Abstract
Nucleophosmin/B23 is a nucleolar phosphoprotein. It has been shown that B23 binds to nucleic acids, digests RNA, and is localized in nucleolar granular components from which preribosomal particles are transported to cytoplasm. The intracellular localization of B23 is significantly changed during the cell cycle. Here, we have examined the cellular localization of B23 proteins and the effect of mitotic phosphorylation of B23.1 on its RNA binding activity. Two splicing variants of B23 proteins, termed B23.1 and B23.2, were complexed both in vivo and in vitro. The RNA binding activity of B23.1 was impaired by hetero-oligomer formation with B23.2. Both subtypes of B23 proteins were phosphorylated during mitosis by cyclin B/cdc2. The RNA binding activity of B23.1 was repressed through cyclin B/cdc2-mediated phosphorylation at specific sites in B23. Thus, the RNA binding activity of B23.1 is stringently modulated by its phosphorylation and subtype association. Interphase B23.1 was mainly localized in nucleoli, whereas B23.2 and mitotic B23.1, those of which were incapable of binding to RNA, were dispersed throughout the nucleoplasm and cytoplasm, respectively. These results suggest that nucleolar localization of B23.1 is mediated by its ability to associate with RNA.
INTRODUCTION
The cell nucleolus is the place for ribosome biogenesis, which is the synthesis and processing of a precursor rRNA (pre-rRNA), and the assembly of ribosomal proteins on rRNAs to form premature ribosome. rRNA is first transcribed by RNA polymerase I as a pre-rRNA of ∼13,500 nucleotides. To produce mature rRNAs (18S, 5.8S, and 28S rRNA), external and internal spacer sequences (ETS and ITS, respectively) in the long pre-rRNA are removed sequentially while nucleotide modifications such as pseudouridylation and 2′-O-methylation occur concurrently (Maden, 1990). For such complex processes, it is suggested that nonribosomal proteins and small nucleolar RNAs are multifunctional (Srivastava and Pollard, 1999; Peculis, 2000). One such nucleolar protein is the well-characterized and abundant nucleophosmin/B23. It is highly conserved in vertebrates. In rat and human cells, at least two isoforms of nucleophosmin/B23, termed B23.1 and B23.2, are expressed (Chang and Olson, 1990; Okuwaki et al., 2001a). These two proteins are identical except that the C-terminal 35 amino acids observed in B23.1 are absent in B23.2. Nucleophosmin/B23 functions to bind nucleic acids (Dumbar et al., 1989), to endonucleolytically cleave RNA preferentially within ITS2 in pre-rRNA (Savkur and Olson, 1998), to suppress the aggregation of proteins (Szebeni and Olson, 1999), and to bind peptides containing nuclear localization signals for their nuclear import (Szebeni et al., 1995). Recently, phosphorylation of nucleophosmin/B23 by cyclin E/cdk2, a G1 cyclin-dependent kinase, was shown to be essential for centrosome duplication in fibroblast cells (Okuda et al., 2000). Furthermore, it is shown that B23 protein binds a wide diversity of proteins, such as nucleolar phosphoproteins, nucleolin and p120 (Durban et al., 1995; Li et al., 1996), HIV1-Rev protein (Fankhauser et al., 1991), retinoblastoma protein (Takemura et al., 1999) to stimulate the DNA polymerase α activity (Umekawa et al., 2001), and hepatitis delta virus delta antigens (Huang et al., 2001). Because the B23-nucleic acid binding and RNase activity domains are mapped in the C-terminal region present only in B23.1 (Hingorani et al., 2000), the C-terminal region of B23.1 is likely to be important for ribosome biogenesis. The function of B23.1 has been well studied and characterized, whereas that of B23.2 is not well understood.
Eukaryotic gene expression is regulated through the cell cycle, partly by the phosphorylation and dephosphorylation of proteins involved in this process (Gottesfeld and Forbes, 1997). In particular, transcription is significantly repressed during mitosis. This also applies to the transcription system mediated by RNA polymerase I (pol I). Several factors involved in pol I transcription are found phosphorylated and inactivated during mitosis. The promoter selectivity factor SL1 is phosphorylated during mitosis, which inactivates it for transcription from the pol I promoter (Heix et al., 1998). During mitosis, not only transcription machinery but also factors involved in mRNA biogenesis (Colgan et al., 1998) and in chromatin remodeling (Sif et al., 1998) are phosphorylated and inactivated. On the contrary, the activity of several proteins is stimulated by phosphorylation during mitosis. For example, a component of the chromosome condensation machinery, condensin, becomes activated through phosphorylation by cdc2 kinase during mitosis (Kimura et al., 1998). Nucleophosmin/B23 is also phosphorylated by cyclin B/cdc2 during mitosis (Peter et al., 1990), thereafter changing its cellular localization drastically. The localization of nucleophosmin/B23 in mitotic cells can be found in three locations; throughout cytoplasm, on the mitotic apparatus, or at nucleolus-derived foci (NDF) (Zatsepina et al., 1997, 1999; Dundr et al., 2000). At NDF, B23 remains associated with pre-rRNA and rRNA processing factors such as nucleolin, fibrillalin, and snoRNP (Dundr and Olson, 1998; Pinol-Roma, 1999). However, it is not known if phosphorylation of B23 proteins during mitosis has any effect on its function in ribosome biogenesis.
We previously identified nucleophosmin/B23 to be a major component of template activating factor (TAF)-III (Okuwaki et al., 2001a). TAF-III can stimulate cell-free replication from the adenovirus genome complexed with viral basic core proteins, thus forming a chromatin-like structure. Because nucleophosmin/B23 binds directly to core histones and mediates assembly of nucleosome in vitro, it is suggested that nucleophosmin/B23 functions as a histone chaperone (Okuwaki et al., 2001b). Here, we have shown that the B23 subtypes were complexed with each other in vivo and in vitro, and that the RNA binding activity of B23.1 was modulated through its complex formation with B23.2. Green fluorescent protein (GFP)-tagged B23.2 was dispersed throughout nucleoplasm, whereas a major fraction of GFP-tagged B23.1 was concentrated in nucleoli as endogenous B23.1. In addition, B23.2 was extracted with detergent, whereas B23.1 remained tightly associated with nuclear structure in the presence of detergent. These in vitro and in vivo properties of both subtypes of B23 proteins suggest that B23.2 could modulate the ribosome biogenesis function of B23.1 as well as its intracellular localization. Furthermore, the RNA binding activity of B23.1 was completely disrupted by cyclin B/cdc2-mediated phosphorylation. A mutant B23.1 in which four threonines for phosphorylation were substituted by alanines was not phosphorylated by cyclin B/cdc2, and remained associated with RNA after phosphorylation reaction in the presence of cyclin B/cdc2. This result strongly suggests that the RNA binding activity of B23.1 is regulated partly by phosphorylation and dephosphorylation. Thus, it is proposed that at least two mechanisms, hetero-oligomerization with a modulator protein (e.g., B23.2) and phosphorylation/de-phosphorylation of B23.1, contribute to regulating the involvement of B23 in ribosome biogenesis.
MATERIALS AND METHODS
Plasmid Construction
For expression of N-terminal HA-epitope tag–fused hB23.1 in HeLa cells, the pCHA vector (Nagata et al., 1998), which is driven by the cytomegalovirus immediate early gene enhancer, and chicken β-actin promoter was used. pCHA-hB23.1 was prepared as described (Okuwaki et al., 2001a). To generate pCHA-hB23.2, a fragment of hB23.2 cDNA was obtained by digestion of pET14b-hB23.2 (Okuwaki et al., 2001a) with NdeI and BamHI. The fragment was then cloned in-frame into BstEII- and BglII-digested pCHA. To generate mutant B23.1 proteins, an appropriate set of oligonucleotide primers were used for site-directed amino acid substitutions (threonine to alanine). To generate a T199A expression vector, cDNA fragments containing a mutation at T199 were amplified by polymerase chain reaction (PCR) using the N-terminal primer for B23.1 and 5′-tttggctggagcatctcgtat-3′, or C-terminal primer for B23.1 and 5′-atacgagatgctccagccaaa-3′, with pET14b-hB23.1 as a template. Two kinds of amplified cDNA fragments were purified and used as templates for the second PCR to amplify a full-length cDNA fragment for T199A using N- and C-terminal primers of B23.1. The same protocol was applied to construct T219A and T234/237A expression vectors. Primer sets for T219A and T234/237A were 5′-ccatcatcagcaccaagatca-3′ and 5′-tgatcttggtgctgatgatgg-3′, and 5′-caggaaaaagctcctaaagcaccaaaagga-3′ and 5′-tccttttggtgctttaggagctttttcctg-3′, respectively. The mutated cDNA fragments were then digested with NdeI and BamHI, and then cloned into NdeI- and BamHI-digested pET 14b (Novagen, Madison, WI). Triple mutant proteins T199/234/237A and T219/234/237A were prepared by PCR using the appropriate primer sets described above and pET14b-T234/237A as a template. Recombinant proteins were expressed and purified as described by Okuwaki et al. (2001a). For expression of Flag-tagged wild-type and mutant B23.1 proteins in HeLa cells, each cDNA fragment digested with NdeI and BamHI was subcloned into NdeI- and BamHI-digested pBS-Falg vector to generate pBS-Flag-B23.1, -T199A, -T219/23 4/237A, and -T4A. Each cDNA attached by the Flag-tag at its 5′ terminus was cut out from pBS-Flag vector by digestion with BamHI, and then subcloned into BglII-digested pCAGGS vector. To express GFP-tagged B23.1 or B23.2, cDNA fragments attached by the Flag-tag were cut out from pBS-Flag vector by BamHI, and then subcloned into BamHI-digested pEGFPC1 vector (CLONTECH, Palo Alto, CA). Transient transfection of each plasmid to HeLa cells was performed by the calcium phosphate precipitation method (Graham and Eb, 1973).
Immunoprecipitation Analysis
HeLa cells expressing HA-tagged hB23.1 or hB23.2 were lysed in buffer A (50 mM Tris, pH 7.9, 0.1 mM EDTA, and 0.1% Nonidet P40) containing 150 mM NaCl on ice for 10 min and were then disrupted by extensive sonication. Cell extracts recovered by centrifugation were mixed with anti-HA antibody (3F10; Roche, Indianapolis, IN) and incubated on ice for 30 min. Then, protein A Sepharose beads (5 μl of resin; Amersham Pharmacia, Piscataway, NJ) were added and further incubated for 2 h with gentle agitation. Proteins bound to resins were eluted by an SDS sample buffer, boiled, separated on a 10% SDS-PAGE, and transferred to a polyvinylidene difluoride (PVDF) membrane. Endogenous B23.1 was detected by western blotting with anti-B23.1 antibody (C19; Santa Cruz Biotechnology, Santa Cruz, CA).
In Vitro Phosphorylation of B23 Proteins by Cyclin-dependent Kinases
To prepare cyclin E/cdk2, NIH3T3 cells were first grown in DMEM supplemented with 0.1% calf serum for 48 h. The medium was changed to DMEM containing 10% calf serum, and cells were further incubated for 14 h. Cells were lysed in buffer A containing 150 mM NaCl, 10 mM β-d-glycerophosphate, 1 mM NaF, 1 mM NaVO 4, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and disrupted by extensive sonication. Cell extracts (200 μg) recovered by centrifugation were mixed with anti-cyclin E antibody (M20; Santa Cruz Biotechnolgy). The cyclin E-associated kinase bound to the antibody was purified using protein A Sepharose beads. The beads were washed extensively with buffer A containing 150 mM NaCl, and were suspended in a kinase reaction buffer (20 mM Tris, pH 7.9, 10 mM MgCl 2, 10 mM β-d-glycerophosphate, 1 mM NaF, 1 mM NaVO 4, and 0.5 mM PMSF). For preparation of cyclin B/cdc2, HeLa cells were arrested at prometaphase by incubation with nocodazole, a microtubule destabilizing reagent, at a final concentration of 50 ng/ml for 12 h. Mitotic cells were collected by shaking dishes gently. Cell extracts were prepared as described (Pinol-Roma, 1999). In standard experiments, cyclin B/cdc2 kinase was purified from 500 μg of mitotic extracts using anti-cyclin B antibody (GNS1; Santa Cruz Biotechnology, 500 ng). The cyclin B/cdc2-immobilized protein A beads were then suspended in 100 μl of buffer H (20 mM HEPES-NaOH, 0.5 mM EDTA, 50 mM NaCl, 10% glycerol, and 0.5 mM PMSF) and used as a kinase. The phosphorylation reaction was performed as follows: Recombinant B23 proteins (5 μg) were mixed with 20 μl of a kinase fraction in the presence of 1 mM ATP in 20 mM Tris, pH 7.4, 10 mM MgCl 2, 50 mM NaCl, 5 mM β-d-glycerophosphate, and 0.5 mM PMSF. The mixture was incubated at 30°C for 30 min. Under these reaction conditions, 5 μg of B23.1 was completely phosphorylated. Phosphorylated B23.1 was separable from nonphosphorylated protein by SDS-PAGE (see Figure 7A). Note that for results shown in reactions of Figures 6, C through E, and 8B, ATP concentration was adjusted to 1 μM, and 1 μCi of [γ-32P]ATP (Amersham Pharmacia) was supplemented.
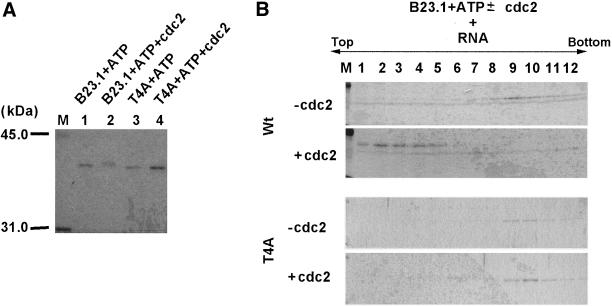
Figure 7.
Inactivation of the RNA binding activity of B23.1 by cyclin B/cdc2-mediated phosphorylation. (A) Phosphorylation of rB23.1 proteins by purified cyclin B/cdc2 kinase. rB23.1 (lanes 1 and 2) or T4A mutant (lanes 3 and 4) proteins (5 μg each) were incubated in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of purified cyclin B/cdc2 kinase. A part of the protein solution (200 ng) was separated on a 10% SDS-PAGE and stained with Coomassie Brilliant Blue (CBB). Lane M indicates molecular weight markers. (B) RNA binding activity of phosphorylated B23.1. Wild-type (upper two panels) or T4A mutant B23.1 (bottom two panels) proteins phosphorylated in the absence (first and third panels) or presence (second and fourth panels) of cyclin B/cdc2 kinase were examined for the RNA binding activity by a sucrose gradient sedimentation analysis as in Figure 4. Proteins in each fraction collected from the top were separated on a 10% SDS-PAGE and stained with CBB.
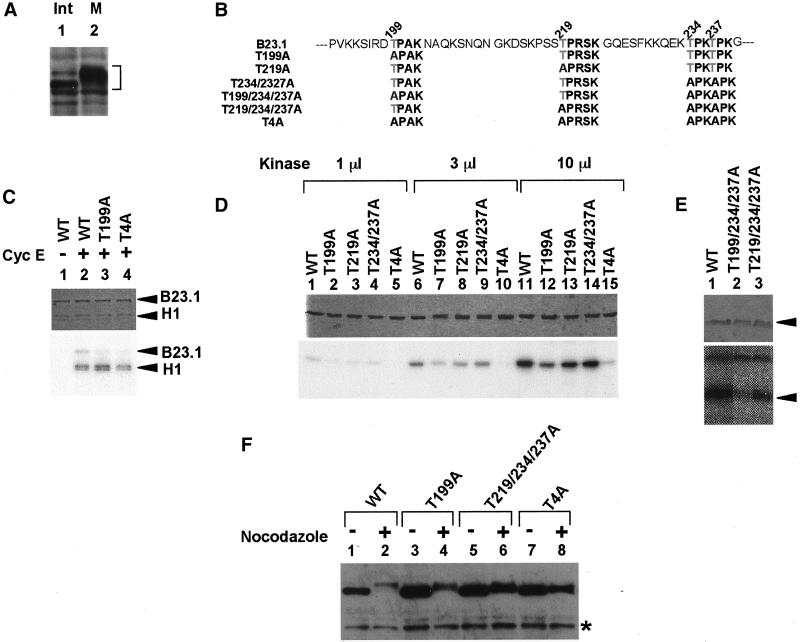
Figure 6.
Phosphorylation sites of B23.1 by cyclin B/cdc2. (A) Hyperphosphorylation of B23.1 during mitosis. Cell extracts (20 μg) prepared from interphase (lane 1) or prometaphase (lane 2) HeLa cells were separated on a 10% SDS-PAGE, and B23.1 was detected by western blotting with anti-B23.1 antibody (C19). Positions of B23.1 are indicated with a bracket at the right side of the panel. (B) Schematic representation of mutant proteins used in this study. Amino acids are indicated with single letter abbreviations. Threonine residues (T) replaced by alanine (A) are indicated by gray letters. (C) Phosphorylation of B23.1 by cyclin E/cdk2. Cyclin E/cdk2 was purified from NIH3T3 cells without (lane 1) or with (lanes 2–4) anticyclin E antibody (M20). Wild-type B23.1 (lanes 1 and 2), T199A (lane 3), and T4A (lane 4) (200 ng) were phosphorylated by purified cyclin E/cdk2 in the presence of histone H1 (100 ng) and [γ-32P]ATP at 30°C for 30 min. (D) In vitro phosphorylation of wild-type and mutant B23.1 proteins by cyclin B/cdc2 kinase. Cyclin B/cdc2 was purified from mitotic HeLa cell extracts. B23.1 and point mutant proteins (200 ng) as indicated above each lane were phosphorylated by 1 (lanes 1–5), 3 (lanes 6–10), and 10 (lanes 11–15) μl of cyclin B/cdc2 kinase (see “Materials and Methods”) in the presence of [γ-32P]ATP at 30°C for 30 min. (E) Threonine residues phosphorylated by cyclin B/cdc2. Wild-type (WT, lane 1) and mutant B23.1 (T199/234/237A and T219/234/237A, lanes 2 and 3, respectively) were phosphorylated by cyclin B/cdc2 and separated on a 10% SDS-PAGE. In C through E, proteins were stained with CBB, and phosphorylated proteins were detected by autoradiography (upper and bottom panels, respectively). (F) Phosphorylation of Flag-tagged wild-type and mutant B23.1 proteins in vivo. Cell extracts (30 μg) prepared from exponentially growing (lanes 1, 3, 5, and 7) or nocodazole-arrested (lanes 2, 4, 6, and 8) HeLa cells expressing either Flag-tagged B23.1 (lanes 1 and 2), T199A (lanes 3 and 4), T219/234/237A (lanes 5 and 6), or T4A (lanes 7 and 8) were separated on a 8% SDS-PAGE and subjected to western blotting with anti-Flag antibody. Protein bands shown by an asterisk are nonspecific background bands that reacted with the secondary antibody (biotinylated anti-mouse IgG).
Cell Fractionation Experiments
Cell fractionation experiments were performed essentially as described (He et al., 1990). HeLa cells grown in Eagle's minimum essential medium supplemented with 10% fetal bovine serum were washed with ice-cold phosphate-buffered saline, and then lysed on ice for 5 min in cytoskeletal (CSK) buffer containing 0.5% Triton X-100. Then, supernatant was collected and the cell pellet was washed with the CSK buffer. The cell pellet that contained the chromatin DNA was digested by 2 units/μl RNase-free DNase I (Invitrogen, Carlsbad, CA) at 37°C for 15 min and was extracted with (NH 4)2SO 4 at a final concentration of 0.25 M in CSK buffer. The supernatant was collected while the pellet was further extracted on ice for 5 min with 2 M NaCl in CSK buffer. Remaining insoluble fractions were dissolved in 8 M urea. For Figure 5, a part of Triton X-100–extracted cell pellet was digested with 0.1 mg/ml RNase A instead of DNase I. Extracted proteins at each step were separated on a 10% SDS-PAGE and were subjected to western blotting using anti-B23 polyclonal (C19) or monoclonal (NB23) antibodies, antitopoisomerase IIα (8D2), and anticyclin B (GNS1) antibodies. To detect HA- and Flag-tagged B23 proteins, anti-HA (12CA5; Roche) and anti-Flag (M2; Sigma, St. Louis, MO) antibodies, respectively, were used. For experiments in Figure 8, HeLa cells were arrested at prometaphase in the presence of 50 ng/ml nocodazole for 12 h, and the mitotic cells were then collected. Mitotic cells were subjected to cell fractionation experiment as described above.
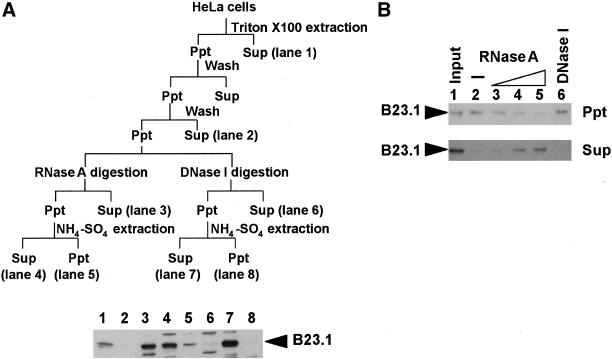
Figure 5.
B23.1 is released from nuclear structure by RNase treatment. (A) Cell fractionation experiment using RNase A or DNase I. The experimental scheme is shown in the upper panel. Exponentially growing HeLa cells were first treated with 0.5% Triton X-100 in CSK buffer and were then subjected to digestion with DNase I (3 unit/μl) or RNase A (200 μg/ml). Released proteins by nuclease treatment were recovered by centrifugation as supernatant fraction, and the cell pellet was further extracted with 0.25 M ammonium sulfate on ice for 5 min. Remaining insoluble proteins were dissolved in 8 M urea. Proteins in each fraction were separated on a 10% SDS-PAGE, and B23 proteins were detected by western blotting with anti-B23 antibody (NB23). (B) RNA-dependent nuclear retention of B23.1. Cells treated with Triton X-100 were digested with increasing amounts of RNase A (0, 2, 20, and 200 μg/ml for lanes 2–5, respectively) or DNase I (3 units/μl for lane 6) at 37°C for 20 min. Proteins were separated from insoluble materials by centrifugation, and the remaining pellet was dissolved in 8 M urea. Insoluble (Ppt, upper panel) and released (Sup, bottom panel) proteins were separated on a 10% SDS-PAGE, and B23 proteins were detected by western blotting. Insoluble proteins after treatment with Triton X-100 are shown in lane 1 (Input). Positions of B23.1 are indicated by arrowheads at the left sides of panels.
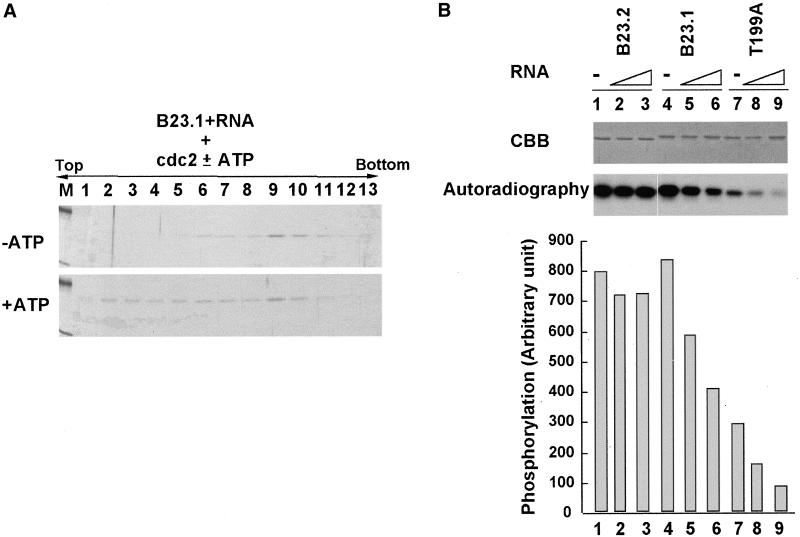
Figure 8.
Phosphorylation of B23.1-RNA complex in vitro. (A) Release of B23.1 from RNA by cyclin B/cdc2-mediated phosphorylation. rB23.1 (5 μg) preincubated with RNA (5 μg) was phosphorylated by cyclin B/cdc2 kinase in the absence (upper panel) or presence (bottom panel) of ATP, and was then subjected to sucrose gradient sedimentation analyses as shown in Figures 4 and 7. Proteins in each fraction were separated on a 10% SDS-PAGE and stained with CBB. (B) Inhibition of cyclin B/cdc2-mediated phosphorylation of B23.1 through interaction with RNA. B23.2 (lanes 1–3), B23.1 (lanes 4–6), and T199A (lanes 7–9) (200 ng each) were preincubated in the absence (lanes 1, 4, and 7) or presence of 50 ng (lanes 2, 5, and 8) or 200 ng (lanes 3, 6, and 9) of RNA purified from HeLa cells and were phosphorylated by cyclin B/cdc2 in the presence of [γ- 32P]ATP. After the phosphorylation reaction, proteins and phosphorylated proteins were separated on a 10% SDS-PAGE and stained with CBB (upper panel) or autoradiographed (bottom panel), respectively. The phosphorylation efficiency was quantified using a FUJIX BAS2000 image analyzer system and is shown in the bottom graph.
RNA Binding Assay
RNA was extracted from HeLa cells using guanidium thiocyanate. Recombinant B23 proteins (5 μg) were mixed with 5 μg of RNA, incubated at room temperature for 30 min, and then loaded onto a 15–40% sucrose gradient in 20 mM Tris, pH 7.4, 50 mM NaCl, 0.5 mM PMSF, and 1 mM dithiothreitol. Samples were centrifuged at 54,000 rpm for 2 h in a TLS55 rotor (Beckman Instruments, Fullerton, CA), and fractions (150 μl) were collected from the top. For analysis of phosphorylated proteins, 10 mM β-d-glycerophosphate, 1 mM NaF, and 0.1 mM NaVO 4 were added to inhibit the potentially contaminating phosphatase activity. Proteins in each fraction were separated by SDS-PAGE and were visualized by staining with CBB.
For nitrocellulose filter binding assay, RNA (5 μg) extracted from HeLa cells were end-labeled by polynucleotide kinase (Toyobo) in the presence of 30 μCi of [γ-32P]ATP (3000 Ci/mmol). To form B23 complexes in which the ratio of B23.1 to B23.2 was 1:0, 1:0.2, 1:0.5, 1:1, or 1:2, B23 proteins were mixed and subjected to the denature-renature protocol (Hager and Burgess, 1980). [32P]-labeled RNA (100 ng, specific activity was ∼500 cpm/ng) was incubated in the presence or absence of B23 complexes at 30°C for 30 min in 20 mM HEPES-NaOH, pH 7.9, 150 mM NaCl, and 0.5 mM EDTA. The RNA–protein complexes were filtrated through a nitrocellulose filter (Hybond N; Amersham Pharmacia). The radioactivity retained on the filter was measured by a FUJIX BAS 2000 image analyzing system.
Indirect Immunofluorescence Analysis
HeLa cells grown on coverslips were transiently transfected with pCAGGS-Flag B23.1 (5 μg) by the calcium phosphate precipitation method. Twenty-four hours after transfection, cells were fixed by ice-cold acetone-methanol (1:1) at −30° for 10 min, and Flag-tagged B23.1 was bound by the primary anti-Flag (M2) antibody and detected by a secondary antibody consisting of anti-mouse IgG conjugated with Alexa564 (Molecular Probes, Eugene, OR).
RESULTS
Homo- and Hetero-Oligomerization and Cellular Distribution of Splicing Variants of Nucleophosmin/B23
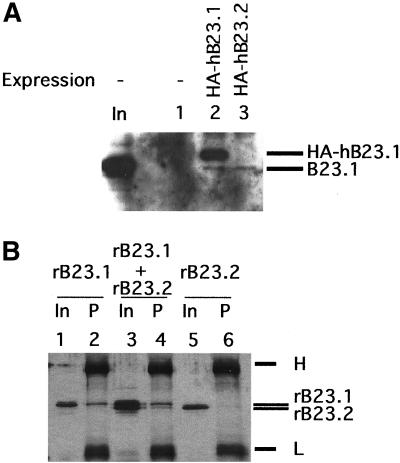
The C-terminal region specific to B23.1 is suggested to be involved in several important functions of B23 in ribosome biogenesis, whereras the function of B23.2, which lacks this C-terminal tail, is totally unknown. B23.1 and B23.2 cofractionated after five steps of column chromatography, including ion-exchange and gel filtration columns (Okuwaki et al., 2001a). In addition, several previous reports have suggested that the N-terminal portion of B23 protein is involved in oligomer formation (Yung and Chan, 1987; Herrera et al., 1996; Dutta et al., 2001). Therefore, B23.1 and B23.2 could be complexed in vivo. We first decided to examine this possibility by immunoprecipitation and by looking at cellular localization of both proteins. We used anti-HA antibody to see if endogenous B23.1 from cell extracts coimmunoprecipitates transiently expressed HA-tagged B23.1 or B23.2. Cell extracts prepared from HeLa cells expressing either HA-tagged B23.1 or B23.2 were prepared and subjected to immunoprecipitation analyses. Coimmunoprecipitation of endogenous B23.1 by anti-HA antibody was detected by western blotting with anti-B23.1 antibody (Figure 1A). Endogenous B23.1 was coimmunoprecipitated with both HA-tagged B23.1 and B23.2 (Figure 1A, lanes 2 and 3), indicating that both subtypes are complexed in vivo. To confirm that B23.1 and B23.2 directly interact with each other, recombinant B23.1 and B23.2 proteins were mixed and subjected to immunoprecipitation analysis using a B23.1-specific antibody. To reconstitute the complex, recombinant His-tagged B23.1 and B23.2 were mixed and codenatured/renatured before immunoprecipitation analysis. Recombinant B23.1 indeed was precipitated with anti-B23.1 antibody, whereas B23.2 alone was not (Figure 1B, compare lanes 2 and 6). On the other hand, when B23.2 was corenatured with B23.1, it was coimmunoprecipitated with B23.1 with the same anti-B23.1 antibody (Figure 1B, lane 4). From these results, we concluded that the B23 subtypes directly interact in vivo and in vitro. This is also consistent with a previous report that B23 forms an oligomer in solution (Yung and Chan, 1987).
Figure 1.
B23.1 and B23.2 are complexed in vivo and in vitro. (A) Immunoprecipitation of HA-tagged B23 proteins from cell extracts. Cell extracts prepared from HeLa cells transfected with pCHA (lane 1), pCHA-hB23.1 (lane 2), or pCHA-hB23.2 (lane 3) were subjected to immunoprecipitation analysis using anti-HA (3F10) antibody. Proteins associated with anti-HA were separated on a 10% SDS-PAGE, transferred to a PVDF membrane, and endogenous and HA-tagged B23.1 proteins were detected with anti-B23.1 antibody (C19). Positions of HA-tagged and endogenous B23.1 are indicated by arrowheads at the right side of the panel. (B) Direct interaction between B23.1 and B23.2. Either recombinant (r) B23.1 alone (2 μg) (lanes 1 and 2), rB23.2 alone (2 μg) (lanes 5 and 6), and a mixture of rB23.1 and rB23.2 (2 μg each) (lanes 3 and 4) were subjected to the denature-renature protocol (Hager and Burgess, 1980). Renatured proteins were immunoprecipitated with the anti-B23.1 (C19) antibody that specifically recognizes the C-terminal region of B23.1. Immunoprecipitated proteins (“P,”, lanes 2, 4, and 6) were separated on a 10% SDS-PAGE and visualized via silver staining. Positions of rB23.1 and rB23.2 are indicated by arrowheads at the right side of the panel. “In” indicates 10% of input proteins (lanes 1, 3, and 5). Protein bands indicated by “H” and “L” shown at the right side of the panel correspond to heavy and light chains of immunoglobulin, respectively.
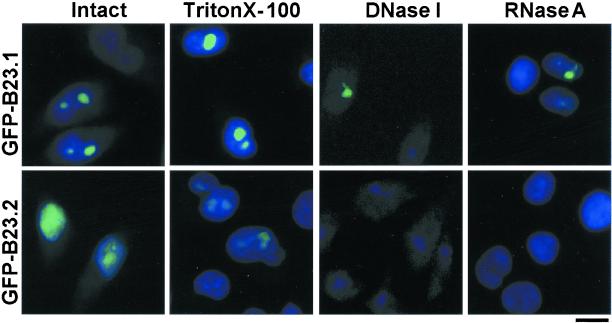
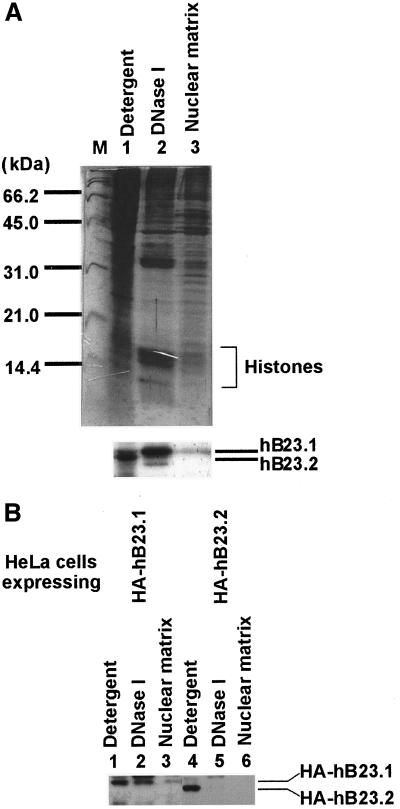
Next, the subcellular localization of these proteins was examined. B23 proteins fused with GFP-tag at their N termini were expressed in HeLa cells. The localization of GFP-tagged proteins was visualized under a fluorescent microscope (Figure 2). GFP-tagged B23.1 was mainly localized in nucleoli in HeLa cells. This localization pattern is identical to that of endogenous B23.1 (unpublished results). In contrast, GFP-tagged B23.2 was localized not only in nucleoli, but was also dispersed throughout the nucleoplasm. These observations support the previous reports that the C-terminal region specific to B23.1 plays an important role in nucleolar localization (Peculis and Gall, 1992; Wang et al., 1993; Zirwes et al., 1997a). Next, cells on coverslips were sequentially extracted in situ as follows. Cells on coverslips were exposed to detergent (0.5% Triton X-100, Figure 2, second column), and then digested with DNase I followed by extraction with 0.25 M ammonium sulfate (Figure 2, third column) or RNase A (Figure 2, fourth column) followed by extraction with 0.25 M ammonium sulfate. The majority of B23.1 was resistant to detergent extraction and a fraction of B23.1 was detected in nuclei even after treatment with DNase I or RNase A. Thus, it is possible that a fraction of B23.1 is associated with protein(s) that remains in nuclease-treated nuclei. On this line, it is reported that B23.1 is a nuclear matrix–associated protein (Feuerstein and Mond, 1987). On the other hand, nucleoplasmic GFP-tagged B23.2 was released by detergent extraction, whereas a trace amount of B23.2 remained in nucleoli (Figure 2, second column in bottom panels). GFP-tagged B23.2 was not qualitatively detected after nuclease treatment (Figure 2, third and fourth columns in bottom panels). To further examine these properties of B23 subtypes, protein composition in the fractions collected after each treatment was analyzed with SDS-PAGE followed by western blotting (Figure 3). Core histones (major chromatin proteins) fractionated mainly in the “DNase I” fraction (Figure 3A, upper panel), whereas TAF-I, a nuclear protein involved in chromatin assembly and remodeling (Nagata et al., 1995), was mainly detected in the “Detergent” fraction (unpublished results). The majority of B23.1 was recovered in both “DNase I” and “Detergent” fractions. A low but distinct amount of B23.1 was in the “Nuclear Matrix” fractions. In contrast, B23.2 mainly fractionated in the “Detergent” fraction, whereas only a part of B23.2 was detected in the “DNase I” fraction. To further confirm these B23 protein properties, HA-tagged B23.1 or B23.2 were expressed in HeLa cells and the cells were then subjected to fractionation as described above. HA-tagged B23.1 was recovered mainly in the “Detergent” and “DNase I” fractions as endogenous protein, whereas HA-tagged B23.2 is mainly in the “Detergent” fraction (Figure 3B). Figures 2 and 3 suggest that B23.1, but not B23.2, is a nuclear matrix–associated protein, and B23.2 is less tightly associated with nuclear structures such as chromatin and nuclear matrix relative to B23.1.
Figure 2.
Cellular localization of B23 splicing variants. HeLa cells grown on coverslips were transfected with pEGFPC1-hB23.1 (upper panels) or -hB23.2 (bottom panels). Thirty hours after transfection, cells on coverslips were fixed with 3% paraformaldehyde (first column) or were treated with 0.5% Triton X-10 0 followed by fixation with 3% paraformaldehyde (second column). Triton X-100–treated cells were further digested with DNase I (third column) or RNase A (fourth column) followed by extraction with 0.25 M ammonium sulfate before fixation with paraformaldehyde. DNA stained with Hoechst 33258 (blue) and GFP-tagged proteins (green) were visualized under a fluorescent microscope. The bar at the right bottom indicates 10 μm.
Figure 3.
Cell fractionation and diverse distribution of B23 subtypes. (A) Cell fractionation experiments. Exponentially growing HeLa cells were fractionated sequentially as described in the “Materials and Methods.” Proteins in “Detergent,” “DNase I,” and “nuclear matrix” (lanes 1–3, respectively) fractions (1 × 105 cells) were separated on a 12.5% SDS-PAGE and visualized with CBB staining (upper panel) or transferred to a PVDF membrane and subjected to western blotting with anti-B23 (NB23 that recognizes both subtypes of B23 proteins) (bottom panel). Positions of the proteins are indicated on the right side of the panels. Lane M in the upper panel shows standard molecular weight markers. Positions of core histones are also indicated with a bracket. (B) Fractionation of HeLa cells expressing HA-tagged B23 proteins. HeLa cells were transiently transfected with 500 ng of pCHA-hB23.1 (lanes 1–3) or -hB23.2 (lanes 4–6) and were incubated at 37°C for 30 h. Cell fractionation experiments were carried out as described in A. Positions of HA-tagged B23.1 and B23.2 are indicated at the right side of the panel.
RNA Binding Activity of B23.1 Is Modulated by Hetero-Oligomer Formation with B23.2
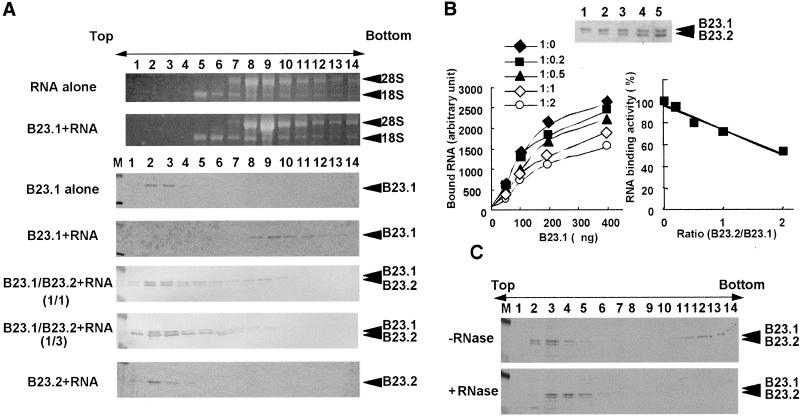
The observations that both subtypes of B23 proteins are directly complexed in vivo and that their localization patterns are different from each other led us to hypothesize that hetero-oligomer formation of B23.1 and B23.2 may impair the C-terminal–specific function of B23.1 such as nucleic acid binding and RNase function. To address this, the RNA binding activity of B23 proteins was examined in a sucrose gradient sedimentation assay. B23 complex was mixed with total RNA purified from HeLa cells, incubated, and then loaded onto the sucrose gradient. B23.1 or RNA alone was recovered in relatively low and high density fractions (Figure 4A, lanes 2–4 or lanes 8–10), respectively. B23.2 alone was also recovered in the low density fractions (unpublished results). In sharp contrast, B23.1 preincubated with RNA was sedimented in the high density fractions (Figure 4A, lanes 8–11) and cofractionated with RNA containing 18S and 28S rRNA (Figure 4A, fourth panel), whereas B23.2 was detected in low density fractions even after mixing with RNA (Figure 4A, seventh panel). This is consistent with the previous observation that rat B23.1 but not B23.2 binds nucleic acids (Wang et al., 1994). Although the peak position of 28S rRNA is slightly shifted from fraction 8 to fraction 9 in the presence of B23.1, we cannot conclude that B23.1 binds directly to 28S rRNA or which RNA is a target of B23.1 at present. To evaluate the effect of B23.2 on the RNA binding activity of B23.1, both proteins were mixed at a ratio of 1:1 or 1:3 and then subjected to the denature-renature protocol to form hetero-oligomer as shown in Figure 1B. Renatured complexes were tested for the RNA binding activity as above. Interestingly, B23.1 complexed with B23.2 was recovered mainly in the low density fractions (Figure 4A, lanes 2–4), whereas only a part of B23.1 sedimented to the RNA containing fractions with a small amount of B23.2 (Figure 4A, lanes 8–10). When the ratio of B23.2 to B23.1 was increased in the complex, the RNA binding activity of B23.1 significantly decreased (Figure 4A, compare fourth, fifth, and sixth panels), suggesting that B23.2 impairs the RNA binding activity of B23.1. To quantitatively estimate the effect of B23.2 on the RNA binding activity of the B23 complex, nitrocellulose filter binding assays were carried out. Total RNA prepared from HeLa cells was end-labeled and used for the filter binding assay. B23.1 were mixed with B23.2 at a ratio (B23.1:B23.2) of 1:0, 1:0.2, 1:0.5, 1:1, or 1:2, and then subjected to the denature-renature protocol (Figure 4B, upper panel). Renatured B23 complexes were incubated with the end-labeled RNA, followed by filtration through a nitrocellulose filter. Radioactive RNA retained on the nitrocellulose filter was quantified (Figure 4B, left plot). RNA alone passed through the filter and the radioactive RNA was not retained. Homo-oligomer of B23.1 efficiently bound to RNA in a B23.1 dose-dependent manner, whereas B23.2 homo-oligomer did not (Figure 4B and unpublished results). This is consistent with the results of sucrose gradient sedimentation analyses. The RNA binding ability of the B23 complex was decreased with the increasing ratio of B23.2 to B23.1 in the complex (Figure 4B, left and right plots). The amount of B23.1 recovered in high density fractions (Figure 4A, fourth panel, lanes 8–11) was significantly decreased with increasing amounts of B23.2 (Figure 4A, fifth and sixth panels) under a sucrose gradient assay condition, whereas under the filter binding assay condition, the RNA binding activity of the 1:2-ratio–B23.1:B23.2 complexes was reduced ∼50%. This difference of the effect of B23.2 on the RNA binding activity of B23 complex could be explained by observing that the B23-RNA complexes recovered in middle density fractions (Figure 4A, fifth and sixth panels, lanes 4–7) were collected on filters.
Figure 4.
The RNA binding activity of B23.1 complexed with B23.2. (A) Sucrose gradient analyses of the B23–RNA complex. rB23.1 (5 μg), rB23.2 (5 μg), and a 1:1 ratio mixture (5 μg each), and a 1:3 mixture (5 μg and 15 μg, respectively) of B23.1 and B23.2 were subjected to the denature-renature protocol (Hager and Burgess, 1980). Renatured proteins were incubated in the absence or presence of RNA purified from HeLa cells as indicated at the left side of the panels and were loaded on a 15–40% sucrose gradient. Proteins in fractions collected from the top were separated on a 10% SDS-PAGE and were visualized by staining with CBB (third through seventh panels). RNA was extracted from each fraction and was separated on a 1% agarose gel containing 6.3% formaldehyde in MOPS buffer [3-(N-morpholino)-propanesulfonic acid] and was visualized by staining with ethidium bromide. (Top and second panels) Positions of 18S and 28S rRNA, and rB23.1 and rB23.2 are indicated at the right side of the panels. Lane M indicates molecular weight markers. (B) Filter binding assay. The B23.1 and B23.2 hetero-oligomers were reconstituted and subjected to nitrocellulose filter RNA binding assays. The reconstituted B23 complexes (B23:1:B23.2 = 1:0, 1:0.2, 1:0.5, 1:1, and 1:2 for lanes 1–5, respectively; each lane contains 100 ng of B23.1) were separated on a 10% SDS-PAGE and stained with CBB (upper panel). The B23 complexes (a ratio of B23.1 to B23.2 = 1:0 (♦), 1:0.2(▪), 1:0.5 (▴), 1:1 (⋄), or 1:2 (○)) containing 50, 100, 200, and 400 ng of B23.1 were mixed and incubated with [32P]-end-labeled RNA (100 ng), followed by filtration through nitrocellulose filters. The radioactivity of RNA retained on the filter was quantified by a FUJIX BAS2000 image analyzing system. The amount of RNA bound on the filter is plotted as a function of the amount of B23. 1 in the B23 complexes (left plot). The efficiency of the RNA binding activity of the B23 complexes as a function of the ratio of B23.2 to B23.1 is calculated and summarized at the right plot. The RNA binding activity of B23.1 homo-oligomer is represented as 100%. Shown are the average of a series of experiments where different amounts of B23.1 (50, 100, 200, and 400 ng) in the B23 complexes were used for filter binding assays. (C) Sucrose density gradient of cell extracts. Cell extracts prepared from HeLa cells as described by Manley et al. (1980) were subjected to sucrose gradient sedimentation assays. Cell extracts (100 μg) treated without (upper panel) or with (bottom panel) RNase A (0.2 mg/ml) were loaded on a 15–40% sucrose density gradient and centrifuged as in A. Fractions (150 μl) were collected from the top, and proteins in each fraction were analyzed by SDS-PAGE. B23 proteins were detected by western blotting with anti-B23 antibody (NB23). Positions of B23.1 and B23.2 are shown at the right side of the panels. Lane M shows molecular weight markers.
Next, to examine the relationship between oligomerization form and the RNA binding activity of B23 proteins in vivo, whole cell extracts prepared from HeLa cells were subjected to the sucrose gradient analysis (Figure 4C). Fractionation patterns of B23.1 and B23.2 were analyzed by western blotting using anti-B23 antibody, which recognizes both subtypes. B23.1 sedimented in low (Figure 4C, fractions 2–4) and high (Figure 4C, fractions 11–14) density fractions, whereas B23.2 mainly fractionated in low density fractions (Figure 4C, fractions 2–4). These fractionation patterns are quite similar to a mixed pattern of in vitro reconstituted B23–RNA complexes. When cell extracts were treated with RNase A before loading on a sucrose gradient, B23.1 in the high density fractions disappeared (Figure 4C, bottom panel). Thus, it is likely that B23.1 recovered in the high density fractions is associated with RNA. This observation supports the idea that B23.2 modulates the RNA binding activity of B23.1 in a cell. In addition, B23.1 present in cell extracts was recovered in higher density fractions than the in vitro reconstituted B23.1–RNA complex. Therefore, B23.1 would be integrated into large complex containing RNA and other proteins associated with RNA and/or B23 in cell extracts. From results in Figure 4, it is strongly suggested that the ratio of B23.2 to B23.1 in B23 oligomer is important in determining the RNA binding activity and that B23.2 modulates the RNA binding ability of B23.1 by hetero-oligomer formation.
The different intracellular localization and the RNA binding ability of each B23 subtype allowed us to hypothesize that nucleolar localization of B23.1 is partly mediated by its RNA binding activity. Because there are abundant rRNAs and small nucleolar RNAs in nucleoli, it is possible that B23.1 is recruited to nucleoli by association with these RNA molecules. To address this, exponentially growing HeLa cells were subjected to cell fractionation experiments (summarized in Figure 5A, upper panel). Cells were extracted by Triton X-100, followed by digestion with RNase A or DNase I. B23.1 was not released from nucleoli by DNase I digestion alone (Figure 5, lane 6). To release B23.1 from DNase I-digested nuclear structure, extraction of DNase I-treated nuclei with ammonium sulfate was required (Figure 5A, lane 7). This process was also applied for experiments in Figures 2 and 3. In contrast, B23.1 was released only by RNase A digestion of nuclei (Figure 5A, lane 3). The amounts of B23.1 increased proportionally to the increasing amounts of RNase A (Figure 5B, lanes 2–5), whereas a small but distinct level of B23.1 remained on nuclear structure after RNase A treatment (Figure 5A, lane 5 and 5B, lane 5). These observations support that nucleolar localization of B23.1 is partly mediated by its RNA binding. However, it should be noted that a trace but detectable amount of B23.1 remains in RNase A-treated nuclei (Figures 2A and 5A). This could be interpreted that a part of B23.1 is tightly associated with nuclear (nucleolar) structure through protein–protein interaction, although a majority of B23.1 is retained in nuclei (nucleoli) by association with RNA molecule.
Mitotic Phosphorylation of B23.1
B23 protein is hyperphosphorylated during mitosis (Figure 6A). A candidate kinase responsible for B23 phosphorylation is reported to be cyclin B/cdc2 kinase (Peter et al., 1990). There are several patches of serine (S) or threonine (T) residues similar to the consensus sequences that are potentially targeted by CDK (Figure 6B). It has been shown previously that threonine(s) is a B23 phosphorylation target during mitosis (Peter et al., 1990). In fact, the 234th and 237th threonine residues of glutathione S-transferase (GST)-tagged B 23.1 were found to be phosphorylated by cyclin B/cdc2 kinase in vitro (Tokuyama et al., 2001). To confirm these observations, we examined the in vitro level of cyclin B/cdc2-derived phosphorylation using a series of C-terminal deletion mutants of B23.1. Cyclin B/cdc2 kinase was purified from mitotic HeLa cell extracts using anti-cyclin B antibody as described in “Materials and Methods.” Wild-type full-length B23.1 was efficiently phosphorylated (same levels in Figure 6 and unpublished results). B23.1(1–257), which contains 257 amino acids from the N-terminus of B23.1, was phosphorylated as efficiently as the full-length B23.1, whereas the more truncated B23.1(1–160) was not efficiently phosphorylated by cyclin B/cdc2 in vitro (unpublished results). This indicates that a target residue(s) for phosphorylation by cyclin B/cdc2 resides between the 160th and 257th amino acids in the B23.1 protein sequence. We constructed point mutant proteins that contained amino acid substitutions for potential cyclin B/cdc2 kinase target sites as shown in Figure 6B in an effort to determine the precise phosphorylation site(s) by cyclin B/cdc2 kinase. His-tagged wild-type and mutant B23.1 proteins, WT, T199A, T219A, T234/237A, T199/234/234/A, T219/234/237A, and T4A (see Figure 6B) were generated in Escherichia coli and purified. Wild-type and mutant B23.1 proteins were subjected to phosphorylation by cyclin E/cdk2 or cyclin B/cdc2 kinase in the presence of [γ-32P]ATP. Phosphorylated proteins were separated by SDS-PAGE and were detected by autoradiography. Before examining phosphorylation by cyclin B/cdc2 kinase in detail, we performed phosphorylation reactions using purified cyclin E/cdk2 to confirm that T199 is a phosphorylation target of cyclin E/cdk2 (Tokuyama et al., 2001). Wild-type B23.1 and histone H1 were efficiently phosphorylated by cyclin E/cdk2 purified from NIH3T3 cells (Figure 6C, lane 2), although T199A and T4A, in which known cyclin E/cdk2-target threonine residues are substituted by alanine (Figure 6B), were not efficiently phosphorylated (Figure 6C, lanes 3 and 4). This is consistent with the previous report that T199 in B23.1 is phosphorylated by cyclin E/cdk2 (Tokuyama et al., 2001). Wild-type B23.1 was efficiently phosphorylated by cyclin B/cdc2 in a kinase dose-dependent manner (Figure 6D, lanes 1, 6, and 11). Although Tokuyama et al. (2001) reported that a GST-tagged T234/237A mutant is nonphosphorylatable by cyclin B/cdc2, His-tagged T234/237A mutant protein was phosphorylated as efficiently as wild-type protein by cyclin B/cdc2 in our study. On the other hand, the T199A mutant showed decreased phosphorylation efficiency compared with wild-type protein, and the T4A mutant was hardly phosphorylated by cyclin B/cdc2 in vitro. To confirm that T199 is one of the most accessible targets of cyclin B/cdc2, the mutant proteins T199/234/237A and T219/234/237A, in which three threonine residues were substituted by alanine, were also tested for phosphorylation susceptibility (Figure 6E). As expected, the phosphorylation efficiency of the mutant protein T199/234/237A (i.e., T219 intact) was significantly decreased, whereas the T219/234/237A (i.e., T199 intact) is efficiently phosphorylated. The T199/234/237A was inefficiently but distinctly phosphorylated by cyclin B/cdc2 in vitro, whereas the T4A mutant was hardly phosphorylated. Therefore, T219 may also be a potential target of cyclin B/cdc2, but with less accessibility than the T199. To examine the phosphorylation site(s) of B23.1 proteins in vivo, Flag-tagged wild-type and mutant B23.1 proteins were transiently expressed in HeLa cells. Cell extracts prepared from cells grown in the presence or absence of nocodazole were subjected to western blotting using anti-Flag antibody. Separation by SDS-PAGE shows that relative to endogenous B23.1 from interphase cells, B23.1 from mitotic cell extracts migrated slower due to hyperphosphorylation (Figure 6A). The band position of transiently expressed mitotic Flag-tagged B23.1 on SDS-PAGE is shifted from that of the interphase protein, suggesting that Flag-tagged B23.1 is phosphorylated during mitosis (Figure 6F, lanes 1 and 2). Because phosphatase-treated mitotic Flag-tagged B23.1 migrated faster than mock-treated protein on SDS-PAGE, the band shift of mitotic Flag-tagged B23.1 on SDS-PAGE was confirmed to be due to phosphorylation (unpublished results). Migration positions of Flag-tagged T199A and T219/234/237A are also shifted from those of interphase proteins, although the band shift of these mutant proteins in mitotic extracts was less than that of the wild type. In addition, the band shift of T4A mutant in mitotic extracts on SDS-PAGE was decreased significantly, indicating that this mutant protein is not efficiently phosphorylated during mitosis. These observations support our results of the in vitro phosphorylation assay using purified cyclin B/cdc2. From these in vitro and in vivo observations, it is strongly suggested that at least four threonine residues, T199, T219, T234, and T237, are the potential phosphorylation targets during mitosis possibly by cyclin B/cdc2.
Mitotic Inactivation of the RNA Binding Activity of B23.1
Next, we examined if phosphorylation of B23 protein during mitosis affects its RNA binding activity. Recombinant B23.1 preincubated in the presence or absence of cyclin B/cdc2 and ATP was mixed with RNA and was then subjected to a sucrose gradient sedimentation assay. As shown in Figure 7A, phosphorylated B23.1 migrated slower than nonphosphorylated B23.1 on SDS-PAGE (Figure 7A, compare lanes 1 and 2), suggesting that recombinant B23.1 is completely phosphorylated by cyclin B/cdc2. Nonphosphorylated B23.1 bound to RNA and was recovered in high density fractions (Figure 7B, first panel, lanes 8–11) as in Figure 8. In contrast, B23.1 phosphorylated by cyclin B/cdc2 was recovered in low density fractions (Figure 7B, second panel, lanes 2–5), even after incubation with RNA, indicating that phosphorylated B23.1 lost the ability to bind RNA. To confirm that B23.1 phosphorylation is critical for inactivation of the RNA binding activity, T4A mutant protein (a nonphosphorylatable mutant protein by cyclin B/cdc2) was subjected to the RNA binding assay. Before conducting this assay, it was first established that T4A mutant bound RNA similarly to wild-type protein (Figure 7B, third panel), meaning that the T to A substitutions in mutant B23.1 have no effect on its RNA binding activity. Although the phosphorylation reaction was performed under the same conditions as for wild-type protein, a band shift of T4A mutant was not detected (Figure 7A, compare lanes 3 and 4). In addition, nonphosphorylatable B23.1 mutant T4A protein remained associated with RNA even after the phosphorylation reaction before the RNA binding assay (Figure 7B, fourth panel). These results strongly support the concept that the RNA binding activity of B23.1 is inactivated by cyclin B/cdc2-mediated phosphorylation.
This concept lead us to propose that B23.1 is associated with RNA in interphase cells, whereas it is released from RNA by phosphorylation when cells enter mitosis. In fact, B23 proteins are mainly dispersed throughout the cytoplasm during mitosis (see Figure 9A and “Discussion” below), whereas some are reported to be concentrated in NDF and complexed with pre-rRNA and rRNA processing factors during mitosis (Dundr and Olson, 1998). Therefore, we supposed that a fraction of B23.1 remains associated with RNA as a nonphosphorylated form. We tested if the preformed B23.1–RNA complex is disrupted by cyclin B/cdc2-mediated phosphorylation in vitro (Figure 8). B23.1 was mixed and incubated with RNA to form a complex, followed by phosphorylation with cyclin B/cdc2 kinase in the absence or presence of ATP. The phosphorylation conditions used here were essentially the same as in Figure 7, where the free form of B23.1 was completely phosphorylated by cyclin B/cdc2. The RNA binding status of B23.1 was monitored by sucrose gradient sedimentation. The B23.1–RNA complex incubated with cyclin B/cdc2 in the absence of ATP remained associated with RNA as indicated by being recovered in high density fractions (Figure 8A, upper panel). In contrast, when the B23.1–RNA complex was incubated with cyclin B/cdc2 in the presence of ATP, some of B23.1 was released from RNA and was found in low density fractions (Figure 8A, bottom panel). These results suggest that the interaction between B23.1 and RNA is somewhat disrupted during mitosis by cyclin B/cdc2-mediated phosphorylation, whereas a distinct level of B23.1 remains associated with RNA by escaping from phosphorylation. Because completely phosphorylated B23.1 looses its ability to associate with RNA (Figure 7), it is speculated that phosphorylation of RNA-associated B23.1 is inhibited through its interaction with RNA. To examine this, the phosphorylation efficiency of the B23.1–RNA complex by cyclin B/cdc2 kinase was quantified relative to those of B23.2 and T199A protein–RNA complexes. Recombinant B2 3.1, B23.2, and T199A were mixed with increasing amounts of RNA, and were then phosphorylated by a fixed amount of cyclin B/cdc2 kinase in the presence of [γ- 2P]ATP (Figure 8B). As predicted, the phosphorylation efficiency of B23.1 decreased when B23.1 was preincubated with increasing amounts of RNA, whereas B23.2, which lacks RNA binding activity, was efficiently phosphorylated, even in the presence of RNA. These results indicate that phosphorylation of B23.1 is negatively regulated by its complex formation with RNA. Phosphorylation of the mutant B23.1 protein, T199A, wherein phosphorylation accessibility was much less than that of wild-type protein, showed a pattern similar to B23.1 (Figure 8B, lanes 7–9). Results shown here suggest that the B23.1 protein associated with RNA is resistant to phosphorylation and remains associated with RNA, whereas protein free from RNA is phosphorylated and thus restricted in accession of target RNA during mitosis. Because partially phosphorylated B23.1 is capable of associating with RNA (unpublished results), the phosphorylation status of B23.1 oligomers such as the ratio of nonphosphorylated to phosphorylated B23.1 may be a key determinant in regulation of the RNA binding activity of B23.1 during mitosis.
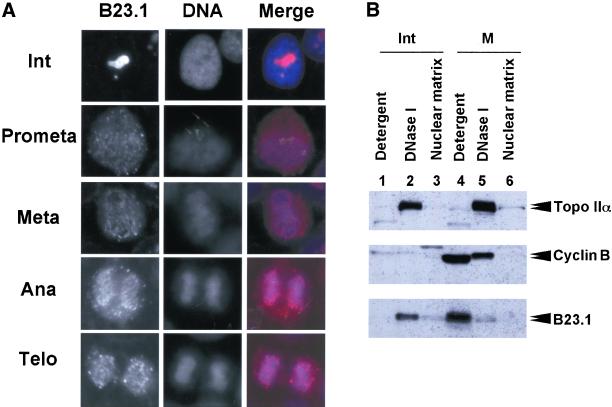
Figure 9.
Dissociation of B23 from nuclear structure during mitosis. (A) Cellular localization of Flag-tagged B23.1. Asynchronous HeLa cells expressing Flag-tagged B23.1 were fixed with an acetone:methanol (1:1) solution at −30°C for 10 min, and were then immunostained with anti-Flag antibody. Flag-tagged proteins were visualized by secondary antibody against mouse IgG conjugated with Alexa564. Images of B23.1 in a HeLa cell in interphase, prometaphase, metaphase, anaphase, and telophase on a coverslip are arranged as indicated. DNA was stained with Hoechst33258. The bar corresponds to 10 μm. (B) Cell fractionation of mitotic HeLa cells. Exponentially growing (lanes 1–3) or nocodazole-arrested (lanes 4–6) HeLa cells were subjected to cell fractionation experiments as shown in Figure 3. Proteins in each fraction (1 × 10 5 cells equivalent) were separated on a 8% SDS-PAGE followed by western blotting using anti-Topo IIα (8D2) (upper panel), anticyclin B (GNS1) (middle panel), or anti-B23.1 (C19) (bottom panel) antibodies. Positions of proteins are indicated at the right side of the panels.
To confirm the localization of B23.1 during mitosis, exponentially growing HeLa cells expressing Flag-tagged B23.1 were subjected to indirect immunofluorescence analysis using anti-Flag antibody. As shown in Figure 9A, Flag-tagged B23.1 was localized in nucleoli of interphase cells as the GFP-tagged protein (Figure 2). In contrast, mitotic Flag-tagged B23.1 was dispersed throughout cells, and partially concentrated in the periphery of chromosomes and NDF (Figure 9A). Before the end of mitosis, NDF is assembled into nuclei and nucleoli are reconstituted. This localization pattern of B23 during mitosis supports previous reports (Zatsepina et al., 1997, 1999; Dundr and Olson, 1998; Dundr et al., 2000). To further confirm that B23 is released from nuclear structure during mitosis by phosphorylation, cell fractionation experiments were performed using exponentially growing or mitotic HeLa cells (Figure 9B). The expression level of cyclin B in mitotic cells was much higher than that in asynchronous cells, and cyclin B was recovered in the “Detergent” and “DNase I” fractions (Figure 9B, middle panel, compare lanes 1 and 2, and 4 and 5). This is in good agreement with the fact that cyclin B accumulates during G2/M phase and is translocated to nuclei at the beginning of mitosis (Pines and Hunter, 1991). It is noted that the migration of cyclin B detected in the “DNase I” fraction is slower than that in the “Detergent” fraction (Figure 9B, lanes 4 and 5). Because cyclin B is phosphorylated by a polo-like kinase and translocated to nuclei (Toyoshima-Morimoto et al., 2001), cyclin B recovered in the “DNase I” fraction should be phosphorylated. Topoisomerase IIα, which is also phosphorylated during mitosis (Heck et al., 1989; Kimura et al., 1996) and binds tightly to chromatin, was detected mainly in the “DNase I” fraction (Figure 9B, upper panel). The migration position of mitotic topoisomerase IIα was also slightly shifted from that of interphase protein (Figure 9B, compare lanes 2 and 5). Interestingly, interphase B23.1 fractionated mainly in the “DNase I” fraction, whereas the majority of mitotic B23.1, which was supposed to be phosphorylated, fractionated in the “Detergent” fraction (Figure 9B, bottom panel). A minor potion of mitotic B23.1 was also detected in the “DNase I” fraction, but the B23.1 in this fraction showed faster mobility on SDS-PAGE. From these observations, it is suggested that B23.1 in interphase cells associates with nuclear structures with the help of RNA, whereas B23.1 phosphorylated by cyclin B/cdc2 during mitosis looses its RNA binding activity and is released from nuclei.
DISCUSSION
As evidenced through the RNA binding and cellular localization of ribosome biogenesis factor B23, we have shown its cell cycle-related regulation mechanism. Both splicing variants of B23 proteins, B23.1 and B23.2, are complexed in vivo (Figure 1), and the RNA binding activity of B23.1 is modulated by hetero-oligomerization with B23.2 (Figure 4). Furthermore, phosphorylation of B23.1 by cyclin B/cdc2 inactivates its RNA binding activity (Figure 7). It is possible that these regulation mechanisms of the RNA binding activity of B23.1 are important for regulation of ribosome biogenesis and possibly cell cycle progression.
In rat and human cells, both subtypes of B23 proteins are expressed, although the expression level of B23.2 is much lower than that of B23.1. As shown here, the cellular localizations of B23.1 and B23.2 are not exactly identical (Figure 2). B23.1 is mainly localized in nucleoli, whereas B23.2 is localized not only in nucleoli but also throughout nucleoplasm. From these observations, it is presumed that B23.1 is targeted to rRNA rich-nucleoli, possibly due to its RNA binding activity. This is supported by the facts that B23.2, which is incapable of associating with RNA, fails to localize nucleoli (Figure 2), that a majority of B23.1 is released by RNase A digestion of nuclei (Figure 5), and that mitotic B23.1 phosphorylated by cyclin B/cdc2, which is incapable of binding to RNA, is dispersed throughout cells (Figure 9). In addition, B23.2 and mitotic B23.1 easily leak out of nuclei after detergent treatment (Figures 2, 3, and 9), indicating that these proteins are less tightly associated with nuclear structures compared with interphase B23.1. However, when cells overexpressing HA-tagged B23.2 were subjected to cell fractionation experiments as in Figures 2 and 3, distinct levels of HA-tagged B23.2 were detected in detergent- or nuclease-treated cell nuclei (Figure 3 and unpublished results), although GFP-tagged B23.2 was not detected after DNase I or RNase A digestion (Figure 2). B23.2 may have a potential ability to associate with nuclear structure. Because B23.2 complexed with B23.1 can bind to RNA in vitro (Figure 4), a detergent- and nuclease resistant-nucleolar B23.2 is probably complexed with B23.1. This is supported by the fact that HA-tagged B23.2 in detergent-extracted HeLa cells coprecipitates with endogenous B23.1. When the ratio of B23.2 to B23.1 is increased, the RNA binding activity of the complex decreased even in the presence of constant amounts of B23.1 (Figure 4). Therefore, the ratio of B23.1 to B23.2 is likely important in determining the RNA binding activity of the B23 complex. B23 proteins are suggested to form hexamer or pentamer/decamer in solution (Yung and Chan, 1987; Dutta et al., 2001). Therefore, there are several different combinations of B23.1/B23.2 complexes that could be formed. Hetero-oligomer formation of B23.1 and B23.2 is a possible RNA binding modulation mechanism that may be involved in nucleolar targeting and ribosome biogenesis function. The expression level of B23.1 is closely correlated with the cell growth rate, whereas that of B23.2 is relatively constant (Wang et al., 1993). Therefore, it is possible that the ribosome biogenesis function of B23.1 in slowly growing cells could be impaired through two distinct pathways: 1) expression of B23.1 is down-regulated, and 2) the low level of B23.1 is trapped by B23.2 and its RNA binding activity is thus suppressed, thereby resulting in inhibition of ribosome biogenesis, including rRNA processing and ribosome assembly. This could act to control the rate of ribosome biogenesis.
Although the N-terminal portion of B23 proteins is shown to be involved in oligomer formation (Yung and Chan, 1987; Herrera et al., 1996; Dutta et al., 2001), the interaction force for oligomer formation is unknown. Cellular B23 oligomer can be seen even in the presence of SDS or a reducing agent such as dithiothreitol, but is disrupted by extensive boiling (unpublished results). The N-terminal region is highly conserved in nucleoplasmin family proteins (Schmidt-Zachmann et al., 1987), including nucleoplasmin, B23/NO38, NO29 (Zirwes et al., 1997b), and mitotic apparatus protein p62 (Ye and Sloboda, 1997). In B23 proteins, oligomer formation is critical for its molecular chaperone activity (Hingorani et al., 2000) and RNA binding activity (in this study). However, it is shown that oligomer formation is dispensable for B23 RNase activity (Hingorani et al., 2000), and that RNase activity of B23.2 is about one-half of B23.1. Because the C-terminal region of B23.1 is essential to tether B23.1 protein to the target RNA, hetero-oligomer formation of B23.1 and B23.2 (which interferes the RNA binding activity of B23.1) would obstruct RNase function. Again, it is emphasized that B23.2 modulates the ribosome biogenesis function of B23.1.
B23 protein is phosphorylated by cyclin B/cdc2 during mitosis possibly at the 199th, 219th, 234th, or 237th threonine residues. Previously, T234 and T237 were shown to be phosphorylation sites (Tokuyama et al., 2001). However, under the experimental conditions used here, the T199 residue was the most likely candidate to be phosphorylated by cyclin B/cdc2 (Figure 6). This difference could be explained by the structure of recombinant proteins used as a substrate. A long N-terminal extra GST polypeptide may block the proper structure of B23 proteins and change the target residue of cyclin B/cdc2, although we cannot exclude the possibility that the histidine-tag containing 20 amino acid residues attached at the N-terminus may also have altered the structure of B23 proteins. As shown here, because the T199A, T234/237A, and even T199/234/237A mutants are phosphorylated by cyclin B/cdc2 kinase in vitro, four threonine residues, T99, T219, T234, and T237, are possibly phosphorylated. Furthermore, T4A mutant is distinctly phosphorylated by cyclin B/cdc2 in vivo and in vitro but at significantly low level (Figure 6, D and F). Thus, there could be another potential target residue phosphorylated by cyclin B/cdc2 in B23. Nevertheless, an important point is that phosphorylation of B23.1 by cyclin B/cdc2 leads to inactivation of its RNA binding activity. Two distinct mechanisms, hetero-oligomer formation and phosphorylation by cyclin B/cdc2, regulate the RNA binding activity of B23.1. Not only the RNA binding activity of B23.1, but also other functions specific to the C-terminal region such as protein binding (Takemura et al., 1999) and RNase could be similarly regulated by cyclin B/cdc2-mediated phosphorylation and hetero-oligomer formation with B23.2. On this line, it is likely that B23 localizes to centrosome through the C-terminal tail of B23.1, because B23 is released from centrosome by the phosphorylation at T199 with cyclin E/cdk2 (Tokuyama et al., 2001). Because release of B23.1 from centrosome by phosphorylation is indispensable for centrosome duplication, it is important to identify the target protein of B23.1 that localizes to centrosome in clarifying the mechanism of centrosome duplication. Furthermore, a fraction of B23.1 is tightly associated with nuclear structures, possibly so-called nuclear matrix, whereas B23.2 is not (Figures 2 and 3). Because B23.1 associates with nuclear structure after RNase treatment (Figures 2 and 5), it is presumed that B23.1 associates with nuclear matrix or residual nuclear materials through protein–protein interaction between an unidentified nuclear structural protein(s) and the C-terminal tail of B23.1. This interaction is also impaired by the phosphorylation during mitosis and hetero-oligomer formation (Figure 9).
In mitotic cells, a part of B23 is associated with a large complex containing pre-rRNA, fibrillarin, nucleolin, and U3 small nucleolar RNA (Pinol-Roma, 1999), and is localized in cytoplasmic dots termed NDF (Dundr et al., 2000; Dundr and Olson, 1998; Zatsepina et al., 1999; Zatsepina et al., 1997). It remains unclear how NDF is released from chromosomes during mitosis and is reassembled to form nucleoli after cell division. As shown in Figure 8, B23.1 complexed with RNA was not efficiently phosphorylated, and a fraction of B23.1 remains associated with RNA even after phosphorylation. Because NDF is suggested to be important to form nucleoli and restart ribosome biogenesis after cell division, it is possible that the phosphorylation restriction of non-rRNA binding proteins is essential to maintain NDF during mitosis. Protection from mitotic phosphorylation of RNA binding proteins is one possible mechanism for preserving a “core” nucleolar structure for the next cell cycle. Nucleolin, a major RNA binding nucleolar phosphoprotein, shows similar behavior to B23; nucleolin is also phosphorylated during mitosis by cyclin B/cdc2 kinase and is dispersed throughout cells, and some nucleolin can be found in NDF (Pinol-Roma, 1999; Srivastava and Pollard, 1999). Therefore, mitotic inactivation of the RNA binding activity by phosphorylation may also be significant to nucleolin. Alternatively, it is possible that a factor(s) that connects NDFs or NDFs and chromosomes may exist whereby the interaction between the putative factor and the NDF or chromosome may be broken down when cells enter mitosis. Because both B23 and nucleolin were shown to bind chromatin (Olson and Thompson, 1983; Okuwaki et al., 2001a), these nonribosomal proteins may function as linker proteins between NDFs or between NDFs and chromosomes.
During mitosis, several cellular functions such as transcription and translation are silenced so that genetic material can be correctly condensed and segregated into daughter cells. Mitotic inactivation of these cellular processes is partly mediated through phosphorylation by mitotic kinases including cyclin B/cdc2. Most of transcription and chromatin binding factors are phosphorylated and released from chromosomes, thereby silencing transcription (Gottesfeld and Forbes, 1997). Mitotic release also functions in pre-rRNA processing components, whereas the pol I transcription machinery, including RNA polymerase I, UBF, and DNA topoisomerase I, remains associated with chromosomes during mitosis (Weisenberger and Scheer, 1995; Roussel et al., 1996). Our findings demonstrate the first case where a factor involved in ribosome biogenesis is regulated through phosphorylation and dephosphorylation during the cell cycle.
ACKNOWLEDGMENTS
We thank Dr. H. Umekawa (Mie University, Japan) for mAb for B23 (NB23), Dr. A. Kikuchi (Nagoya University, Japan) for antitopoisomerase II antibody (8D2), and Dr. K. Matsumoto (RIKEN, Japan) for useful discussion and critical comments on the manuscript. We also thank K. Pepin for proofreading of the manuscript. This work was supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by a grant for Bioarchitect Research Program from RIKEN (to K.N.). M.O. is a Special Postdoctoral Researcher of RIKEN.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–03–0036. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–03–0036.
REFERENCES
- Chang JH, Olson MO. Structure of the gene for rat nucleolar protein B23. J Biol Chem. 1990;265:18227–18233. [PubMed] [Google Scholar]
- Colgan DF, Murthy KG, Zhao W, Prives C, Manley JL. Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of multiple consensus and non-consensus sites. EMBO J. 1998;17:1053–1062. doi: 10.1093/emboj/17.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbar TS, Gentry GA, Olson MO. Interaction of nucleolar phosphoprotein B23 with nucleic acids. Biochemistry. 1989;28:9495–9501. doi: 10.1021/bi00450a037. [DOI] [PubMed] [Google Scholar]
- Dundr M, Misteli T, Olson MO. The dynamics of postmitotic reassembly of the nucleolus. J Cell Biol. 2000;150:433–446. doi: 10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Olson MO. Partially processed pre-rRNA is preserved in association with processing components in nucleolus-derived foci during mitosis. Mol Biol Cell. 1998;9:2407–2422. doi: 10.1091/mbc.9.9.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durban E, Valdez BC, Gustafson WC, Taylor CW, Cardellini E, Busch H. Functional domains of nucleolar phosphoprotein p120. Physiol Chem Phys Med NMR. 1995;27:303–311. [PubMed] [Google Scholar]
- Dutta S, Akey IV, Dingwall C, Hartman KL, Laue T, Nolte RT, Head JF, Akey CW. The crystal structure of nucleoplasmin-core. Implications for histone binding and nucleosome assembly. Mol Cell. 2001;8:841–853. doi: 10.1016/s1097-2765(01)00354-9. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Izaurralde E, Adachi Y, Wingfield P, Laemmli UK. Specific complex of human immunodeficiency virus type 1 rev and nucleolar B23 proteins: dissociation by the Rev response element. Mol Cell Biol. 1991;11:2567–2575. doi: 10.1128/mcb.11.5.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerstein N, Mond JJ. “Numatrin,” a nuclear matrix protein associated with induction of proliferation in B lymphocytes. J Biol Chem. 1987;262:11389–11397. [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Graham FL, Eb AJvd. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hager DA, Burgess RR. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980;109:76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- He DC, Nickerson JA, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck MM, Hittelman WN, Earnshaw WC. In vivo phosphorylation of the 170-kDa form of eukaryotic DNA topoisomerase. II. Cell cycle analysis. J Biol Chem. 1989;264:15161–15164. [PubMed] [Google Scholar]
- Heix J, Vente A, Voit R, Budde A, Michaelidis TM, Grummt I. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera JE, Correia JJ, Jones AE, Olson MO. Sedimentation analyses of the salt- and divalent metal ion-induced oligomerization of nucleolar protein B23. Biochemistry. 1996;35:2668–2673. doi: 10.1021/bi9523320. [DOI] [PubMed] [Google Scholar]
- Hingorani K, Szebeni A, Olson MO. Mapping the functional domains of nucleolar protein B23. J Biol Chem. 2000;275:24451–24457. doi: 10.1074/jbc.M003278200. [DOI] [PubMed] [Google Scholar]
- Huang WH, Yung BY, Syu WJ, Lee YH. The nucleolar phosphoprotein B23 interacts with hepatitis delta antigens, and modulates the hepatitis delta virus RNA replication. J Biol Chem. 2001;276:25166–25175. doi: 10.1074/jbc.M010087200. [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano M, Kobayashi R, Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- Kimura K, Nozaki N, Enomoto T, Tanaka M, Kikuchi A. Analysis of M phase-specific phosphorylation of DNA topoisomerase II. J Biol Chem. 1996;271:21439–21445. doi: 10.1074/jbc.271.35.21439. [DOI] [PubMed] [Google Scholar]
- Li YP, Busch RK, Valdez BC, Busch H. C23 interacts with B23, a putative nucleolar-localization-signal-binding protein. Eur J Biochem. 1996;237:153–158. doi: 10.1111/j.1432-1033.1996.0153n.x. [DOI] [PubMed] [Google Scholar]
- Maden BE. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1990;39:24. doi: 10.1016/s0079-6603(08)60629-7. 1–303. [DOI] [PubMed] [Google Scholar]
- Manley JL, Fire A, Cano A, Sharp PA, Gefter ML. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci USA. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K, Kawase H, Handa H, Yano K, Yamasaki M, Ishimi Y, Okuda A, Kikuchi A, Matsumoto K. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc Natl Acad Sci USA. 1995;92:4279–4283. doi: 10.1073/pnas.92.10.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K, Saito S, Okuwaki M, Kawase H, Furuya A, Kusano A, Hanai N, Okuda A, Kikuchi A. Cellular localization and expression of template-activating factor I in different cell types. Exp Cell Res. 1998;240:274–281. doi: 10.1006/excr.1997.3930. [DOI] [PubMed] [Google Scholar]
- Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, Fukasawa K. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Okuwaki M, Iwamatsu A, Tsujimoto M, Nagata K. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J Mol Biol. 2001a;311:41–55. doi: 10.1006/jmbi.2001.4812. [DOI] [PubMed] [Google Scholar]
- Okuwaki M, Matsumoto K, Tsujimoto M, Nagata K. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 2001b;506:272–276. doi: 10.1016/s0014-5793(01)02939-8. [DOI] [PubMed] [Google Scholar]
- Olson MO, Thompson BA. Distribution of proteins among chromatin components of nucleoli. Biochemistry. 1983;22:3187–3193. doi: 10.1021/bi00282a023. [DOI] [PubMed] [Google Scholar]
- Peculis BA. RNA-binding proteins: if it looks like a sn(o)RNA. Curr Biol. 2000;10:R916–R918. doi: 10.1016/s0960-9822(00)00851-4. [DOI] [PubMed] [Google Scholar]
- Peculis BA, Gall JG. Localization of the nucleolar protein NO38 in amphibian oocytes. J Cell Biol. 1992;116:1–14. doi: 10.1083/jcb.116.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Nakagawa J, Doree M, Labbe JC, Nigg EA. Identification of major nucleolar proteins as candidate mitotic substrates of cdc2 kinase. Cell. 1990;60:791–801. doi: 10.1016/0092-8674(90)90093-t. [DOI] [PubMed] [Google Scholar]
- Pines J, Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S. Association of nonribosomal nucleolar proteins in ribonucleoprotein complexes during interphase and mitosis. Mol Biol Cell. 1999;10:77–90. doi: 10.1091/mbc.10.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel P, Andre C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs, and absent in inactive NORs. J Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur RS, Olson MO. Preferential cleavage in pre-ribosomal RNA byprotein B23 endoribonuclease. Nucleic Acids Res. 1998;26:4508–4515. doi: 10.1093/nar/26.19.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Zachmann MS, Hugle-Dorr B, Franke WW. A constitutive nucleolar protein identified as a member of the nucleoplasmin family. EMBO J. 1987;6:1881–1890. doi: 10.1002/j.1460-2075.1987.tb02447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Pollard HB. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 1999;13:1911–1922. [PubMed] [Google Scholar]
- Szebeni A, Herrera JE, Olson MO. Interaction of nucleolar protein B23 with peptides related to nuclear localization signals. Biochemistry. 1995;34:8037–8042. doi: 10.1021/bi00025a009. [DOI] [PubMed] [Google Scholar]
- Szebeni A, Olson MO. Nucleolar protein B23 has molecular chaperone activities. Protein Sci. 1999;8:905–912. doi: 10.1110/ps.8.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura M, Sato K, Nishio M, Akiyama T, Umekawa H, Yoshida S. Nucleolar protein B23.1 binds to retinoblastoma protein and synergistically stimulates DNA polymerase α activity. J Biochem. 1999;125:904–909. doi: 10.1093/oxfordjournals.jbchem.a022367. [DOI] [PubMed] [Google Scholar]
- Tokuyama Y, Horn HF, Kawamura K, Tarapore P, Fukasawa K. Specific phosphorylation of nucleophosmin on Thr (199) by cyclin-dependent kinase 2-cyclin E, and its role in centrosome duplication. J Biol Chem. 2001;276:21529–21537. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- Toyoshima-Morimoto F, Taniguchi E, Shinya N, Iwamatsu A, Nishida E. Polo-like kinase 1 phosphorylates cyclin B1, and targets it to the nucleus during prophase. Nature. 2001;410:215–220. doi: 10.1038/35065617. [DOI] [PubMed] [Google Scholar]
- Umekawa H, Sato K, Takemura M, Watanabe Y, Usui S, Takahashi T, Yoshida S, Olson MO, Furuichi Y. The carboxyl terminal sequence of nucleolar protein B23.1 is important in its DNA polymerase α-stimulatory activity. J Biochem. 2001;130:199–205. doi: 10.1093/oxfordjournals.jbchem.a002973. [DOI] [PubMed] [Google Scholar]
- Wang D, Baumann A, Szebeni A, Olson MO. The nucleic acid binding activity of nucleolar protein B23.1 resides in its carboxyl-terminal end. J Biol Chem. 1994;269:30994–30998. [PubMed] [Google Scholar]
- Wang D, Umekawa H, Olson MO. Expression and subcellular locations of two forms of nucleolar protein B23 in rat tissues and cells. Cell Mol Biol Res. 1993;39:33–42. [PubMed] [Google Scholar]
- Weisenberger D, Scheer U. A possible mechanism for the inhibition of ribosomal RNA gene transcription during mitosis. J Cell Biol. 1995;129:561–575. doi: 10.1083/jcb.129.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Sloboda RD. Molecular characterization of p62, a mitotic apparatus protein required for mitotic progression. J Biol Chem. 1997;272:3606–3614. doi: 10.1074/jbc.272.6.3606. [DOI] [PubMed] [Google Scholar]
- Yung BY, Chan PK. Identification and characterization of a hexameric form of nucleolar phosphoprotein B23. Biochim Biophys Acta, 1987;925:74–82. doi: 10.1016/0304-4165(87)90149-8. [DOI] [PubMed] [Google Scholar]
- Zatsepina OV, Rousselet A, Chan PK, Olson MO, Jordan EG, Bornens M. The nucleolar phosphoprotein B23 redistributes in part to the spindle poles during mitosis. J Cell Sci. 1999;112:455–466. doi: 10.1242/jcs.112.4.455. [DOI] [PubMed] [Google Scholar]
- Zatsepina OV, Todorov IT, Philipova RN, Krachmarov CP, Trendelenburg MF, Jordan EG. Cell cycle-dependent translocations of a major nucleolar phosphoprotein, B23, and some characteristics of its variants. Eur J Cell Biol. 1997;73:58–70. [PubMed] [Google Scholar]
- Zirwes RF, Kouzmenko AP, Peters JM, Franke WW, Schmidt-Zachmann MS. Topogenesis of a nucleolar protein: determination of molecular segments directing nucleolar association. Mol Biol Cell. 1997a;8:231–248. doi: 10.1091/mbc.8.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirwes RF, Schmidt-Zachmann MS, Franke WW. Identification of a small, very acidic constitutive nucleolar protein (NO29) as a member of the nucleoplasmin family. Proc Natl Acad Sci USA. 1997b;94:11387–11392. doi: 10.1073/pnas.94.21.11387. [DOI] [PMC free article] [PubMed] [Google Scholar]