Abstract
Cyclin-dependent kinases (CDKs) are important for both mitotic and meiotic cell cycles. In fission yeast, the major CDK, Cdc2p is involved in premeiotic DNA replication and in meiosis II. One of its partners, the mitotic cyclin Cdc13p is known to be required for meiosis, whereas there are no studies on the G1/S cyclin Cig2p. In this article, we have studied the regulation of the Cdc2p/Cdc13p and Cdc2p/Cig2p complexes during synchronous meiosis. We observed that Cdc2p/Cig2p kinase is activated in an unexpected biphasic manner, first at onset of premeiotic S phase and again during meiotic nuclear divisions. The role of Cig2p during meiosis was investigated using cig2-deleted strains that exhibit delays in onset of both S phase and meiotic divisions as well as an inefficient completion of MII. Furthermore, analysis of cig2 transcripts revealed a meiosis-specific regulation of cig2 expression during MI/MII dependent upon the Mei4p transcription factor leading to a different transcription start site at this stage of meiosis.
INTRODUCTION
Meiosis generates haploid gametes from diploid parental cells. After premeiotic S phase, cells undergo successive reductional (MI) and equational (MII) divisions without a further intervening round of DNA replication. In the fission yeast Schizosaccharomyces pombe, cells initiate sexual development when they are starved of nitrogen. This can occur either when two haploid cells of opposite mating types mate or in diploid cells at the end of the vegetative growth and results in four nuclei each within an ascospore. The pathway controlling entry into meiosis is well understood in fission yeast (for review Yamamoto, 1996); the pat1 gene encodes a protein kinase (McLeod and Beach, 1986) that negatively regulates entry into meiosis, and the temperature-sensitive pat1–114 allele initiates meiosis at high temperature (Iino and Yamamoto, 1985; Nurse, 1985). Nitrogen starvation induces the expression of specific genes required for mating and meiosis (Shimoda et al., 1985; reviewed in Yamamoto, 1996), including mei2, which encodes an RNA-binding protein inactivated by Pat1p (Watanabe and Yamamoto, 1994; Watanabe et al., 1997), mei3, a Pat1p inactivator (McLeod and Beach, 1988), mei4, which encodes a transcription factor (Horie et al., 1998) essential for entry into meiosis I, and mes1, which is essential for the second meiotic division (Shimoda et al., 1985).
The cyclin-dependent kinases (CDKs), with their regulatory partners the cyclins, regulate the major cell cycle transitions in eukaryotes and are involved in both mitotic and meiotic cell cycles. In fission yeast mitotic cycles, the Cig2p and Cig1p B-cyclins control S phase (Connolly and Beach, 1994; Obara-Ishihara and Okayama, 1994; Martin-Castellanos et al., 1996), and Cdc13p is the M-phase cyclin (Booher and Beach, 1987; Hagan et al., 1988). These cyclins all form complexes with Cdc2p. In a cig2Δ strain, Cdc13p substitutes for Cig2p to bring about DNA replication after a short delay (Fisher and Nurse, 1996; Mondesert et al., 1996). Cig2p cooperates with Cig1p to promote progression through DNA replication (Connolly and Beach, 1994). It has been proposed that Cig2p and Cdc13p share overlapping functions in mitosis, although they are not functionally redundant (Bueno and Russell, 1993). Cdc2p in Cdc2p/Cdc13p complexes is regulated by phosphorylation on its Y15 inhibitory residue (Gould and Nurse, 1989) brought about by the inhibitory kinases Wee1p (Featherstone and Russell, 1991; Parker et al., 1992) and Mik1p (Lundgren et al., 1991; Lee et al., 1994) and is removed by the activating phosphatase Cdc25p (Millar et al., 1991). Cdc2p is required for premeiotic DNA replication (Iino et al., 1995) and the second meiotic division (Grallert and Sipiczki, 1989, 1990, 1991; Iino et al., 1995), but its role in meiosis I is unclear. Y15 phosphorylation of Cdc2p appears after premeiotic DNA replication as Wee1p levels increase (Daya-Makin et al., 1992), and cdc25 is required for both meiotic divisions (Iino et al., 1995). The cdc13 gene is essential for both meiosis I and II (Iino et al., 1995) and high Cdc2p/Cdc13p kinase activity has been reported during meiotic divisions (Murakami and Nurse, 1999). Deletion of cig2 (cyc17) enhances conjugation (Obara-Ishihara and Okayama, 1994), and so Cig2p is likely to be a negative regulator of the initiation of conjugation and meiosis. However, nothing is known concerning the regulation and role of Cig2p during meiosis.
Cig2 expression fluctuates during vegetative growth peaking in G1 and S phases and is dependent on the Cdc10p transcriptional machinery (Obara-Ishihara and Okayama, 1994; Aytéet al., 2001). Cig2 mRNA is induced during conjugation and nitrogen starvation, an induction dependent on Cdc10p/Res2p, but there is no report on cig2 expression during meiosis itself. Expression of cdc13 mRNA is induced during meiosis and is reported to be reduced in mei4Δ strains (Iino et al., 1995). Mei4p is a transcription factor essential for MI entry and has a forkhead DNA-binding domain that in humans binds genes containing the heptamer core, GTAAAYA (Horie et al., 1998). Other Mei4p-dependent genes include spo6, which is required for meiosis II and sporulation, and the mde genes (Abe and Shimoda, 2000). In this article, we study the regulation and kinase activities of the Cdc2p/Cdc13p and Cdc2p/Cig2p complexes during meiosis. We establish that Cig2p has a role during meiosis and demonstrate that there is a specific regulation of cig2 expression in MI.
MATERIALS AND METHODS
Fission Yeast Strains and Methods
Strains used were constructed using standard procedures (Moreno et al., 1991) and are given in Table 1. The diploid strains homozygous for the mating-type locus were constructed by normal h+ × h− crosses (usually using the ade6-M210 and ade6-M216 markers) at 25°C on YEPD plates (1% yeast extract YE, 2% peptone, 2% glucose). After crosses, cells were grown on minimal medium with glutamate as a nitrogen source, and stable diploid colonies homozygous for the mating-type locus were selected as nonstaining colonies after 5–10 min exposure to iodine. Synchronous meiosis was induced in liquid culture using haploid or diploid pat1ts mutants (Bähler et al., 1991; Murakami and Nurse, 1999). Cells were grown at 25°C in YES to stationary phase and diluted in MM–NH4Cl–glucose medium supplemented with 5 mg/ml NH4Cl, 1% glucose, and 100 μg/ml leucine to 2 × 107 cells/ml at 25°C. Cells were then filtered through a Millipore membrane, washed with MM–NH4Cl, resuspended at a concentration of 5 × 106 cells/ml in MM–NH4Cl–glucose supplemented with 1% glucose and 50 μg/ml leucine and incubated for 16 h at 25°C for G1 arrest. Meiosis was induced by shifting the temperature to 34°C after addition of 500 μg/ml NH4Cl and 50 μg/ml leucine to the culture medium.
Table 1.
Strains constructed and/or used during this study
| Number | Genotype |
|---|---|
| PN1 | 972 h− |
| PN2 | 968 h90 |
| PN4 | 975 h+ |
| PN1675 | pat1-114 leu1-32 h− |
| PN1926 | cig2∷ura4 ura4-D18 h+ |
| PN1942 | cig2∷ura4 ura4-D18 h− |
| PN2158 | pat1-114/pat1-114 leu1-32/leu1-32 ade6-M210/ade6-M216 h−/h− |
| PN2298 | pat1-114 cig2∷ura4 ura4-D18 leu1-32 h− |
| PN2448 | pat1-114 mei4∷ura4 ura4-D18 leu1-32 ade6-M210 h+ |
| PN2468 | pat1-114/pat1-114 mes1∷LEU2/mes1∷LEU2 leu1-32/leu1-32 ade6-M210/ade6-M216 h−/h− |
| PN2758 | pat1-114 cig2∷ura4 ura4-D18 leu1-32 ade6-M210 h+ |
| PN2991 | pat1-114/pat1-114 cig2∷ura4/cig2∷ura4 ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-M210/ade6-M216 h+/h+ |
| PN3796 | cig2∷ura4 ura4-D18 h90 |
| HM1391 | pat1-114 cig1∷ura4 cig2∷ura4 ura4-D18 leu1-32 h+ |
DAPI Staining
For each meiotic time point, cells were fixed in 70% ethanol and stored. After washing the cells with water, DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; Moreno et al., 1991). The number of nuclei of at least 200 cells was counted under the microscope.
Flow Cytometric Analysis
We used a Becton Dickinson FACScan (fluorescence-activated cell sorter; Mountain View, CA) for flow cytometric analysis. Fixed cells were washed with 50 mM Na citrate and incubated overnight at 37°C with 0.1 mg/ml Rnase A in Na citrate. Then, DNA were stained with propidium iodide (Sazer and Sherwood, 1990).
Dissection and Random Spore Analysis
h+ and h− strains were crossed or h90 plated on YEPD plates and incubated at 25°C for 2 d. Asci were dissected and spores analyzed by FACS to determine their ploidy. For random spore analysis, asci were treated with Helicase (Helix pomatia juice; IBF Biotechnics, Columbia, MD) to break down the ascus wall and kill vegetative cells (Moreno et al., 1991), and the spores were grown at 25°C on YES and YEP (YE + phloxin B) plates for 2 d. The presence of diploid colonies was measured by incorporation of phloxin B (for details see Moreno et al., 1991) and confirmed by microscopic examination (larger cells).
Protein Extraction
Boiled and native extracts were prepared as described in Borgne and Nurse (2000).
Immunoprecipitation and Histone H1 Kinase Assay
One milligram of native extract was incubated at 4°C for 45 min on a rotating wheel with the polyclonal anti-Cdc13p (SP4, 2.5 μl/ml; Correa-Bordes and Nurse, 1995) or the anti-Cig2p (Moc6, 6 μl/ml; O'Connell and Correa-Bordes, unpublished data) antibodies. Then, 30 μl of preequilibrated protein A-Sepharose beads (Amersham Pharmacia Biotech Inc., Piscataway, NJ) was added for further 45 min. Beads were washed three times with HB buffer (25 mM MOPS, pH 7.2, 60 mM β-glycerophosphate, 15 mM p-nitrophenyl phosphate, 15 mM MgCl2, 15 mM EGTA, 1 mM DTT, 0.1 mM sodium vanadate, 1% Triton X-100, 1 mM PMSF, protease inhibitor cocktail), and the immunoprecipitated complex was analyzed for its histone H1 kinase activity. Beads were incubated for 20 min at 30°C with 20 μl of KIN buffer (1 mg/ml histone H1 [Calbiochem] and 200 μM [γ-32P] ATP [Amersham Pharmacia Biotech Inc.] in HB buffer) and boiled for 3 min in 20 μl of 2× Laemmli sample buffer before SDS-PAGE. Phosphorylated histone H1 was detected by autoradiography and quantitated using a PhosphorImager.
Western Blotting
Samples were run in 12% SDS-polyacrylamide gels (Laemmli, 1970). Proteins were then blotted to Immobilon-P membrane (Millipore, Bedford, MA) and detected using ECL (Amersham Pharmacia Biotech Inc.). The following antibodies were used: the polyclonal anti-Cdc13p (SP4, 1:1000; Correa-Bordes and Nurse, 1995) and anti-PY15 (1:1000; New England Biolabs, Beverly, MA) antibodies and the monoclonal anti-Cdc2p (Y63–2, 1:500) and anti-Cig2p (5E3–4, 1:2000) antibodies (gifts of Dr. H. Yamano).
RNA Preparation and Northern Blot Analysis
Fifty milliliters of cells was taken every half an hour during synchronous meiotic time courses then washed in ice-cold Stop buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3, pH 8), frozen in liquid nitrogen, and kept at −70°C. RNA was prepared as already described (Baum et al., 1997). The [α-32P]dATP probes were prepared by random oligo priming using a Prime-It kit (Stratagene, La Jolla, CA). The template DNA for the probes were as follows: an XhoI-EcoRV cdc13 fragment (1.9 kb) from pSAB1 cdc13 plasmid (lab), a BamHI-SacI cig1 fragment (0.96 kb) from pREP1 cig1 plasmid (lab), and for cig2, an NdeI-EcoRV fragment (0.91 kb in ORF), a HpaII-NdeI fragment (1 kb in 5′ untranslated region), and a SacI-BamHI fragment (0.8 kb in 3′ untranslated region) from a genomic cig2 clone in pAL-SK (Sergio Moreno). A DraI-EcoRV his3 fragment was used as loading control.
RNA Preparation and Primer Extension
RNA from synchronous meiotic culture (2- and 4-h time points) was prepared on a CsCl gradient. Cells were broken with acid-washed glass beads (Sigma, St. Louis, MO) with the FastPrep system (BIO 101) in 4 M guanidine thiocyanate, 0.25 mM Na citrate, pH 7.0, 0.5% sarkosyl, and 0.1 M β-mercaptoethanol. After 10 min centrifugation at 13,000 rpm, the supernatant was centrifuged in presence of 5.7 M CsCl for 16 h at 26,000 rpm at 20°C. RNA was then phenol/chloroform extracted and EtOH precipitated. To determine the cig2 transcriptional start sites, primer extension was carried out using synthetic oligonucleotides called A, B, C, D, E, and F, where A is 5′-CTTCAAGGTGGATCCAACCTTTGG-3′ (+84 to +107) and F is 5′-GGTAAATTAATCTCAATTGACAAG-3′ (+1172 to +1195). The oligonucleotides (0.1 μg) were end-labeled with [γ-32P]ATP using the T4 polynucleotide kinase (4 U) for 30 min at 37°C. After purification on a ProbeQuant G-50 microcolumn (Amersham Pharmacia Biotech Inc.), the oligonucleotides were mixed to 14 μg RNA, heated at 95°C for 5 min, hybridized for 1 h at 67°C (for primer A) or 63°C (for primer F) in the presence of 120 mM NaCl, and slowly cooled down to room temperature. The extension reaction was performed using MuLV reverse transcriptase (25 U) in the presence of 100 mM Tris, pH 8.7, 12 mM MgOAc, 20 mM DTT, and 5 mM each dNTP at 45°C for 1 h. After incubation at 37°C for 15 min with RNase A, phenol/chloroform extraction and EtOH precipitation were performed. The primer-extended products were boiled in 2× formamide loading dye and separated on a 8.3 M urea, 8% polyacrylamide gel using the Sequagel sequencing system. The gel was exposed overnight to BioMax film. The sizes of the resulting labeled primer-extended products were inferred from their position relative to φx174 DNA/HinfI markers (Promega) labeled with [γ-32P]ATP.
RESULTS
Cdc2p Regulation during pat1-induced Meiosis
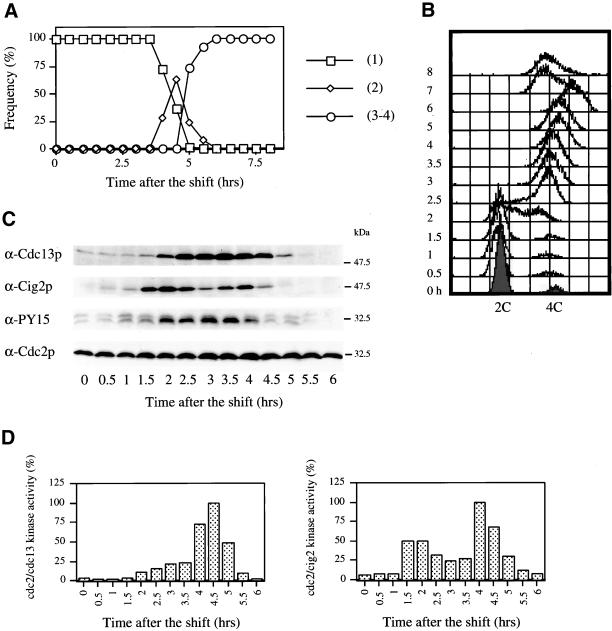
We monitored Cdc2p regulation during the meiotic cell cycle by inducing fission yeast pat1ts/pat1ts cells to undergo meiosis by nitrogen starvation and shifting the temperature from 25 to 34°C (Figure 1). Meiotic progression was monitored by counting nuclei number (Figure 1A) and by measuring DNA content (Figure 1B). Premeiotic DNA replication started at 2 h and was completed by 2.5 h (Figure 1B), meiosis I (2 nuclei) occurred at 4 h , and meiosis II (3–4 nuclei) at 5 h (Figure 1A). Spore walls formed at 7 h, resulting in a leftwards shift of the FACS profile. We investigated Cdc2p regulation by following the level of Cig2p and Cdc13p B-type cyclins and Cdc2p Y15 phosphorylation using Western blotting (Figure 1C). As cells underwent premeiotic DNA replication (2.5 h), the G2/M cyclin Cdc13p level increased and remained high until cells underwent meiosis II. The G1/S cyclin Cig2p appeared before premeiotic DNA replication (1.5 h) and declined in level immediately after completion of replication. Unexpectedly, a second peak of Cig2p was observed at 4 h despite the absence of DNA replication at this time. The level of Cdc2p was constant throughout the meiotic cell cycle, and Cdc2p was phosphorylated on its Y15 inhibitory residue from 2 to 4 h, corresponding to the period between DNA replication and the initiation of MI. Cdc2p/Cdc13p protein kinase activity was maintained at a low level until 4 h, probably because of Y15 phosphorylation, and at entry into MI rapidly increased to a peak around 4.5 h before declining by 5.5 h. Cdc2p/Cig2p kinase activity increased at 1.5 h just before premeiotic DNA replication, reduced between 2.5 and 3.5 h, and finally increased at 4 h to a second peak before declining by 5.5 h.
Figure 1.
Cyclin-B protein levels and Cdc2p-associated kinase activities during pat1-induced meiosis. (A) pat1ts/pat1ts diploid cells (strain PN2158) were grown to late log-phase, washed, transferred to −N media for 16 h at 25°C and shifted to 34°C at time 0 h. Samples were taken every 0.5 h until completion of MII. The number of nuclei were counted under the microscope after DAPI staining and is given as a percentage of total cells. (□), Cells with 1 nucleus; (⋄). cells that completed meiosis I (2 nuclei); and (○), cells that completed meiosis II (3–4 nuclei). (B) Cells progressing into meiosis were analyzed by FACS. (C) Boiled samples were analyzed by Western blotting with anti-Cdc13p (SP4), anti-Cig2p (5E3–4), anti-PY15, and anti-Cdc2p (Y63–2) antibodies. (D) Cdc2p/cyclin B complexes from native extracts have been immunoprecipitated using antibodies directed against Cdc13p (polyclonal SP4) and Cig2p (polyclonal Moc8) and analyzed for their histone H1 kinase activities. [32P]phosphate incorporation in histone H1 was measured on PhosphorImager after SDS-PAGE. The maximum of Cdc2p kinase activity (100%) was determined for Cdc13p and Cig2p and served as a reference for the other experiments (Figures 2, 4, and 5).
We conclude that the major mitotic cell cycle G2/M CDK Cdc2p/Cdc13p becomes activated to a high level at the onset of MI and remains high until exit from MII. A lower level of activation to 20–25% of this high level is present as cells proceed through premeiotic S phase and correlates with the appearance of the Cdc13p cyclin. Cdc2p Y15 phosphorylation probably restrains full CDK activation at this time. In contrast, the major mitotic G1/S CDK Cdc2p/Cig2p is activated in an unexpected biphasic manner, first at the onset of premeiotic S phase, and then again as cells proceed through MI and MII.
Cig2p Absence Delays Meiotic Progression
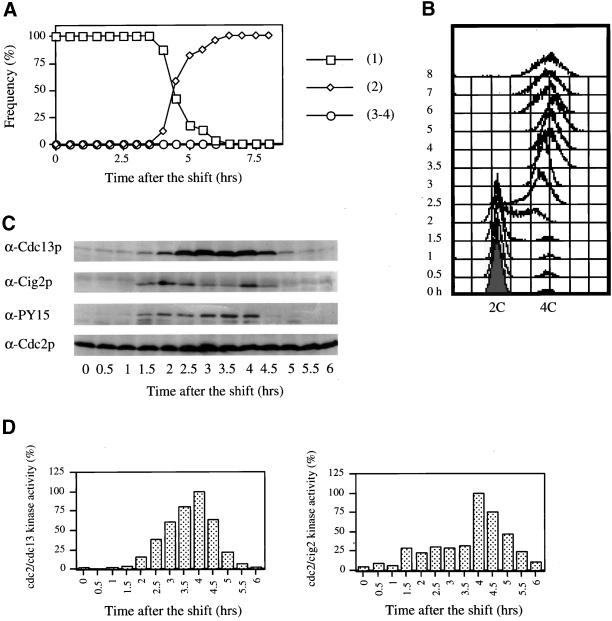
No role has been described for Cig2p during the meiotic cell cycle. Given that Cig2p levels and Cdc2p/Cig2p protein kinase activities show a biphasic pattern we monitored meiotic progression in cells deleted for cig2 using a diploid pat1ts/pat1ts cig2Δ/cig2Δ strain (Figure 2), with the cig2 ORF replaced by ura4+ (Obara-Ishihara and Okayama, 1994). Premeiotic DNA replication was delayed by ∼0.5–1 h compared with a pat1ts/pat1ts control (see Figure 1), starting at 2.5 h with completion at 3.5 h (Figure 2B). The binucleated cells (MI) reached a peak at 5 h (Figure 2A), ∼0.5 h later than the control. The number of MII cells reached a final plateau at 7.5 h , rather than 6 h as in wild-type in several different experiments. It appears that the timing between S and MII in the mutant is extended by around 1 h. Cdc13p accumulation began 0.5 h later than cells with cig2, and its disappearance was delayed by 0.5–1 h. A similar delay was observed for both Cdc2p Y15 phosphorylation (Figure 2C) and Cdc2p/Cdc13p kinase activity (Figure 2D).
Figure 2.
Cig2p absence delays meiotic progression. pat1ts/pat1ts cig2Δ/cig2Δ cells (strain PN2991) were induced to undergo meiosis as in Figure 1. Cells were analyzed by DAPI staining (A) and FACS (B). Protein levels were analyzed by Western blotting (C) and Cdc2p/Cdc13p kinase activity was measured (D) as in Figure 1.
We conclude that the presence of Cig2p is required for normal meiotic progression, and in the absence of Cig2p both the onset of premeiotic S phase and entry into the meiotic nuclear divisions are delayed by 0.5–1 h.
To address the redundancy between cig1 and cig2 in meiosis, we monitored the double cig2Δ cig1Δ mutant in a pat1ts background (Figure 3). We compared time courses in haploid pat1ts (Figure 3A), pat1ts cig2Δ (Figure 3B), and pat1ts cig2Δ cig1Δ (Figure 3C) cells (strains PN1675, PN2298, and HM1391, respectively). There was a 0.5-h delay in premeiotic DNA replication between the pat1ts control (Figure 3A, right panel) and the pat1ts/pat1ts in Figure 1. Premeiotic DNA replication in the pat1ts cig2Δ cig1Δ cells (Figure 3C, right panel) was only slightly delayed compared with the pat1ts cig2Δ cells (Figure 3B, right panel). Furthermore, the cig2Δ and cig2Δ cig1Δ strains behaved identically a 0.5-h delay in the onset of meiotic divisions compared with the wild-type strain (Figure 3, left panels). We conclude that Cig1p either has no role or only a minor one during meiosis.
Figure 3.
Cig1p has a minor role during meiosis. Meiosis was induced in pat1ts (A), pat1ts cig2Δ (B), and pat1ts cig2Δ cig1Δ (C) cells (strains PN1675, PN2298, and HM1391, respectively) as in Figure 1. Cells were analyzed by DAPI staining (left panels) and FACS (right panels).
Cig2p Involvement in Meiotic Nuclear Divisions
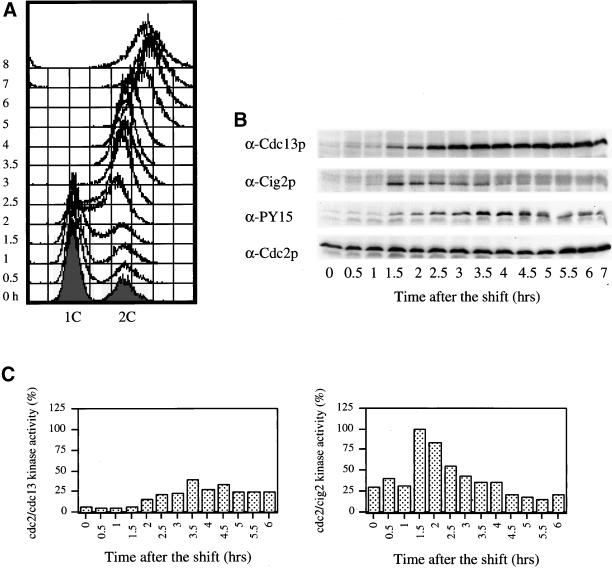
We next investigated whether the Cig2p peak in protein level was related to MI or MII. Because MI and MII occur very rapidly, we used a haploid pat1ts mei4Δ strain (Figure 4; diploids were not constructable) that blocks before MI. In this mutant, cells underwent DNA replication (Figure 4A) around 2–2.5 h and became arrested with one nucleus before MI. We found that in these arrested cells Cdc13p was at a high level (Figure 4B), but the kinase activity associated with Cdc13p remained low (Figure 4C), probably because of the maintenance of Cdc2p Y15 phosphorylation (Figure 4B). Cig2p was present during premeiotic DNA replication, decreased, and did not reappear (Figure 4B). Cdc2p/Cig2p kinase activity increased on schedule before premeiotic S phase (Figure 4C) to a level that was twice that observed during normal meiosis. After DNA replication, Cdc2p/Cig2p kinase activity decreased and remained low, consistent with the absence of a second Cig2p peak. We propose that the mei4Δ mutant arrests in G2 because of inhibitory phosphorylation of the Cdc2p/Cdc13p complex and because of the absence of the Cdc2p/Cig2p complex.
Figure 4.
The second Cig2p peak is not present in cells blocked before meiosis I. Meiosis was induced in pat1ts mei4Δ cells (strain PN2448) as in Figure 1 and was followed during 8 h. (A) DNA content was measured by FACS analysis. Western blots (B) and kinase assays (C) were performed as in Figure 1.
We next used the pat1ts/pat1ts mes1Δ/mes1Δ mutant (Figure 5) which arrests just before meiosis II. These cells underwent premeiotic DNA replication and MI (Figure 5, A and B) at the same time as the pat1ts/pat1ts control. The changes in Cdc13p and Cig2p levels were similar to the pat1ts/pat1ts strain, showing that the drop in Cdc13p and the second peak of Cig2p did not require entry into MII. The only difference was a small advancement of <0.5 h in the decline of Cdc13p level (Figure 5C). Cdc13p/Cdc2p kinase activity (Figure 5D) was also found to decline in the mes1Δ cells arrested before MII. These results demonstrate that Cdc13p and its associated kinase activity are able to decline after MI, although normally the onset of MII is so rapid that such a fall is not observed. Like the Cig2p level, the Cig2p-associated kinase activity was biphasic (Figure 5D), although the first peak remained lower than in the control. Because the second peak of Cig2 protein and activity are present in cells not undergoing MII, we conclude that Cig2p is associated either with progression through MI or the interphase period between MI and MII.
Figure 5.
The second Cig2p peak is detected in meiosis II arrested cells. pat1ts/pat1ts mes1Δ/mes1Δ cells (strain PN2468) were induced to undergo meiosis as in Figure 1. (A) The number of nuclei was counted indicating a block at a two-nuclei stage. (B) DNA content was analyzed by FACS. Western blots (C) and kinase assays (D) were performed as in Figure 1.
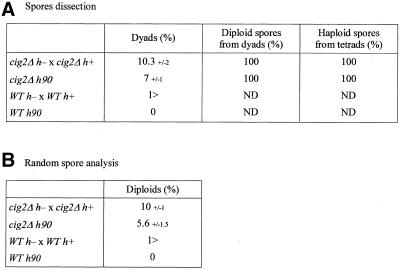
Because a delay in MII was observed for some cells in a cig2Δ strain (Figure 2A), we examined whether there was any drop in the efficiency of haploid spore formation during meiosis and sporulation. Two cig2Δ strains of opposite mating type were crossed and a cig2Δ h90 strain plated on YEPD medium (Figure 6). Wild-type strains were also crossed as control. The asci were dissected under the microscope (Figure 6A) or analyzed by random spore (Figure 6B). Dapi staining and observation by phase contrast microscopy revealed a small increase in the number of dyad asci in the absence of cig2: 10.3% in a cig2Δ cross and 7% in a cig2Δ h90 strain (Figure 6A; column 2), compared with <1% in wild-type controls. Twenty dyads were dissected, and after germination the exponentially growing cells were analyzed by FACS, which indicated they were all diploids (Figure 6A; column 3). Tetrads were also dissected and shown to generate only haploid spores (Figure 6A; column 4). The viability of the spores from the cig2Δ asci was similar to wild type at ∼95%. Random spore analysis demonstrated that 10% diploid colonies were formed in the cig2Δ cross and 5.6% using a cig2Δ h90 strain (Figure 6B; column 2), with <1% diploids in the wild-type controls. For unknown reasons, the score of 10% diploids is higher than would be expected, given the 10.3% dyads (and 89.7% tetrads). In Figure 2, 100% of the pat1ts cig2Δ cells completed MII, giving only haploid spores (4 nuclei per asci). This difference is presumably due to the physiology of the pat1ts strain, which undergoes meiosis more readily than a wild-type strain. However, pat1ts cig2Δ mutant (Figure 2) cells reached the MII plateau 1.5 h later than the control, suggesting a delay in MII (Figure 1). FACS analysis established that the diploids were only formed in the dyads. These observations suggest that the cig2Δ diploid cells are likely to arise as a consequence of a failure to proceed into MII. Thus, in addition to its role in contributing to the onset of premeiotic S phase, the G1/S CDK Cdc2p/Cig2p has a second minor role in the meiotic cell cycle to ensure efficient completion of MII.
Figure 6.
In the absence of cig2, meiosis generates dyads and diploid cells. Strains deleted (PN1926 h+, PN1942 h−, PN3796 h90) or not (PN1 h−, PN4 h+, PN2 h90) for cig2 were crossed (h+ × h−) or spread (h90) on YEPD plates and incubated at 25°C for 2 d. (A) The asci were observed under the microscope to determine the percentage of dyads and then dissected using the Singer-MSM micromanipulator and analyzed by FACS to determine the ploidy of spores coming from dyads and tetrads (ND: non determined). (B) Another group of asci was used to perform random spore analysis. Asci were digested with helicase, plated on YES and YEP and incubated at 25°C to follow the formation of diploid colonies. The percentage of dyads (A) and diploid colonies (B) reported are an average from three different experiments. The SDs are also reported.
Meiosis-specific Regulation of cig2 Expression
Given the requirement of the Cdc13 and Cig2 B-type cyclins for proper meiotic progression, we investigated the transcriptional regulation of B-type cyclin genes using Northern blot analysis. Vegetatively cycling cells have a 2.5-kb cdc13 mRNA (Hagan et al., 1988). In the pat1ts/pat1ts mutant (Figure 7; left panels), cdc13 was present in two transcripts (2.5 and 2.2 kb), as already observed by Iino et al. (1995). Both transcripts began to accumulate during S phase and rose to a high level by MI and MII before decreasing. Cig2 mRNA increased to a high level at 1 h, just before S phase, and both the poly(A)+ 3.2 kb- and poly(A)− 3.0-kb transcripts present in exponentially growing cells (Obara-Ishihara and Okayama, 1994) were detected. These transcripts were maintained until completion of DNA replication but then disappeared, to be replaced by a smaller cig2 transcript of 2.15 kb, which peaked at 4 h and decreased after MII. These levels correlated with the changes in protein level observed in Figure 1C. The cyclin B Cig1 mRNA was also monitored and was observed to accumulate just after premeiotic DNA replication and to decline during MI and MII.
Figure 7.
cig2 presents a smaller transcript later in meiosis, which is dependent on mei4. RNA was extracted from cells taken during pat1ts/pat1ts (strain PN2158, left) and pat1ts mei4Δ (strain PN2448, right) progression into meiosis and analyzed by Northern blotting using cdc13, cig2, cig1, and his3 (loading control) probes. Each probes are DNA fragments located in the ORF of the genes.
Mei4p is a transcription factor specific for meiosis (Horie et al., 1998), and so we investigated its effect on B-type cyclin transcription, analyzing RNA from pat1ts mei4Δ cells undergoing meiosis (Figure 7; right panels). In the absence of mei4 the larger cdc13 mRNA increased like that of controls, but the small mRNA did not rise to the high level seen during MI and MII of a normal meiosis. Cig2 mRNA levels increased just before S phase like that of controls, but the smaller 2.15-kb transcript did not appear (Figure 7). Cig1 mRNA levels increased ∼2 h later than normal and did not decline during the time course of the experiment. Therefore, in the absence of the Mei4p transcription factor, the transcription patterns of all three B-type cyclins are modified, with the most dramatic effect observed for cig2 2.15-kb transcript, which completely failed to appear.
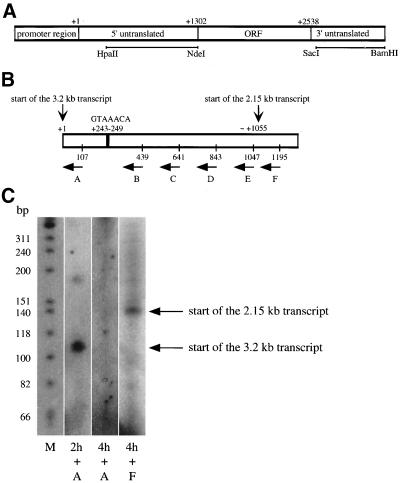
To elucidate the effects on cig2 mRNA further we tested if there were modifications of the 5′ or the 3′ untranslated regions of the gene. Northern blot analysis (Figure 8) was performed using DNA fragments located in the 5′ and 3′ untranslated regions of cig2 as probes (indicated in Figure 9A). The 3.2- and 3.0-kb transcripts were detected by both probes, but only the 3′ fragment detected the 2.15-kb cig2 mRNA, suggesting differences in the 5′-region of cig2 mRNA during the second half of meiosis. Examination of the cig2 genomic sequence (EMBL SPAPB2B4, S. pombe, chromosome 1, region 3000–7000, where 3713 is nucleotide +1) did not reveal any introns in the 5′ region, and thus the 5′ region is likely to be untranslated sequence (Figure 9A). Our analysis revealed an Mei4p-binding site consensus sequence (GTAAACA; Horie et al., 1998) in the 5′ untranslated region of cig2 (indicated in Figure 9B). To further examine this region, we determined the start site of transcription for both transcripts by primer extension using primers corresponding to different parts of the 5′ untranslated region (Figure 9B). Primer A was hybridized to total RNA from early meiotic cells (at 2 h; Figure 9C, second lane) and was extended to ∼110 base pairs, consistent with transcription start site at the nucleotide +1 for the long 3.2-kb transcript as shown by other primer extension data (our unpublished results). The primers A–F were hybridized to total RNA from 4-h late meiotic cells. No extension product was detected for primers A–E (third lane, primer A shown). Primer F was extended to ∼140 base pairs, indicating the presence of a second transcription start site for the smaller 2.15-kb cig2 transcript around nucleotide 1055. We conclude that the small cig2 transcript present later in meiosis has a different transcription start site, explaining its reduction in length to 2.15 kb (Figures 7 and 8) and that it may be regulated by the meiosis-specific transcription factor Mei4p.
Figure 8.
The cig2 transcript is modified in its 5′ untranslated region in MI. RNA from pat1ts/pat1ts (strain PN2158) meiotic time course was analyzed by Northern blotting using DNA probes from the 5′ untranslated (top) and 3′ untranslated (bottom) regions of cig2.
Figure 9.
Cig2 has two different transcription start sites. (A) Diagram of the cig2 gene with probes used in Figure 8 indicated. (B) Detailed diagram of the 5′ untranslated region and position of primers A–F used to perform the primer extension experiment. A putative Mei4p-binding site (GTAAACA) is also indicated. (C) Primer extension experiment was performed by hybridizing primers A–F with RNA extracted from cells taken at 2 or 4 h of a pat1-induced meiosis (strain PN2158). The results obtained for primers A and F are shown. To determine the size of the extended products labeled φx174 DNA/HinfI markers (M) were loaded (first lane). The polyacrylamide gel was exposed to a BioMax film.
DISCUSSION
In this article we have investigated the role and regulation of CDKs during meiosis in fission yeast. We have found that the G1/S CDK Cdc2p/Cig2p is activated in a biphasic manner during meiosis, at the onset of premeiotic S phase, and at the MI/MII transition. Analysis of a cig2 deleted strain revealed that Cig2p is required both for the normal timing of meiotic events and for the efficient completion of the second meiotic division. We have also shown that there is a meiosis-specific regulation of cig2 transcription during MI/MII that is dependent on the Mei4p transcription factor and involves a different transcription start site for cig2.
During pat1ts-induced synchronous meiosis, Cdc13p accumulates as cells proceed through premeiotic S phase. This is accompanied by an increase in Cdc2p/Cdc13p kinase activity that remains high from onset of MI to exit from MII because Cdc2p is dephosphorylated on Y15 (Figure 1, C and D). This is consistent with the requirement of the activating phosphatase Cdc25p for both the first and the second meiotic divisions (Grallert and Sipiczki, 1989, 1991; Iino et al., 1995). Studies using oocytes (Kobayashi et al., 1991; Hunt et al., 1992) have revealed degradation of the B-type cyclins at the MI/MII transition. In pat1ts strains undergoing synchronous meiosis, we observed no decrease in Cdc13p level between MI and MII. However, Cdc13p level is not maintained in cells arrested before MII (Figure 5), suggesting there may be some proteolysis of Cdc13p at MI/MII transition that is too transient to be detected in our synchronized meiosis.
Cig2p is present and Cdc2p/Cig2p is active during premeiotic S phase (Figure 1, C and D). In the absence of Cig2p, premeiotic DNA replication is delayed for about half an hour (Figure 2B), revealing a role for the mitotic G1/S CDK in meiotic S phase. In Saccharomyces cerevisiae, Clb5p and Clb6p act as the premeiotic S phase B-type cyclins. The clb5 mutant shows a more severe delay in premeiotic DNA replication (Stuart and Wittenberg, 1998) than the cig2 mutant. As in mitosis (Fisher and Nurse, 1996; Mondesert et al., 1996), in the absence of Cig2p, Cdc13p appears to substitute for Cig2p although there is a short delay in the onset of S phase. Our study also revealed an unexpected involvement of Cig2p in meiotic divisions. Cig2 protein level reaches a peak at 4 h, 2 h after DNA replication (Figure 1C), when the associated Cdc2p kinase activity is twice that of the S-phase Cdc2p/Cig2p activity (Figure 1D). This second peak is dependent on entry into MI because it is not detected in mei4Δ cells, which are blocked before MI (Figure 4). However, the second Cig2p peak is still present in mes1Δ cells blocked before MII (Figure 5), demonstrating that it is associated either with progression through MI or the interphase period between MI and MII. This result also establishes that Cdc2p is likely to have a function in MI when in association with Cig2p. This is important because a Cdc2p requirement during meiosis I has not been clearly shown before because of the experimental difficulty of separating MI from premeiotic S phase (Iino et al., 1995). In the mes1Δ mutant the Cdc2p/Cig2p kinase activity present during premeiotic DNA replication is four times lower than the maximal activity detected at 4 h (Figure 5D). In normal meiosis (Figure 1) it appears that premeiotic S phase requires a lower Cdc2p kinase activity than do the meiotic divisions. This can be compared with the mitotic cell cycle, where Cdc13p/Cdc2p is able to execute both S and M phases in the absence of Cig2p and does so with a single oscillation of kinase activity (Fisher and Nurse, 1996). Activity is much lower during S phase establishing that a low Cdc2p kinase activity is sufficient to induce mitotic S phase. Like Cig2p in S. pombe, Clb5p in S. cerevisiae also shows a biphasic kinetic during meiosis, peaking in S and M phases (Grether and Herskowitz, 1999), suggesting that a similar mechanism may be operative in both these organisms.
Studies in Xenopus and starfish oocytes (Furuno et al., 1994; Picard et al., 1996) suggest a role for B-type cyclins in the suppression of DNA replication between MI and MII. However, this is unlikely be the role of Cig2p during fission yeast meiosis. The formation of dyads producing diploid cells observed in cig2Δ crosses (Figure 6) means that some cig2Δ cells fail to proceed into one of the meiotic divisions. MII cells plateau at 7.5 h rather than 6 h in wild-type (Figure 1), indicating that entry into MII is delayed and suggesting that cig2Δ dyads are likely to be produced because of a failure of MII, although eventually pat1ts cig2Δ cells are able to complete MII. This could be confirmed by analyzing the segregation of Cen1-markers. One explanation for a failure in MII completion is that Cig2p may be required for efficient sister chromatid separation. In the absence of Cig2p, separation does not occur before spore wall formation generating dyads containing two diploid spores. We also cannot exclude a redundant role for Cig2p in suppression of S phase between MI and MII, although this cannot explain the formation of dyad diploid spores in the cig2Δ strain.
Given the involvement of cig2 in meiosis, we examined the meiotic expression of cig2 and other B-type cyclin genes (Figure 7). Cdc13 expression rises to a high level during MI and MII (Figure 7), as already described by Iino et al. (1995). Cdc13 mRNA is still detected in the absence of the meiotic transcription factor Mei4p, at least for the larger transcript, indicating that Mei4p is not absolutely required for induction of cdc13 expression, as suggested by Iino et al. (1995). Mei4p seems to have a stronger influence on cig1 expression, which reaches a peak with a 2-h delay in mei4Δ compared with normal meiosis (Figure 7), although Cig1p seems to play a minor role in meiosis (Figure 3). Cig2 mRNA produces three transcripts during meiosis, and only the larger two transcripts are detected in the mei4Δ mutant. The longer transcripts present during DNA replication are likely to be dependent on the Cdc10p transcription factor, as is the case in mitosis (Obara-Ishihara and Okayama, 1994), whereas the smaller cig2 mRNA never detected in the mitotic cell cycle clearly depends on the meiotic transcription factor Mei4p (Figure 7). It is interesting to note that Clb5 mRNA levels in S. cerevisiae are also controlled by a meiosis-specific transcription factor, in this case Ndt80p (Chu and Herskowitz, 1998). One of the best known Mei4p targets is spo6, which is required for meiosis II and sporulation (Horie et al., 1998). The mde genes are other Mei4p targets but generally have not been functionally defined (Abe and Shimoda, 2000). All these genes contain the Mei4p-binding sequence GTAAAYA. Our finding that cig2 expression during the meiotic divisions is dependent on Mei4p is supported by the fact that cig2 also contains a putative Mei4p-binding sequence (GTAAACA) in its 5′ untranslated region (Figure 9, A and B). Primer extension experiments (Figure 9C) and the absence of putative introns in the 5′ untranslated region indicate the existence of two different transcription start sites for cig2, one for the longer transcripts present during premeiotic DNA replication and a second for the shorter 2.15-kb transcript present later in the meiotic cell cycle. All these transcripts appear to generate the same Cig2 protein of 47.3 kDa. The second transcription start site is located approximately at nucleotide 1055 (Figure 9, B and C), close to the ORF that begins at nucleotide 1302 (Figure 9, A and B). Analysis of the cig2 sequence revealed the presence of TATA boxes at 84 and 209 base pairs upstream of the start site, whereas the putative Mei4p-binding sequence is located 812 nucleotides upstream of the start site. Mei4p is absolutely required for cells to undergo MI, and it is possible that Cig2p has an important role in meiosis as one of the first Mei4p target during meiotic process. The absence of the second Cig2p peak in mei4Δ mutant possibly contributes to the subsequent meiotic arrest.
We conclude that the mitotic G1/S cyclin Cig2p is involved in premeiotic DNA replication in S. pombe and is regulated at the transcriptional level as in the mitotic cell cycle. However, Cig2p also plays a role during the meiotic divisions, being required for the normal timing of the divisions and for efficient completion of MII. Thus Cig2p is important to ensure genome stability during meiosis. Finally, we have shown that cig2 transcription is regulated differently during the MI/MII transition, by the meiosis-specific transcription factor Mei4p.
ACKNOWLEDGMENTS
The authors thank all members of the Cell Cycle Laboratory, especially Heidi Browning, Satoko Yamaguchi, and Damien Hermand for encouragement and helpful discussions; Stephanie Yanow, Anabelle Decottignies, and Satoko Yamaguchi for critical reading of the manuscript; Ralf Behrens and Trevor Duhig for technical advice on RNA experiments; Nigel Peat for assistance in computational analysis; Vaughan C. Howells and Liz Eaton for their help with the manuscript; and H. Yamano for the monoclonal anti-Cdc2p and anti-Cig2p antibodies.
Footnotes
DOI: 10.1091/mbc.01–10–0507.
REFERENCES
- Abe H, Shimoda C. Autoregulated expression of Schizosaccharomyces pombe meiosis-specific transcription factor Mei4 and a genome-wide search for its target genes. Genetics. 2000;154:1497–1508. doi: 10.1093/genetics/154.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayté J, Schweitzer C, Zarzov P, Nurse P, DeCaprio JA. Feedback regulation of the MBF transcription factor by cyclin Cig2. Nat Cell Biol. 2001;3:1043–1050. doi: 10.1038/ncb1201-1043. [DOI] [PubMed] [Google Scholar]
- Bähler J, Schuchert P, Grimm C, Kohli J. Synchronized meiosis and recombination in fission yeast: observations with pat1–114 diploid cells. Curr Genet. 1991;19:445–451. doi: 10.1007/BF00312735. [DOI] [PubMed] [Google Scholar]
- Baum B, Wuarin J, Nurse P. Control of S-phase periodic transcription in the fission yeast mitotic cycle. EMBO J. 1997;16:4676–4688. doi: 10.1093/emboj/16.15.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher R, Beach D. Interaction between cdc13+ and cdc2+ in the control of mitosis in fission yeast; dissociation of the G1 and G2 roles of the cdc2+ protein kinase. EMBO J. 1987;6:3441–3447. doi: 10.1002/j.1460-2075.1987.tb02667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgne A, Nurse P. The Spd1p S phase inhibitor can activate the DNA replication checkpoint pathway in fission yeast. J Cell Sci. 2000;113:4341–4350. doi: 10.1242/jcs.113.23.4341. [DOI] [PubMed] [Google Scholar]
- Bueno A, Russell P. Two fission yeast B-type cyclins, Cig2 and Cdc13, have different functions in mitosis. Mol Cell Biol. 1993;13:2286–2297. doi: 10.1128/mcb.13.4.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- Connolly T, Beach D. Interaction between the Cig1 and Cig2 B-type cyclins in the fission yeast cell cycle. Mol Cell Biol. 1994;14:768–776. doi: 10.1128/mcb.14.1.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Bordes J, Nurse P. p25rum1 orders S phase and mitosis by acting as an inhibitor of the p34cdc2 mitotic kinase. Cell. 1995;83:1001–1009. doi: 10.1016/0092-8674(95)90215-5. [DOI] [PubMed] [Google Scholar]
- Daya-Makin M, Szankasi P, Tang L, MacRae D, Pelech SL. Regulation of p105wee1 and p34cdc2 during meiosis in Schizosaccharomyces pombe. Biochem Cell Biol. 1992;70:1088–1096. doi: 10.1139/o92-154. [DOI] [PubMed] [Google Scholar]
- Featherstone C, Russell P. Fission yeast p107wee1 mitotic inhibitor is a tyrosine/serine kinase. Nature. 1991;349:808–811. doi: 10.1038/349808a0. [DOI] [PubMed] [Google Scholar]
- Fisher DL, Nurse P. A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 1996;15:850–860. [PMC free article] [PubMed] [Google Scholar]
- Furuno N, Nishizawa M, Okazaki K, Tanaka H, Iwashita J, Nakajo N, Ogawa Y, Sagata N. Suppression of DNA replication via Mos function during meiotic divisions in Xenopus oocytes. EMBO J. 1994;13:2399–2410. doi: 10.1002/j.1460-2075.1994.tb06524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- Grallert B, Sipiczki M. Initiation of the second meiotic division in Schizosaccharomyces pombe shares common functions with that of mitosis. Curr Genet. 1989;15:231–233. [Google Scholar]
- Grallert B, Sipiczki M. Dissociation of meiotic and mitotic roles of the fission yeast cdc2 gene. Mol Gen Genet. 1990;222:473–475. doi: 10.1007/BF00633860. [DOI] [PubMed] [Google Scholar]
- Grallert B, Sipiczki M. Common genes and pathways in the regulation of the mitotic and meiotic cell cycles of Schizosaccharomyces pombe. Curr Genet. 1991;20:199–204. doi: 10.1007/BF00326233. [DOI] [PubMed] [Google Scholar]
- Grether ME, Herskowitz I. Genetic and biochemical characterization of the yeast Spo12 protein. Mol Biol Cell. 1999;10:3689–3703. doi: 10.1091/mbc.10.11.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Hayles J, Nurse P. Cloning and sequencing of the cyclin-related cdc13+ gene and a cytological study of its role in fission yeast mitosis. J Cell Sci. 1988;91:587–595. doi: 10.1242/jcs.91.4.587. [DOI] [PubMed] [Google Scholar]
- Horie S, Watanabe Y, Tanaka K, Nishiwaki S, Fujioka H, Abe H, Yamamoto M, Shimoda C. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol Cell Biol. 1998;18:2118–2129. doi: 10.1128/mcb.18.4.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T, Luca FC, Ruderman JV. The requirements for protein synthesis and degradation, and the control of destruction of cyclins A and B in the meiotic and mitotic cycles of the clam embryo. J Cell Biol. 1992;116:707–724. doi: 10.1083/jcb.116.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino Y, Hiramine Y, Yamamoto M. The role of cdc2 and other genes in meiosis in Schizosaccharomyces pombe. Genetics. 1995;140:1235–1245. doi: 10.1093/genetics/140.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino Y, Yamamoto M. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol Gen Genet. 1985;198:416–421. doi: 10.1007/BF00332932. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Minshull J, Ford C, Golsteyn R, Poon R, Hunt T. On the synthesis and destruction of A- and B-type cyclins during oogenesis and meiotic maturation in Xenopus laevis. J Cell Biol. 1991;114:755–765. doi: 10.1083/jcb.114.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee MS, Enoch T, Piwnica-Worms H. Mik1+ encodes a tyrosine kinase that phosphorylates p34cdc2 on tyrosine 15. J Biol Chem. 1994;269:30530–30537. [PubMed] [Google Scholar]
- Lundgren K, Walworth N, Booher R, Dembski M, Kirschner M, Beach D. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell. 1991;64:1111–1122. doi: 10.1016/0092-8674(91)90266-2. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, Labib K, Moreno S. B-type cyclins regulate G1 progression in fission yeast in opposition to the p25rum1 cdk inhibitor. EMBO J. 1996;15:839–849. [PMC free article] [PubMed] [Google Scholar]
- McLeod M, Beach D. Homology between the ran1+ gene of fission yeast and protein kinases. EMBO J. 1986;5:3665–3671. doi: 10.1002/j.1460-2075.1986.tb04697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod M, Beach D. A specific inhibitor of the ran1+ protein kinase regulates entry into meiosis in Schizosaccharomyces pombe. Nature. 1988;332:509–514. doi: 10.1038/332509a0. [DOI] [PubMed] [Google Scholar]
- Millar J, McGowan C, Lenaers G, Jones R, Russell P. p80cdc25 mitotic inducer is the tyrosine phosphatase that activates p34cdc2 kinase in fission yeast. EMBO J. 1991;10:4301–4309. doi: 10.1002/j.1460-2075.1991.tb05008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondesert O, McGowan CH, Russell P. Cig2, a B-type cyclin, promotes the onset of S in Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:1527–1533. doi: 10.1128/mcb.16.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Murakami H, Nurse P. Meiotic DNA replication checkpoint control in fission yeast. Genes Dev. 1999;13:2581–2593. doi: 10.1101/gad.13.19.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. Mutants of the fission yeast Schizosaccharomyces pombe which alter the shift between cell proliferation and sporulation. Mol Gen Genet. 1985;198:497–502. [Google Scholar]
- Obara-Ishihara T, Okayama H. A B-type cyclin negatively regulates conjugation via interacting with cell cycle ‘start’genes in fission yeast. EMBO J. 1994;13:1863–1872. doi: 10.1002/j.1460-2075.1994.tb06455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker LL, Atherton-Fessler S, Piwnica-Worms H. p107wee1 is a dual-specificity kinase that phosphorylates p34cdc2 on tyrosine 15. Proc Natl Acad Sci USA. 1992;89:2917–2921. doi: 10.1073/pnas.89.7.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard A, Galas S, Peaucellier G, Dorée M. Newly assembled cyclin B-cdc2 kinase is required to suppress DNA replication between meiosis I and meiosis II in starfish oocytes. EMBO J. 1996;15:3590–3598. [PMC free article] [PubMed] [Google Scholar]
- Sazer S, Sherwood SW. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J Cell Sci. 1990;97:509–516. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- Shimoda C, Hirata A, Kishida M, Hashida T, Tanaka K. Characterization of meiosis-deficient mutants by electron microscopy and mapping of four essential genes in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1985;200:252–257. doi: 10.1007/BF00425432. [DOI] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 1998;12:2698–2710. doi: 10.1101/gad.12.17.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Shinozaki-Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature. 1997;13:187–190. doi: 10.1038/386187a0. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yamamoto M. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell. 1994;78:487–498. doi: 10.1016/0092-8674(94)90426-x. [DOI] [PubMed] [Google Scholar]
- Yamamoto M. The molecular control mechanisms of meiosis in fission yeast. Trends Biochem Sci. 1996;21:18–22. [PubMed] [Google Scholar]