Abstract
Background
Seed-associated microorganisms play crucial roles in maintaining plant health by providing nutrients and resistance to biotic and abiotic stresses. However, their functions in seed germination and disease resistance remain poorly understood. In this study, we investigated the microbial community assembly features and functional profiles of the spermosphere and endosphere microbiomes related to germinated and ungerminated seeds of Astragalus mongholicus by using amplicon and shotgun metagenome sequencing techniques. Additionally, we aimed to elucidate the relationship between beneficial microorganisms and seed germination through both in vitro and in vivo pot experiments.
Results
Our findings revealed that germination significantly enhances the diversity of microbial communities associated with seeds. This increase in diversity is driven through environmental ecological niche differentiation, leading to the enrichment of potentially beneficial probiotic bacteria such as Pseudomonas and Pantoea. Conversely, Fusarium was consistently enriched in ungerminated seeds. The co-occurrence network patterns revealed that the microbial communities within germinated and ungerminated seeds presented distinct structures. Notably, germinated seeds exhibit more complex and interconnected networks, particularly for bacterial communities and their interactions with fungi. Metagenome analysis showed that germinated seed spermosphere soil had more functions related to pathogen inhibition and cellulose degradation. Through a combination of culture-dependent and germination experiments, we identified Fusarium solani as the pathogen. Consistent with the metagenome analysis, germination experiments further demonstrated that bacteria associated with pathogen inhibition and cellulose degradation could promote seed germination and vigor. Specifically, Paenibacillus sp. significantly enhanced A. mongholicus seed germination and plant growth.
Conclusions
Our study revealed the dynamics of seed-associated microorganisms during seed germination and confirmed their ecological role in promoting A. mongholicus seed germination by suppressing pathogens and degrading cellulose. This study offers a mechanistic understanding of how seed microorganisms facilitate successful seed germination, highlighting the potential for leveraging these microbial communities to increase plant health.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-024-02014-5.
Keywords: Community assembly, Disease suppression, Plant growth-promoting bacteria, Seed germination, Seed microbiome
Introduction
Plant-associated microbial communities profoundly influence various plant traits, including growth promotion [1], drought tolerance [2], pathogen resistance, and overall plant health maintenance [3]. Emerging studies have demonstrated that the microbiomes residing in the rhizosphere and phyllosphere play pivotal roles in enhancing plant adaptations, particularly in bolstering resistance against pathogens [4, 5]. For example, rhizosphere microbes such as Trichoderma and Bacillus could help plants resist a range of diseases, including tomato wilt [6, 7]. Despite their crucial role as a primary source of the plant microbiome, particularly in shaping early rhizosphere microbial communities, plant seed-associated microorganisms remain largely underexplored regarding their beneficial functions for plant health and adaptability [8].
Seeds represent the beginning and end of the plant life cycle [9], providing a unique habitat (ecological niche) for numerous microorganisms. Bacterial isolates from plant seeds perform diverse functions, including nitrogen fixation, phosphorus solubilization, phytohormone synthesis, and antimicrobial compound production, potentially contributing to plant fitness [10–12]. Importantly, seed microbiomes can be vertically transmitted between generations via seeds [13, 14], suggesting that the conserved traits of seed microorganisms offer new opportunities for agricultural production. The spermosphere, the immediate interface for seed-microbe interactions, represents the most proximal ecological niche to the seed [15]. Microbes in the spermosphere community are derived primarily from the surrounding soil and are recruited through seed exudation during germination [16], and their assembly is dependent on the soil type and varying seed exudation characteristics. Although germination-driven dynamics of seed endophytic microorganisms have been demonstrated [17–19], the relationship between germination and the assembly of seed-associated microbial communities, particularly regarding the structural and functional characteristics of spermosphere microorganisms, remains inadequately understood.
Successful seed germination is influenced by both beneficial endophytic microorganisms within the seeds and environmental factors, as well as seed-borne pathogens. Seed-borne pathogens, which reside within seeds and are difficult to detect and identify, can spread globally due to animal and human activities, posing a significant threat to plant health. Some pathogens, such as Fusarium, can produce toxins that pose serious health risks to animals and humans [20]. Studies have documented the transgenerational transmission of various pathogens through seeds, such as Fusarium [21] and Burkholderia [12]. These pathogens can lead to seedling wilt or germination failure, potentially causing substantial crop losses. However, natural soil microorganisms can assist plants against seed-borne Fusarium [21]. For example, microbial communities recruited by seeds can inhibit pathogenic fungi by preventing their colonization of the seed surface [22]. Additionally, seed-borne bacteria are crucial for seed germination and growth. Studies have shown that rice seeds treated with bacterial antibiotics significantly reduced seed germination rates, while reinoculation with Enterobacter asburiae and Pantoea dispersa restored seedling growth and development [23]. Moreover, Sphingomonas melonis can suppress the seed-borne pathogen Burkholderia plantarii by producing metabolites and promoting the growth of seedlings [12]. These findings suggest that seed-associated microorganisms are involved in important processes related to seed germination and plant development.
Astragalus membranaceus Bge. var. mongholicus (Bge.) Hsiao (hereafter, Astragalus), a perennial legume with high economic and medicinal value in traditional Chinese medicine, faces challenges due to its low germination rate under natural conditions [24]. The factors that affect seed germination of Astragalus include insufficient seed vitality and external environmental conditions. Specifically, physical dormancy, resulting from mechanical obstruction of the seed coat, and physiological dormancy, induced by chemical signals, are significant factors that impede the germination of Astragalus seeds. Traditional mechanical treatments and chemical agents exert adverse effects on the growth and health of Astragalus in subsequent developmental stages. Harnessing microbial engineering to promote Astragalus seed germination and protect plant health may be another alternative and effective strategy for the future [25]. Although studies have shown that rhizosphere microorganisms can assist Astragalus in resisting Fusarium wilt disease [26, 27], the assembly, interactions and functions of seed-associated microbial communities during Astragalus seed germination and their impact on plant health remain to be elucidated.
In this study, we employed amplicon sequencing and shotgun metagenomic sequencing to explore the microbial community assembly and functional adaptation of seed in germinated and ungerminated Astragalus seeds. We further validated the link between seed microorganisms, seed germination, and disease resistance through culture-dependent methods. Specifically, we aimed to address the following questions: (1) How does seed germination affect the spermosphere and endophytic microbiome signatures of seeds? (2) Which beneficial microorganisms have been harnessed by the seed? and (3) How do these microorganisms facilitate seed germination and protect plant health?
Results
Seed-associated microbial community structure and assembly
To investigate the impact of germination on the assembly of seed-associated microbial communities, we analyzed bacterial 16S rRNA genes and fungal ITS sequences from germinated and ungerminated seeds at days 4 and 10 across different ecological niches (endosphere and spermosphere) (Fig. 1). Germination was defined as the emergence of the radicle through the seed coat on day 4 or 10, while ungerminated seeds showed no radicle emergence on day 4 or 10. Our results indicated that the germination rate of seeds in the soil was 23.33%, the germination force was 5%, and the germination index was 1.78. To mitigate the influence of seed vitality on germination, we measured the hard seed rate (70.67%) of Astragalus seed and selected seeds uniform in size that had imbibed successfully after soaking at 25 °C for 24 h. Successful imbibition indicated that the seeds have broken the obstacle of high hard seed rate and possessed high vitality. From 32 samples, a total of 3,198,515 high-quality bacterial 16S rRNA genes and 131,540 fungal ITS sequences were obtained (Table S1). Following rarefaction, we identified 11,943 bacterial and 1187 fungal amplicon sequencing variants (ASVs).
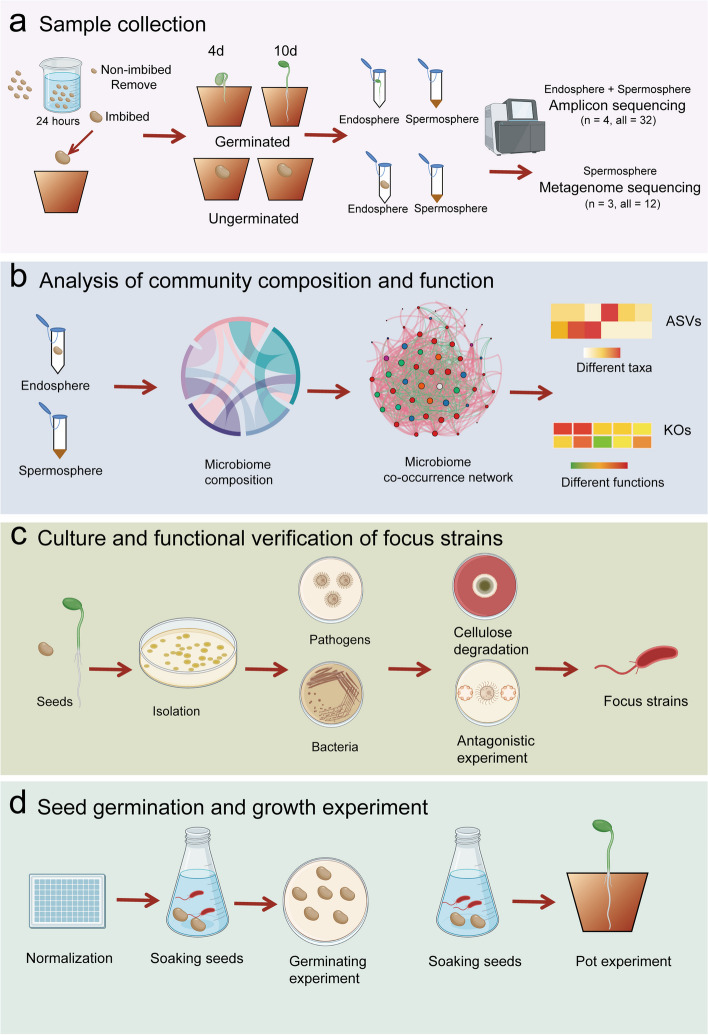
Fig. 1.
Experimental design for the collection, processing, and analysis of microbial communities associated with Astragalus seeds. a Collection of microbial samples from seeds. Imbibed seeds were planted in soil, and endosphere and spermosphere soil samples of germinated and ungerminated seed were collected on day 4 and day 10, respectively. b Analysis of microbial communities. The composition and functional analysis of microbial communities associated with germinated and ungerminated seeds are performed to identify representative taxonomic and functional information. c Isolation, cultivation, and functional screening of seed-associated microorganisms. Potential pathogens and microorganisms are isolated and cultured from the seeds. The ability of seed-associated microorganisms to degrade cellulose and inhibit pathogens was measured. d Pot validation of target strains. Seeds inoculated with the strains were grown in plates and pots for verification
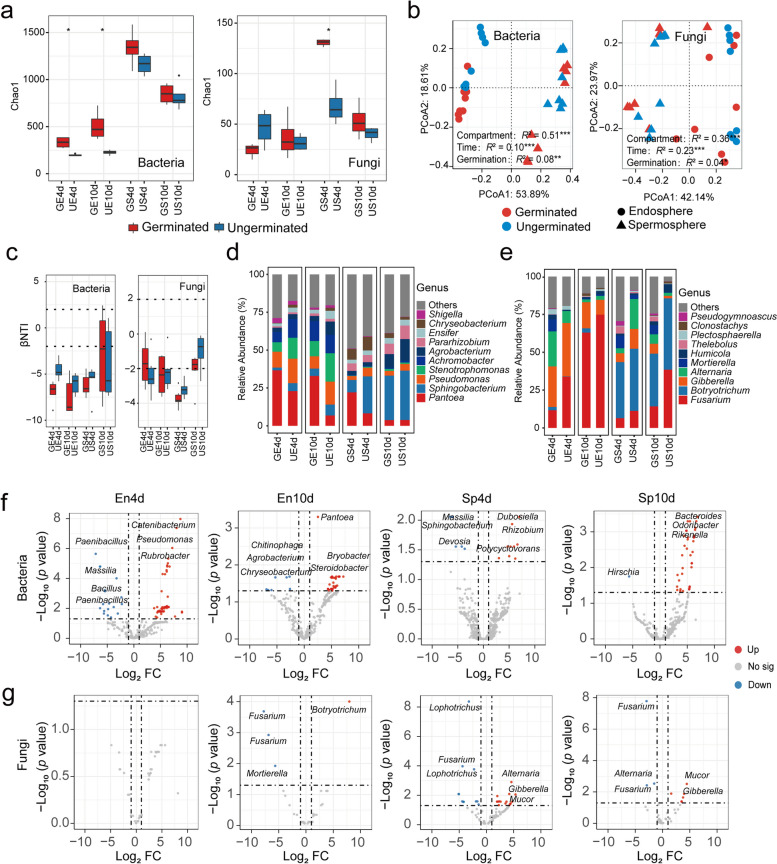
The Chao1 index revealed greater microbial diversity in germinated seeds than in ungerminated seeds, particularly for endophytic bacteria (Fig. 2a and Fig. S1; Table S2), indicating greater diversity of endophytic bacteria in germinated seeds. The PERMANOVA indicated that the compartments had the strongest influence on the structure of the seed-associated bacterial (R2 = 0.51; p < 0.001) and fungal (R2 = 0.36; p < 0.001) communities (Fig. 2b). Notably, germination significantly affected the community structure of seed-associated bacteria (R2 = 0.08; p < 0.001) and fungi (R2 = 0.04; p < 0.05). Under different compartmental niches and germination times, germination had the greatest effect on the community structure of the seed endophytic bacteria on day 10 (R2 = 0.66; p < 0.05) and the spermosphere fungal community on day 4 (R2 = 0.54; p < 0.05) (Fig. S2a-c; Table S3). Beta dispersion analysis revealed that the structure of the microbial community associated with germinated seeds presented greater variation than did that associated with ungerminated seeds (Fig. S2d and Fig. S3).
Fig. 2.
Diversity and assembly of Astragalus seed-associated microbiome. a Chao1 diversity index of bacterial and fungal communities in germinated and ungerminated seeds across different time and compartments. b Principal coordinate analysis (PCoA) of seed-associated bacterial and fungal communities based on the Bray–Curtis distances, with PERMANOVA to test differences. c βNTI value of bacterial and fungal communities of germinated and ungerminated seeds across the different time and compartments. d Composition of seed-associated bacterial community at genus levels. e Composition of seed-associated fungal community at genus levels. Volcano plots indicate taxa (ASV) with differences in bacterial (f) and fungal (g) communities associated with germinated and ungerminated seeds at different time and compartments. GE4d: Germinated seed endosphere on Day 4, UE4d: Ungerminated seed endosphere on Day 4, GE10d: Germinated seed endosphere on Day 10, UE10d: Ungerminated seed endosphere on Day 10, GS4d: Germinated seed spermosphere on Day 4, US4d: Ungerminated seed spermosphere on Day 4, GS10d: Germinated seed spermosphere on Day 10, US10d: Ungerminated seed spermosphere on Day 10. Differences were analyzed using Wilcoxon rank-sum test. Asterisks represent distinct significant differences (*, p < 0.05; **, p < 0.01; ***, p < 0.001)
We further investigated the impact of germination on the assembly process of seed microbial community. Neutral community model (NCM) analysis indicated that microbial communities associated with germinated seeds were generally less affected by neutral processes (Fig. S4). β-nearest taxon index (βNTI) based on null-model results indicated that the assembly of all seed-associated bacterial communities was predominantly governed by deterministic (homogeneous selection) processes, with germinated bacteria being more influenced by deterministic processes than ungerminated seed bacteria were (Fig. 2c; Table S4). In contrast, the assembly of seed-associated fungal communities was dominated by stochastic processes.
Our results showed that the relative abundance of Pantoea in germinated seeds was higher than that in ungerminated seeds, especially in the spermosphere on day 4 and seed endosphere on day 10 (Fig. 2d; p < 0.01). In addition, the relative abundance of Fusarium in ungerminated seed was higher than germinated seeds (Fig. 2e). Enriched taxon analysis of germinated and ungerminated seed-associated microbiomes revealed that germinated seeds enriched more microbial taxa in endosphere and spermosphere soils (Fig. 2f, g; Table S5). Specifically, the endophytes of germinated seeds were enriched with potential growth-promoting microorganisms such as Pseudomonas and Pantoea (Fig. 2f; Table S5), as well as fungi such as Mucor (Fig. 2g; Table S5). Notably, both the endophytes and the spermosphere of the ungerminated seeds were enriched with Fusarium (Fig. 2e and g). Interestingly, many potential biocontrol bacteria, such as Paenibacillus, Bacillus, Massilia, and Chitinophaga, were significantly enriched in the endophytes of ungerminated seeds, particularly Paenibacillus, on day 4 (Fig. 2f; Table S5).
Seed-associated microbial co-occurrence network
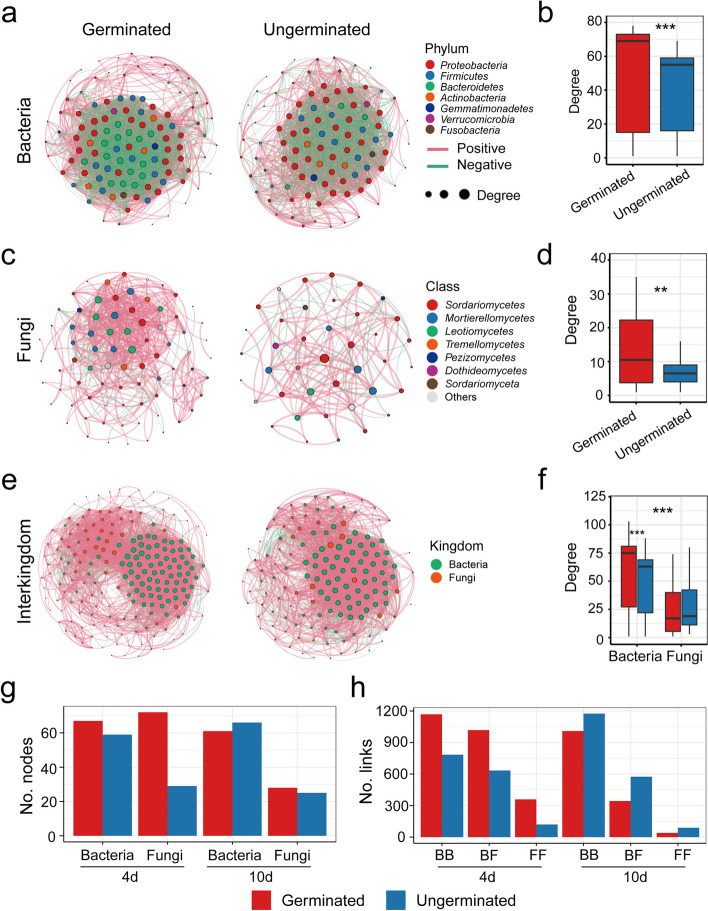
To explore the co-occurrence network patterns of the microbiome associated with seed germination, we constructed and analyzed networks for bacteria–bacteria, fungi–fungi, and bacteria–fungi interactions. Our results revealed distinct microbial co-occurrence network patterns between germinated and ungerminated seeds (Fig. 3; Table S6). The bacterial network of germinated seeds (nodes/edges: 113/2838) was more complex than that of ungerminated seeds (nodes/edges: 99/1956) (Fig. 3a, b; Table S6), with a greater degree across different seed compartments (endophyte and spermosphere) (Fig. S5-6) and different times (day 4 and day 10) (Fig. S7-8). Similarly, the fungal community co-occurrence network also exhibited greater complexity in germinated seeds (Fig. 3c, d), with numerous (3/10) ASVs, such as Fusarium, identified as keystone taxa (Table S7).
Fig. 3.
Microbiome co-occurrence networks of germinated and ungerminated seeds. a Bacterial community co-occurrence networks of germinated and ungerminated seeds. b Comparison of the network degree of bacterial communities associated with germinated and ungerminated seeds. c Fungal community co-occurrence networks in germinated and ungerminated seeds. d Comparison of the network degree of fungal communities associated with germinated and ungerminated seeds. e Interkingdom co-occurrence networks of germinated and ungerminated seeds. f Comparison of the microbiome interkingdom network degree in germinated and ungerminated seeds. g Number of bacterial and fungal nodes in the co-occurrence network at different germination times. h Number of bacterial and fungal links in the co-occurrence network at different germination times. Nodes color represents different phyla/kingdom, and node size represents the size of the degree. Red edge represents a positive correlation, and the green edge represents a negative correlation. Differences were analyzed using Wilcoxon rank-sum test. Asterisks represent distinct significant differences (**, p < 0.01; ***, p < 0.001)
The interkingdom association network further revealed greater complexity in the germinated seeds (nodes/edges: 197/4290) than in the ungerminated seeds (nodes/edges: 143/2943) (Fig. 3e). Notably, the bacterial taxa exhibited a greater degree in the germinated seed microbiome network (Fig. 3f), whereas Fusarium was a hub taxon in the ungerminated seed microbiome network (Table S7). The dynamics of seed germination revealed a temporal shift in network dominance: fungi dominated the seed microbial community co-occurrence network at early germination (day 4), whereas bacteria dominated at the later stage (day 10) (Fig. 3g). Additionally, fungi displayed increased interactions with other taxa in early germinating seeds (Fig. 3h), highlighting their potential role in initiating the germination process.
Seed-associated microbial function profiles
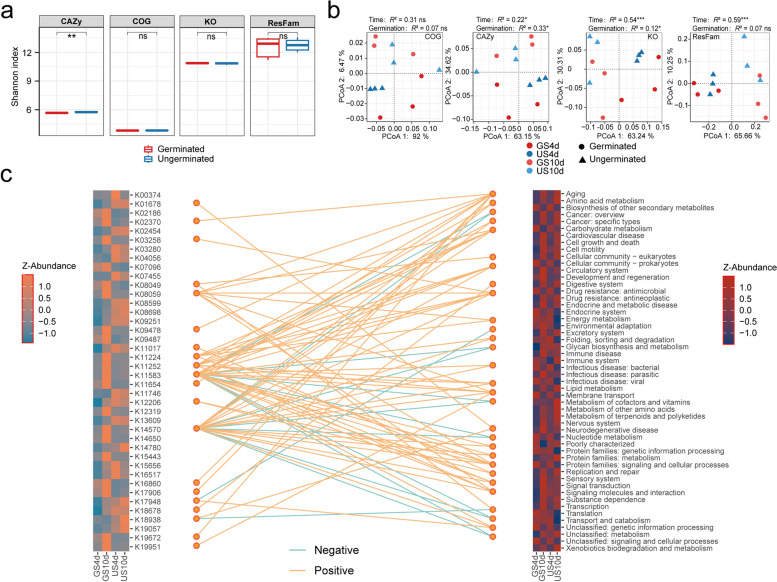
We investigated the impact of germination on the potential function of the seed-associated microbial community. Metagenomic sequencing revealed distinct functional profiles between microbial communities inhabiting germinated and ungerminated Astragalus spermosphere soil. The germinated spermosphere soil had lower functional diversity, particularly for carbohydrate-active enzymes (CAZy; p < 0.01; Fig. 4a). Moreover, with increasing germination time, the functional diversity of the microorganisms increased (Fig. S9a). PERMANOVA confirmed a significant difference in functional composition (CAZy and KO) between the germinated and ungerminated spermosphere (p < 0.05; Fig. 4b; Table S8). Differential functional analyses indicated that germinated seeds were enriched with more KOs (Fig. S9; Table S9). In the early stages (day 4) of seed germination, there was notable enrichment of pathways related to carbohydrate metabolism, drug resistance, environmental adaptation, energy metabolism, membrane transport, nucleotide metabolism, and translation. Moreover, pathways such as cell growth and death, cell motility, and signal transduction were enriched in the later stages (day 10) of seed germination. More specifically, growth hormone synthesis, secretion, and action (K11224), along with membrane trafficking (K17906 and K19951), are prominent among the enriched pathways in the germinated spermosphere. Conversely, the ungerminated spermosphere is enriched in metabolic pathways involving cofactors, vitamins, amino acids, terpenoids, and polyketides (Fig. 4c and Fig. S10).
Fig. 4.
Differences in the microbiome function between germinated and ungerminated spermosphere soil. a Shannon diversity index of functional genes in germinated and ungerminated seed spermosphere under different time. b PCoA of spermosphere soil microbiome function (KO, COG, CAZy, and ResFam) based on the Bray–Curtis distances, and PERMANOVA to test for differences between germinated and ungerminated seeds. c Paired heatmaps of the diverse differentially expressed KEGG Orthology (KO) functional categories (top 40) between groups, and the relationships between differentially functional genes and pathways. The connections in the middle were calculated through the co-occurrence network, and only connections with |r|> 0.7 and p < 0.01 were retained. The orange line indicates a positive correlation and the light blue line indicates a negative correlation. (*, p < 0.05; **, p < 0.01; ***, p < 0.001); ns indicates no significant difference
Linear discriminant analysis (LDA) of the CAZy profiles revealed enrichment of pathogen-inhibiting enzymes in the germinated spermosphere microbiome (Fig. S11; Table S10), including chitin-binding enzymes (CBM50), chitin-degrading enzymes (GH23), and rhamnolipid synthesis enzymes (GT2, GT41 and GT4), associated with detoxifying fungal metabolites. The cellulose degradation (GH94 and AA3) was enriched in the germinated spermosphere. In addition, GH94 was significantly and positively correlated with Pantoea, Lactobacillus, and Paenibacillus (Fig. S11b, p < 0.05). Temporal analysis revealed distinct functional patterns during germination. In the early stage of seed germination (day 4), the germinated seed spermosphere was enriched in functions such as CH1, GH6, GH8, and GT2, whereas the similar functions CH1, GH3, CH5, and GH6 were enriched in the ungerminated seed spermosphere in the later stages (Fig. S12; Table S10). These results demonstrate that seed germination drives significant shifts in the functional profiles of the spermosphere microbiome, with enhanced capacities for cellulose degradation and pathogen resistance.
Ability of seed microorganisms to inhibit pathogens, degrade cellulose, and promote germination
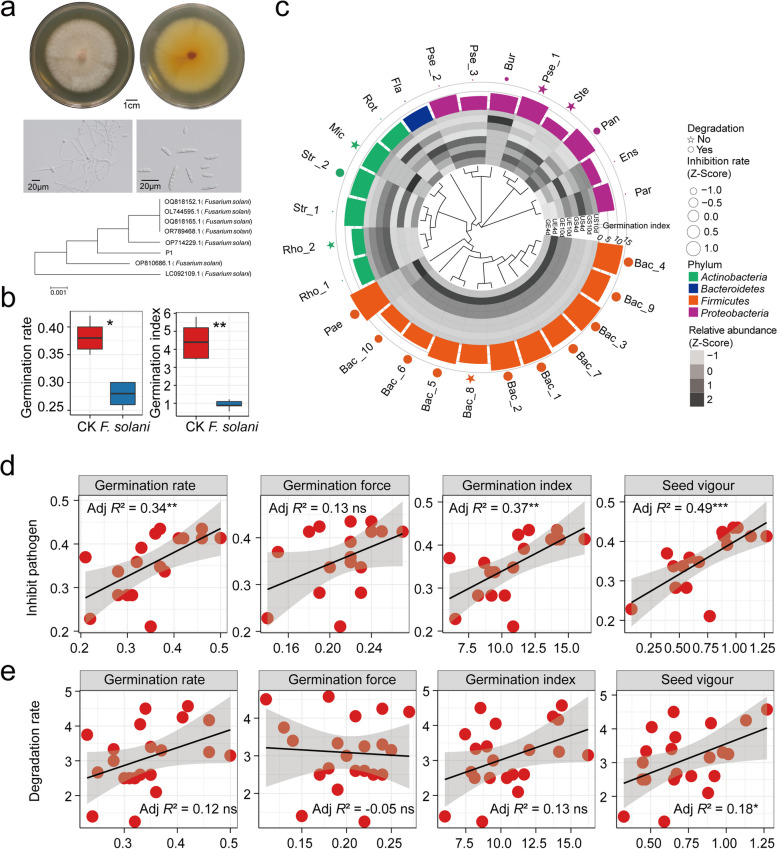
The analysis of amplicon data revealed that Fusarium was enriched in ungerminated seeds, leading to the hypothesis that Fusarium inhibits Astragalus seed germination. To test this hypothesis, a potentially pathogenic fungus, Fusarium solani strain (P1), was isolated from ungerminated seeds and identified on the basis of morphological and molecular ecological comparisons (Fig. 5a). Inoculation of P1 conidia suspension (107 spores/mL) with Astragalus seeds significantly reduced the seed germination rate (p < 0.05) and germination index (p < 0.01) (Fig. 5b), supporting the hypothesis that Fusarium plays a key inhibitory role.
Fig. 5.
Seed-associated microbiome of promote seed germination. a Morphological characteristics (macroscopic and microscopic) and phylogenetic tree of isolated pathogenic fungi. b The effect of Fusarium solani on seed germination rate and germination index, differences were analyzed using Wilcoxon rank-sum test. c Phylogenetic tree of isolated strains, ability to inhibit pathogens, whether to degrade cellulose and its effect on seed germination of Astragalus. The heatmap in the inner circle represents the relative abundance (normalized) of the genera of the strains in the amplicon sample. The outermost circle represents the germination index of the strain, and different colors represent phyla. Circles in the phylogenetic tree represent degradable cellulose, and star represent non-degradable cellulose. The size of the shape represents the ability to inhibit pathogenic fungi (normalized). Phylogenetic tree construction based on Maximum Likelihood method. d Relationship between the ability of strains to suppress pathogenic fungi and Astragalus seed germination parameters (germination rate, germination force, germination index, and seed vigor). e Relationship between the ability of strains to degrade cellulose and Astragalus seed germination parameters (germination rate, germination force, germination index, and seed vigor). Asterisks represent significant differences (*, p < 0.05; **, p < 0.01; ***, p < 0.001); ns indicates no significant difference
To further confirm the positive effect of seed-associated microorganisms on seed germination. A total of 26 bacterial strains, representing Firmicutes (11 strains), Proteobacteria (8), Actinobacteria (6), and Bacteroidetes (1), were isolated from germinating seeds and spermosphere, and their abundances differed at the genus level (Fig. 5c; Table S11). Functional screening revealed that 21 of these strains had the ability to degrade cellulose, whereas 16 strains inhibited the pathogenic fungus P1 (Fig. 5c; Table S11). Among these strains, Bac_3 (Bacillus sp. DLAE23) presented the strongest cellulose-degrading ability, whereas Bac_7 (Bacillus sp. DLAE19) presented the strongest inhibition rate for Fusarium solani strain (P1). Further experiments revealed that 12 of the 26 seed-associated bacterial strains promoted the germination of Astragalus seeds under sterile conditions (Fig. 5c; Table S11), with Pae (Paenibacillus sp. DLAE12) showing the greatest ability to improve seed germination by 17% compared with that of the control. In addition, 60% of the isolated Bacillus exhibited positive effects on seed germination. Importantly, the ability of the bacterial strains to inhibit pathogenic fungi was positively correlated with the germination rate, germination index, and vigor of Astragalus seeds (p < 0.05, Fig. 5d). Additionally, the capacity for cellulose degradation was significantly correlated with seed vigor (p < 0.05, Fig. 5e).
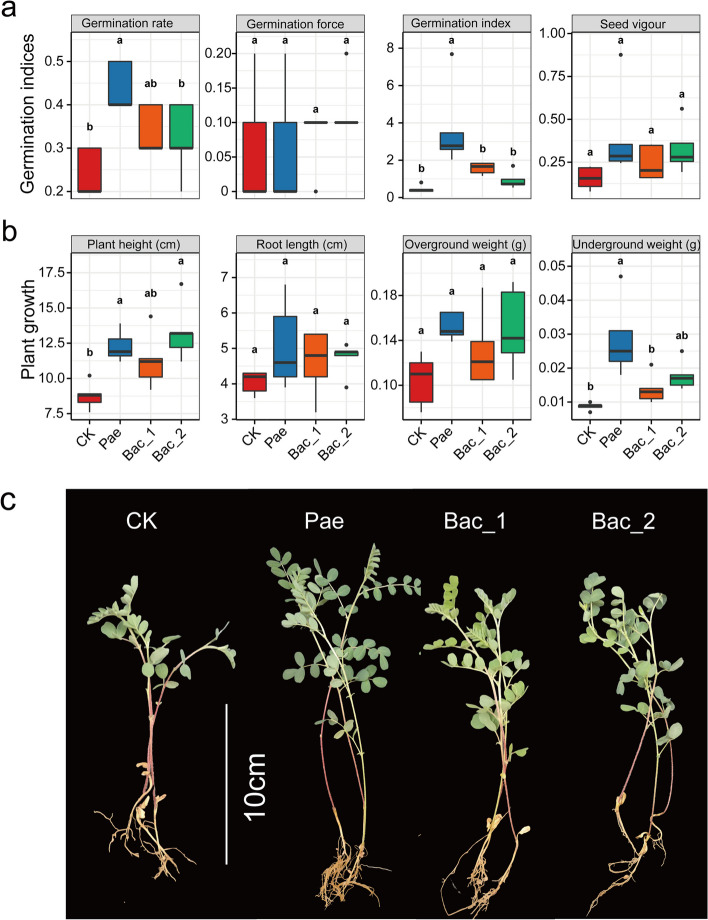
To further investigate the effects of the isolated bacteria on the seed germination and plant health of Astragalus in soil, the top three seed germination-promoting strains under sterile conditions, including Pae, Bac_1 (Bacillus sp. DLAE26) and Bac_2 (Bacillus sp. DLAE24), were selected for the pot planting experiments. The results demonstrated that Pae significantly promoted the germination rate, germination index, plant height, and underground biomass of Astragalus (Fig. 6a–c). Bac_2 significantly promoted the height of Astragalus plants. These results indicate that seed microorganisms can promote seed germination by inhibiting pathogenic fungi and degrading cellulose.
Fig. 6.
Seed-associated microbiomes promote Astragalus seed germination and plant growth. a Effects of inoculation of growth-promoting bacteria on Astragalus seed germination (germination rate, germination force, germination index, and seed vigor). b Effects of inoculation of growth-promoting bacteria on Astragalus plant growth (plant height, root length, overground weight, and underground weight). According to Tukey’s HSD test, different lowercase letters indicate statistically significant differences (p < 0.05). c Impact of inoculated strains on Astragalus growth
Discussion
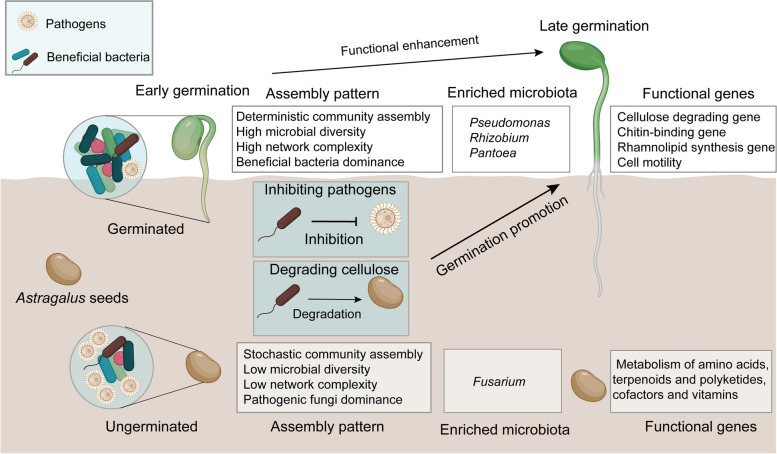
The intricate mutualistic relationship between seed microorganisms and plants is a fascinating area in the field of plant-soil-microbe interactions, particularly in the early stages of plant life [28]. During seed germination, beneficial microorganisms protect against successful seed germination by producing phytohormones and alleviating biotic and abiotic stresses [9]. However, it remains unclear whether germination failure is attributed to inherent seed vitality issues or interactions with microbes. Using imbibed seeds as research material allowed us to more effectively demonstrate the process of seed microbiome-mediated seed germination by partially excluding the influence of insufficient seed vitality. Furthermore, the dynamics of the overall composition and functional adaptations of the microbial community during seed germination remain elusive. Our investigation revealed that the diversity of endophytes and spermosphere soil microbial communities associated with germinated seeds was significantly greater than that associated with ungerminated seeds. The bacterial communities associated with germinated seeds presented a relatively high abundance of taxa with potential growth-promoting characteristics. Conversely, the enrichment of pathogens serves as a significant limiting factor for seed germination in Astragalus. Analysis of the metagenomic data revealed that the microbiome present in the spermosphere of germinated seeds performs functions involving pathogen inhibition and cellulose degradation. Further experiments with inoculated strains revealed that seed-associated microorganisms promoted seed germination and enhanced seed vitality by inhibiting pathogenic fungi and degrading cellulose (Fig. 7). These findings provide fundamental evidence for the potential of manipulating seed-associated microbial communities to increase seed germination and protect plant health.
Fig. 7.
Conceptual diagram of microbiome-mediated germination of Astragalus seeds
Germination leads to a shift in seed microbial communities from fungal to bacterial dominance
The germination process profoundly reshapes the assembly of seed-associated microbial communities. Previous studies conducted under sterile soil conditions indicated a reduction in the diversity of seed endophytic bacteria during germination [18, 19]. However, our findings in natural soil reveal a contrasting pattern. We observed a significant increase in microbial diversity associated with germinating seeds. This heightened diversity is likely driven by the secretion of a complex blend of exudates from the germinating seeds, creating a nutrient-rich spermosphere that attracts and facilitates the colonization of a wider range of microorganisms [29]. The microbial communities colonizing germinating seeds presented greater complexity, which was particularly evident in the early stages of seed germination. This high complexity may be due to the diverse exudates offering multiple carbon sources and promoting microbial interactions [30].
Our observations revealed a succession of microbial communities during seed germination, with fungi initially dominating the microbial network. This early fungal dominance is likely due to their rapid colonization of available niches and ability to utilize these recalcitrant carbon sources [31]. Interestingly, we found that the potential pathogen Fusarium was enriched in early ungerminated seeds, whereas in the later stages of germination, beneficial Firmicutes and Basidiomycota became increasingly abundant. For example, some Basidiomycota are well known for forming mycorrhizal fungi with plants, increasing nutrient uptake [32]. Similarly, Firmicutes, particularly Bacillus species, are recognized for promoting plant growth and disease resistance [21]. This succession from a pathogenic fungal-dominated community to one rich in beneficial bacteria and symbiotic fungi highlights the intricate ecological choreography unfolding during seed germination. Our findings suggest that as seeds transition to seedlings, they actively shape their microbial environment, potentially selecting for microorganisms that contribute to their growth and health.
Seeds recruit beneficial microorganisms to assist in seed germination
Many Fusarium species are globally distributed seed-borne pathogens that hinder seed germination and threaten plant health, often leading to severe crop yield losses in various species, including soybean, maize, and wheat [21, 33, 34]. However, the impact of seed-borne Fusarium species on seed germination in Astragalus remains unknown. Our results revealed an enrichment of Fusarium in ungerminated Astragalus seeds, with this genus emerging as a keystone species in the co-occurrence network of the ungerminated seed microbiome, indicating its dominant role in shaping the microbial community within ungerminated seeds. Pathogenicity experiments demonstrated that Fusarium solani significantly inhibited Astragalus seed germination. The mechanism underlying the detrimental effects of Fusarium is attributed to its production of toxins and enzymes that disrupt crucial cellular processes. For example, Fusarium graminearum produces deoxynivalenol, a mycotoxin that disrupts normal cellular functions by inhibiting protein synthesis [35]. Similarly, Fusarium solani secretes cell wall-degrading enzymes, including cutinase, which contribute to plant tissue decay and pathogenicity [33, 36]. In addition, a study has shown that Fusarium solani infection can increase alanine and fatty acids during seed germination, thereby reducing the seed germination rate [37].
In contrast to ungerminated seeds, germination activates metabolic processes that recruit and enrich a greater number of microorganisms in the endosphere and spermosphere through the release of seed exudates [15, 38]. We observed an enrichment of potentially beneficial microorganisms, including Pseudomonas and Pantoea, which are known to antagonize pathogenic fungi and promote plant growth [39, 40]. Our experiments confirmed the ability of Pseudomonas and Pantoea to promote seed germination. The presence of pathogens, such as tomato Fusarium wilt [41], wheat Fusarium head blight [42] and Diaporthe citri melanose pathogens [4], alters the composition of plant-associated microbial communities. The symbiotic relationship between the microbiome and the plant host, which functions as a “holobiont,” serves as a defence mechanism against pathogen stress [43]. Plants exhibit a remarkable ability to recruit beneficial microbes in response to pathogen-induced stress [4, 44, 45].
Intriguingly, our findings revealed that ungerminated seeds in the early stages harbored diverse potentially beneficial microorganisms, including Paenibacillus and Bacillus. However, not all isolated Bacillus species have a positive effect on seed germination. Thus, considering the variations in their metabolic diversity and functions [46], the diversity of Bacillus species and strains may play a significant role in seed germination and plant growth. Subsequent disease suppression assays demonstrated the efficacy of these bacteria in inhibiting the growth of pathogenic fungi and increasing Astragalus seed germination rates (Fig. 5c; Tale S11). Furthermore, under pathogen stress, plants may mitigate this stress by recruiting beneficial microbes [3]. This “cry-for-help” strategy may partially elucidate the observed enrichment of Paenibacillus and Bacillus in ungerminated seed. Nevertheless, the exact mechanism remains unclear, which is a limitation of our study.
Specific functions of seed microorganisms stimulating Astragalus seed germination
The microbial communities associated with germinated seeds present diverse functions crucial for promoting germination, particularly pathogen inhibition and cellulose degradation. Our analysis revealed fungal cell wall degradation in the spermosphere, including an increased abundance of genes encoding CBM50 (chitin or peptidoglycan binding) and CH23 (chitin degradation). This finding is consistent with previous research linking chitinase activity to disease suppression in sugar beet infected with Rhizoctonia solani [47]. Furthermore, we observed enrichment of rhamnosidase synthesis-related functions (GT2 and GT41 glycosyltransferase families), which are often associated with Paenibacillus species known to mediate plant–microbe interactions and confer pathogen resistance [48]. Notably, the microbial cell motility functional pathway was enriched in the germinated seeds, potentially enabling Pseudomonas and Bacillus to effectively colonize the seed environment [49]. This targeted recruitment of beneficial microbes with specific functions underscores the active role of plants in shaping their microbiome to promote germination success.
Another key mechanism by which microbes support seed germination is microbial cellulose degradation, which releases nutrients that directly nourish germinating seedlings [50]. In our study, we identified numerous cellulose-degrading enzymes, such as GH94 and AA3, within the spermosphere, emphasizing the importance of this process. Moreover, the positive correlation between Pantoea and Paenibacillus and GH94 was confirmed in cellulose degradation experiments, supporting the view that seed-associated microorganisms promote germination via degrading cellulose. This aligns with previous studies showing that Pantoea, Bacillus, and Paenibacillus, which are common seed endophytes, have robust cellulose-degrading abilities, effectively converting cellulose into readily available carbon sources [51–53]. The presence of these specific bacteria and their associated cellulose degradation enzymes further highlights their critical role in enhancing seed vigor and germination success.
Conclusion
In summary, our study provides compelling evidence for the structural and functional contributions of seed-associated microorganisms in facilitating seed germination and early plant growth. We demonstrated their crucial role in pathogen suppression and cellulose degradation, thereby enhancing seed vigor. The observed shift from fungal dominance to bacterial dominance may represent an adaptive strategy, allowing plants to benefit from different microbial partners at various stages of their early development. Our findings suggest that seeds may actively recruit beneficial microorganisms during germination to combat pathogenic fungi, and add to our understanding of how plants “cry for help” during early life stages, but the mechanism needs to be explored in more detail. Importantly, inoculation with Paenibacillus sp. effectively enhanced the germination and stimulated the growth of Astragalus seeds. These findings highlight the immense potential of seed-associated microorganisms in improving seed germination and plant growth, underscoring the importance of these inconspicuous microbial resources within seeds.
Methods
Experimental design and sample collection
Soil and seeds were collected from annual Astragalus fields located in Yuzhong County, Gansu Province (36° 01′ 38″ N, 104° 22′ 06″ E). The study area is characterized by a temperate continental climate with an average annual temperature of 7.5 °C, a precipitation of 428 mm, a sunshine duration of 2562.5 h, and an altitude of 2280 m. The soil properties are as follows: moisture content, 32.76%; pH, 8.02; organic matter, 13.99 g/kg; total nitrogen, 0.96 g/kg; total phosphorus, 0.95 g/kg; nitrate-nitrogen (NO3), 66.43 mg/kg; ammonium-nitrogen (NH4), 158.11 mg/kg; and available phosphorus, 62.90 mg/kg. The collected soil was sieved and kept in a cool and dark place, while the seeds were stored in a refrigerator at 4 °C.
In the Astragalus seed germination pot experiment, we used 10 cm × 10 cm pots filled with 150 g of fresh soil at 60% water-holding capacity. To mitigate the influence of seed vigor on the microbial-mediated germination of Astragalus, we assessed the hard seed rate and selected seeds capable of imbibition. Uniformly sized Astragalus seeds were randomly selected and surface sterilized by soaking in a 5% sodium hypochlorite solution for 30 s, followed by five rinses with sterile distilled water. The seeds were subsequently immersed in 75% ethanol for 2 min and rinsed five times with sterile water. Subsequently, 100 seeds were immersed in sterile water at 25 °C for 24 h, and the numbers of imbibed and non-imbibed seeds were recorded, with the procedure repeated five times. We calculated the hard seed rate using the formula: hard seed rate (%) = (number of hard seeds / total number of seeds tested) × 100% [54, 55]. The hard seed rate was determined to be 70.67%. Seeds capable of imbibition were selected and again surface sterilized. Ten of these seeds were evenly sown in each pot, and a total of 300 pots were placed in a greenhouse and maintained at 25 °C for 16 h (light) / 20 °C for 8 h (dark), with approximately 40% relative humidity.
At days 4 and 10 after sowing, we collected endosphere and spermosphere (compartment) samples from both germinated and ungerminated seeds were collected. Germination was defined by the emergence of the radicle through the seed coat. A germinated seed had the radicle emerging without signs of wilting or pathogen infection in the seedling, while an ungerminated seed showed no radicle emergence. Day 4 represents the determination time of seed germination force, and day 10 represents the end time of seed germination. Each sample contained 100 seeds (Fig. 1). In total, 32 samples were collected (n = 4 for each type of sample), including germinated seed endophytes (GE4d) and the spermosphere (GS4d) on day 4; ungerminated seed endophytes (UE4d) and the spermosphere (US4d) on day 4; and corresponding samples on day 10 (GE10d, GS10d, UE10d, and US10d).
For sample collection, sterile 50-mL centrifuge tubes containing 35 mL of phosphate buffer solution were used to submerge the seed samples. The tubes were vortexed for 15 s, and the seed washing solution was then homogenized and evenly distributed into two separate 50-mL centrifuge tubes. These samples were subsequently centrifuged at 6000 × g for 5 min. After centrifugation, the resulting seed wash mixture was once again mixed and divided into two additional 50-mL centrifuge tubes, which underwent a second round of centrifugation at 6000 × g for another 5 min. The soil sediment obtained is referred to as spermosphere soil. The seed samples were sonicated twice in sterile water at a frequency of 20 kHz for 30 s each time and then rinsed with sterile water three times. The samples were subsequently lyophilized via liquid nitrogen on an ultraclean bench and pulverized with a sterile mortar. All the samples were stored in a − 80 °C freezer until DNA extraction.
DNA extraction, amplification, and sequencing
DNA was extracted from 0.5 g of seeds or spermosphere soil using the FastDNA® SPIN for soil kit (MP Biomedicals, Solon, USA), and its concentration and purity were determined. The V4 region of the 16S rRNA gene was amplified using the universal primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) [56]. Fungal internal transcribed spacer region 1 (ITS1) was amplified using the primers ITS1 (CTTGGTCATTTAGAGGAAGTAA)/ITS4 (GCTGCGTTCTTCATCGATGC) [57]. The raw sequencing data were processed using the DADA2 pipeline [58], which includes denoising, trimming, and chimaera removal. The obtained amplicon sequence variants (ASV) were classified and identified using the SILVA database v132 [59] for bacteria and the UNITE database (version 1.12.2017) [60] for fungi.
Metagenomic assembly and annotation
Metagenomic sequencing was performed on germinated and ungerminated spermosphere soil samples at days 4 and 10 (n = 3, totalling 12 samples) to measure microbial function during seed germination. Paired-end sequencing (150 bp) was conducted on an Illumina NovaSeq 6000 platform, yielding a minimum of 12 GB of raw data per sample. Reads were assembled using MEGAHIT v1.2.9 [61], followed by gene prediction using Prokka v1.14.5 [62] and clustering at a 0.95 similarity threshold using CD-HIT v4.8.1 [63] to generate a nonredundant gene catalog. Functional annotation was performed using DIAMOND comparison [64] of eggNOG-mapper v1.0.3 with the eggNOG database (v5.0) [65]. The annotation results were reorganized into Kyoto Encyclopedia of Genes and Genomes (KEGG) homology (KO) profiles [66], clusters of homologous proteomes (COG) functional categories [67], and CAZymes (CAZy) [68]. Furthermore, antibiotic resistance genes were detected and reconstituted by using ResFams [69].
Isolation and identification of culturable seed bacterial and pathogen isolates
For bacterial isolation, 5 mL of seed grinding solution was mixed with 45 mL of sterilized distilled water in a 100-mL triangular flask and shaken at 180 rpm for 30 min at 28 °C, and the resulting mixture was serially diluted from 10−1 to 10−6. One hundred microlitres of suspension was applied to LB medium. Single colonies were purified and lysed in 50 μL of 1% sodium hydroxide at 72 °C. The 16S rRNA gene of each strain was amplified with double barcoded universal primers (27F 5′-GAGTTTGATCCTGGCTCAG-3′; 1492R 5′-TACCTTGTTACGACTT-3′) [42]. For pathogenic fungal isolation, 100 μL of the diluted suspension was plated on PDA media. Fungal DNA was extracted and amplified using the primers ITS1/ITS4 (TCCGTAGGTGAACCTGCGG; TCCTCCGCTTATTGATATGC) and tef−1α (EF1 FATGGGTAAGGARGACAAGAC; EF2 RGGARGTACCAGTSATCATGTT) [70].
Approximately 100 seeds were soaked in 20 mL of a spore suspension of 107/mL and/or sterile water for 12 h, dried with aseptic filter paper and placed in water, after which the germination rate and germination index were calculated.
Evaluation of bacterial isolates for fungal pathogen suppression and cellulose degradation
Potential probiotic bacteria were activated in LB medium prior to testing. To assess fungal pathogen suppression, 2 μL of test bacterial suspension (OD600 = 0.7) was inoculated onto 3-day-old pathogen cultures. Inhibition rates were calculated by measuring the growth radii of pathogenic colonies in the presence and absence of antagonistic bacteria:
where Rc is the radius of the pathogenic fungal colony in the control group and Rt is the radius in the treatment group.
To evaluate the cellulose degradation capabilities, bacterial strains were inoculated on sodium carboxymethylcellulose (CMC-Na) media and incubated at 30 °C for 5 days. Plates were then flooded with 1 mg/mL Congo red solution for 1 h, followed by destaining with 1 M NaCl until clear hydrolysis zones appeared. Strains exhibiting distinct degradation halos were selected for further analysis, and both the halo and colony diameter were recorded.
In vitro and in vivo methods for measuring seed germination
Two conditions were used for the germination experiment. For the in vitro experiments, the seeds were immersed in 5% sodium hypochlorite (NaClO) for 1 min, rinsed with sterile water 5 times, immersed in 75% alcohol for 2 min, and rinsed with sterile water 5 times. The seeds were soaked in the suspension of the strain to be tested (OD600 = 0.2 or 107 spores/mL) for 12 h. The seeds were removed and immersed in alcohol for 30 s, rinsed 3–5 times with sterile water, and then the surface was blotted dry with sterile blotting paper. Twenty seeds were placed in sterile water agar media for germination assessment.
For the in vivo experiments, the seeds were treated as described above, and 5 seeds were sown in pots with a total of 150 g of soil in each pot. A total of 20 mL of sterile water was added before sowing, and the germination rate, germination force, germination speed, germination index, and seed vigor were measured:
Where:
Gt = number of germinated seeds on day t
Gt−1 = number of germinated seeds on day t − 1
Dt = number of days since sowing
Data statistics and analysis
All the statistical analyses were performed in the R environment (v4.1.1; http://www.r-project.org/). The richness, Shannon index, and phylogenetic diversity of the seed microbial community were calculated by the “vegan” [71] and “picante” packages [72]. Bray–Curtis distances were used to calculate community similarity, and principal coordinate ranking analysis (PCoA) was performed with the “vegan” package. A permutation multivariate analysis of variance (PERMANOVA) was performed using the “adonis” function in the “vegan” package. Differential abundance analysis between germinated and ungerminated seed microbiota was performed via the EdgeR generalized linear model (GLM) method in the “edgeR” R package [73]. Linear discriminant analysis (LDA) in the “microeco” package was used to explore the differences between germinated and ungerminated seed microbial functions [74]. Phylogenetic trees were constructed using MEGA7 [75] and visualized by the “ggtreeExtra” and “ggplot2” packages [76].
To quantify the relative importance of stochastic processes in community assembly, we used the Sloan neutral community model to predict the relationship between ASV frequency detection and relative abundance [77]. In this model, Nm is defined as the metacommunity size multiplied by the migration value, and the migration rate (m) represents the estimated intercommunity dispersal rate. The metacommunity size (N) is the total number of reads per sample. The parameter R2 represents the overall fit to the neutral model. The 95% confidence intervals for all fitted statistics were calculated by 1000 bootstrap replicates [78].
The microbial community assembly process in different subgroups was assessed using the “picante” null model package and taxonomic β diversity indicators (βNTI and RCbray) [72]. The βNTI quantifies the magnitude and direction of deviation between the observed βMNTD values and the values calculated by the null model [79]. Values of |βNTI|> 2 indicate that deterministic processes dominate community assembly. Specifically, βNTI > 2 reflects that the community assembly process is dominated by variable selection (i.e., environmental sorting), whereas βNTI < − 2 indicates that community assembly is dominated by homogeneous selection. |βNTI|< 2 indicates that community assembly is dominated by stochastic processes. The deviation between the observed Bray–Curtis and null distributions was used to further delineate the stochastic processes. An RCbray > 0.95 indicates dispersal limitation, and an RCbray < − 0.95 indicates homogeneous dispersal-driven community assembly. When |βNTI|< 2 and |RCbray|< 0.95, community assembly was driven by undominant processes (i.e., weak selection, weak dispersal, diversification, or ecological drift).
The co-occurrence patterns of seed-associated microorganisms were explored by constructing separate networks for the germinated and ungerminated seed microbial communities. Microbial taxa present in less than 50% of the samples were excluded, and the topological properties of the network were calculated via Spearman correlation and significance p using the “igraph” and “Hmisc” packages [80]. Subsequently, FDR correction was performed on the p-values, and only the nodes and edges with a correlation exceeding 0.6 and p less than 0.05 were retained and visualized using Gephi (v0.92, https://gephi.org). Keystone taxa were identified on the basis of higher degree values in the network [45, 81]. All schematic diagrams were modified from the BioRender (https://www.biorender.com/).
Supplementary Information
Acknowledgements
We thank Yaoyu Liu for assistance with the soil and seed sampling.
Abbreviations
- ITS
Internal transcribed spacer
- ASVs
Amplicon sequencing variants
- PERMANOVA
Permutational multivariate analysis of variance
- NCM
Neutral community model
- βNTI
β-Nearest taxon index
- CAZy
Carbohydrate-Active enZYmes
- KO
KEGG ORTHOLOGY
- LDA
Linear discriminant analysis
- GLM
Generalized linear model
- CMC-Na
Carboxymethylcellulose sodium
- NaClO
Sodium hypochlorite
Authors' contributions
D. L. conducted the experiments. W. L., H. Z. and Y. L. contributed to the data analysis and visualization. D. L. analyzed the microbial and plant data, and wrote the corresponding manuscript sections. W. C. conceived and designed the experiments and revised the manuscript. G. W. and D. S. designed the experiments. All authors reviewed the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (grant numbers: 42177106 and 31870476).
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA018766, CRA018767, CRA018768) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. The data visualization code can be found in https://github.com/lidas666/seed-microbiome.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/14/2025
The original online version of this article was revised: Video Abstract has been updated.
Contributor Information
Weimin Chen, Email: chenwm029@nwsuaf.edu.cn.
Duntao Shu, Email: Donald.shu@nwafu.edu.cn.
Gehong Wei, Email: weigehong@nwafu.edu.cn.
References
- 1.Zai X, Luo W, Bai W, Li Y, Xiao X, Gao X, et al. Effect of root diameter on the selection and network interactions of root-associated bacterial microbiomes in Robinia pseudoacacia L. Microb Ecol. 2021;82:391–402. [DOI] [PubMed] [Google Scholar]
- 2.Xu L, Dong Z, Chiniquy D, Pierroz G, Deng S, Gao C, et al. Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics. Nat Commun. 2021;12:3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu H, Li J, Carvalhais LC, Percy CD, Prakash Verma J, Schenk PM, et al. Evidence for the plant recruitment of beneficial microbes to suppress soil-borne pathogens. New Phytol. 2021;229:2873–85. [DOI] [PubMed] [Google Scholar]
- 4.Li P, Zhu Z, Zhang Y, Xu J, Wang H, Wang Z, et al. The phyllosphere microbiome shifts toward combating melanose pathogen. Microbiome. 2022;10:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiao Y, Wang Z, Sun H, Guo H, Song Y, Zhang H, Ruan Y, Xu Q, Huang Q, Shen Q, et al. Synthetic community derived from grafted watermelon rhizosphere provides protection for ungrafted watermelon against Fusarium oxysporum via microbial synergistic effects. Microbiome. 2024;12:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao C, Wang Z, Liu S, Lv N, Deng X, Xiong W, et al. Additive fungal interactions drive biocontrol of Fusarium wilt disease. New Phytol. 2023;238:1198–214. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Tang S, Gao H, Ren J, Xu P, Dong W, et al. Plant growth-promoting rhizobacterium Bacillus cereus AR156 induced systemic resistance against multiple pathogens by priming of camalexin synthesis. Plant Cell Environ. 2024;47:337–53. [DOI] [PubMed] [Google Scholar]
- 8.Tabassum N, Ahmed HI, Parween S, Sheikh AH, Saad MM, Krattinger SG, et al. Host genotype, soil composition, and geo-climatic factors shape the fonio seed microbiome. Microbiome. 2024;12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg G, Raaijmakers JM. Saving seed microbiomes. ISME J. 2018;12:1167–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santoyo G, Moreno-Hagelsieb G, del Carmen O-M, Glick BR. Plant growth-promoting bacterial endophytes. Microbiol Res. 2016;183:92–9. [DOI] [PubMed] [Google Scholar]
- 11.Bergna A, Cernava T, Rändler M, Grosch R, Zachow C, Berg G. Tomato seeds preferably transmit plant beneficial endophytes. Phytobiomes J. 2018;2:183–93. [Google Scholar]
- 12.Matsumoto H, Fan X, Wang Y, Kusstatscher P, Duan J, Wu S, et al. Bacterial seed endophyte shapes disease resistance in rice. Nat Plants. 2021;7:60–72. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Ma Y, Wang X, Liao K, He S, Zhao X, et al. Dynamics of rice microbiomes reveal core vertically transmitted seed endophytes. Microbiome. 2022;10:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sulesky-Grieb A, Simonin M, Bintarti AF, Marolleau B, Barret M, Shade A. Stable, multigenerational transmission of the bean seed microbiome despite abiotic stress. mSystems. 2024;9:e00951-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiltz S, Gaillard I, Pawlicki-Jullian N, Thiombiano B, Mesnard F, Gontier E. A review: what is the spermosphere and how can it be studied? J Appl Microbiol. 2015;119:1467–81. [DOI] [PubMed] [Google Scholar]
- 16.Pitzschke A. Molecular dynamics in germinating, endophyte-colonized quinoa seeds. Plant Soil. 2018;422:135–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh CM, Becker-Uncapher I, Carlson M, Fierer N. Variable influences of soil and seed-associated bacterial communities on the assembly of seedling microbiomes. ISME J. 2021;15:2748–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barret M, Briand M, Bonneau S, Préveaux A, Valière S, Bouchez O, et al. Emergence shapes the structure of the seed microbiota. Appl Environ Microbiol. 2015;81:1257–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres-Cortés G, Bonneau S, Bouchez O, Genthon C, Briand M, Jacques MA, et al. Functional microbial features driving community assembly during seed germination and emergence. Front Plant Sci. 2018;9: 902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang G, Wang Y, Ji F, Xu L, Yu M, Shi J, et al. Biodegradation of deoxynivalenol and its derivatives by Devosia insulae A16. Food Chem. 2019;276:436–42. [DOI] [PubMed] [Google Scholar]
- 21.Xun W, Ren Y, Yan H, Ma A, Liu Z, Wang L, et al. Sustained inhibition of maize seed-borne Fusarium using a Bacillus-dominated rhizospheric stable core microbiota with unique cooperative patterns. Adv Sci. 2023;10:2205215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jack ALH, Nelson EB. A seed-recruited microbiome protects developing seedlings from disease by altering homing responses of Pythium aphanidermatum zoospores. Plant Soil. 2018;422:209–22. [Google Scholar]
- 23.Verma SK, Kingsley K, Irizarry I, Bergen M, Kharwar RN, White JF Jr. Seed-vectored endophytic bacteria modulate development of rice seedlings. J Appl Microbiol. 2017;122:1680–91. [DOI] [PubMed] [Google Scholar]
- 24.Yang M, Li Z, Liu L, Bo A, Zhang C, Li M. Ecological niche modeling of Astragalus membranaceus var. mongholicus medicinal plants in Inner Mongolia, China. Sci Rep. 2020;10:12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poppeliers SWM, Sánchez-Gil JJ, de Jonge R. Microbes to support plant health: understanding bioinoculant success in complex conditions. Curr Opin in Microbiol. 2023;73: 102286. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Yang Y, Wu T, Zhang H, Wei G, Li Z. Rhizosphere bacterial and fungal spatial distribution and network pattern of Astragalus mongholicus in representative planting sites differ the bulk soil. Appl Soil Ecol. 2021;168: 104114. [Google Scholar]
- 27.Li Z, Bai X, Jiao S, Li Y, Li P, Yang Y, et al. A simplified synthetic community rescues Astragalus mongholicus from root rot disease by activating plant-induced systemic resistance. Microbiome. 2021;9:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong H, Song G, Ryu CM. Inheritance of seed and rhizosphere microbial communities through plant–soil feedback and soil memory. Env Microbiol Rep. 2019;11:479–86. [DOI] [PubMed] [Google Scholar]
- 29.Olofintila OE, Noel ZA. Soybean and cotton spermosphere soil microbiome shows dominance of soilborne copiotrophs. Microbiol Spectr. 2023;11:e00377–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge M, Wei X. Spermosphere bacterial community at different germination stages of Ormosia henryi and its relationship with seed germination. Sci Hortic. 2024;324: 112608. [Google Scholar]
- 31.Wang C, Kuzyakov Y. Mechanisms and implications of bacterial–fungal competition for soil resources. ISME J. 2024;18:wrae073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyauchi S, Kiss E, Kuo A, Drula E, Kohler A, Sánchez-García M, et al. Large-scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits. Nat Commun. 2020;11:5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiotta ML, Alaniz Zanon MS, Palazzini JM, Scandiani MM, Formento AN, Barros GG, et al. Pathogenicity of Fusarium graminearum and F. meridionale on soybean pod blight and trichothecene accumulation. Plant Pathol. 2016;65:1492–7. [Google Scholar]
- 34.Khaledi N, Taheri P, Falahati Rastegar M. Identification, virulence factors characterization, pathogenicity and aggressiveness analysis of Fusarium spp., causing wheat head blight in Iran. Eur J Plant Pathol. 2017;147:897–18. [Google Scholar]
- 35.Yu J, Bai G, Zhou W, Dong Y, Kolb F. Quantitative trait loci for Fusarium head blight resistance in a recombinant inbred population of Wangshuibai/Wheaton. Phytopathology. 2008;98:87–94. [DOI] [PubMed] [Google Scholar]
- 36.Coleman JJ. The Fusarium solani species complex: ubiquitous pathogens of agricultural importance. Mol Plant Pathol. 2016;17:146–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tahmasebi A, Roach T, Shin SY, Lee CW. Fusarium solani infection disrupts metabolism during the germination of roselle (Hibiscus sabdariffa L.) seeds. Front Plant Sci. 2023;14:1225426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson EB. The seed microbiome: Origins, interactions, and impacts. Plant Soil. 2018;422:7–34. [Google Scholar]
- 39.Yang R, Shi Q, Huang T, Yan Y, Li S, Fang Y, et al. The natural pyrazolotriazine pseudoiodinine from Pseudomonas mosselii 923 inhibits plant bacterial and fungal pathogens. Nat Commun. 2023;14:734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu S, Liu YX, Cernava T, Wang H, Zhou Y, Xia T, et al. Fusarium fruiting body microbiome member Pantoea agglomerans inhibits fungal pathogenesis by targeting lipid rafts. Nat Microbiol. 2022;7:831–43. [DOI] [PubMed] [Google Scholar]
- 41.Zhou X, Wang J, Liu F, Liang J, Zhao P, Tsui CKM, et al. Cross-kingdom synthetic microbiota supports tomato suppression of Fusarium wilt disease. Nat Commun. 2022;13:7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Wang J, Yang N, Wen Z, Sun X, Chai Y, et al. Wheat microbiome bacteria can reduce virulence of a plant pathogenic fungus by altering histone acetylation. Nat Commun. 2018;9:3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A. The importance of the microbiome of the plant holobiont. New Phytol. 2015;206:1196–206. [DOI] [PubMed] [Google Scholar]
- 44.Bakker PAHM, Pieterse CMJ, de Jonge R, Berendsen RL. The Soil-Borne Legacy. Cell. 2018;172:1178–80. [DOI] [PubMed] [Google Scholar]
- 45.Gao M, Xiong C, Gao C, Tsui CKM, Wang MM, Zhou X, et al. Disease-induced changes in plant microbiome assembly and functional adaptation. Microbiome. 2021;9:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia L, Miao Y, Cao A, Liu Y, Liu Z, Sun X, Xue Y, Xu Z, Xun W, Shen Q, et al. Biosynthetic gene cluster profiling predicts the positive association between antagonism and phylogeny in Bacillus. Nat Commun. 2022;13:1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrión VJ, Perez-Jaramillo J, Cordovez V, Tracanna V, de Hollander M, Ruiz-Buck D, et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science. 2019;366:606–12. [DOI] [PubMed] [Google Scholar]
- 48.Mendes LW, Raaijmakers JM, de Hollander M, Mendes R, Tsai SM. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 2018;12:212–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Xu Z, Chen L, Xun W, Shu X, Chen Y, et al. Root colonization by beneficial rhizobacteria. FEMS Microbiol Rev. 2023;48:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Artzi L, Bayer E, Moraïs S. Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat Rev Microbiol. 2017;15:83–95. [DOI] [PubMed] [Google Scholar]
- 51.Tao J, Chen Q, Chen S, Lu P, Chen Y, Jin J, Li XuY, He W, Long T, et al. Metagenomic insight into the microbial degradation of organic compounds in fermented plant leaves. Environ Res. 2022;214:113902. [DOI] [PubMed] [Google Scholar]
- 52.Jain D, Ravina, Bhojiya AA, et al. Polyphasic characterization of plant growth promoting cellulose degrading bacteria isolated from organic manures. Curr Microbiol. 2021;78:739–48. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Lei L, Zheng L, et al. Genome sequencing of gut symbiotic Bacillus velezensis LC1 for bioethanol production from bamboo shoots. Biotechnol Biofuels. 2020;13:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janská A, Pecková E, Sczepaniak B, Smýkal P, Soukup A. The role of the testa during the establishment of physical dormancy in the pea seed. Ann Bot. 2018;123:815–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun L, Miao Z, Cai C, Zhang D, Zhao M, Wu Y, Zhang X, Swarm SA, Zhou L, Zhang ZJ, et al. GmHs1-1, encoding a calcineurin-like protein, controls hard-seededness in soybean. Nat Genet. 2015;47:939–43. [DOI] [PubMed] [Google Scholar]
- 56.Kim H, Lee KK, Jeon J, Harris WA, Lee Y-H. Domestication of Oryza species eco-evolutionarily shapes bacterial and fungal communities in rice seed. Microbiome. 2020;8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuang J, Han S, Chen Y, et al. Root-associated fungal community reflects host spatial co-occurrence patterns in a subtropical forest. ISME Commun. 2021;1:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abarenkov K, Nilsson RH, Larsson K-H, Taylor Andy FS, May Tom W, Frøslev TG, et al. The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: sequences, taxa and classifications reconsidered. Nucleic Acids Res. 2023;52:791–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li D, Luo R, Liu C, Leung C, Ting H, Sadakane K, et al. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. [DOI] [PubMed] [Google Scholar]
- 62.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. [DOI] [PubMed] [Google Scholar]
- 63.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. [DOI] [PubMed] [Google Scholar]
- 65.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2018;47:309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:445–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gibson MK, Forsberg KJ, Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 2015;9:207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan X, Hong S, Xiong W, Raza W, Shen Z, Wang B, et al. Development of fungal-mediated soil suppressiveness against Fusarium wilt disease via plant residue manipulation. Microbiome. 2021;9:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–30. [Google Scholar]
- 72.Kembel S, Cowan P, Helmus M, Cornwell W, Morlon H, Ackerly D, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–4. [DOI] [PubMed] [Google Scholar]
- 73.Robinson M, McCarthy D, Smyth G. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu C, Cui Y, Li X, Yao M. microeco: an R package for data mining in microbial community ecology. FEMS Microbiol Ecol. 2020;97:255. [DOI] [PubMed] [Google Scholar]
- 75.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu S, Dai Z, Guo P, Fu X, Liu S, Zhou L, et al. ggtreeExtra: Compact visualization of richly annotated phylogenetic data. Mol Biol Evol. 2021;38:4039–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sloan WT, Woodcock S, Lunn M, Head IM, Curtis TP. Modeling taxa-abundance distributions in microbial communities using environmental sequence data. Microb Ecol. 2007;53:443–55. [DOI] [PubMed] [Google Scholar]
- 78.Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, et al. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2016;10:655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stegen JC, Lin X, Konopka AE, Fredrickson JK. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012;6:1653–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Toju H, Peay KG, Yamamichi M, Narisawa K, Hiruma K, Naito K, et al. Core microbiomes for sustainable agroecosystems. Nat Plants. 2018;4:247–57. [DOI] [PubMed] [Google Scholar]
- 81.Hu Q, Tan L, Gu S, Xiao Y, Xiong X, Zeng W, et al. Network analysis infers the wilt pathogen invasion associated with non-detrimental bacteria. npj Biofilms and Microbi. 2020;6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA018766, CRA018767, CRA018768) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. The data visualization code can be found in https://github.com/lidas666/seed-microbiome.