to 37°C for 3 h (C and D). Cellular DNA was visualized by DAPI staining (A and C) and microtubules by immunofluorescence (B and D). (E–H) stu1–5 stain expressing Spc42-GFP (CUY1158) was grown at 22°C (E and F) and shifted to 37°C for 3 h (G and H). (E and G) DIC. (F and H) Spc42-GFP. (I) DNA content of stu1–5 cells (CUY1000) grown at 22°C (0 h) and after a shift to 37°C for 2, 4, and 6 h. The DNA content of individual cells was determined by flow cytometry (Haase and Lew, 1997).
Immunofluorescence staining of microtubules revealed that stu1–5–arrested cells lack normal preanaphase spindles at 37°C (Figure 1D). Instead of the ∼1.5-μm bar typically observed in G2/M cells (Figure 1B, bottom cell), stu1–5 cells contained a bright dot or two closely apposed bright dots of staining coincident with the nuclear DNA. Extending from these brightly staining regions were cytoplasmic microtubules that appeared unusually long and numerous compared with the cytoplasmic microtubules in STU1 cells.
To determine the degree of spindle pole separation, we examined live cells expressing GFP-tagged Spc42p, an integral component of the yeast spindle pole body (SPB). For growing cultures, we measured the distance between Spc42-GFP dots in preanaphase cells, which we defined as those that contained two distinguishable dots with a separation ≤2.0 μm. We observed cells grown at 22 and 30°C and cells grown at 22°C and then shifted to 37°C for 3 h. The average distance of SPB separation in preanaphase STU1 cells at 22°C was 1.45 ± 0.31 μm, and this distance did not change significantly at 30°C or 37°C. In preanaphase stu1–5 cells at 22°C, the average SPB separation was 1.19 ± 0.35 μm (Figure 1, E and F, top cell), or ∼80% of the value for STU1 cells (p < 0.001). At 30°C, the average distance in stu1–5 cells was 0.91 ± 0.25 μm, ∼65% of the distance observed in STU1 cells at this temperature (p < 0.001). For stu1–5 cells arrested at 37°C, we examined SPB separation in all of the large-budded arrested cells (Figure 1, G and H). In more than half of these cells, we could distinguish only one dot of Spc42-GFP, indicating that the SPBs resided very close to each other. We estimate that the minimal SPB separation we can distinguish is 0.25–0.5 μm, depending on the orientation of the spindle relative to the plane of the field of view. In cells in which we could distinguish two dots, the average distance of separation was only 0.57 ± 0.25 μm, ∼ 40% of the distance seen in STU1 cells (p < 0.001). Overall, the average separation of SPBs, assuming a value of zero for cells with one dot, was 0.24 ± 0.33 μm. Thus, SPB separation is significantly reduced in stu1–5 cells and decreases substantially with increasing temperature.
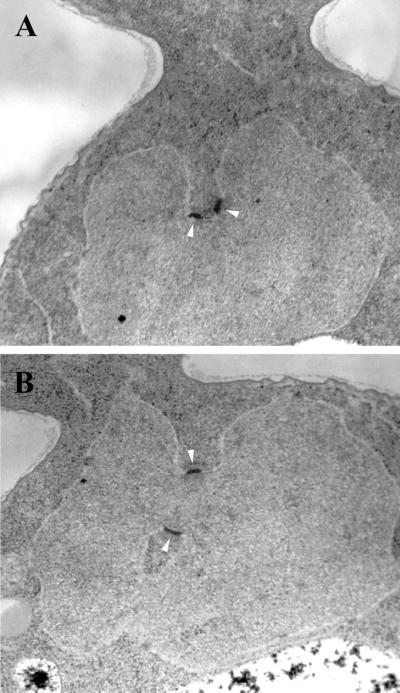
We also used electron microscopy to examine SPB separation in stu1–5 cells that had been shifted to 37°C for 3 h. We observed 10 cells in which both SPBs were contained in a single thin section. Six cells contained side-by-side SPBs. In four of these six cells, the two SPBs were located on deep invaginations of the nuclear envelope, and the bridge between them appeared bent (Figure 2A). In the other four cells, the SPBs were separated from each other on the surface of the nuclear envelope but located on invaginations of the nuclear envelope, which brought them into close proximity (Figure 2B). The separation between these pairs of SPBs ranged from 0.25 to 0.40 μm. The requirement that a single thin section contain both SPBs creates a bias favoring the observation of SPBs that are close together and will probably overestimate the percentage of cells that contain side-by-side SPBs. Cell sections containing a single SPB were observed more often than those containing two SPBs. These former cells probably contain SPBs that have separated. In nearly all cases, the single SPB was found on an invagination of the nuclear envelope.
Figure 2.
SPB separation and nuclear morphology in stu1–5 cells. stu1–5 cells (CUY1001) were grown at 22°C, shifted to 37°C for 3 h, and viewed by thin-section electron microscopy. Arrowheads indicate SPBs.
Overall, these results show that Stu1p is required for the pole separation that normally accompanies bipolar spindle formation.
Stu1p Is Required for Spindle Assembly and Maintenance
stu1–5 cells do not contain normal preanaphase spindles at the restrictive temperature. We wanted to determine whether these cells were unable to assemble spindles, unable to maintain spindles once they had assembled, or both. To determine whether stu1–5 cells are able to assemble spindles, we assessed their ability to separate SPBs at the restrictive temperature. Cells were synchronized at a stage in the cell cycle before SPB duplication by treatment with α-factor at 22°C. Arrested cells were shifted to 37°C for 1.5 h to provide time for the mutation to take effect and then were washed out of α-factor and allowed to proceed through the cell cycle at 37°C. After 1.5 h, both STU1 (CUY1139) and stu1–5 (CUY1158) cultures contained ∼80% large-budded cells. SPB separation was assessed on this population of cells by viewing Spc42-GFP. In 95% of STU1 large-budded cells, SPBs were separated by > 5 μm, indicating that they had entered anaphase. In the stu1–5 culture, half of the cells contained only one discernible dot of staining. The other half of the cells contained two dots, with an average separation of 0.62 ± 0.19. Thus, the stu1–5 mutation does not completely block SPB separation but does keep them from separating to their normal distance in preanaphase cells.
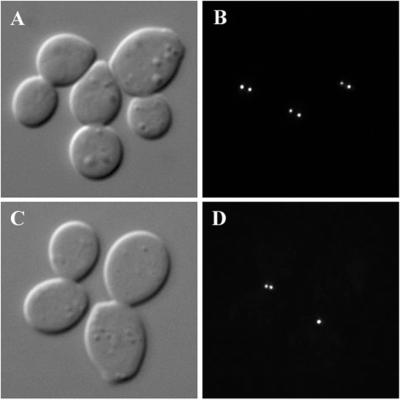
To determine whether Stu1p is necessary to maintain spindle integrity, we synchronized cells before anaphase by treatment with hydroxyurea at 22°C and then shifted them to 37°C in the presence of hydroxyurea. Samples were taken at 30-min intervals, and the separation between Spc42-GFP dots was measured in the large-budded arrested cells. The average distance of SPB separation in STU1 cells was 1.37 ± 0.4 μm at 22°C and 1.33 ± 0.31 after 1 h at 37°C. In stu1–5 cells at 22°C, SPB separation averaged 0.70 ± 0.19 μm (Figure 3, A and B). After 30 min at 37°C, 67% of the stu1–5 cells contained a only single dot of Spc42; the average SPB separation in cells with two distinguishable dots was 0.55 ± 0.17 μm (Figure 3, C and D). Overall, the average distance of SPB separation was 0.18 ± 0.28 μm. This value did not change significantly after 1 h at 37°C. These results demonstrate that the stu1–5 mutation causes the collapse of preformed mitotic spindles at the restrictive temperature.
Figure 3.
Stu1p is necessary for spindle maintenance. stu1–5 cells expressing Spc42-GFP (CUY1158) were arrested with hydroxyurea at 22°C (A and B) and shifted to 37°C for 30 min in the continued presence of hydroxyurea (C and D). (A and C) DIC. (B and D) Spc42-GFP.
stu1–5 Causes a Checkpoint-Independent Block in Spindle Elongation
We do not observe anaphase spindles in stu1–5 cells. Because these cells cannot assemble normal preanaphase spindles, it seemed likely that the stu1–5 mutation causes an inherent defect in spindle structure that prevents formation of an anaphase spindle. However, we could not rule out the possibility that the anaphase block was being caused by activation of the spindle checkpoint. To determine whether the spindle checkpoint is responsible, we deleted MAD2, a gene encoding a component of the spindle checkpoint, in the stu1–5 strain. STU1 (CUY25), stu1–5 (CUY1000), and stu1–5 mad2Δ (CUY1150) strains were synchronized in G1 by α-factor treatment, shifted to 37°C for 1 h, and then released into α-factor–free medium at 37°C. STU1 cells entered anaphase and elongated their spindles at ∼90 min after release from α-factor, as determined by immunofluorescence staining of microtubules. In contrast, we did not observe anaphase spindles in stu1–5 or stu1–5 mad2Δ cells at any time up to 3 h after release. Because this phenotype is observed in the absence of a functional spindle checkpoint, we conclude that the stu1–5 mutation causes an inherent defect that inhibits spindle assembly and elongation.
Stu1p Localizes to the Spindle Midregion
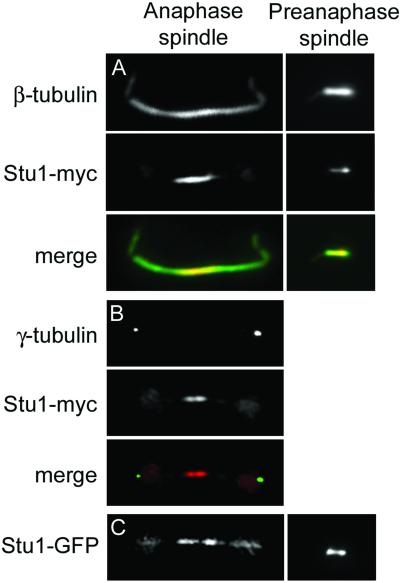
Interpolar microtubules play a role in maintaining spindle structure by supporting the outward forces necessary to separate spindle poles. Because Stu1p is required for pole separation, we wanted to see whether Stu1p associates with this class of microtubules. We previously localized Stu1p to the mitotic spindle by immunofluorescence microscopy using an HA epitope–tagged version of Stu1p expressed from a plasmid (Pasqualone and Huffaker, 1994). This analysis was limited because the signal was weak and was observed only in a fraction of cells. We attempted to improve this approach by attaching 13 copies of the myc epitope tag to the C terminus of chromosomally encoded Stu1p. This tag had no adverse effects on the growth of cells and could be observed by immunofluorescence microscopy in virtually all cells that contained spindles.
The haploid S. cerevisiae spindle contains 16 kinetochore microtubules originating from each of the SPBs, 1 for each of the 16 sets of duplicated chromosomes (Winey et al., 1995). Each SPB also nucleates approximately four interpolar microtubules that interdigitate with their counterparts from the opposite pole. Kinetochore microtubules are concentrated near the spindle poles and, by fluorescence microscopy, appear as tufts that occupy ∼60% of the length of the 1.5-μm preanaphase spindle (Maddox et al., 2000). Stu1-myc is concentrated near the poles of preanaphase spindles, suggesting that it associates with kinetochore microtubules in these cells (Figure 4A). Because kinetochore microtubules occupy much of the length of the preanaphase spindle and outnumber the interpolar microtubules by several-fold, it is difficult to determine whether Stu1-myc also associates with interpolar microtubules of preanaphase spindles. However, interpolar microtubules are readily distinguished in anaphase cells. As cells enter anaphase, kinetochore microtubules remain closely associated with the poles, so that the central region of the spindle is composed entirely of interpolar microtubules. Stu1-myc is concentrated in the midregion of anaphase spindles (Figure 4, A and B), the region of interpolar microtubule overlap. In a number of cases, this signal is split into two, suggesting that Stu1-myc may be more concentrated near the plus ends of overlapping antiparallel interpolar microtubules (Figure 4B).
Figure 4.
Stu1p localizes to the midregion of anaphase spindles. Double labeling of CUY1386 cells, expressing Stu1-myc, by use of (A) anti-β-tubulin and anti-myc antibodies and (B) anti-γ-tubulin and anti-myc antibodies. (C) GFP fluorescence in living CUY1293 cells expressing Stu1-GFP from the GAL1 promoter.
We also constructed a Stu1-GFP fusion to examine Stu1p localization in living cells. When GFP was fused to the chromosomal copy of STU1, we were unable to detect any GFP fluorescence in cells, even although this fusion allowed the cells to grow in the absence of STU1. Next, we integrated a copy of Stu1-GFP expressed from the inducible GAL1 promoter. These cells, which lack the wild-type STU1 gene, are able to grow on galactose-containing medium and provide visible GFP fluorescence in live cells. Although Stu1-GFP is overexpressed in these cells, its localization pattern is similar to that of Stu1-myc (Figure 4C). In particular, it is concentrated at the midregion of anaphase spindles.
Stu1p Binds Microtubules In Vitro
Genetic evidence suggests that Stu1p associates with tubulin in vivo (Pasqualone and Huffaker, 1994). To determine whether Stu1p binds microtubules in vitro, we performed a microtubule-cosedimentation assay. 35S-labeled Stu1p was synthesized by in vitro transcription and translation and incubated with a large excess of taxol-stabilized bovine brain microtubules. After sedimentation of microtubules by centrifugation, both the pellet and the supernatant were analyzed for the presence of 35S-Stu1p. When microtubules were added to a final concentration of 5 μM tubulin, >80% of the Stu1p pelleted with the microtubules. Conversely, >80% of the Stu1p remained in the supernatant in the absence of microtubules.
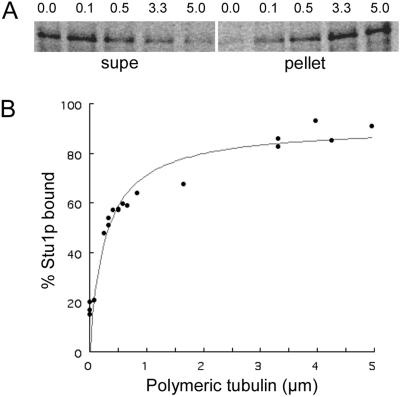
To determine the binding affinity of Stu1p for microtubules, we incubated a constant amount of 35S-labeled Stu1p with various amounts of microtubules. The binding of Stu1p to microtubules is concentration dependent and saturable (Figure 5). The apparent dissociation constant, Kd, equal to the concentration of polymerized tubulin necessary to cosediment half of the Stu1p is 3.2 × 10−7 M.
Figure 5.
Measurement of the dissociation constant (Kd) for full-length Stu1p. A constant amount of 35S-labeled in vitro–translated Stu1p was incubated with various amounts of taxol-stabilized bovine brain microtubules. Microtubules were pelleted by centrifugation, and both the bound Stu1p (in pellets) and unbound Stu1p (in supernatants) were subjected to SDS-PAGE analysis. Band intensities were quantified by use of a phosphoimager, and the percentage of Stu1p bound was calculated. (A) Phosphoimager scans of a gel showing Stu1p in supernatants (supe) and the corresponding pellets at various tubulin concentrations. The polymeric tubulin concentration ranging from 0 to 5 μM is shown above each lane. (B) The binding curve that includes data from the gel shown above and additional gels not shown. The Kd is equal to the concentration of polymerized tubulin necessary to cosediment 50% of the Stu1p in the reaction.
The Microtubule-Binding Domain of Stu1p
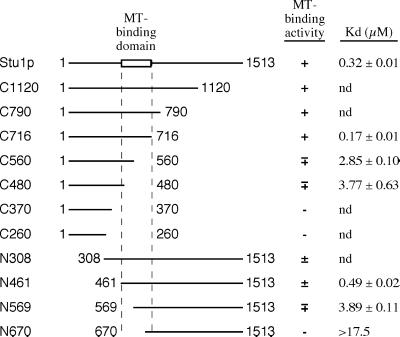
To define the microtubule-binding domain of Stu1p, we measured the relative microtubule-binding affinities of a series of N-terminal and C-terminal truncation constructs (Figure 6). Initially, we measured the fraction of Stu1p polypeptides that cosedimented with microtubules at a single tubulin concentration (5 μM). Deletions of up to ∼800 amino acids (C1120, C790, and C716 in Figure 6) from the C terminus of Stu1p have little effect on microtubule binding. However, deletions of ∼950 amino acids or more from the C terminus (C560, C480, C370, and C260) sharply diminish microtubule binding. Truncations of up to 461 amino acids from the N-terminus of Stu1p (N308 and N461) have microtubule-binding affinities that are comparable to that of full-length Stu1p. In contrast, deletions of 569 and 670 amino acids from the N-terminus eliminate most binding activity.
Figure 6.
Identification of the Stu1p microtubule-binding domain. Various truncated Stu1p polypeptides were translated in vitro and tested for microtubule-binding activity by the cosedimentation assay using a constant amount of microtubules (5 μm tubulin). The percentage of Stu1p that pelleted with microtubules is indicated by (+), >80%; (±), 60–80%; (∓), 40–60%; and (−), <20%. The Kds of some of these Stu1p polypeptides were determined as shown for full-length Stu1p in Figure 4. The Kds shown are the average values from two separate assays, and the range is indicated. The top line represents the full-length Stu1p. The numbers adjacent to the end points represent the positions of the corresponding amino acids.
To obtain more quantitative binding data for some of these constructs, we measured the fraction of polypeptide bound to microtubules over a range of microtubule concentrations and calculated the apparent Kd as described for the full-length Stu1p above (Figure 6). The Kd for the C716 and N461 polypeptides are ∼2-fold lower and ∼1.5-fold higher, respectively, than the Kd for full-length Stu1p, confirming that the ∼800 C-terminal and 461 amino-terminal amino acids do not contribute significantly to microtubule binding. In fact, the C-terminal ∼800 amino acids actually inhibit the microtubule binding of Stu1p to some degree. Deletions that remove either the amino-terminal (N569) or the C-terminal (C560) portions of the 461–716 domain have Kds that are ∼10-fold higher than that for the full-length protein. The N670 polypeptide, which lacks most of the 461–700 domain, has a Kd that is >17-fold higher than that of Stu1p. We could not determine an accurate Kd value for C370 because a significant fraction of this polypeptide pelleted in the absence of microtubules.
These results localize the microtubule-binding domain of Stu1p to the 256-amino-acid region between amino acids 461 and 716. This domain is highly basic, with a predicted isoelectric point of 9.9 and a positive charge of 16.5 at pH 7.0. It includes a 103-amino-acid sequence (574–676) that is particularly serine-rich (28% serine residues) and has a predicted isoelectric point of 10.5.
Stu1p Interacts with Itself In Vivo
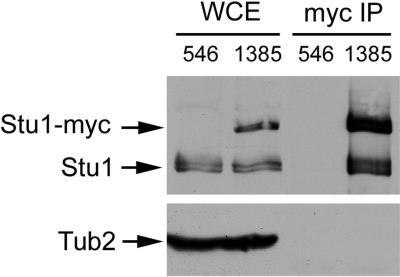
BimC kinesin–related proteins are homotetramers. Their self-association produces a complex that contains microtubule motor domains at each end and enables them to cross-link microtubules. Because Stu1p is localized to a region of microtubule overlap in anaphase spindles, we investigated whether it too had the ability to self-associate in vivo. We constructed a yeast strain that expresses both Stu1p and Stu1-myc. Stu1-myc was immunoprecipitated from extracts of this strain by use of anti-myc antibody, and the immunoprecipitated material was analyzed for the presence of Stu1p by immunoblotting with anti-Stu1p antibodies. As shown in Figure 7, Stu1p coimmunoprecipitated with Stu1-myc. Tub2p was not found in the immunoprecipitated material, indicating that the Stu1p–Stu1p interaction is not mediated by tubulin.
Figure 7.
Stu1p associates with itself in vivo. Whole-cell extracts from yeast strains CUY546 (lacking Stu1-myc) and CUY1385 (containing Stu1-myc) were immunoprecipitated with anti-myc antibody. Whole-cell extracts (WCE) and the immunoprecipitates (myc IP) were subjected to SDS-PAGE. The upper half of the gel was immunoblotted with anti-Stu1p antibody; the lower half of the gel was immunoblotted with anti-Tub2p antibody.
Interaction of Stu1p with β-Tubulin
The above experiments demonstrate that Stu1p interacts with microtubules. To determine whether this interaction is mediated by Stu1p binding to α-tubulin or β-tubulin or both, we used a two-hybrid assay. A portion of STU1 encoding its microtubule-binding domain (amino acids 308–718) was fused to the GAL4 activation domain. TUB1 (encoding α-tubulin) and TUB2 (encoding β-tubulin) were fused to the GAL4 DNA-binding domain on two independent plasmids. Because overexpression of TUB2 is lethal and co-overexpression of TUB1 is known to suppress this lethality, the plasmid containing GAL4BD-TUB2 also contained a copy of TUB1 expressed from the ADH1 promoter. Stu1p was able to interact with Tub2p, but not Tub1p, in this assay system.
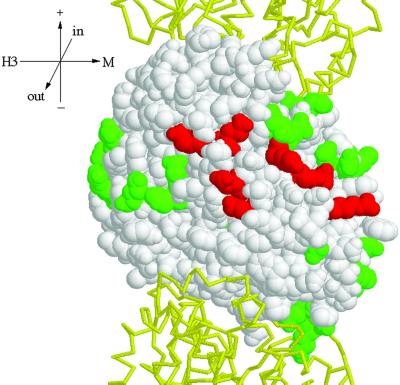
Next, we determined the residues of β-tubulin involved in the interaction with Stu1p, taking advantage of a set of tub2 alanine-scanning alleles (Reijo et al., 1994). In each allele, one to three closely spaced charged amino acids are changed to alanine. Charged residues were targeted in this set because they are likely to reside on the protein surface and be involved in protein–protein interactions. Together, these mutations span the entire length of the TUB2 gene. Fifty tub2 alleles were fused to the GAL4 DNA-binding domain and tested for their ability to interact with Stu1p. Twenty-five alleles failed to interact with Stu1p. However, 21 of these also failed to interact with a number of other microtubule-binding proteins that do interact with the wild-type Tub2p, suggesting that they might affect protein binding in a nonspecific manner. The other four alleles that failed to interact specifically with Stu1p are tub2–440 (D304A, R306A), tub2–449 (E376A, K379A), tub2–455 (E410A, E412A), and tub2–456 (D417A, E421A). The eight amino acids affected by these mutations have been mapped onto the structure of yeast β-tubulin (Richards et al., 2000) in Figure 8. These residues (304, 306, 376, 379, 410, 412, 417, and 421) form a patch on the surface of β-tubulin that faces the outside of the microtubule.
Figure 8.
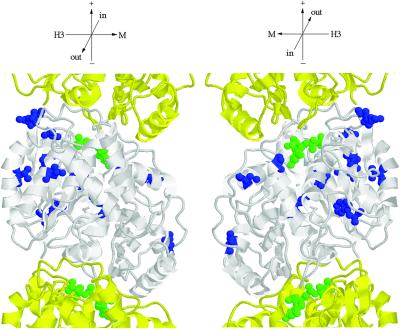
Two-hybrid interaction footprint of Stu1p on Tub2p. Tub2p is shown as a space-fill model with its GTP-binding site at the top (Richards et al., 2000). The Tub1ps that reside above and below Tub2p in the protofilament are show as backbone traces in yellow. This view shows the outside of the microtubule. Stu1p was tested for interaction with each of the Tub2p mutant proteins by use of the two-hybrid system. Those amino acids whose substitution abolished the interaction are shown in red; those that did not interfere with the interaction are shown in green. Tub2p mutations that abolished interactions with all β-tubulin–binding proteins tested are not shown. Orientation axes: + and − show microtubule orientation; H3 and M show lateral sides marked by the H3 α-helix and M loop, respectively; in and out represent the inside and outside of the microtubule. Structure drawn with RASMOL (Sayle and Milner-White, 1995).
tub2 Mutations Suppress stu1–5
Alleles of STU1 were originally identified by their ability to suppress the cold sensitivity of the tub2–406 mutation. If Stu1p interacts directly with β-tubulin in vivo, then it seemed likely that we would also be able to obtain mutations in TUB2 that suppress the temperature sensitivity of stu1–5. We isolated spontaneous suppressors of the temperature sensitivity of the stu1–5 mutant. Twenty-eight strains that could grow at 37°C were obtained from 2 × 108 stu1–5 cells. Seven of these suppressors are linked to STU1 and are presumably intragenic alleles. Of the 21 extragenic suppressors, 20 are linked to TUB2 and 1 is linked to TUB3. The 20 tub2 suppressor alleles were sequenced. Each contains a single nucleotide substitution that results in a single amino acid change (Figure 9; amino acid substitutions are listed in the legend). All of these amino acids except for Gln-94, Ser-95, and Gly-223 are buried within the β-tubulin structure. Gln-94, Ser-95, and Gly-223 are located on the surface of β-tubulin that lies inside the microtubule. Therefore, it is unlikely that any of these amino acids directly contact Stu1p.
Figure 9.
Location of amino acid substitutions in Tub2p suppressors. Tub2p is shown in white; adjacent Tub1p monomers of the protofilament are shown in yellow (Richards et al., 2000). Amino acids changed in Tub2p suppressors are indicated in blue. GTP is shown in green. The following amino acid substitutions in Tub2p were found to suppress stu1–5: G17C, G17D, W59L, V66F, Q94K, S95R, A102S, V116A, H137N, T149 M, C211F, G223R, F265I, and E288A. C211F was obtained four times; Q94R, S95R, and H137N were each obtained twice; all others were obtained once. Orientation axes: + and − show microtubule orientation; H3 and M show lateral sides marked by the H3 α-helix and M loop, respectively; in and out represent the inside and outside of the microtubule. Structure drawn with RASMOL (Sayle and Milner-White, 1995).
Overexpression of CIN8 Suppresses stu1–5
To identify proteins that functionally interact with Stu1p, we looked for overexpression suppressors of stu1–5. Initially, we transformed stu1–5 cells with a yeast genomic DNA library carried on a high-copy-number YEp plasmid. The only gene we found that could suppress the temperature sensitivity of stu1–5 was STU1 itself. Because a number of genes are lethal on high-copy plasmids, we also transformed stu1–5 cells with a yeast genomic DNA library carried on a low-copy-number YCp plasmid. We found one gene, CIN8, that allowed growth of stu1–5 cells at 35°C but not 37°C (Table 2). CIN8 also suppresses the temperature sensitivity of stu1–7 but not stu1–8, stu1–9, or stu1–12. CIN8 could not restore the growth defect caused by deletion of STU1. Although YCp plasmids are generally carried in cells at a low copy number, they can be present in more than one copy per cell. To see whether only one extra copy of CIN8 is sufficient to suppress stu1–5, we integrated a second copy of CIN8 at the CIN8 locus. This strain, containing stu1–5 and two copies of CIN8, also grew at 35°C.
Table 2.
Suppression of stu1 alleles by CIN8
| 30°C | 35°C | 37°C |
CIN8
YCp
|
CIN8∷CIN8
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| 30°C | 35°C | 37°C | 30°C | 35°C | 37°C | ||||
| stu1-5 | + | − | − | + | + | − | + | + | − |
| stu1-7 | + | − | − | + | + | − | |||
| stu1-8 | + | − | − | + | − | − | |||
| stu1-9 | + | + | − | + | + | − | |||
| stu1-12 | + | + | − | + | + | − | |||
| stu1Δ | − | − | |||||||
DISCUSSION
Role of Stu1p in Spindle Assembly
Stu1p performs a role that is essential for mitotic spindle function. It is necessary for complete SPB separation during assembly of the bipolar spindle. After the spindle is assembled, its activity is also necessary to prevent spindle collapse. Therefore, it is reasonable to conclude that Stu1p aids in producing a pole-separating force.
The phenotype of stu1–5 resembles that of cin8 and kip1 mutations (Hoyt et al., 1992; Saunders and Hoyt, 1992). Cin8p and Kip1p are BimC kinesin-related proteins that localize to the yeast spindle. Like other BimC family members, Cin8p and Kip1p are believed to cross-link overlapping polar microtubules and slide them past one another to generate an outward force on the spindle. In the absence of Cin8p and Kip1p, SPBs fail to separate and the SPBs of preformed spindles collapse back to the side-by-side configuration. The facts that Stu1p and Cin8p/Kip1p are both required for spindle pole separation and that a single extra copy of CIN8 can suppress a stu1 mutation suggests that these proteins have overlapping roles in the cell. This view is also supported by the observation that cells containing the stu1–5 mutation and a deletion of cin8 are inviable (our unpublished observations). Although the sequence of Stu1p does not indicate that it is a motor protein, its ability to bind microtubules and its localization to the midregion of anaphase spindles are consistent with Stu1p playing a structural role in the spindle through its interaction with antiparallel interpolar microtubules. Stu1p self-association may produce homodimers that are capable cross-linking these antiparallel microtubules. As far as we are aware, Stu1p is the first example of a nonmotor microtubule-binding protein that is necessary for bipolar spindle formation in yeast.
A phenotypic comparison can also be made between stu1–5 yeast and mast/orbit Drosophila mutants (Inoue et al., 2000; Lemos et al., 2000). The mast/orbit mutations lead to high levels of cells with polyploid chromosomes in the larval CNS. Such chromosomes are frequently associated with multipolar spindles. In addition, these mutations cause the appearance of circular mitotic figures in which the major chromosomes are arranged in a circle with their centromeres inward and arms oriented toward the periphery. In general, these cells contain a reduced number of microtubule-organizing centers relative to their chromosome content, but their microtubule-organizing centers frequently contain multiple centrosomes. These results suggest that mast/orbit cells cannot separate centrosomes properly or cannot maintain centrosome separation once it has occurred. This model is supported by the observation that the mast/orbit phenotype closely resembles the phenotype caused by mutations in KLP61F, a Drosophila kinesin–like protein in the BimC family, which is necessary to maintain spindle pole separation (Heck et al., 1993; Sharp et al., 1999b). Thus, it appears likely that Stu1p and Mast/Orbit play similar roles in spindle assembly and maintenance in yeast and Drosophila, respectively.
Nature of the Stu1p–β-Tubulin Interaction
In this report, we show that Stu1p binds to microtubules in vitro and identify the domains on both Stu1p and β-tubulin necessary for this interaction. Stu1p binds to microtubules with an apparent Kd of 0.32 μM, a value that is approximately two times greater than that obtained for the neuronal microtubule-associated protein tau by use of the same assay (Goode and Feinstein, 1994). In vitro binding assays using truncations of Stu1p have localized the microtubule-binding region to a 256-amino-acid sequence in the amino-terminal half of the protein. This region is highly basic and contains a 103-amino-acid serine-rich stretch. Two of the Stu1p homologues, the Drosophila Orbit/Mast and the human CLASP proteins, have also been shown to bind microtubules (Inoue et al., 2000; Lemos et al., 2000; Akhmanova et al., 2001). Although the specific regions of these proteins that mediate their interactions with microtubules have not been defined, they do contain a sequence that is similar to the microtubule-binding domain of the microtubule-associated protein MAP4. This sequence is located just inside the amino-terminal half of these proteins, as is the microtubule-binding region of Stu1p.
Stu1p binds to β-tubulin, but not α-tubulin, in the two-hybrid assay. To map the Stu1p-binding site on β-tubulin, we used an approach that is similar to one previously used for mapping binding sites of other proteins on α-tubulin (Feierbach et al., 1999; Richards et al., 2000) and actin (Wertman et al., 1992; Holtzman et al., 1994). We made use of a set of clustered charge–to-alanine mutations in β-tubulin (Reijo et al., 1994) and the three-dimensional structure of yeast β-tubulin constructed by modeling its sequence onto the mammalian β-tubulin structure (Richards et al., 2000). We reasoned that the mutations in β-tubulin that disrupt the interaction with Stu1p in the two-hybrid assay would identify side chains that make up the interacting surface. Four tub2 alleles, comprising eight amino acid substitutions, specifically disrupted Stu1p binding. These residues form a patch on the surface of β-tubulin that would be exposed to the cytoplasm when tubulin is assembled into a microtubule and most likely define the domain on tubulin that mediates the interaction between Stu1p and microtubules. Two of the tub2 alleles (tub2–455 and tub2–456) that disrupt the interaction with Stu1p are recessive lethal mutations in yeast. Their lethality could result from their inability to interact productively with Stu1p.
STU1 was initially identified by allele-specific suppression of a cold-sensitive tub2 mutation. Here, we report that tub2 mutations can also suppress the temperature sensitivity of stu1–5. Such mutual suppression has been taken as strong evidence of a direct in vivo interaction (Adams and Botstein, 1989). One view of the mechanism by which this type of suppression occurs is that an alteration in one protein causes a decrease in binding affinity, which is restored by a compensating alteration in the second protein. We have shown that the tub2 suppressor mutations change residues predicted to lie in internal regions of β-tubulin or on the inside of the microtubule, which is inconsistent with their direct participation in the Stu1p-binding interface. Thus, if these mutations increase the affinity of β-tubulin for Stu1-5p, they must do so by longer-range actions, such as altering the conformation of β-tubulin in a way that exposes novel residues or changes the orientation of existing surface residues. Such conformational changes have been proposed to account for the suppression of actin alleles by mutations in Sac6p (Goldsmith et al., 1997; Sandrock et al., 1997). A second mechanism by which suppression could occur is that the tub2 suppressors change the properties of microtubules so that they do not require the wild-type level of Stu1p activity. For example, an alteration in β-tubulin may increase its affinity for another protein whose activity overlaps with that of Stu1p.
ACKNOWLEDGMENTS
We thank Justin Warner and Cindy Tian for help in constructing strains; Steve Elledge, John Kilmartin, and Andy Hoyt for providing yeast strains; and Frank Solomon and Tim Stearns for providing antibodies. This work was supported by a grant from the National Institutes of Health (GM-40479) to T.C.H.
Abbreviations used:
- bp
base pairs
- GFP
green fluorescent protein
- SPB
spindle pole body
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–09–0458. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–09–0458.
REFERENCES
- Adams AEM, Botstein D. Dominant suppressors of yeast actin mutations that are reciprocally suppressed. Genetics. 1989;121:675–684. doi: 10.1093/genetics/121.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, Galjart N. Clasps are clip-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Bloom K. It's a kar9ochore to capture microtubules [news] Nat Cell Biol. 2000;2:E96–E98. doi: 10.1038/35014089. [DOI] [PubMed] [Google Scholar]
- Boeke J, LaCroute F, Fink G. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Busson S, Dujardin D, Moreau A, Dompierre J, De Mey JR. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr Biol. 1998;8:541–544. doi: 10.1016/s0960-9822(98)70208-8. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Preparation of yeast cells for thin-section electron microscopy. Methods Enzymol. 1991;194:602–607. doi: 10.1016/0076-6879(91)94044-d. [DOI] [PubMed] [Google Scholar]
- Cadwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- Chen XP, Yin H, Huffaker TC. The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J Cell Biol. 1998;141:1169–1179. doi: 10.1083/jcb.141.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Bertram G, Egerton M, Gow NA, Falkow S, Brown AJ. Yeast-enhanced green fluorescent protein (yEGFP) a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feierbach B, Nogales E, Downing KH, Stearns T. Alf1p, a CLIP-170 domain-containing protein, is functionally and physically associated with alpha-tubulin. J Cell Biol. 1999;144:113–124. doi: 10.1083/jcb.144.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith SC, Pokala N, Shen W, Fedorov AA, Matsudaira P, Almo SC. The structure of an actin-crosslinking domain from human fimbrin [letter] Nat Struct Biol. 1997;4:708–712. doi: 10.1038/nsb0997-708. [DOI] [PubMed] [Google Scholar]
- Goode BL, Feinstein SC. Identification of a novel microtubule binding and assembly domain in the developmentally regulated inter-repeat region of tau. J Cell Biol. 1994;124:769–782. doi: 10.1083/jcb.124.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Lew DJ. Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol. 1997;283:322–332. doi: 10.1016/s0076-6879(97)83026-1. [DOI] [PubMed] [Google Scholar]
- Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DA, Wertman KF, Drubin DG. Mapping actin surfaces required for functional interactions in vivo. J Cell Biol. 1994;126:423–432. doi: 10.1083/jcb.126.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue YH, do Carmo Avides M, Shiraki M, Deak P, Yamaguchi M, Nishimoto Y, Matsukage A, Glover DM. Orbit, a novel microtubule-associated protein essential for mitosis in Drosophila melanogaster. J Cell Biol. 2000;149:153–166. doi: 10.1083/jcb.149.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos CL, Sampaio P, Maiato H, Costa M, Omel'yanchuk LV, Liberal V, Sunkel CE. Mast, a conserved microtubule-associated protein required for bipolar mitotic spindle organization [In Process Citation] EMBO J. 2000;19:3668–3682. doi: 10.1093/emboj/19.14.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini D J, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KL, O'Toole ET, Mastronarde DN, McIntosh JR. Kinetochore microtubules in Ptk cells. J Cell Biol. 1992;118:369–383. doi: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J Cell Biol. 1997;137:1567–1580. doi: 10.1083/jcb.137.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Pasqualone D, Huffaker TC. STU1, a suppressor of a β-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J Cell Biol. 1994;127:1973–1984. doi: 10.1083/jcb.127.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijo RA, Cooper EM, Beagle GJ, Huffaker TC. Systematic mutational analysis of the yeast β-tubulin gene. Mol Biol Cell. 1994;5:29–43. doi: 10.1091/mbc.5.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KL, Anders KR, Nogales E, Schwartz K, Downing KH, Botstein D. Structure-function relationships in yeast tubulins. Mol Biol Cell. 2000;11:1887–1903. doi: 10.1091/mbc.11.5.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrock TM, O'Dell JL, Adams AE. Allele-specific suppression by formation of new protein-protein interactions in yeast. Genetics. 1997;147:1635–1642. doi: 10.1093/genetics/147.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The bipolar kinesin, K.L.P61F., cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999a;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Yu KR, Sisson JC, Sullivan W, Scholey JM. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat Cell Biol. 1999b;1:51–54. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop AR, White JG. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr Biol. 1998;8:1110–1116. doi: 10.1016/s0960-9822(98)70465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T. Motoring to the finish: kinesin and dynein work together to orient the yeast mitotic spindle [comment] J Cell Biol. 1997;138:957–960. doi: 10.1083/jcb.138.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg HA, Goetsch L, Byers B, Davis TN. Role of calmodulin and Spc110p interaction in the proper assembly of spindle pole body components. J Cell Biol. 1996;133:111–124. doi: 10.1083/jcb.133.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PJ, Huffaker TC. Stu2p: A microtubule-binding protein that is an essential component of the yeast spindle pole body. J Cell Biol. 1997;139:1271–1280. doi: 10.1083/jcb.139.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertman KF, Drubin DG, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Hyman A, Desai A. The spindle: a dynamic assembly of microtubules and motors. Nat Cell Biol. 2001;3:E28–E34. doi: 10.1038/35050669. [DOI] [PubMed] [Google Scholar]