Abstract
Gin4, a Nim1-related kinase, is required in budding yeast for localization of the septins and for proper control of daughter cell growth during G2/M. Gin4 becomes hyperphosphorylated when cells enter mitosis, leading to activation of Gin4 kinase activity. In this study, we have used immunoaffinity chromatography to identify proteins that associate with Gin4 during mitosis, with the goal of finding targets of Gin4 kinase activity and proteins that play a role in Gin4 activation. We show that during mitosis Gin4 is assembled into a multiprotein complex that includes Nap1, Bni5, the septins, and at least two molecules of Gin4. The associated Gin4 molecules present in this complex phosphorylate each other, leading to Gin4 hyperphosphorylation. Furthermore, the Shs1 septin present in the complex undergoes Gin4-dependent phosphorylation during mitosis and appears to be a substrate of Gin4 in vitro, suggesting that it is a target of Gin4 kinase activity in vivo. Genetic data support the idea that Shs1 is an important target of Gin4 kinase activity. Association of Gin4 with the septins during mitosis requires Shs1, Nap1, Cla4, Elm1, and the kinase activities of Gin4 and Cdc28. Self-association of Gin4 molecules requires Shs1 but not Cla4 or Nap1. Previous work has suggested that the septins function together as a tight complex, and we found that the majority of the Shs1 in the cell is tightly bound to the other septins Cdc3, Cdc10, Cdc11, and Cdc12. Interestingly, however, Shs1 can bind to Gin4 and induce Gin4 oligomerization under conditions in which the Cdc11 septin does not bind to Gin4, suggesting that Shs1 can function independently of the other septins. Taken together, these findings suggest that highly regulated protein-binding events ensure that the Gin4 kinase is activated only during mitosis and only in association with Shs1, a likely in vivo substrate of Gin4. In addition, these results provide clues to how Gin4 may regulate the localization or function of the septins.
INTRODUCTION
The septins are a conserved family of GTP-binding proteins that were first identified genetically in screens for genes that play a role in cell cycle progression in budding yeast (Hartwell, 1971). Loss of septin function causes budding yeast cells to arrest at G2/M while continuing to undergo cell growth, leading to the formation of highly elongated cells. Loss of septin function also causes defects in cytokinesis. There are five members of the septin family expressed in vegetatively growing yeast cells: Cdc12, Cdc11, Cdc10, Cdc3, and Shs1/Sep7. All of these are localized to the bud neck, and biochemical experiments show that Cdc12, Cdc11, Cdc10, and Cdc3 form a tight complex, consistent with genetic studies showing that loss of function of one septin can cause mislocalization of the others (Haarer and Pringle, 1987; Ford and Pringle, 1991). In animal cells, the septins play a role in cytokinesis but are also expressed in nondividing neuronal tissues and have been implicated in vesicle fusion events at the plasma membrane, indicating that their functions are not restricted to cytokinesis (Neufeld and Rubin, 1994; Fares, 1995; Field et al., 1996; Kinoshita et al., 1997; Hsu et al., 1998; Beites et al., 1999; Trimble, 1999; Nguyen et al., 2000).
Recent work has demonstrated that an intricate signaling network is required in budding yeast for septin localization, proper control of bud growth, and cell cycle progression during G2/M (Kellogg and Murray, 1995; Ma et al., 1996; Altman and Kellogg, 1997; Carroll et al., 1998; Longtine et al., 1998; McMillan et al., 1998; Tjandra et al., 1998; Barral et al., 1999; Shulewitz et al., 1999; Sreenivasan and Kellogg, 1999; Longtine et al., 2000). This signaling network includes the protein kinases Gin4, Elm1, Cla4, and Hsl1, as well as a number of proteins that are required for proper regulation and localization of these kinases, including Nap1, Cdc42, and Hsl7. Inactivation of any of these proteins can cause a prolonged G2/M delay and an elongated cell phenotype similar to the phenotype caused by loss of function of the septins. In addition, many of the proteins that function in this signaling network interact genetically with the septins and are required in vivo for proper septin localization. Finally, Gin4 and Hsl1 bind to the septins and undergo septin-dependent hyperphosphorylation during mitosis (Altman and Kellogg, 1997; Carroll et al., 1998; Barral et al., 1999). These observations suggest that there is a close functional relationship between the septins and the signaling network that includes the Gin4, Hsl1, Cla4, and Elm1 kinases. The nature of this relationship, however, remains poorly understood.
In this study, we have focused on learning more about the functional and regulatory relationships between the Gin4 kinase and the septins. Our results demonstrate that Gin4 is assembled into a multiprotein complex during mitosis that includes Nap1, the septins, Bni5, and at least two molecules of Gin4. Assembly of the Gin4-septin complex requires mitotic CDK activity. The Gin4 molecules present in the complex phosphorylate each other, leading to hyperphosphorylation of Gin4. In addition, the Shs1 septin plays an important role in Gin4 activation but also appears to be an important target of Gin4 kinase activity. These results provide important clues to the molecular mechanisms that control Gin4 activity during mitosis and suggest that Gin4 may exert its effects on septin organization via phosphorylation of Shs1.
MATERIALS AND METHODS
Strains and Culture Conditions
Except where noted, all strains were grown in yeast extract/peptone/dextrose (YPD) media. All strains are in the W303 background (leu2-3112 ura3-52 can1-100 ade2-1 his3-11 trp1-11). The glutathione S-transferase (GST)-Gin4–integrating plasmid was a gift of M. Longtine (Longtine et al., 1998). Additional features of the strains used in this study are as follows. DK186: Mata Δbar1; RA24: Mata Δbar1 Δshs1::URA3 (Carroll et al., 1998); HT160: Mata Δbar1 Δcla4::HIS5; DK272: Mata Δbar1 GIN4-3×HA::URA3 (pDK63B); DK273: Mata Δbar1 gin4K48A (Altman and Kellogg, 1997); DK274: Mata/α Δbar1/Δbar1 gin4K48A -3×HA/GIN4; CC10: Mata Δbar1 SHS1-3×HA::URA3 (Carroll et al., 1998); AS18: Mata Δbar1 Δswe1::TRP1; AS20: Mata Δbar1 Δelm1::URA3; AS36: Mata Δbar1 Δcla4::LEU2 Δswe1::URA3; DK244: Mata Δbar1 Δnap1::URA3; DK280: Mata Δbar1 GIN4-HA::URA3 (pDK64); RA19: Mata Δbar1 Δgin4::LEU2; EM9: Mata Δbar1 GIN4-GST::URA3 GIN4-3×HA::TRP1 (pEM103); EM13: Mata Δbar1 GIN4-GST::URA3 GIN4-3×HA::TRP1 (pEM103) Δnap1::LEU2; EM14: Mata Δbar1 GIN4-GST::URA3 GIN4-3×HA::TRP1 (pEM103) Δcla4::HIS5; EM19: Mata Δbar1 GIN4-GST::URA3 GIN4-3×HA::TRP1 (pEM103) Δshs1::HIS5; DK219: Mata Δbar1 cdc28-4; DK351: Mata Δbar1 cdc28-as1; DK254: Mata Δbar1 Δcln1::hisG Δcln2 Δcln3 gal1-CLN3.
Plasmid Construction and Antibody Generation
To create a 3× hemagglutinin (HA)-tagged version of Gin4, the 3′ end of the GIN4 open reading frame was amplified by PCR (oligonucleotides: GCGTCTAGAAAATATCAATCATTTGGAGG and CGCGGTACCTTTTTGTAGAACGCCTTCCTT) and cloned into the XbaI and BamHI sites of pDK51 to generate an in-frame fusion with the 3×HA tag. This plasmid (pDK63B) was cut with BglII to target integration at Gin4. A 1×HA-tagged version of Gin4 was created by amplifying the 3′ end of Gin4 by PCR (oligonucleotides: CTCAAACATCCGCTATTAC and CGGGATCCCTAGCCCGCATAGTCAG-GAACATCGTATGGGTAGCCCGCATATTTTTGTAGAACGCCTTCC). The fragment was cut with EcoRI and BamHI, utilizing an internal EcoRI site, and cloned into yIPlac211 to generate an in-frame fusion with the amino acid sequence YAGYPYDVPDYAG (this is the last three amino acids of the HA sequence followed by the full HA sequence). This plasmid (pDK64) was cut with ClaI to target integration at the GIN4 gene. To create pEM103, the Gin4 fragment from pDK63B was excised and cloned into pDK53 to generate pEM103. This plasmid was cut with BglII to integrate at the GIN4 gene.
To generate anti-Shs1 antibodies, a fusion protein including full-length Shs1 fused to maltose-binding protein was purified from Escherichia coli and used to immunize rabbits. Antibodies that recognize Shs1 were affinity purified using a column containing GST-Shs1 as described previously (Kellogg and Alberts, 1992).
Cell Cycle Arrests and Treatment with 1NM-PP1
Strains were arrested in G1 by addition of 1 μg/ml α factor to log phase cultures, followed by growth at room temperature for 3 h. Mitotic arrests were carried out by resuspending log phase cells in YPD media containing 30 μg/ml benomyl followed by growth at room temperature for 2.5–3 h. Mitotic arrests for experiments with the cdc28-as1 strain were carried out by the adding 10 μg/ml nocodazole to log phase cells in YPD media followed by growth at room temperature for 2.5 h. Cells were then treated with either 50 nM 1NM-PP1 from a 12 μM stock in dimethyl sulfoxide (DMSO) or mock treated with an equivalent amount of DMSO.
Coimmunoprecipitation of Gin4, Nap1, Cdc11, and Shs1
Immunoaffinity beads for the precipitation of Gin4 were made by binding affinity-purified anti-Gin4 polyclonal antibodies to protein A beads (Bio-Rad, Hercules, CA) overnight at 4°C on a rotator. Anti-Gin4 antibodies were prepared as previously described (Altman and Kellogg, 1997). For each immunoprecipitation, 5 μg of anti-Gin4 antibody was bound to 20 μl of protein A beads in the presence of phosphate-buffered saline containing 500 mM NaCl and 0.1% Tween-20. Control beads were prepared in the same way using affinity-purified anti-GST or anti-MBP antibodies.
To prepare cells for immunoprecipitation experiments, 50 ml of cells at OD0.7 were pelleted, resuspended in 3 ml of 50 mM HEPES-KOH, pH 7.6, and aliquoted into two 1.6-ml screw-top tubes, pelleted again, and frozen on liquid nitrogen, typically yielding two cell pellets of ∼150 μl each. For immunoprecipitations performed during a time course, a 500-ml culture of cells was grown overnight to OD0.7 and arrested in G1 by the addition of α factor. The cells were then released from the arrest by washing three times with 1 l of fresh YPD. After the final wash, the cells were resuspended in 400 ml of YPD and allowed to proceed synchronously through the cell cycle. At each time point, a 50-ml sample of culture was taken and the cells were pelleted, resuspended in 1.6 ml of 50 mM HEPES-KOH, pH 7.6, and then pelleted again in a 1.6-ml screw-top tube. After removing the supernatant, the cell pellet was frozen on liquid nitrogen.
Extracts for immunoprecipitations were made by adding 300 μl of acid-washed glass beads to the frozen pellets, followed by 300 μl of ice-cold lysis buffer (50 mM HEPES-KOH, pH 7.6, 150 mM KCl, 100 mM β-glycerol phosphate, 25 mM NaF, 1 mM EGTA, 1 mM MgCl2, 0.15% Tween-20, 1 mM phenylmethylsulfonyl fluoride [PMSF]). The tubes were placed immediately into a Biospec Multibeater-8 and beaten at top speed for 30 s. The tubes were then placed in an ice-water bath for 30 s before a 5-min spin in a microfuge at top speed. Supernatant (300 μl) was removed and replaced with 200 μl of lysis buffer and the tubes were beaten again for 30 s. The supernatants were pooled and then added to the immunoaffinity beads equilibrated in lysis buffer. Samples of the extract (10 μl) were taken before and after treatment with antibody and frozen in liquid nitrogen for analysis by Western blotting. The tubes were rotated gently end over end at 4°C for 1 h and 45 min and then washed three times with 500 μl of lysis buffer containing 10% glycerol and no PMSF. At the end of these washes, the beads were transferred to a fresh tube and washed once more. Gin4-associated proteins were eluted from the beads by the addition of 200 μl of elution buffer (50 mM HEPES-KOH, pH 7.6, 1 M KCl, 1 mM EGTA, 1 mM MgCl2, 10% glycerol). After addition of the elution buffer, the beads were pipeted up and down several times and pelleted in a microfuge; then 150 μl of the supernatant was removed, taking care to avoid the antibody-containing beads. This process was repeated once more and the supernatants were pooled together and precipitated by the addition of trichloroacetic acid to 10%. The resulting pellet was resuspended in 20 μl of 1× protein sample buffer and one-half of this was loaded onto a 10% polyacrylamide gel and used for a Western blot. After the elution of associated proteins, the anti-Gin4 beads were pelleted, washed once in 50 mM HEPES-KOH, pH 7.6, resuspended in 100 μl of 1× protein sample buffer, and boiled to release the bound Gin4 from the beads. Samples (10 μl) were loaded onto a 10% gel to demonstrate that equal amounts of Gin4 were precipitated from each sample. For Western blotting of the crude extracts, 100 μl of 1× protein sample buffer was added to 10 μl of crude extract, the samples were incubated at 100°C, and 10 μl was used for PAGE.
Immunoaffinity Purification and Mass Spectrometry Analysis of Gin4 Complexes
We raised an anti-HA polyclonal antibody by immunizing rabbits with an HA peptide (peptide sequence: CPDYAGYPYDVPDYAG) conjugated to keyhole limpet hemocyanin. We raised the anti-HA antibodies against a partial HA peptide followed by a full-length peptide to obtain antibodies that recognize the junction between two HA peptides, as well as against the HA peptide. The peptide was conjugated to keyhole limpet hemocyanin via an amino-terminal cysteine and via primary amines. The anti-HA antibody was affinity purified on a column constructed with a GST-HA fusion protein.
Anti-HA beads were prepared by binding 500 μg of affinity-purified anti-HA antibody to 500 μl of protein A agarose in a 1.5-ml tube in the presence of phosphate-buffered saline containing 0.5 M NaCl, 0.1% Tween-20. Binding was carried out on a rotator for 1–2 h at room temperature or overnight at 4°C. A control column was constructed with an affinity-purified anti-GST antibody in the same manner. After binding the antibody, the beads were transferred to a 15-ml tube and washed once with lysis buffer (50 mM HEPES-KOH, pH 7.6, 175 mM KCl, 75 mM NaF, 1 mM EGTA, 1 mM MgCl2, 0.45% Tween-20, 5% glycerol).
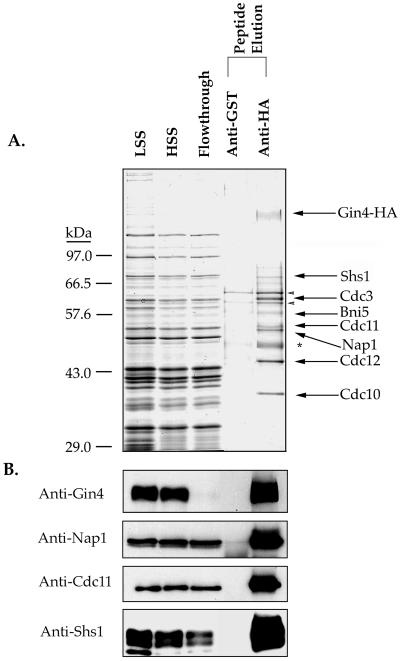
Cells containing an HA-tagged copy of Gin4 were grown overnight at 30°C to OD0.7 and then arrested in mitosis for 2.5 h at room temperature in YPD media containing 30 μg/ml benomyl. The cells were pelleted, resuspended in 50 mM HEPES-KOH, pH 7.6, transferred to a 50-ml tube, pelleted again, and frozen in liquid nitrogen. The cell pellet was liberated from the tube by smashing with a hammer and transferred to a prechilled mortar and pestle. The pellet was then ground for 30 min under liquid nitrogen until a fine powder the consistency of flour was obtained. This powder (25 g) was transferred to a beaker prechilled with liquid nitrogen. Just as the powder began to thaw around the edges of the beaker, 30 ml of ice-cold lysis buffer containing 1 mM PMSF was added and the powder was rapidly resuspended by mixing with a metal spatula. All subsequent steps were carried out at 4°C. After most of the powder was solubilized in the buffer solution, a stir bar was added and the extract was stirred until all chunks were in solution (15–20 min). The extract was then centrifuged for 5 min at 10,000 × g, followed by 100,000 × g for 45 min. Samples taken from this first spin were denoted “LSS” (Figure 3), whereas those from the second spin were denoted “HSS.” After the final spin, the clarified extract was removed carefully to avoid taking any of the pellet and was divided equally between the anti-HA and anti-GST beads. The protein concentration of these extracts was typically 10–15 mg/ml.
Figure 3.
Purification of a Gin4 multiprotein complex from cells arrested in mitosis. (A) Yeast cells carrying an HA-tagged GIN4 were arrested in mitosis, and a crude extract from the cells was incubated with anti-HA beads or anti-GST control beads. After washing, both columns were eluted with an excess of HA dipeptide. Samples from each step of the purification procedure were loaded onto a 10% SDS-polyacrylamide gel, which was stained with Coomassie blue. The lanes marked “LSS” and “HSS” are the low- and high-speed supernatants, respectively, and the lane marked “flowthrough” represents the unbound proteins from the anti-HA immunoaffinity column. Asterisk denotes the antibody heavy chain. The arrowheads denote two background proteins that we consistently see in our purifications. Mass spectrometry analysis identifies these as Ssa1 and Ssa2. (B) The same fractions shown in A were loaded onto a 10% SDS-polyacrylamide gel and transferred to nitrocellulose. Fractions were probed with anti-Gin4-HA, anti-Shs1, anti-Cdc11, and anti-Nap1 antibodies.
The extracts and immunoaffinity beads were gently rotated end over end in the cold room for 2 h at 4°C. The beads were then pelleted by brief centrifugation, and a sample of the supernatant was taken. The beads were washed twice with 15 ml of lysis buffer and then transferred to a 1.5-ml Biospin column (Bio-Rad). The columns were washed with 5 ml of lysis buffer by pipeting 1-ml aliquots of buffer on top of the column, allowing the buffer to flow through by gravity. After the final wash, the columns were washed once with 1 ml of elution buffer (50 mM HEPES-KOH, pH 7.6, 200 mM KCl, 1 mM EGTA, 1 mM MgCl2, 10% glycerol). The columns were then transferred to room temperature and 250 μl of elution buffer containing 0.5 mg/ml HA dipeptide (amino acid sequence: YPYDVPDYAGYPYDVPDYAG) was added and the flow-through fraction was collected. After a 15-min incubation, another aliquot of elution buffer was added. This was repeated for a total of six fractions. Fractions 2–5 were pooled and one-sixth of this pool was precipitated with trichloroacetic acid, resuspended in 20 μl of protein gel sample buffer, incubated at 100°C for 3 min, and loaded onto a 10% polyacrylamide gel for staining with Coomassie blue. The 10-μl fractions from each step of the purification were boiled in 100 μl of 1× protein sample buffer and 10 μl were loaded on the gel. For Western blots, the same amount of each extract sample was loaded onto a 10% gel, whereas one-sixtieth of the dipeptide elution fractions was used. We estimated that our total yield was ∼5 μg. We carried out this procedure with both single HA-tagged Gin4 (tag sequence: YAGYPYDVPDYAG) and with 3×HA-tagged Gin4, with equivalent results, except that the yield of protein tended to be higher with the 3×HA-tagged Gin4.
To analyze Gin4 complexes by mass spectrometry, one-fifth of the total elution from an anti-HA or anti-GST control column was precipitated with methanol/chloroform and analyzed by mass spectrometry as previously described (Carroll et al., 1998; Link et al., 1999).
Immunoaffinity Purification of Septin Complexes or Gin4 in the Presence of High Salt
All steps used for the purification of the septins or Gin4 in the presence of high salt were identical to those described above for purification of Gin4 complexes, except that the lysis buffer used both for lysis and washes included 50 mM HEPES-KOH, pH 7.6, 1 M KCl, 100 mM β-glycerol phosphate, 5 mM MgCl2, 1 mM EGTA, 0.45% Tween-20, 5% glycerol. After elution, fractions were pooled and one-sixth of the pool was precipitated with trichloroacetic acid and analyzed on a 10% polyacrylamide gel. The remainder of the pool was supplemented with 10 μg bovine serum albumin and then concentrated to ∼100 μl in a Microcon-10 spin filtration device (Amicon, Beverly, MA). Activated Gin4 was purified from cells arrested in mitosis by treatment with 30 μg/ml benomyl.
Treatment of Shs1 with Phosphatase
For treatment of Shs1 with phosphatase, we used anti-Shs1 antibodies to immunoprecipitate Shs1 from wild-type cells arrested in mitosis, using the same techniques described for immunoprecipitation of Gin4, except that the immunoprecipitation and wash buffers contained 1 M KCl. After the final wash, the beads were washed three times with phosphatase buffer (50 mM Tris-HCl, pH 7.5, 5 mM dithiothreitol, 2 mM MnCl2, 100 μg/ml bovine serum albumin). After the final wash in phosphatase buffer the beads were resuspended in 50 μl of phosphatase buffer and aliquoted equally to two tubes. λ-Phosphatase (1.5 μl) was added to one tube, and both tubes were incubated at 30°C for 30 min with gentle mixing every 10 min. The reaction was stopped by the addition of 125 μl of 1× protein sample buffer, the samples were incubated in a boiling water bath, and 10 μl of each sample was run out on an 11% polyacrylamide gel.
Kinase Assays
To demonstrate phosphorylation of Shs1 by Gin4, ∼0.25 μg of purified active Gin4 was added to tubes containing 20 μl of kinase assay buffer (50 mM HEPES-KOH, pH 7.6, 1 mM EGTA, 2 mM MgCl2, 0.1% Tween-20, 10% glycerol, 1 mM dithiothreitol, 0.25 mM ATP, 0.1 mCi/ml [γ32P]ATP) with or without 1 μg of purified septin complex. Reactions were incubated at 30°C for 30 min with gentle mixing every 10 min and terminated by the addition of 10 μl of 4× sample buffer. After incubation of the samples at 100°C for 30 s, one-fourth of the reaction was loaded onto a 10% SDS polyacrylamide gel, which was stained with Coomassie blue, dried, and placed on film to visualize phosphorylated proteins.
To demonstrate that the phosphorylated band corresponded to Shs1, a duplicate reaction was set up that contained Gin4 and the septin complex. At the end of the reaction, SDS was added to 1% and the sample was boiled for 2 min to disrupt the septin complex. RIPA buffer (150 mM NaCl, 1.0% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0; 500 μl) was then added to the reaction, the beads were spun down, and the supernatant was split equally between two tubes, one containing anti-Shs1 beads and the other containing anti-GST control beads. These tubes were rotated end over end at room temperature for 2 h and washed three times in RIPA buffer. After the third wash, 20 μl of 2× protein sample buffer was added to the beads, and 10 μl was loaded onto a 10% SDS polyacrylamide gel.
PAGE, Western Blotting, and Sample Preparation
PAGE and Western blotting were carried out as previously described (Anderson et al., 1973; Harlow and Lane, 1988). For routine Western blotting, 1.6-ml samples of culture were pelleted in a 1.6-ml screw-top tube and the cells were frozen on liquid nitrogen. Acid-washed glass beads (300 μl) were added to the frozen pellets, followed by 200 μl of 1× protein sample buffer containing 2 mM PMSF, 50 mM NaF, and 50 mM β-glycerol phosphate. The cells were lysed by beating in a Biospec Multibeater-8 at top speed for 90 s at 4°C. After incubating in a boiling water bath for 5 min, the tubes were centrifuged for 2 min and 10 μl was loaded onto a polyacrylamide gel. Gin4 phosphorylation forms were resolved as described previously (Altman and Kellogg, 1997), whereas Shs1 isoforms were resolved on an 11% polyacrylamide gel for 2 h at 170 V.
RESULTS
Gin4 Hyperphosphorylation Involves Cross-Phosphorylation between Gin4 Molecules
In previous work, we demonstrated that the Gin4 kinase is hyperphosphorylated and activated during mitosis and that hyperphosphorylation is required for Gin4 kinase activity (Altman and Kellogg, 1997). In addition, we found that a catalytically inactive version of Gin4 (gin4K48A) fails to undergo hyperphosphorylation, suggesting that hyperphosphorylation of Gin4 during mitosis involves autophosphorylation (Altman and Kellogg, 1997). To further understand Gin4 hyperphosphorylation, we determined whether hyperphosphorylation is due to an intra- or intermolecular event. To do this, we tested whether gin4K48A undergoes hyperphosphorylation when placed in trans to a wild-type copy of GIN4. We constructed a diploid strain carrying the gin4K48A allele tagged with three copies of the HA epitope and an untagged copy of wild-type GIN4. As controls we used strains carrying single copies of either wild-type GIN4-3×HA or gin4K48A-3×HA. We arrested these strains in mitosis by treatment with the microtubule-depolymerizing drug benomyl and then used Western blotting to determine whether gin4K48A-3×HA undergoes a hyperphosphorylation-induced shift in electrophoretic mobility. In the control strains, we observed that wild-type GIN4-3×HA undergoes hyperphosphorylation, whereas the gin4K48A-3×HA protein does not, as expected (Figure 1). In the presence of wild-type GIN4, however, the gin4K48A-3×HA protein undergoes hyperphosphorylation. These results suggest that Gin4 hyperphosphorylation involves intermolecular cross-phosphorylation events between Gin4 molecules but do not exclude the possibility that additional kinases play roles in Gin4 hyperphosphorylation.
Figure 1.
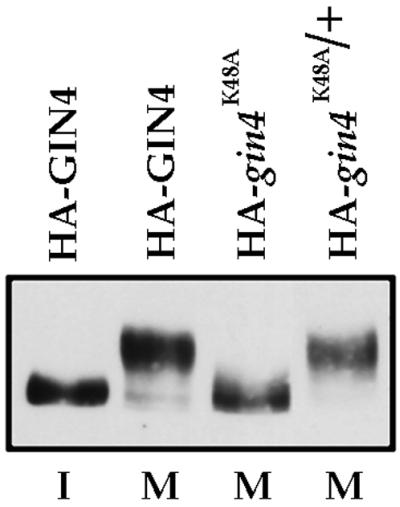
Gin4 autophosphorylation is due to an intermolecular event. Haploid or diploid cells expressing 3×HA-tagged versions of wild-type GIN4 and/or a catalytically inactive GIN4 (gin4K48A) were arrested in mitosis with the microtubule-depolymerizing drug benomyl, and the electrophoretic mobility of 3×HA-GIN4 was monitored by Western blotting with an anti-HA antibody. A strain carrying wild-type 3×HA-GIN4 arrested in G1 with α factor is included for comparison. The genotype of each strain is indicated at the top of the figure, and the cell cycle arrest point is indicated below (I, interphase; M, mitosis).
Gin4 Is Found in a Complex with Itself during Mitosis
Hyperphosphorylation of Gin4 during mitosis appears to be due to intermolecular events. We therefore hypothesized that Gin4 is found in a complex with itself during mitosis, leading to cross-phosphorylation between Gin4 molecules. We tested this idea by determining whether Gin4-Gin4 interactions can be detected during mitosis. We generated a haploid strain carrying one copy of Gin4 tagged with the 3×HA epitope and another copy tagged with GST. Previous experiments have demonstrated that Gin4 tagged with 3×HA or GST is fully functional (Longtine et al., 1998). We arrested this strain either in G1 by treatment with the mating pheromone α factor or in mitosis by treatment with benomyl. We then immunoprecipitated Gin4-GST and used Western blotting with an anti-HA antibody to determine whether the Gin4-3×HA coprecipitates with Gin4-GST. We found that Gin4-3×HA coprecipitates with Gin4-GST but only in extracts made from cells arrested in mitosis, consistent with a model in which Gin4-Gin4 interactions during mitosis result in cross-phosphorylation between Gin4 molecules (Figure 2). These results do not discern whether Gin4 interacts directly with itself or whether bridging proteins are required.
Figure 2.
Gin4 oligomerization is mitosis specific. A strain carrying 3×HA-GIN4 and GST-GIN4 (strain EM9) was arrested in interphase or mitosis, and GST-GIN4 was immunoprecipitated with anti-GST antibodies. The coprecipitation of 3×HA-Gin4 was detected by Western blotting of the precipitates with anti-HA antibody. A control immunoprecipitation (IP) was carried out using equal amounts of a nonspecific antibody. Western blotting of the crude extracts revealed that 3×HA-Gin4 is present at equal levels in interphase and mitotic cells, and anti-GST Western blots of the beads used for immunoprecipitation demonstrated that equal amounts of GST-Gin4 were precipitated from interphase and mitotic cells. The cell cycle arrest point is indicated above each panel (I, interphase; M, mitosis). Note that the gels used for these Western blots were not run long enough to give full resolution of Gin4 isoforms and therefore appear somewhat different from the Western blots shown in Figure 1.
Purification of Endogenous Gin4 Complexes by Immunoaffinity Chromatography
We next wanted to identify proteins that play a role in inducing the oligomerization of Gin4. We reasoned that such proteins would be likely to associate with Gin4 specifically during mitosis. In previous work we used protein affinity chromatography to demonstrate that Gin4 associates with Nap1 and the septins (Altman and Kellogg, 1997; Carroll et al., 1998). However, these experiments utilized affinity columns constructed with fusion proteins purified from bacteria, which precluded the identification of protein-protein interactions that require cell cycle-dependent posttranslational modifications.
To circumvent this problem, we developed a method that allows rapid and specific purification of endogenous multiprotein complexes. For this approach, the protein of interest is tagged with the HA epitope. A crude extract made from cells carrying the HA-tagged protein is then loaded onto an immunoaffinity column made with an affinity-purified anti-HA polyclonal antibody. After washing the column extensively with buffer, protein complexes are competitively eluted with an excess of a peptide consisting of a tandem repeat of the HA epitope, which provides highly specific elution under gentle conditions. We have used this approach successfully to purify both single HA-tagged proteins and 3×HA-tagged proteins (see below). Use of an HA dipeptide is essential for obtaining efficient elution of 3×HA-tagged proteins, most likely because of a high avidity interaction between 3×HA and the anti-HA antibody. As a control, we use either an untagged strain or an identical column made with a nonspecific antibody. We have used this approach to purify six different multiprotein complexes from yeast, indicating that it is generally useful (our unpublished data). In addition, previous work in other systems has shown that immunoaffinity chromatography is a powerful means of purifying endogenous multiprotein complexes under native conditions (Kellogg and Alberts, 1992; Zheng et al., 1995; Field et al., 1996; Frazier et al., 1998).
To purify endogenous Gin4 complexes, we first arrested GIN4-HA cells in mitosis by treatment with the microtubule polymerization inhibitor benomyl. We then loaded a crude extract made from the arrested cells onto an anti-HA column and an anti-GST control column. Figure 3A shows a Coomassie blue-stained gel of the affinity-purified Gin4 complex obtained by this approach. The purified HA-Gin4 appears as a disperse band representing differently phosphorylated forms of Gin4 (arrow, Figure 3A). In addition to Gin4, we observe a number of other proteins that specifically copurify with Gin4. We used mass spectrometry to identify these proteins, as well as the proteins that elute from the anti-GST control column. We found that Cdc3, Cdc10, Cdc11, Cdc12, Shs1, Nap1, and Bni5 are all retained on the anti-HA-Gin4 column but not on the anti-GST control column (Table 1). We verified the presence of Gin4, Cdc11, Shs1, and Nap1 by Western blotting (Figure 3B), and verified the presence of Cdc3, Cdc10, and Cdc12 by comigration with purified septin complexes (not shown). None of these proteins are present in other multiprotein complexes that we have purified using the same approach under the same conditions. In addition, we have obtained equivalent results using an untagged strain as the control. Note that we are unable to detect any Gin4 in the extract after antibody treatment, indicating that Gin4 is quantitatively depleted from the extract. However, only a fraction of the total septins present in the extract is found in the Gin4 complex.
Table 1.
Analysis of the Gin4 complex by mass spectrometry
| Gene name | Peptides |
|---|---|
| Identification of proteins that bind to the anti-HA column | |
| GIN4 | 18 |
| SEP7 | 7 |
| CDC10 | 2 |
| CDC11 | 1 |
| CDC12 | 8 |
| CDC3 | 6 |
| NAP1 | 6 |
| BNI5 | 3 |
| CTF7 | 1 |
| YJR083C | 1 |
| YLL054C | 1 |
| HSP26 | 4 |
| SSA1 | 8 |
| SSA2 | 2 |
| SSB1 | 3 |
| SSE1 | 1 |
| Identification of proteins that bind to the anti-GST control column | |
| SSA1 | 5 |
| SSA2 | 1 |
| RPL27B | 1 |
In addition to the proteins described above, we identified single peptides for Ctf7, YJR083C, and YLL054C in the elution from the anti-HA column but not the control column. However, we were unable to clearly identify Coomassie blue-stained bands corresponding to these proteins, and we have not yet carried out further experiments to confirm these interactions. We routinely identify Ssa1, Ssa2, Ssb1, and Hsp26 on both anti-HA and control columns.
These results demonstrate the existence of an endogenous Gin4 complex present in cells arrested in mitosis and identify proteins that are likely to play a direct role in the function and regulation of Gin4. Further characterization of the functional and physical interactions of these proteins with Gin4 is presented below. Interestingly, we did not detect Cla4, Elm1, Clb2, or Cdc28 in the Gin4 complex by mass spectrometry or by Western blotting, although previous results suggest they play an important role in the regulation of Gin4 (Altman and Kellogg, 1997; Carroll et al., 1998; Tjandra et al., 1998; Sreenivasan and Kellogg, 1999).
Assembly of the Gin4 Complex Occurs as Cells Enter Mitosis
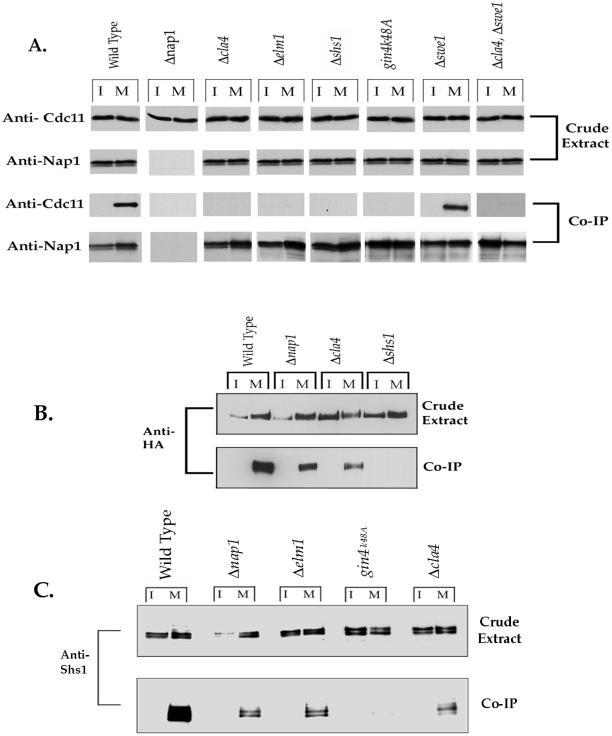
We next wished to determine whether Nap1 and the septins form a complex with Gin4 during mitosis, as would be expected if these proteins play a role in the activation of Gin4 during mitosis. To define when the Gin4 complex is assembled, we first arrested cells in G1 with α factor, released the cells from the arrest, and then took samples every 15 min as the cells proceeded through the cell cycle. Gin4 was then immunoprecipitated from each sample and the coprecipitation of Cdc11 and Nap1 was assayed by Western blotting. In addition, we used Western blotting to follow the behavior of Cdc11, Nap1, and Clb2 in the crude extracts used for the immunoprecipitations (Figure 4A). We found that the Gin4-Cdc11 complex is detected only during mitosis, whereas Gin4 and Nap1 are bound to each other throughout the cell cycle. Additionally, the assembly of the Gin4-Cdc11 complex is exactly correlated with levels of the mitotic cyclin Clb2. Western blotting of the crude extracts showed that Cdc11 and Nap1 protein levels do not change during the cell cycle (Figure 4A), and previous work has demonstrated that Gin4 levels remain constant throughout the cell cycle (Altman and Kellogg, 1997).
Figure 4.
Gin4 associates with Cdc11 in a mitosis-specific manner. (A) Log phase cells were synchronized in G1 by treatment with α factor, and samples were taken every 15 min after release from the arrest. Gin4 was immunoprecipitated (IP) from each sample, and the coprecipitation of Cdc11 and Nap1 was assayed by Western blotting. In addition, the levels of Nap1, Cdc11, and Clb2 present in the crude extracts were assayed by Western blotting. Previous work has shown that levels of Gin4 remain constant during the cell cycle (Altman and Kellogg, 1997). (B) Wild-type and cdc28-4 cells were grown to log phase at room temperature and then shifted to 37°C The association of Gin4 with Cdc11 was detected by coprecipitation. (C) Log phase cells containing Cln3 under the control of the Gal promoter were synchronized in G1 by the addition of dextrose-containing media and then released from the arrest. Samples were taken every 30 min and the coprecipitation of Cdc11 and Nap1 was assayed as before.
To demonstrate that the cell cycle dependence of the Gin4-Cdc11 interaction is not an artifact caused by pheromone-induced arrest, we repeated the experiment using several other means of synchronizing cells. First, we demonstrated that the Gin4-Cdc11 interaction cannot be detected in cells arrested in G1 by shifting the cdc28-4 temperature-sensitive allele to the restrictive temperature, ruling out the possibility that failure to detect the Gin4-Cdc11 interaction during G1 is an artifact of the pheromone arrest (Figure 4B). In addition, we arrested cells in G1 by depletion of the G1 cyclins in a Δcln1 Δcln2 Δcln3 gal1-CLN3 strain. We then released the cells from the arrest by turning on expression of Cln3 and assayed the Gin4-Cdc11 interaction and Clb2 levels at 30-min intervals. Again, we found that the appearance of the Gin4-Cdc11 complex correlated with levels of the Clb2 cyclin as cells entered mitosis (Figure 4C).
The finding that the Gin4-Cdc11 interaction is correlated with Clb2 levels suggested that the interaction should be dependent on mitotic Cdc28 activity. To test whether this is the case, we utilized an analogue-sensitive allele of CDC28 (cdc28-as1), which allows rapid and specific inhibition of Cdc28 kinase activity in vivo by addition of the inhibitor compound 1NM-PP1 (Bishop et al., 2000). We arrested cdc28-as1 cells in mitosis by treatment with the microtubule-destabilizing drug nocodazole, added 1NM-PP1, and assayed Gin4 hyperphosphorylation and Gin4 complex formation 15 min later. We found that inhibition of Cdc28 activity caused loss of Gin4 hyperphosphorylation (Figure 5A). In addition, the interaction between Gin4 and Cdc11 or Shs1 was completely eliminated, whereas the interaction with Nap1 was unaffected (Figure 5B). Controls showed that 1NM-PP1 has no affect on complex formation in cells carrying wild-type CDC28 (not shown). Clb2 levels remained high after treatment with 1NM-PP1, suggesting that disruption of the complex was not due simply to exit from mitosis (Figure 5C).
Figure 5.
Mitotic Cdc28 kinase activity is required for Gin4 hyperphosphorylation and complex assembly. (A) Cells containing the cdc28-as1 allele were arrested in mitosis and then treated with 1NM-PP1 or mock treated with DMSO for 15 min. The behavior of the Gin4 protein was assayed by Western blotting. (B) Gin4 was immunoprecipitated (IP) from the indicated samples and coprecipitation of Nap1, Cdc11, and Shs1 was monitored by Western blotting. (C) Western blotting of the same samples shown in A revealed that Clb2 levels remain high in cells treated with 1NM-PP1.
In Vivo Requirements for Interaction of Gin4 with Nap1, Cdc11, and Gin4
To learn more about the molecular mechanisms leading to formation of the Gin4 complex, we further examined the in vivo requirements for complex assembly. To do this, we assayed whether Gin4 interacts with Cdc11 or Nap1 in Δcla4, Δelm1, Δnap1, Δshs1, or gin4K48A cells. We found that the Gin4-Cdc11 interaction fails to occur in each of these genetic backgrounds, whereas the Gin4-Nap1 interaction is unaffected (Figure 6A). We also tested whether the Gin4-Gin4 interaction occurs normally in Δcla4, Δnap1, and Δshs1 cells. We found that neither Cla4 nor Nap1 are required for the Gin4-Gin4 interaction, suggesting that they do not affect Gin4 activation by inducing binding of Gin4 to itself. In contrast, Shs1 is absolutely required for the Gin4-Gin4 interaction (Figure 6B).
Figure 6.
In vivo requirements for complex assembly. (A) Gin4 was immunoprecipitated (IP) from the indicated strains, and coprecipitation of Cdc11 and Nap1 was assayed by Western blotting. Western blots of the crude extracts were carried out to verify equal protein levels in the crude extracts. (B) The Gin4-Gin4 interaction was monitored in the indicated strains by immunoprecipitation of GST-Gin4 and assaying for coprecipitation of 3×HA-Gin4. Note that the gels used for these Western blots were not run long enough to give full resolution of Gin4 isoforms and therefore appear somewhat different from the Western blots shown in Figure 1. (C) Gin4 was immunoprecipitated from the indicated strains arrested in either interphase (I) or mitosis (M), and coprecipitation of Shs1 was monitored by Western blotting.
Deletion of the SWE1 gene has been shown to partially rescue the elongated cell phenotype observed in Δgin4 and Δcla4 cells (Longtine et al., 2000). This observation has led to the suggestion that the elongated cell phenotype may be due to activation of an Swe1-dependent checkpoint that inactivates Cdc28 (Longtine et al., 2000). To determine whether the failure in the Gin4-Cdc11 interaction in Δcla4 cells is due to Swe1-dependent inhibition of Cdc28 activity, we tested whether Δswe1 could restore the Gin4-Cdc11 interaction in Δcla4 cells. We found, however, that Δswe1 does not restore the Gin4-Cdc11 interaction in Δcla4 cells, demonstrating that the failure in the Gin4-Cdc11 interaction is not due to activation of an Swe1-dependent checkpoint (Figure 6A).
Gin4 Can Interact with Shs1 Independently of Cdc11
Shs1 is required for the Gin4-Gin4 interaction, whereas Cla4 and Nap1 are not. However, Shs1, Cla4, and Nap1 are all required for the Gin4-Cdc11 interaction. Taken together, these results suggest that Shs1 can induce the Gin4-Gin4 interaction under conditions where there is no detectable Cdc11 associated with Gin4. These results are surprising because previous work has shown that septins are found in tight complexes with each other, and we have found that Shs1 is present in a tight complex that includes Cdc11 (see below). The most simple explanation for these observations is that Shs1 can associate with Gin4 and facilitate the Gin4-Gin4 interaction independently of Cdc11. To test this idea, we immunoprecipitated Gin4 under conditions in which we do not detect an interaction between Gin4 and Cdc11. Gin4-Shs1 coimmunoprecipitations were performed in Δcla4, Δnap1, Δelm1, and in cells carrying the gin4K48A allele. We found that a fraction of Shs1 is able to interact with Gin4 independently of Cdc11 in all backgrounds tested, with the exception of the gin4K48A allele (Figure 6C). Additionally, we found that Cdc28 kinase activity is also required for the Gin4-Shs1 interaction because we were unable to detect Shs1 binding to Gin4 after treatment of cdc28-as1 cells with 1NM-PP1 (Figure 5B). We used a similar coimmunoprecipitation assay to confirm that Shs1 is still able to interact with Cdc11 in Δcla4 cells (Mortensen and Kellogg, unpublished results). These results suggest that, although the majority of Shs1 is found in a complex that includes Cdc11, a fraction of Shs1 is able to interact with Gin4 in the absence of detectable Cdc11 and in a manner that is dependent on Cdc28 and Gin4 kinase activity.
Shs1 Undergoes Gin4-dependent Phosphorylation
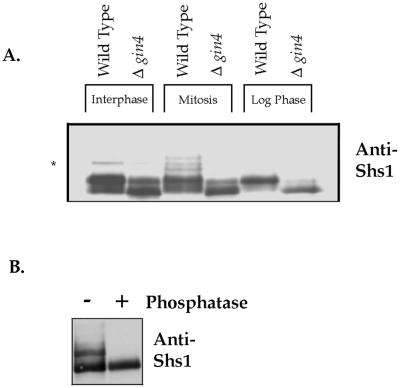
Because the Gin4-septin interaction is cell cycle-dependent, and Gin4 is bound to the septins at a time when it is activated, we were interested in determining whether any of the septins undergo Gin4-dependent phosphorylation in vivo. We therefore used Western blotting to analyze the behavior of Shs1 in Δgin4 cells, in cells arrested in interphase by α factor, in cells arrested in mitosis by nocodazole, and in asynchronous cells undergoing rapid growth (Figure 7A). We found that Shs1 exhibits multiple isoforms during both interphase and mitosis; however, Shs1 undergoes unique modification during mitosis. The mitotic isoforms of Shs1 largely disappear in Δgin4 cells, indicating that they are dependent on Gin4 in vivo. Loss of Gin4 function also causes a dramatic change in the modification state of Shs1 in rapidly growing cells (Figure 7A). To determine whether Shs1 isoforms are due to phosphorylation, we immunoprecipitated Shs1 from cells arrested in mitosis and incubated the precipitated Shs1 with phosphatase. This caused the isoforms to collapse into a single band, demonstrating that they are due to phosphorylation (Figure 7B). These data demonstrate that Gin4 is required in vivo for the mitosis-specific modification of Shs1.
Figure 7.
Shs1 is a phosphoprotein that undergoes Gin4-dependent phosphorylation during mitosis. (A) The indicated strains were arrested in interphase with α factor, or in mitosis with nocodazole, and the behavior of Shs1 was monitored by Western blotting with an anti-Shs1 antibody. The behavior of Shs1 in rapidly growing cells was also analyzed. The asterisk denotes a variable background band that we observed in an Shs1 deletion. (B) Shs1 was immunoprecipitated from cells arrested in mitosis with anti-Shs1 antibody, and the precipitated Shs1 was incubated in the presence (+) or absence (−) of λ-phosphatase. After treatment with phosphatase, differently phosphorylated forms of Shs1 were detected by Western blotting.
Phosphorylation of Shs1 by Purified Gin4 Kinase In Vitro
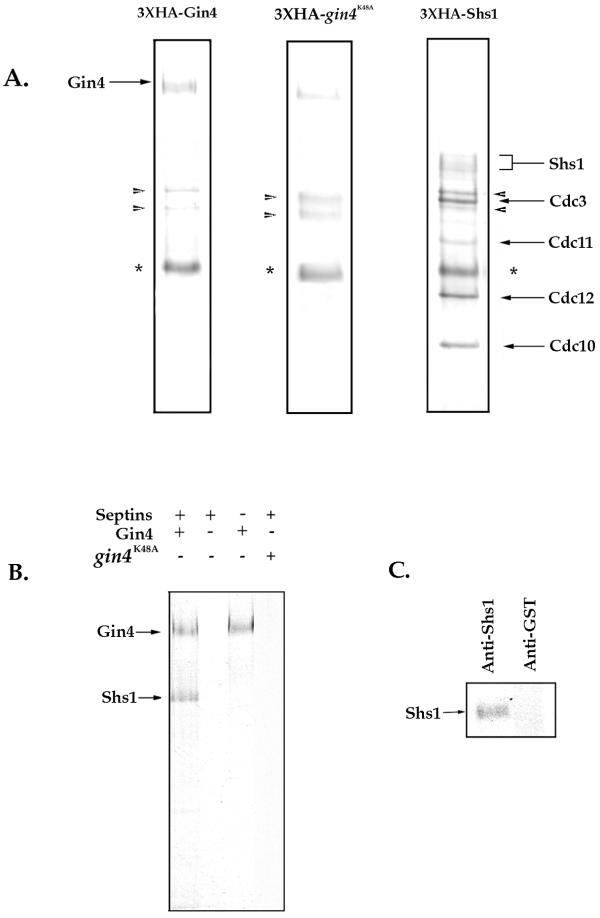
To determine whether Shs1 might be a direct substrate of Gin4, we used immunoaffinity chromatography to purify HA-tagged versions of Gin4 and Shs1 and then tested whether Gin4 can phosphorylate Shs1 in vitro. We purified 3×HA-tagged Gin4 and 3×HA-tagged Shs1 using the same procedure described above for purification of the Gin4 complex, except that we carried out the purification in the presence of 1 M KCl to remove associated proteins. Gin4 was purified from cells arrested in mitosis to obtain the hyperphosphorylated active form of Gin4. As a control, we also purified a 3×HA-tagged version of gin4K48A from benomyl-arrested cells. The purified proteins are shown in Figure 8A. We found that Shs1 copurifies with Cdc3, Cdc10, Cdc11, and Cdc12 as determined by Western blotting and mass spectrometry (not shown), indicating that it forms a tight complex with these septins that is stable in 1 M KCl (see MATERIALS AND METHODS). We next tested whether purified Gin4 can phosphorylate any of the proteins present in the purified septin complex. We found that a protein of the same molecular weight as Shs1 is phosphorylated upon the addition of activated Gin4 to the septins (Figure 8B). To confirm that the phosphorylated band corresponds to Shs1, we disrupted the septin complex by boiling the reaction in the presence of 1% SDS and then reprecipitated Shs1 using an anti-Shs1 antibody. We found that the phosphorylated band can be precipitated by anti-Shs1 antibodies but not by anti-GST antibodies, suggesting that it corresponds to Shs1 (Figure 8C). Additionally, gin4K48A was unable to phosphorylate Shs1 (Figure 8B), suggesting that this event is dependent on catalytically active Gin4. These experiments suggest that Gin4 directly phosphorylates Shs1 in vitro.
Figure 8.
Shs1 is a member of the core septin complex and is an in vitro substrate for the Gin4 kinase. (A) 3×HA-Shs1, 3×HA-Gin4, and 3×HA-gin4K48A were purified by immunoaffinity chromatography in the presence of 1 M KCl, and the purified proteins were separated on an SDS-polyacrylamide gel. The gel was stained with Coomassie blue. The 3×HA-Gin4 was purified from cells arrested in mitosis to obtain the active hyperphosphorylated form of Gin4. The asterisk denotes the antibody heavy chain. The arrowheads denote two background bands corresponding to chaperone proteins that we consistently see in our purifications. (B) Purified Gin4 and purified septin complexes were incubated together or alone in the presence of [γ32P]ATP, and the reactions were loaded onto a 10% SDS-polyacrylamide gel. (C) After incubating Gin4 and septin complexes with [γ32P]ATP, protein complexes were disrupted by the addition of 1% SDS and incubation at 100°C. Samples were then incubated with either anti-Shs1 or anti-GST control beads. After incubation, samples were loaded onto a 10% SDS-polyacrylamide gel.
DISCUSSION
The function and regulation of the septins is poorly understood at the molecular level. Biochemical and genetic experiments demonstrate that the Gin4 kinase is likely to play an important role in mediating septin function or regulation. An understanding of the mechanisms that lead to activation of Gin4 kinase activity, and identification of Gin4 targets, is therefore likely to provide clues to septin function and regulation. In addition, because Gin4 is hyperphosphorylated and activated during mitosis, an understanding of these mechanisms may also provide clues to how mitotic events are controlled at the molecular level. In this study, we have initiated a characterization of the molecular mechanisms underlying Gin4 function and regulation.
Catalytically Inactive Gin4 Is Hyperphosphorylated In Trans to Wild-Type Gin4
In previous work, we demonstrated that hyperphosphorylation of Gin4 involves at least one autophosphorylation event, because a catalytically inactive Gin4 fails to undergo hyperphosphorylation (Altman and Kellogg, 1997). In this study, we have shown that Gin4 autophosphorylation is likely to be due to cross-phosphorylation between associated Gin4 molecules, because a catalytically inactive version of Gin4 (gin4K48A) becomes hyperphosphorylated when a wild-type copy of Gin4 is supplied in trans. It remains possible that additional kinases are involved in the phosphorylation of Gin4. For example, Gin4 could activate another kinase that then phosphorylates the catalytically inactive allele of Gin4. Alternatively, cross-phosphorylation between associated Gin4 molecules could trigger further phosphorylation of Gin4 by another kinase or phosphorylation of Gin4 by another kinase could trigger Gin4 oligomerization and cross-phosphorylation. Cla4, Elm1, and Cdc28 are all candidates for kinases that could directly phosphorylate Gin4. Gin4 activation is reminiscent of the activation of receptor tyrosine kinases, in which binding of a ligand induces dimerization and subsequent cross-phosphorylation of associated kinase monomers (Weiss and Schlessinger, 1998). In these cases, ligand binding induces a conformational change that allows the two kinase monomers to associate or provides a bridge upon which the two monomers are linked. Subsequent hyperphosphorylation of kinase monomers initiates the phosphorylation-dependent assembly of a signaling complex that includes downstream substrates. Shs1-induced oligomerization of Gin4 may function in an analogous manner.
Gin4 Assembles into a Multiprotein Complex as Cells Enter Mitosis
To identify proteins that play a role in the regulation and function of Gin4 during mitosis, we developed an immunoaffinity chromatography approach to purify endogenous Gin4 complexes from cells arrested in mitosis. Our results demonstrate that Gin4 is assembled into a multiprotein complex during mitosis that includes Nap1, 5 members of the septin family, Bni5, and at least two molecules of Gin4. We were able to purify the Gin4 complex under stringent salt and washing conditions, suggesting that the proteins in the complex interact with relatively high affinity and are likely to function together as a complex within the cell. Previous studies have shown that Nap1 and the septins are required in vivo for hyperphosphorylation of Gin4, consistent with the biochemical associations we have observed. Similarly, Bni5 was first identified genetically in a screen for mutations that cause synthetic lethality in combination with septin mutations (M. Longtine, Saccharomyces Genome Database).
Coimmunoprecipitation assays demonstrated that Gin4 associates with the septins during mitosis, whereas Gin4 and Nap1 associate during both mitosis and interphase. The fact that the complex is assembled during mitosis coincident with the activation of the Gin4 kinase suggests that complex assembly plays an important role in the activation and/or activity of Gin4 during mitosis. By assaying protein interactions in mutant strains, we found that the Gin4-Gin4 association requires Shs1, but not Cla4 or Nap1, whereas the Gin4-Cdc11 association requires Shs1, Nap1, Cla4, Elm1, and Gin4 and Cdc28 kinase activity. The Gin4-Shs1 interaction requires the kinase activity of Gin4 and Cdc28 and appears to occur in the absence of detectable levels of Cdc11. The interaction of Nap1 with Gin4 does not require Cla4, Elm1, Shs1, Cdc28, or Gin4 kinase activity. These dependency relationships are summarized in Table 2 and begin to define the in vivo requirements for assembly of the Gin4 complex.
Table 2.
Summary of in vivo requirements for events leading to Gin4 hyperphosphorylation
| Event: | In vivo requirements
|
||||||
|---|---|---|---|---|---|---|---|
| Entry into mitosis | Nap1 | Cla4 | Elm1 | Shs1 | Gin4 kinase activity | Cdc28 kinase activity | |
| Gin4-Nap1 Interaction | No | — | No | No | No | No | No |
| Gin4-Gin4 Interaction | Yes | No | No | N.D. | Yes | N.D. | N.D. |
| Gin4-Shs1 Interaction | Yes | No | No | No | — | Yes | Yes |
| Gin4-Cdc11 Interaction | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Gin4 hyperphosphorylation | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
It is noteworthy that we did not detect Cdc28, Clb2, Cla4, or Elm1 in the Gin4 complex by mass spectrometry or by Western blotting; yet all appear to play important roles in Gin4 activation (Altman and Kellogg, 1997; Tjandra et al., 1998; Sreenivasan and Kellogg, 1999). These proteins may play a role in transducing signals required for complex formation or they may interact with Gin4 transiently during early steps in complex assembly and therefore cannot be detected in a Gin4 complex purified from cells arrested in mitosis. Another possibility is that these proteins affect Gin4 activation indirectly, perhaps by contributing to proper septin architecture (Longtine et al., 2000).
In previous work we found that the septins bind to a Gin4 affinity column constructed with GST-Gin4 purified from bacteria, suggesting that yeast-specific posttranslational modification of Gin4 is not required for interaction with the septins (Carroll et al., 1998). However, in the experiments described here we observed a strict cell cycle dependence of the Gin4-Cdc11 interaction, suggesting that it is dependent on posttranslational modifications. There are a number of possible explanations for these observations. One possibility is that only the septins need to be modified in order for the Gin4-septin interaction to occur and that septins in a yeast crude extract carry the necessary modifications. Another possibility is that GST-Gin4 expressed to high levels in bacteria undergoes autophosphorylation and thereby becomes competent to interact with the septins. GST has been reported to dimerize, which could facilitate Gin4 autophosphorylation in the absence of other factors normally required for autophosphorylation in yeast cells (Walker et al., 1993). A third possibility is that septins in a yeast extract interact with truncated forms of the GST-Gin4 purified from bacteria. Such truncated forms of Gin4 are present because of partial proteolysis and may lack autoinhibitory domains that prevent inappropriate interactions (Carroll et al., 1998). Finally, Gin4 may show a weak affinity for Gin4 and the septins that is independent of posttranslational modification, and this can be detected on an affinity column that contains high concentrations of Gin4 but not in an immunoaffinity experiment in which one is purifying relatively small quantities of Gin4 from extracts. The formation of a more robust Gin4 complex could be facilitated in vivo by cell cycle-dependent phosphorylations of Gin4 or the septins.
Catalytically inactive Gin4 is localized to the bud neck, even though it is not hyperphosphorylated and cannot be detected binding to the septins in coimmunoprecipitation assays (Figure 6; Longtine et al., 1998). In addition, Gin4 colocalizes with the septins early in the cell cycle at the site of bud emergence, apparently before we detect an interaction with the septins. These results suggest that Gin4 may bind to a non-septin component of the bud neck that is not detected in the purified Gin4 complex. An alternative possibility is that inactive Gin4 may have a low affinity for the septins that can localize Gin4 to the bud neck in vivo but is not strong enough to maintain an interaction with the septins through a coimmunoprecipitation assay. According to this model, low-affinity interactions between Gin4 and the septins during interphase could be strengthened by mitotic signaling events and complex assembly.
The Role of Nap1 in Gin4 Activation
The finding that Nap1 interacts with Gin4 during both interphase and mitosis rules out simple models in which the cell cycle-regulated binding of Nap1 plays a role in inducing Gin4 activation. These results are perhaps consistent, however, with previous experiments showing that Nap1 is required both for activation of Gin4 during mitosis and for keeping Gin4 inactive during interphase (Altman and Kellogg, 1997). The molecular mechanisms by which Nap1 regulates Gin4 activity remain unclear. Nap1 also binds to the Clb2 cyclin, suggesting a link between Clb2/Cdc28 activity and Gin4 activation (Kellogg et al., 1995). In addition, Nap1 interacts with a highly conserved protein called Sda1, which carries out functions required for passage through Start (Zimmerman and Kellogg, 2001). The Nap1-Sda1 complex does not appear to include Gin4 or Clb2, suggesting that Nap1 functions with Sda1 independently of these proteins. Nap1 may therefore carry out a basic molecular function that is required in multiple contexts.
Shs1 Plays an Important Role in Complex Assembly
The septins were first identified in budding yeast by temperature-sensitive mutants that cause cells to arrest at G2/M with elongated buds and defects in cytokinesis and were subsequently found to be colocalized with 10-nm filaments at the bud neck (Hartwell, 1971; Byers, 1976; Haarer and Pringle, 1987). More recent work has demonstrated that septins form a tight complex consisting of Cdc12, Cdc11, Cdc10, and Cdc3 (Frazier et al., 1998), and our studies have demonstrated that Shs1 also forms a tight complex with these septins that is stable to 1 M KCl.
The fact that the septins bind to Gin4 during mitosis and are required in vivo for Gin4 activation suggests that they play an important and direct role in the activation of Gin4. It is particularly interesting that the Shs1 septin appears to play a unique and important role in Gin4 activation and can bind to Gin4 and induce Gin4 oligomerization under conditions in which no Cdc11 can be detected binding to Gin4. These observations suggest that septins may carry out functions independently of the core septin complex and that different septins may have unique functions. Recent experiments in Drosophila support the idea that septins can function independently of a core septin complex (Adam et al., 2000). Three septins have been characterized in Drosophila: Pnut, Sep1, and Sep2. Biochemical studies have shown that these septins form a tight complex, but in vivo experiments show that Sep2 can localize to the cellularization front in the absence of Sep1 in Pnut-deficient cells, suggesting that it can at least localize properly in the absence of Sep1 or Pnut (Field et al., 1996; Adam et al., 2000).
Because the septins are GTPases, it is possible that the GTPase activity of Shs1 plays a role in complex formation. For example, the GTP-bound form of Shs1 might bind and partially activate Gin4, leading to phosphorylation of Shs1 by Gin4 and additional events in complex assembly. Further studies exploring the role of GTP binding in the assembly of the Gin4 complex and Gin4 activation should prove to be informative. Recent work has shown that Shs1 is modified with the ubiquitin-like protein Sumo during mitosis, which might also play a role in complex assembly (Johnson and Blobel, 1999). Alternatively, SUMO conjugation to the septins during mitosis could be a consequence of complex assembly and/or signals from the Gin4 kinase. Further work will be required to understand the function and regulation of SUMO conjugation.
The Shs1 Septin Is a Likely In Vivo Target of Gin4 Kinase Activity
Several lines of evidence suggest that Shs1 may be an important substrate for Gin4. First, Shs1 undergoes Gin4-dependent hyperphosphorylation during mitosis in vivo. Second, the Shs1 present in purified septin complexes is a substrate of purified Gin4 in vitro. Third, Shs1 associates with Gin4 specifically during mitosis, when Gin4 is active. Fourth, Gin4 kinase activity is required for the Gin4-Shs1 association. Finally, Δgin4 and Δshs1 cells show similar phenotypes, supporting the idea that Shs1 mediates the effects of Gin4 (Carroll et al., 1998). The finding that Gin4 kinase activity is required for association of Gin4 with Shs1 suggests that phosphorylation of Shs1 by Gin4 is a key step in formation of the Gin4-septin complex. The phosphorylation of Shs1 by Gin4 might also cause changes in Shs1 function that lead to changes in septin organization or function. Alternatively, Shs1 phosphorylation may serve only to activate and recruit Gin4 to the septin ring, where Gin4 then phosphorylates other substrates that play roles in septin function.
Gin4 kinase activity is required for association of Gin4 with Shs1; however, Gin4 can associate with Shs1 in Δcla4, Δnap1, and Δelm1 cells, where Gin4 does not become fully hyperphosphorylated or activated. These observations could be explained by the presence of a basal level of Gin4 kinase activity in these mutant cells. In support of this, in vitro kinase assays have demonstrated that Gin4 is partially active in Δnap1 cells (Altman and Kellogg, 1997). This low activity may be sufficient to facilitate an association with Shs1.
Shs1 phosphorylation is not completely eliminated in Δgin4 cells, suggesting that additional kinases phosphorylate Shs1. Because Hsl1 and Kcc4 are related to the Gin4 kinase and appear to play a role in septin organization, they represent candidates for kinases that phosphorylate Shs1 (Barral et al., 1999). However, we assayed Shs1 phosphorylation in Δkcc4 Δhsl1 Δgin4 cells but observed no differences between the triple mutant and the Δgin4 single mutant, suggesting that another kinase is responsible for Shs1 phosphorylation in Δgin4 cells (Mortensen and Kellogg, unpublished results).
Mechanisms Leading to the Activation of Gin4 during Mitosis
At this point it is difficult to discern whether Gin4 complex formation is a requirement for Gin4 hyperphosphorylation, a consequence of Gin4 hyperphosphorylation, or both. A number of in vivo dependency relationships, however, suggest that Gin4 activation is not due to a simple linear sequence of events and that activation is both a requirement for and a consequence of complex formation. For example, Gin4 self-association, hyperphosphorylation, activation, and localization are all dependent on the septins (Carroll et al., 1998; Longtine et al., 1998). Conversely, septin localization and Shs1 phosphorylation are dependent on Gin4, and Gin4 hyperphosphorylation and Gin4 association with Shs1 and Cdc11 are dependent on Gin4 kinase activity (Altman and Kellogg, 1997; Longtine et al., 1998). These dependency relationships seem to suggest that assembly of the Gin4 complex requires bidirectional signaling events between the septins and Gin4. One model that might explain these observations is that assembly of the complex is initially mediated by low-affinity Gin4-Gin4 or Gin4-Shs1 interactions. These low-affinity interactions could then trigger phosphorylation of Shs1 and/or Gin4, leading to formation of a high-affinity complex, association with the other septins, and full hyperphosphorylation of Gin4.
The finding that Gin4 activation involves a complex series of events may reflect mechanisms that ensure that activation occurs only in the correct context. In addition, if Gin4 activation requires multiple independent events and is highly cooperative, then activation should occur in an all or nothing manner (Ferrell and Machleder, 1998). The finding that Shs1 appears to be required for Gin4 activation and may also be a target of Gin4 kinase activity is particularly interesting. This kind of target-dependent activation would ensure that activation of a kinase occurs only in the correct context and only when it is bound to the protein that it acts upon.
What are the signals that trigger formation of the Gin4-septin complex? A number of observations suggest that Clb2/Cdc28 activity sends the signal that initiates assembly of the Gin4 complex, as might be expected for an event that occurs during mitosis. For example, Gin4 hyperphosphorylation, activation, and association with the septins all occur with a timing that is exactly correlated with the rise in Clb2 levels as cells enter mitosis (Figure 4; Altman and Kellogg, 1997). In addition, Gin4 hyperphosphorylation, and association with the septins, requires mitotic Cdk activity (Figure 5). Finally, the Δgin4 phenotype is significantly more severe in cells that are dependent on Clb2 for survival, suggesting that Gin4 is part of a signaling mechanism initiated by Clb2 (Altman and Kellogg, 1997). The Clb2/Cdc28 kinase complex might play a direct role in signaling formation of the Gin4 complex, possibly by directly phosphorylating Gin4 to initiate complex assembly. Alternatively, Clb2/Cdc28 activity might initiate assembly of the complex by inducing events that are indirectly required for complex assembly. To distinguish these possibilities, we have attempted to reconstitute Gin4 activation in vitro using purified Clb2/Cdc28, septins, and Nap1. Thus far these experiments have not been successful, suggesting that the molecular events surrounding Gin4 activation are more complex than direct phosphorylation by Cdc28. A final possibility is that the Gin4-septin complex is induced by signals that occur in parallel to Clb2/Cdc28 activation but independently of Clb2/Cdc28 activation. In this view, Gin4-septin complex formation would be independent of Clb2/Cdc28 activity, whereas maintenance of the complex would be dependent on signaling from Cdc28. A complete understanding of the molecular mechanisms underlying Gin4 activation during mitosis will require further study.
ACKNOWLEDGMENTS
We thank Grant Hartzog and Topher Carroll, and members of the lab for critical reading of the manuscript. This work was supported by grants from the Pew Charitable Trusts and the National Institutes of Health.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc. 01–10–0500. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–10–0500.
REFERENCES
- Adam JC, Pringle JR, Peifer M. Evidence for functional differentiation among Drosophila septins in cytokinesis and cellularization. Mol Biol Cell. 2000;11:3123–3135. doi: 10.1091/mbc.11.9.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman R, Kellogg DR. Control of mitotic events by Nap1 and the Gin4 kinase. J Cell Biol. 1997;138:119–130. doi: 10.1083/jcb.138.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CW, Baum PR, Gesteland RF. Processing of adenovirus 2-induced proteins. J Virol. 1973;12:241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y, Parra M, Bidlingmaier S, Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beites CL, Xie H, Bowser R, Trimble W. The septin CDC-re1–1 binds syntaxin and inhibits exocytosis. Nat Neurosci. 1999;2:434–439. doi: 10.1038/8100. [DOI] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Byers B. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69:717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C, Altman R, Schieltz D, Yates J, Kellogg DR. The septins are required for the mitosis-specific activation of the Gin4 kinase. J Cell Biol. 1998;143:709–717. doi: 10.1083/jcb.143.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H. Localization and possible functions of Drosopila septins. Mol Cell Biol. 1995;6:1843–1859. doi: 10.1091/mbc.6.12.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- Field CM, al-Awar O, Rosenblatt J, Wong ML, Alberts B, Mitchison TJ. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J Cell Biol. 1996;133:605–616. doi: 10.1083/jcb.133.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford SK, Pringle J. Cellular morphogenesis of the S. cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev Genet. 1991;12:281–292. doi: 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- Frazier JA, Wong ML, Longtine MR, Pringle JR, Mann M, Mitchison TJ, Field C. Polymerization of purified yeast septins: evidence that organized filament arrays may not be needed for septin function. J Cell Biol. 1998;143:737–749. doi: 10.1083/jcb.143.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer BK, Pringle JR. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10-nm filaments in the mother-bud neck. Mol Cell Biol. 1987;7:3678–3687. doi: 10.1128/mcb.7.10.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hartwell L. Genetic control of the cell cycle in yeast: genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Hazuka CD, Roth A, Foletti DL, Heuser J, Scheller RH. Subunit composition, protein interactions, and structures of the mammalian brain sec6/8 complex and septin filaments. Neuron. 1998;20:1111–1122. doi: 10.1016/s0896-6273(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–993. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Alberts BM. Purification of a multiprotein complex containing centrosomal proteins from the Drosophila embryo by chromatography with low-affinity polyclonal antibodies. Mol Biol Cell. 1992;3:1–11. doi: 10.1091/mbc.3.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Kikuchi A, Fujii-Nakata T, Turck CW, Murray A. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J Cell Biol. 1995;130:661–673. doi: 10.1083/jcb.130.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Murray AW. NAP1 acts with Clb2 to perform mitotic functions and suppress polar bud growth in budding yeast. J Cell Biol. 1995;130:675–685. doi: 10.1083/jcb.130.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Kumar S, Mizoguchi A, Ide C, Kinoshita A, Haraguchi T, Hiraok Y, Noda M. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 1997;11:1535–1547. doi: 10.1101/gad.11.12.1535. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR. Direct analysis of protein complexes using mass spectrometry. Nat Biotech. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Fares H, Pringle J. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J Cell Biol. 1998;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, Theesfeld CL, McMillan JN, Weaver E, Pringle JR, Lew DJ. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JX, Lu Q, Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- McMillan JN, Sia RAL, Lew DJ. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J Cell Biol. 1998;142:1487–1499. doi: 10.1083/jcb.142.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T, Rubin G. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. [DOI] [PubMed] [Google Scholar]
- Nguyen TQ, Sawa H, Okano H, White JG. The C. elegans septin genes, unc-59 and unc-61, are required for normal postembryonic cytokinesis and morphogenesis but have no essential function in embryogenesis. J Cell Sci. 2000;113:3825–3837. doi: 10.1242/jcs.113.21.3825. [DOI] [PubMed] [Google Scholar]
- Shulewitz MJ, Inouye CJ, Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan A, Kellogg D. The Elm1 kinase functions in a mitotic signaling network in budding yeast. Mol Cell Biol. 1999;19:7983–7994. doi: 10.1128/mcb.19.12.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra H, Compton J, Kellogg DR. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK kinase family. Curr Biol. 1998;8:991–1000. doi: 10.1016/s0960-9822(07)00419-8. [DOI] [PubMed] [Google Scholar]
- Trimble WS. Septins: a highly conserved family of membrane-associated GTPases with functions in cell division and beyond. J Membr Biol. 1999;169:75–81. doi: 10.1007/s002329900519. [DOI] [PubMed] [Google Scholar]
- Walker J, Crowley P, Moreman AD, Barrett J. Biochemical properties of cloned glutathione S-transferase from Schistosoma mansoni and Schistosoma japonicum. Mol Biochem Parasitol. 1993;61:255–264. doi: 10.1016/0166-6851(93)90071-5. [DOI] [PubMed] [Google Scholar]
- Weiss A, Schlessinger J. Switching signals on or off by receptor dimerization. Cell. 1998;94:277–280. doi: 10.1016/s0092-8674(00)81469-5. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- Zimmerman Z, Kellogg DR. The Sda1 protein is required for passage through Start. Mol Biol Cell. 2001;12:201–219. doi: 10.1091/mbc.12.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]