Abstract
The ESX-1 locus is a region critical for full virulence in Mycobacterium tuberculosis, which encodes two secreted proteins as well as other genes involved in their secretion. The mechanism of secretion of the two proteins, ESAT-6 and CFP-10, and their function remain unknown. Using proteomic methods to search for additional proteins secreted by the ESX-1 locus, we discovered that a protein encoded by a chromosomally unlinked gene, espA, is also secreted by strains that contain the ESX-1 locus but not by strains with ESX-1 deletions. Mutations in individual ESX-1 genes, including those that encode ESAT-6 and CFP-10, were found to block EspA secretion. Surprisingly, mutants that lack espA reciprocally failed to secrete ESAT-6 and CFP-10 and were as attenuated as ESX-1 mutants in virulence assays. The results indicate that secretion of these proteins, which are each critical for virulence of pathogenic mycobacteria, is mutually dependent. The results further suggest that discerning the nature of the interaction and the structure of macromolecular complexes will provide insights into both an alternative mechanism of protein secretion and mycobacterial virulence.
Keywords: ESAT-6 protein, Mycobacterium tuberculosis, virulence factor
Interactions between bacterial pathogens and their hosts occur at the surface of cells. Many pathogenic bacteria secrete proteins involved in virulence (1). These proteins often interfere with normal host defenses and allow bacteria to survive and multiply. Strikingly, many of these virulence factors are secreted by nonclassical secretory systems that, in some cases, facilitate translocation of proteins across host cell membranes (2, 3).
During the evolution of Mycobacterium bovis Bacille Calmette-Guérin (BCG), the attenuated bacillus widely used as vaccine against tuberculosis, a major deletion removed a set of nine ORFs (4). Similar deletions are found in Mycobacterium microti (5) and the Dassie bacillus (6), other relatively avirulent mycobacteria. The BCG deletion, known as RD1, includes two genes that encode the abundantly secreted proteins, ESAT-6 and CFP-10. The RD1 deletion encompasses most but not all of a larger, 15-gene region known as ESX-1 (7), which is hypothesized to encode an alternative secretory system (8, 9).
The proteins encoded by the ESX-1 locus clearly play an important role in virulence. The secreted proteins encoded within the locus are potent T cell antigens (7, 10, 11). More significantly, Mycobacterium tuberculosis strains with RD1 deletions are far less virulent in mice than strains that are otherwise isogenic (12-14). They replicate poorly and cause less pathology in both immunocompetent and immunocompromised mice, although the latter ultimately succumb to infection. RD1-deleted strains of Mycobacterium marinum elicit only very modest macrophage aggregation and granuloma formation in zebrafish (15). Additional work suggests the RD1-encoded proteins may disrupt host cell membranes. In M. tuberculosis, these proteins may be required for lysis of infected cells (13) and cell-to-cell spread (16), whereas, in M. marinum, RD1 is needed for efficient bacterial escape from the phagosome (17).
Several lines of experimental evidence support the idea that the genes in the ESX-1 locus are functionally related. First, several of the ESX-1-encoded proteins have been shown to interact through yeast two-hybrid analysis (14). More strikingly, inactivation of a number of the individual ESX-1 genes disrupts ESAT-6 and CFP-10 secretion and attenuates the bacterium to a similar extent as deletion of the whole locus (13, 16). These data are consistent with the model that ESX-1 encodes an alternative protein secretory apparatus required for the export of ESAT-6 and CFP-10.
However, the specialized type III and IV secretory systems present in Gram-negative pathogens mediate the export of many proteins encoded by genes unlinked to the secretory apparatus (2, 3). These “effector” proteins often play complementary roles in altering host resistance. Here we describe a proteomic search for proteins secreted by the same machinery used for ESAT-6 and CFP-10 secretion. We find that the product of an unlinked gene, Rv3616c, here renamed espA for ESX-1 secretion-associated protein A, has a critical interaction with the genes of the ESX-1 region.
Materials and Methods
Culture of M. tuberculosis and Preparation of Culture Filtrates and Cell Lysates. M. tuberculosis strains were maintained in Middlebrook 7H9 medium supplemented with oleic acid-albumin-dextrose-catalase (Difco) and 0.05% Tween 80. The generation of H37RvΔRD1 and H37RvΔesat6 was described in refs. 12 and 16, and identification and confirmation of the transposon mutants were described in ref. 16.
For analysis of protein expression and secretion, strains were cultured in Sauton's medium supplemented with 0.05% Tween 80 instead of 7H9 medium, in which the large quantity of albumin confounded analysis of secreted bacterial proteins. To analyze bacterial proteins by SDS/PAGE, mid-log phase bacteria were resuspended in fresh medium and cultured overnight at 37°C. Cell pellets were resuspended in protein extraction buffer (50 mM Tris·HCl, pH 7.5/5 mM EDTA/1 mM 2-mercaptoethanol) and disrupted by bead beating. Protein loading buffer was added to cell lysates, and samples were boiled for 20 min before removal from the biosafety level 3 facility and analysis by SDS/PAGE and Western blotting. Culture supernatants were sterilized by double filtration through 0.2-μm filters. Filtrates were concentrated ≈20-fold by using Centricon (Millipore) and Vivacell (Viva-science, Hannover, Germany) filter units and analyzed by SDS/PAGE and Western blotting. The volume of culture filtrate loaded was normalized to the total protein content of the cell pellet as determined by modified Bradford assay (Coomassie Plus, Pierce). For each experiment, equal protein loading for both cell pellets and supernatants was confirmed by visualizing the total protein loaded by Coomassie staining.
For analysis of culture filtrate proteins by LCQ-tandem MS (MS/MS), strains were cultured in Sauton's media containing Tween until mid-log phase as described previously. Strains were then diluted to an OD of ≈0.100 in Sauton's medium without detergent for three further days of culture. Mycobacteria were then resuspended and cultured overnight in modified N salt medium (100 mM Bis/Tris·HCl/5 mM KCl/7.5 mM (NH4)2SO4/0.5 mM K2HSO4/1 mM KH2PO4/10 mM MgCl2/38 mM glycerol, pH 7.0). Culture filtrates were harvested and concentrated as described above. Multiple biologic replicates were performed for each strain involving the independent culture, harvest, and analysis of supernatant proteins.
LCQ-MS/MS. One hundred microliters of concentrated supernatant was diluted with 200 μl of 8 M urea, 100 mM ammonium bicarbonate, and 10 mM DTT and incubated at 37°C for 1 h followed by alkylation of cysteines with iodoacetamide. Samples were diluted with 5 mM CaCl2 to 1.2 ml, and 10 μg of sequencing-grade modified trypsin (Promega) was added. Samples were incubated at 37°C for 20 h, quenched with 10 μl of formic acid, frozen at -80°C, and lyophilized to dryness. Samples were then redissolved in 200 μl of 0.25% formic acid and water.
Samples were run on a LCQ DECA XP plus Proteome X workstation (Thermo, Waltham, MA). Each reconstituted sample was separated on a C18 column running at 2 μl/min flow with a gradient of 5-60% water/0.1% formic acid to acetonitrile/0.1% formic acid over the course of 300 min. Peptide identifications were made by using sequest through bioworks 3.1. Peptide fragment data were searched against the H37Rv database from EMBL using differential carbamidomethyl modified cysteines and oxidized methionines. Peptide score cutoff values were chosen at Xcorr of 1.8 for singly charged ions, 2.5 for doubly charged ions, and 3.0 for triply charged ions, along with ΔCN values of 0.1 and RSP values of 1.
Western Blot Analysis. Antisera reactive to EspA and CFP-10 were generated by antibodies from Fusion Antibodies (Belfast). EspA-reactive serum was obtained by immunizing rabbits with a protein fragment representing amino acids 304-392 of EspA. CFP-10-reactive serum was generated by immunizing rabbits with whole CFP-10. Mouse monoclonal antibody to ESAT-6 was obtained from the Antibody Shop (Gentofte, Denmark). Rabbit antiserum reactive to groEL was obtained from Colorado State University (Fort Collins) through the National Institutes of Health contract for Tuberculosis Research Materials and Vaccine Testing. All antibodies were used at a dilution of 1:1,000 for Western blotting except for anti-groEL, which was used at a dilution of 1:50. Peroxidase-conjugated goat anti-rabbit and goat anti-mouse secondary antibodies were used at concentrations of 1:10,000. Secondary antibodies were detected via chemiluminescence by using SuperSignal (Pierce).
For H37Rv and all of the mutant strains, we confirmed that the secretion differences described were not due to differences in bacterial cytolysis by assessing the amount of groEL, a cytoplasmic chaperonin, in the culture supernatants. In all cases, there was minimal groEL in the supernatant. Blots were over-exposed to compare the amounts of released groEL. No significant differences in cytolysis were observed (data not shown).
Generation of espA Deletion Mutants. The unmarked deletions of espA in H37Rv and H37RvΔRD1 were generated by transformation with sacB counter selection as described in ref. 18. Approximately 1,000 base pairs of upstream flanking sequence (from base 4,056,376 to 4,057,324) and of downstream flanking sequence (from base 4,054,277 to 4,055,194) were cloned into the multiple cloning site of pBluescript to generate p16flank. The abutting flanks were then amplified to add 5′ and 3′ NdeI sites. This PCR product was cloned into the NdeI site of pMP62, a suicide vector carrying hygromycin resistance and sacB, and this construct was validated by sequencing. After transformation into H37Rv and H37RvΔRD1, selection was first performed on 7H10 agar containing hygromycin (50 μg/ml) and then on 3% sucrose. Deletion was confirmed by PCR analysis.
Complementation of RvΔespA. EspA was PCR-amplified from H37Rv genomic DNA with primers that added an N-terminal BamHI site and C-terminal SpeI and HindIII sites. To make the His-Myc tagged construct, forward and reverse oligos encoding a six-residue His tag followed by a Myc tag (protein sequence: HHHHHHMAEQKLISEEDL) were annealed, creating sticky ends compatible with SpeI and HindIII, and then cloned in frame into the C terminus of EspA. These DNAs were cloned downstream of the strong constitutive MOP (mycobacterial optimal promoter) promoter in an integrating plasmid that confers hygromycin resistance (16). To add the native promoter of espA, the MOP promoter was excised from these constructs and replaced with the 502 nucleotides upstream of the espA start codon. All constructs were confirmed by sequencing, and the sequences of these constructs are available through GenBank. The accession number of pALE2 encoding epitope-tagged espA under the MOP promoter is DQ086480, and that of pALE4 encoding epitope-tagged espA under the native promoter is DQ068700.
Mice. C57BL/6 and BALB/c-SCID mice were purchased from The Jackson Laboratory. Mice were infected by i.v. tail vein injection. Infecting doses were confirmed by plating. In single-strain infections, four mice per group were harvested at the given time points. Spleens and lungs were plated for colony-forming units as described in ref. 19. At all time points, the left lung was fixed in 10% formalin for pathology. For survival studies, 10 mice per group were infected with the indicated strains.
For the competition experiments, the indicated strains were mixed at a ratio of 3:1 (unmarked to marked strain). H37Rv and H37RvΔespA are unmarked, and H37RvΔRD1 and H37RvΔRD1ΔespA are kanamycin-resistant. To determine the burden of each strain, spleens and lungs were harvested and plated for colony-forming units on 7H10 agar with and without kanamycin (25 μg/ml).
Results
LCQ-MS/MS Screen Identifies ESX-1 Secreted Proteins. Like the alternative secretion systems of other pathogenic bacteria, the ESX-1 secretion apparatus is likely to secrete proteins in addition to ESAT-6 and CFP-10. Therefore, we analyzed the culture filtrates of wild-type H37Rv and H37RvΔRD1 with the goal of identifying proteins present in wild-type but absent in mutant culture filtrates. To minimize the number of cytoplasmic proteins contaminating the culture filtrates because of background autolysis (20), very-short-term culture supernatants were collected (<24 h of culture). The proteins in these culture filtrates were identified by analyzing HPLC-resolved tryptic fragments by LCQ-MS/MS.
In wild-type culture filtrates, 102 proteins were identified in the three independently obtained samples analyzed. Of these, 53 were present in at least two of three samples. Seventy-five percent of the reproducibly identified proteins have predicted signal sequences, are ESX proteins (8), or have been shown to be secreted in a secA2-dependent fashion (21), indicating that our sampling enriched for secreted proteins. More proteins were identified in the H37RvΔRD1 culture filtrates (178 proteins were identified in three independently obtained samples). In ion-trap MS, abundant peptides can suppress the detection of less abundant species. Peptides from CFP-10 were the most frequently identified peptides in our analyses of wild-type culture filtrates. Thus, we may have identified more proteins in the H37RvΔRD1 culture filtrates because the absence of peptides from CFP-10 and, to a lesser extent, ESAT-6 allowed detection of many peptides present in lower quantities.
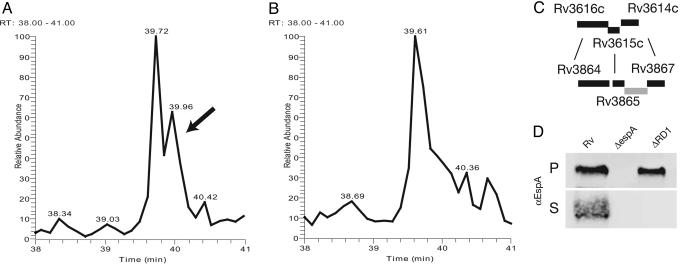
The greater coverage of proteins secreted by the H37RvΔRD1 strain biased our screen against the identification of proteins secreted by wild-type M. tuberculosis but not by the RD1 mutant. Because comparative quantitation by MS is difficult where samples are significantly different (i.e., in the presence or absence of CFP-10 and ESAT-6), we required that proteins be identified in multiple independent samples of the wild-type culture filtrates and not be detected in any of the H37RvΔRD1 culture filtrates. Thus, this screen was specific for differences in proteins that are relatively abundant in wild-type culture filtrates, but it was not comprehensive. By these stringent criteria, we identified four proteins present in wild-type cultures but absent from H37RvΔRD1 culture filtrates. These included three genes encoded within the ESX-1 region: ESAT-6, CFP-10, and PE35, which has not previously been identified in culture filtrates from M. tuberculosis. A single unlinked gene product, encoded by Rv3616c and here named espA, was identified as missing from the mutant culture filtrates (Fig. 1 A and B).
Fig. 1.
EspA is present in H37Rv culture filtrates but not in RvDRD1 culture filtrates. (A and B) EspA identified by MS as present in H37Rv culture filtrates but not in RvΔRD1 culture filtrates. Culture filtrate proteins from H37Rv (A) and H37RvΔRD1 (B) were tryptically digested. In parallel runs, the resultant peptide fragments were resolved through reverse-phase chromatography and analyzed by LCQ-MS/MS. The tracing indicated by the arrow was identified as peptide KYSEGAAAGTEDAERAPVEADAGGGQK from EspA, which was one of four unique peptides identified from EspA. (C) The espA locus shares significant homology to genes upstream of the ESX-1 locus. Rv3616c, here renamed EspA, shares 31.75% identity in a 400-aa overlap with Rv3864. Rv3615c shares 36.25% identity in a 102-aa overlap with Rv3865. Rv3614c shares 54.9% identity in a 173-aa overlap with Rv3867. (D) Presence of EspA in the pellet (P) and supernatant (S) of H37Rv, H37RvΔespA (ΔespA), and H37RvΔRD1 (ΔRD1). Results are representative of more than three experiments.
EspA is a 40-kDa protein without predicted signal sequence or putative transmembrane domains. It is a conserved hypothetical protein of unknown function, but a global transposon analysis of mycobacterial genes required for infection predicted that espA is necessary for the virulence of M. tuberculosis (19). Moreover, the predicted amino acid sequence of EspA is 31% identical to Rv3864, which lies immediately 5′ of the ESX-1 region (Fig. 1C). EspA appears to be in a three-gene operon with Rv3615c and Rv3614c, which show significant homology to Rv3865 and Rv3867, respectively. The homology of the espA operon to genes flanking the ESX-1 locus suggested that EspA might indeed be secreted by or associate with the ESX-1 apparatus.
We confirmed that EspA is not present in H37RvΔRD1 culture filtrates using both epitope-tagged EspA (data not shown) and antibody to native EspA protein. EspA was present in both the cell lysates and supernatants of wild-type mycobacteria. However, the protein was completely absent from the supernatants of the H37RvΔRD1 strain, confirming the LCQ-MS/MS findings (Fig. 1D). In addition, EspA appeared to accumulate in the cell-associated fraction of the H37RvΔRD1 strain, indicating that the protein is produced normally but is retained in the bacterial cell cytoplasm.
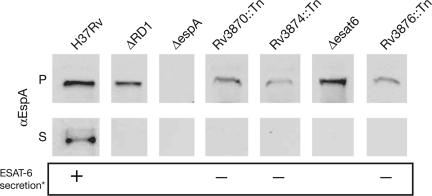
Individual Genes in the ESX-1 Region Are Required for Export of EspA. Mutations in individual genes of the ESX-1 region block secretion of ESAT-6 and CFP-10 and attenuate M. tuberculosis (13, 14, 16). Similarly, mutations in several of the putative structural genes of the ESX-1 apparatus abolish export EspA (Fig. 2). These results are consistent with several models of EspA, ESAT-6, and CFP-10 secretion. EspA and the ESAT-6/CFP-10 complex could be secreted independently by the ESX-1 apparatus. Alternatively, either EspA or ESAT-6 and CFP-10 could be secreted by the apparatus and then form a distal part of the structure, promoting secretion of the other protein(s). To test these models, we assessed the secretion of EspA, ESAT-6, and CFP-10 in the absence of each other.
Fig. 2.
Localization of EspA in wild-type M. tuberculosis and strains carrying deletions of RD1 (ΔRD1), of esat-6 (Δesat-6), and transposon insertions in the indicated genes. Cell pellet (P) and culture supernatant (S) proteins were resolved through SDS/PAGE. EspA was identified by immunoblotting with protein-specific antibody. Results are representative of at least two experiments. *, ESAT-6 secretion as published in ref. 16.
ESAT-6 and CFP-10 form a tight 1:1 complex in vitro and are thought to be secreted as a heterodimer by the ESX-1 apparatus (22). Therefore, we looked at EspA expression and secretion in M. tuberculosis strains carrying a deletion of esat-6 (H37RvΔesat6) or a transposon insertion in cfp-10 (Rv3874::Tn). In H37RvΔesat6, EspA accumulated within the cell pellet but was not found in the culture filtrate (Fig. 2). Likewise, Rv3874::Tn, which expresses but does not secrete ESAT-6, failed to secrete EspA (Fig. 2). Thus, both ESAT-6 and CFP-10 are required for the export of EspA.
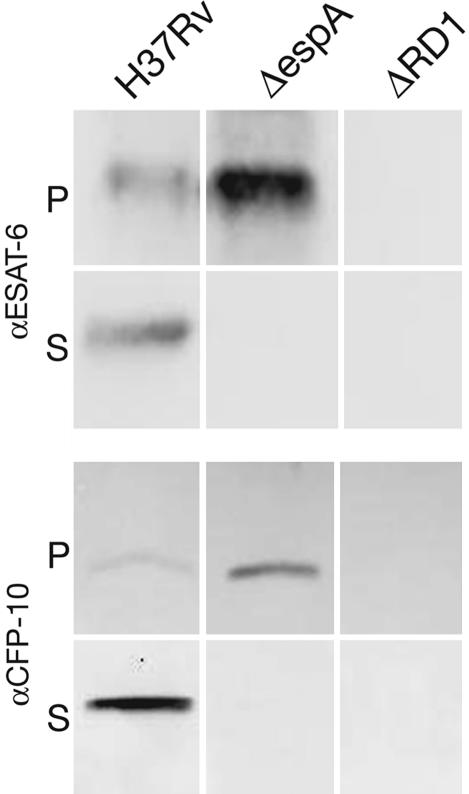
EspA Is Required for Secretion of ESAT-6 and CFP-10. To determine whether EspA is required for ESX-1 function as assessed by ESAT-6 and CFP-10 secretion, we constructed a deletion mutant of espA. In wild-type M. tuberculosis, ESAT-6 was found in both the cell pellet and culture filtrate. However, in H37RvΔespA, ESAT-6 was absent from the culture filtrate and consistently accumulated in the cell pellet (Fig. 3). Thus, EspA is required for ESAT-6 secretion. Likewise, wild-type M. tuberculosis expressed and secreted CPF-10. In H37RvΔespA, secretion of CFP-10 was inhibited and the protein accumulated in the cell pellet (Fig. 3).
Fig. 3.
Localization of ESAT-6 and CFP-10 in the presence or absence of espA. Cell pellet (P) and culture supernatant (S) proteins from H37Rv, H37RvΔRD1 (ΔRD1), and H37RvΔespA (ΔespA) were resolved through SDS/PAGE. ESAT-6 and CFP-10 were identified by immunoblotting with protein-specific antibody. Results are representative of more than three experiments.
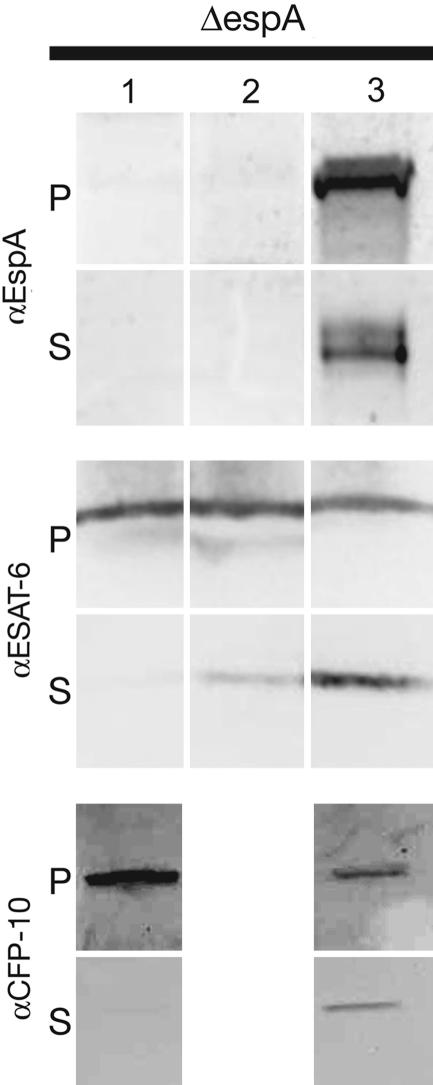
Complementation of H37RvΔespA by integration of a single copy of espA restored ESAT-6 and CFP-10 secretion (Fig. 4). Several complementing constructs were made that expressed and secreted EspA at varying levels. In the presence of these constructs, ESAT-6 secretion was proportional to the amount of EspA expression (Fig. 4).
Fig. 4.
Complementation of H37RvΔespA for ESAT-6 and CFP-10 secretion. H37RvΔespA was complemented by integration of a single copy of a control vector (lane 1), epitope-tagged espA under a synthetic MOP promoter (pALE2) (lane 2) or epitope-tagged espA under its native promoter (pALE4) (lane 3) into the chromosomal attB site. Cell pellet (P) and culture supernatant (S) proteins from the complemented strain or a strain carrying the backbone vector were resolved through SDS/PAGE. EspA, ESAT-6, and CFP-10 were identified by immunoblotting with protein-specific antibody. Pellet-associated EspA expressed from pALE2 could be visualized only by overexposing the blot. Results are representative of two experiments.
RvΔespA Is Attenuated in Mice to a Similar Extent as H37RvΔRD1. Previous work has shown that the ESX-1 region is required for virulence in mice. Deletion of RD1 has been shown to attenuate mycobacterial replication and virulence in immunocompetent mice and to delay mortality in immunocompromised severe combined immunodeficient (SCID) mice. Because our data indicate that EspA is both secreted by the ESX-1 apparatus and necessary for ESX-1 function as measured by ESAT-6 and CFP-10 secretion, it was important to explore whether EspA is required for mycobacterial virulence in these mouse models.
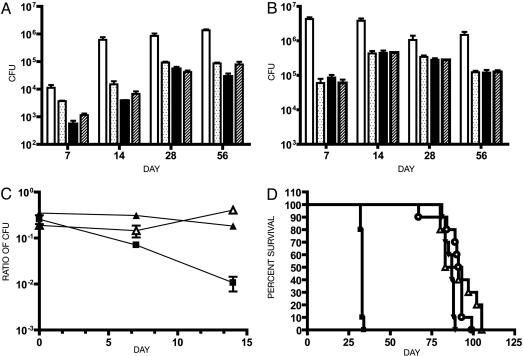
C57BL/6 mice were infected with H37Rv, H37RvΔRD1, H37RvΔespA, or H37RvΔRD1ΔespA. The mycobacterial organ burden in lung and spleen was measured at 1, 2, 4, and 8 weeks. Consistent with data published in ref. 12, H37RvΔRD1 was highly attenuated relative to wild type in both lung and spleen at all time points (Fig. 5 A and B). At all time points, H37RvΔespA or H37RvΔRD1ΔespA was also highly attenuated in both lung and spleen. In both organs they appeared attenuated to an extent similar to or greater than H37RvΔRD1.
Fig. 5.
Deletion of espA attenuates M. tuberculosis to a similar extent as deletion of RD1. Lungs (A) and spleen (B) colony-forming units from C57BL/6 mice infected with the indicated mycobacterial strains. The infecting inocula were determined by plating serial dilutions as follows: open bars, H37Rv, 1.9 × 105; dotted bars, H37RvΔRD1, 1.6 × 105; filled bars, H37RvΔespA, 1.2 × 105; hashed bars, H37RvΔRD1ΔespA, 2.1 × 105. Numbers represent the mean and standard deviation of four mice per time point. Each of the mutants was significantly attenuated compared to H37Rv in both organs at all time points except in the spleen colony-forming units at 28 days (P ≤ 0.01). In addition, the lung burden of RvΔRD1 was slightly but significantly higher than that of H37RvΔespA and H37RvΔRD1ΔespA at 7 days and H37Rv ΔespA at 14 days (P < 0.05). (C) C57BL/6 mice were infected with two strains of M. tuberculosis in competition. Mice were infected with a 1:3 mixture of kanamycin marked to unmarked strains as indicated: ▪, H37RvΔRD1:H37Rv; ▴, H37RvΔRD1:H37RvΔespA; ▵, H37RvΔRD1ΔespA:H37RvΔespA. The organ burden of each strain was determined at the indicated times by plating in the presence and absence of kanamycin. Numbers represent the mean and standard deviation of three mice per time point. H37RvΔRD1 was significantly attenuated relative to H37Rv over time as determined by linear regression analysis (P < 0.05). In the other two competitions, the ratios of the strains did not change significantly over time. (D) SCID mice were infected with a goal inoculum of 300 organisms. The infecting inocula, determined by plating serial dilutions, were as follows: ▪, H37Rv, 3 × 102; ▵, H37RvΔRD1, 7 × 102; ▾, H37RvΔespA, 23 × 102; ○, H37RvΔRD1ΔespA, 7 × 102. Ten mice were infected with each strain. The difference in survival between wild-type and mutant strains was highly statistically significant (P < 0.0001), and the times to death of RvΔespA and RvΔRD1ΔespA were not significantly different from that of RvΔRD1.
Lung histopathology was examined at all time points. At 8 weeks, wild-type M. tuberculosis caused significant disease with focal infiltrates containing foamy macrophages and lymphocytic aggregates (data not shown). In the sections examined, ≈50% of the lung was diseased. In contrast, all of the mutants caused far less disease, although small infiltrates of similar cellular composition to the wild-type infection were evident. At most, 5-10% of the lung was involved in the multiple sections studied (data not shown). There was no clear difference in histopathology between the mutants.
There are several loci that encode ESX-1-related proteins within the mycobacterial genome, which are of unknown significance in vivo. We hypothesized that EspA might be required for the secretion of some of these proteins. If this were true, deletion of EspA might be more attenuating than mutations in the ESX-1 locus. In the infections described above, H37RvΔespA and H37RvΔRD1ΔespA appeared slightly more attenuated than H37RvΔRD1 in the lung at 7 and 14 days. However, it is difficult to make rigorous comparisons of the relative fitness of different strains in single-strain infections; therefore, we performed competition experiments to assess the relative fitness of wild-type H37Rv vs. H37RvΔRD1, H37RvΔRD1 vs. H37RvΔespA, and H37RvΔespA vs. H37RvΔRD1ΔespA (Fig. 5C). H37RVΔRD1 was ≈100-fold less abundant than wild type in the lung at 2 weeks after infection. Deletion of espA attenuated M. tuberculosis to a similar degree as deletion of the RD1 locus. M. tuberculosis carrying both deletions was no more attenuated than the single mutants.
Finally, we examined the relative virulence of H37Rv, H37RvΔRD1, H37RvΔespA, or H37RvΔRD1ΔespA in a SCID mouse model of mycobacterial virulence. SCID mice infected with wild-type H37Rv died at 35 days after infection, whereas those infected with the mutant strains died at ≈90 days after infection. The difference between wild-type and mutant strains was highly statistically significant, whereas the times to death of H37RvΔespA and H37RvΔRD1ΔespA were not significantly different from H37RvΔRD1 (Fig. 5D).
Discussion
The secretion of ESAT-6 and CFP-10, critical to the virulence of M. tuberculosis, appears to be mediated by a secretion apparatus encoded by the surrounding genes (13, 14, 16). We have shown that the same genes are required to secrete an additional protein, EspA. Unexpectedly, the secretion of EspA also depends on the presence of ESAT-6 and CFP-10. Indeed, secretion of EspA, ESAT-6, and CFP-10 is codependent. Each member of the trio of proteins is secreted and is required for the others' secretion. The importance of the association between EspA and the ESX-1-encoded secretory apparatus is confirmed in mice where deletion of espA from both wild-type M. tuberculosis and H37RvΔRD1 results in identical attenuation as the RD1 deletion alone.
M. tuberculosis contains many loci encoding homologues of esat-6 and cfp-10 as well as some of the surrounding genes in the ESX-1 region (8, 23). It is certainly possible that EspA is required for the secretion of other ESAT-6- and CFP-10-like proteins. Indeed, deletion of common, less redundant components of these secretion systems may provide a window into their biology. Although our studies in mice suggest that the primary contribution of EspA to virulence comes from its critical role in ESX-1 function, the mouse model does not fully recapitulate human disease, and it is possible that these other loci have important roles that we fail to detect.
There are alternate hypotheses to explain the observation that secretion of ESAT-6, CFP-10, and EspA is mutually dependent. It is possible that the data could be explained by a lack of protein stability in culture supernatants rather than secretion; however, we feel that this is unlikely, given that the proteins appear individually stable in the cell pellets and that ESAT-6 and CFP-10 can be stably expressed and purified alone (22).
A more likely explanation for our observations is that ESAT-6, CFP-10, and EspA must associate, either directly or indirectly, to be recognized and transported by the ESX-1 secretory apparatus. For example, the proteins may form a heterocomplex in the cytoplasm, raising the possibility that one of these components may act as a chaperone for the others.
Alternatively, ESAT-6, CFP-10, and EspA may be obligate components of the transport machinery itself. Loss of any one of these proteins may alter the integrity of the apparatus and disrupt secretion of all three. If ESAT-6, CFP-10, and/or EspA are structural components of a secretory apparatus, we must account for the large amounts of each protein found in the supernatants of broth-grown mycobacteria. Conceivably, the ESAT-6/CFP-10/EspA portion of the apparatus may be fragile and dissociate when cells are manipulated. Of course, it is possible that ESAT-6, CFP-10, and/or EspA are both structural components of the secretion machinery and direct effectors of virulence. Other secreted proteins behave in multiple ways. For example, the Yersinia effector LcrV is secreted by the type III secretion system and is required for the secretion of other type III effectors (24, 25).
These models predict different mechanisms for how these proteins might affect virulence and suggest a variety of approaches to future investigations. If the proteins must associate before secretion, it will be difficult to study individual proteins in the absence of all components, some of which might not yet be identified. If these proteins form part of the transport apparatus itself, the apparatus may secrete other molecules that are the direct effectors of virulence. Because our LCQ-MS/MS screen was not exhaustive, it does not preclude the possibility that other proteins may be secreted in an ESX-1-dependent manner. However, such effectors may be synthesized or secreted only during infection or may not necessarily be proteins. Finally and most intriguingly, if the proteins remain stably associated after secretion, it suggests that the complex itself might be necessary for biologic activity and virulence. Pursuing the mechanisms of the reciprocal interactions of EspA, ESAT-6, and CFP-10 should provide insights into alternative mechanisms of secretion, their contribution to virulence, and ultimately, the mechanism of ESX-1 mediated virulence itself.
Acknowledgments
We are grateful for the excellent technical help of Ilona Breiterene, Larry Pipkin, and Reiling Liao and the scientific insights of Roderick Bronson and Sloan Siegrist. This work was supported by National Institutes of Health Grants AI48704 and AI51929 (to E.J.R.), AI07118 and AI23545 (to B.R.B.), HL64550 and HL68533 (to D.R.S.), and AI50734 (to S.M.F.). S.M.F. was also supported by a Roche Postdoctoral Fellowship Award from the Infectious Diseases Society of America and a Postdoctoral Research Fellowship for Physicians from the Howard Hughes Medical Institute.
Abbreviations: MS/MS, tandem MS; SCID, immunocompromised severe combined immunodeficient.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ086480 and DQ068700).
References
- 1.Finlay, B. B. & Falkow, S. (1997) Microbiol. Mol. Biol. Rev. 61, 136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galan, J. E. & Collmer, A. (1999) Science 284, 1322-1328. [DOI] [PubMed] [Google Scholar]
- 3.Cascales, E. & Christie, P. J. (2003) Nat. Rev. Microbiol. 1, 137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahairas, G. G., Sabo, P. J., Hickey, M. J., Singh, D. C. & Stover, C. K. (1996) J. Bacteriol. 178, 1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pym, A. S., Brodin, P., Brosch, R., Huerre, M. & Cole, S. T. (2002) Mol. Microbiol. 46, 709-717. [DOI] [PubMed] [Google Scholar]
- 6.Mostowy, S., Cousins, D. & Behr, M. A. (2004) J. Bacteriol. 186, 104-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodin, P., Rosenkrands, I., Andersen, P., Cole, S. T. & Brosch, R. (2004) Trends Microbiol. 12, 500-508. [DOI] [PubMed] [Google Scholar]
- 8.Gey Van Pittius, N. C., Gamieldien, J., Hide, W., Brown, G. D., Siezen, R. J. & Beyers, A. D. (2001) Genome Biol. 2, RESEARCH0044. [DOI] [PMC free article] [PubMed]
- 9.Pallen, M. J. (2002) Trends Microbiol. 10, 209-212. [DOI] [PubMed] [Google Scholar]
- 10.Berthet, F. X., Rasmussen, P. B., Rosenkrands, I., Andersen, P. & Gicquel, B. (1998) Microbiology 144, 3195-3203. [DOI] [PubMed] [Google Scholar]
- 11.Sorensen, A. L., Nagai, S., Houen, G., Andersen, P. & Andersen, A. B. (1995) Infect. Immun. 63, 1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis, K. N., Liao, R., Guinn, K. M., Hickey, M. J., Smith, S., Behr, M. A. & Sherman, D. R. (2003) J. Infect. Dis. 187, 117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu, T., Hingley-Wilson, S. M., Chen, B., Chen, M., Dai, A. Z., Morin, P. M., Marks, C. B., Padiyar, J., Goulding, C., Gingery, M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 12420-12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanley, S. A., Raghavan, S., Hwang, W. W. & Cox, J. S. (2003) Proc. Natl. Acad. Sci. USA 100, 13001-13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volkman, H. E., Clay, H., Beery, D., Chang, J. C., Sherman, D. R. & Ramakrishnan, L. (2004) PloS Biol. 2, e367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guinn, K. M., Hickey, M. J., Mathur, S. K., Zakel, K. L., Grotzke, J. E., Lewinsohn, D. M., Smith, S. & Sherman, D. R. (2004) Mol. Microbiol. 51, 359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, L. Y., Guo, S., McLaughlin, B., Morisaki, H., Engel, J. N. & Brown, E. J. (2004) Mol. Microbiol. 53, 1677-1693. [DOI] [PubMed] [Google Scholar]
- 18.Hondalus, M. K., Bardarov, S., Russell, R., Chan, J., Jacobs, W. R., Jr., & Bloom, B. R. (2000) Infect. Immun. 68, 2888-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sassetti, C. M. & Rubin, E. J. (2003) Proc. Natl. Acad. Sci. USA 100, 12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tullius, M. V., Harth, G. & Horwitz, M. A. (2001) Infect. Immun. 69, 6348-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braunstein, M., Espinosa, B. J., Chan, J., Belisle, J. T. & Jacobs, W. R., Jr. (2003) Mol. Microbiol. 48, 453-464. [DOI] [PubMed] [Google Scholar]
- 22.Renshaw, P. S., Panagiotidou, P., Whelan, A., Gordon, S. V., Hewinson, R. G., Williamson, R. A. & Carr, M. D. (2002) J. Biol. Chem. 277, 21598-21603. [DOI] [PubMed] [Google Scholar]
- 23.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., III, et al. (1998) Nature 393, 537-544. [DOI] [PubMed] [Google Scholar]
- 24.Pettersson, J., Holmstrom, A., Hill, J., Leary, S., Frithz-Lindsten, E., von Euler-Matell, A., Carlsson, E., Titball, R., Forsberg, A. & Wolf-Watz, H. (1999) Mol. Microbiol. 32, 961-976. [DOI] [PubMed] [Google Scholar]
- 25.Sing, A., Rost, D., Tvardovskaia, N., Roggenkamp, A., Wiedemann, A., Kirschning, C. J., Aepfelbacher, M. & Heesemann, J. (2002) J. Exp. Med. 196, 1017-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]