Abstract
Nuclear receptors, including the androgen receptor (AR), regulate target cell transcription through interaction with auxiliary proteins to modify chromatin structure. We describe herein a novel AR-interacting protein, termed ARIP4, that has structural features typical of the SNF2-like protein family. With regard to the Snf2 domain, the closest homolog of ARIP4 is the ATRX protein. ARIP4 is a nuclear protein and comprises 1466 amino acids. It interacts with AR in vitro and in cultured yeast and mammalian cells. ARIP4 can be labeled with 8-azido-[γ-32P]ATP and exhibits DNA-dependent ATPase activity. Like several ATP-dependent chromatin remodeling proteins, ARIP4 generates superhelical torsion within linear DNA fragments in an ATP-dependent manner. With a stably integrated target promoter, ARIP4 elicits a modest enhancement of AR-dependent transactivation. In transient cotransfection assays, ARIP4 modulates AR function in a promoter-dependent manner; it enhances receptor activity on minimal promoters, but does not activate more complex promoters. ARIP4 mutants devoid of ATPase activity fail to alter DNA topology and behave as trans-dominant negative regulators of AR function in transient assays.
INTRODUCTION
The androgen receptor (AR) belongs to the superfamily of nuclear receptors that are ligand-activated transcription factors capable of regulating transcription of genes containing appropriate response elements, usually within or around the proximal promoter regions (Beato et al., 1995; Quigley et al., 1995; Perlmann and Evans, 1997). After hormone binding, the receptors associate with their cognate DNA motifs and modulate transcription initiation. Nuclear receptors may interact directly with the basal transcription factors associated with RNA polymerase II, such as TFIIB (Ing et al., 1992; Blanco et al., 1995; Hadzic et al., 1995), TFIIF (McEwan and Gustafsson, 1997), and TFIIH (Lee et al., 2000), or elicit their actions indirectly via auxiliary regulatory proteins, called coactivators and corepressors (Torchia et al., 1998; Freedman, 1999; McKenna et al., 1999; Glass and Rosenfeld, 2000).
DNA is folded in the nucleus into a tight chromatin structure that often renders important regulatory sequences inaccessible for sequence-specific transcription factors, including steroid receptors (Kingston et al., 1996; Näär et al., 2001). As a consequence, different chromatin-modifying complexes are required to counteract this repressive effect (Björklund et al., 1999; Kingston and Narlikar, 1999; Lemon and Freedman, 1999; Vignali et al., 2000). The protein complexes can be classified into two main categories: 1) ATP-dependent chromatin-remodeling complexes, which use the energy of ATP hydrolysis to alter the association of histones with DNA; and 2) complexes that alter chromatin by covalent modification of its components. These modifications include histone acetylation, methylation, phosphorylation, and ADP-ribosylation. Yeast SWI/SNF was the first ATP-dependent complex shown to facilitate the function of gene regulatory proteins in a chromatin environment (Hirschhorn et al., 1992). Mammalian (hSWI/SNF), Drosophila (Brahma), and yeast (RSC) homologs of this complex have subsequently been characterized (Tamkun et al., 1992; Kwon et al., 1994; Tsuchiya et al., 1998). In yeast, mutations to components of the SWI/SNF complex result in reduced glucocorticoid (GR) and estrogen receptor (ER) activity, indicating the importance of an intact remodeling complex for steroid receptor activity (Yoshinaga et al., 1992). In addition, coexpression of GR and hbrm (a component of the hSWI/SNF complex) in cells depleted of hbrm restores the GR-dependent transcription (Muchardt and Yaniv, 1993).
The yeast SWI2/SNF2 protein was first described as the subunit responsible for the ATPase activity of the SWI/SNF complex (Khavari et al., 1993; Laurent et al., 1993; Cote et al., 1994). SWI2/SNF2 is the founding member of the family of SNF2-like proteins that share in common a 600-amino-acid-long conserved domain (the Snf2 domain) surrounded by nonconserved regions. The SNF2 superfamily of proteins comprises >100 members (Eisen et al., 1995) and, on the basis of the identity of the ATPase subunit, these proteins have been classified into three main groups: 1) the SWI2/SNF2 group, 2) the imitation SWI (ISWI) group, and 3) the Mi-2 group (Vignali et al., 2000). Most of these proteins have no known biological function; however, those members that have been shown to possess well-defined functions all play distinct roles in DNA processing activities, such as replication, repair, and/or transcription (Pazin and Kadonaga, 1997; Kingston and Narlikar, 1999). The Snf2 domain contains seven so-called helicase motifs and a consensus region for binding and hydrolysis of ATP, and the importance of ATPase activity is well established for the function of many SNF2-like proteins (Kingston and Narlikar, 1999). Although no SNF2-like proteins have been found to function in strand displacement assays for DNA helicase activity, the ability of some ATP-dependent chromatin remodelers to generate unconstrained negative superhelical torsion in DNA and chromatin has been described (Havas et al., 2000). Phylogenetic analysis (Eisen et al., 1995) suggests that the nonconserved regions surrounding the Snf2 domain are responsible for targeting the activity of this domain to specific compartments of cells (Peterson and Workman, 2000).
In this work, we have characterized a novel ATPase that belongs to the SNF2-like family of proteins. The protein, termed ARIP4 (for androgen receptor-interacting protein 4), interacts with AR in vivo and in vitro. It generates superhelical torsion within linear DNA fragments in an ATP-dependent manner and modulates AR-mediated transcription. ARIP4 mutants incapable of ATP hydrolysis fail to alter DNA topology and loose the ability to activate AR-dependent transcription. These mutants also behave as trans-dominant negative regulators of AR function when expressed ectopically in transient transfection assays.
MATERIALS AND METHODS
Materials
[α-32P]dCTP, [γ-32P]ATP, and [35S]methionine were purchased from Amersham Biosciences. 8-Azido-[γ-32P]ATP was a product from ICN Biomedicals (Costa Mesa, CA) and M2 anti-FLAG antibody from Eastman Kodak (Rochester, NY). pARE4-tk-LUC, pARE2-TATA-LUC, pPB(−285/+32)-LUC, pSG5rAR, pcDNA3.1-FLAG-AR, and Gal4-AR, VP16 activation domain (AD) fusion to small nuclear RING finger protein (SNURF) have been described previously (Palvimo et al., 1996; Aarnisalo et al., 1998; Moilanen at al., 1998b). The yeast two-hybrid vectors were kindly provided by Dr. Stanley M. Hollenberg (Vollum Institute, Oregon Health Sciences Center, Portland, OR), and the pLexA fusion proteins have been described previously (Moilanen et al., 1998b). PC-3 cell line stably transfected with pcDNA3.1-FLAG-hAR and pPB(−285/+32)-LUC was established in our laboratory and kindly provided by Dr. Taneli Raivio (Biomedicum Helsinki, Helsinki, Finland). The baculovirus transfer vector pVL1393 and BaculoGold Transfection kit were purchased from BD PharMingen (San Jose, CA), and anti-FLAG affinity gel was obtained from Sigma-Aldrich (St. Louis, MO). pCMVβ and mouse E11.5 λgt11 cDNA library were purchased from CLONTECH (Palo Alto, CA). Ni2+-nitrilotriacetic acid resin and pQE-31 vector were products of QIAGEN (Hilden, Germany). Capture-Tec kit for the isolation of transfected eukaryotic cells was from Invitrogen (Carlsbad, CA).
Yeast Two-Hybrid Screening
Partial sequence of ARIP4 was identified by using the yeast two-hybrid assay as described by Moilanen et al. (1998b). Briefly, the human AR zinc-finger region (ZFR) containing the first 20 hinge region residues was fused to the LexA and used as a bait to screen a size-selected mouse E10.5 cDNA library fused to VP16 activation domain (a gift from Dr. S.M. Hollenberg). The positive clones were tested against several control plasmids, such as pLex-a, pLex-lamin, and pLex-WT1ZF (WT1ZF, the zinc-finger region of the Wilms tumor gene product), to eliminate the false positive clones.
cDNA Cloning and Characterization
ARIP4 cDNA clones isolated in the yeast two-hybrid screen were 400–500 nucleotides (nt) in length. To isolate the full-length ARIP4 cDNA, mouse E11.5 λgt11 cDNA library was screened with 32P-labeled ARIP4 cDNA corresponding to amino acids 91–230 (Figure 1A, probe 1) by using standard hybridization conditions (Asubel et al., 1997). The longest insert was ∼4.0 kb in length; it was subcloned into the EcoRI site of pBluescript II S/K to yield pBS46. The most 3′ end of this ARIP4 cDNA was cleaved with EagI and EcoRI and used as a probe to screen again the E11.5 λgt11 cDNA library. The phage clone extending 3′ from pBS46 was isolated, cleaved with EcoRI, and subcloned into pBluescript II S/K to yield pBS56 and pBS57. The former (pBS56) was colinear with the ARIP4 cDNA insert in pBS46 and the latter continued 3′ from pBS56.
Figure 1.
Schematic structure, amino acid sequence, and nuclear localization of ARIP4. (A) Functional domains of ARIP4 include the AR-ID identified in the yeast two-hybrid screen and the regions predicted to encompass ATPase/helicase activity (regions I–VI). The positions of cDNA probes used for library screenings are also shown. (B) ARIP4 amino acid sequence, predicted from the cDNA sequence. The putative functional units are depicted: NLS, nuclear localization signal; numbers I, Ia, and II–IV refer to the corresponding ATPase/helicase motifs. AR-ID, as predicted on the basis of the yeast two-hybrid screening results, is shown in italics and underlined, and the residues potentially important for ATPase activity are boxed. The consensus sequence for SUMO-1 attachment (ΨKXE) is depicted by single underlining and the LXXLL motifs by double underlining. The sequence is deposited into GenBank with accession number AJ132389. (C) Immunoblot analysis of wild-type and mutated ARIP4 proteins. COS-1 cells were transfected with expression vectors encoding FLAG-tagged ARIP4 (lane 1), ARIP4K310A (lane 2), ARIP4DE462AA (lane 3), ARIP4Δ1315-1466 (lane 4), and RIP4Δ1-277 (lane 5), and the cell extract resolved by SDS-PAGE followed by immunoblotting with a polyclonal anti-ARIP4 antibody (K7991). Each lane contains 10 μg of cell protein. Identical bands were detected with M2 monoclonal anti-FLAG antibody (our unpublished data). (D) Localization of ARIP4 in transfected cells. COS-1 cells grown on glass coverslips on 10-cm plastic plates were transfected with 1 μg of pFLAG-ARIP4 as described in MATERIALS AND METHODS. The cells were fixed, permeabilized, and ARIP4 antigen visualized using either anti-FLAG M2 mAb (A) or anti-ARIP4 antiserum (K7991) raised in rabbits against the 280 most carboxyl-terminal amino acid residues of ARIP4 (B).
Plasmid Constructions
To generate full-length FLAG-tagged ARIP4 (pFLAG-ARIP4), the sequences in pBS46 and pBS56 were assembled together. An in-frame EcoRI site was inserted by polymerase chain reaction in front of the first ATG codon in pBS46. The resulting cDNA was digested EcoRI and cloned into pCMV-FLAG-2 vector (Eastman Kodak) to generate pFLAG-ARIP4 containing amino acids 1–1314. pBS56 was then digested with SmaI/EcoRI to yield a fragment containing the rest of the protein coding region (residues 1205–1466) plus ∼500 nt of the 3′-untranslated region of ARIP4 mRNA. This SmaI/EcoRI fragment was inserted into pFLAG-ARIP4(1–1314) that was linearized by a partial SmaI/EcoRI digestion to yield full-length pFLAG-ARIP4. To assemble ARIP4Δ1–277, a fragment corresponding to ARIP4 residues 278–618 was first subcloned into the EcoRI/BglII site of pCMV-FLAG-2. Then, ARIP4 cDNA, subcloned into the HindIII/XbaI site of pBL5CAT, was digested with BglII and the cleaved fragment inserted into the BglII site of pFLAG-ARIP4(278–618) to yield pFLAG-ARIP4Δ1–277. The QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to mutate ARIP4 sequence. K310, D462, and E463 were converted to Ala residues to yield pARIP4K310A and pARIP4DE462AA expression vectors, respectively. The mammalian two-hybrid vector was constructed by fusing the AR interaction domain of ARIP4 (AR-ID, residues 91–230) in-frame to pVP16 (CLONTECH). Human ornithine decarboxylase (ODC) cDNA was cloned between the EcoRI and SalI sites of pVP16 to express VP16-ODC fusion protein.
Purification of C-Terminal Fragment of ARIP4 for Raising Antibodies
A 1.3-kb SacI–PstI cDNA fragment of ARIP4 corresponding to nt 3773–5071 was subcloned into the SacI/PstI site of pQE-31 (QIAGEN). This fragment encodes the very C-terminal 280 amino acids of ARIP4 fused to an N-terminal His-tag. The protein was expressed in Escherichia coli (strain JM109) and extracted from a 250-ml bacterial culture by suspension in 10 ml of buffer containing 8 M urea, 0.1 M sodium phosphate, 0.01 M Tris-HCl pH 8.0, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and 10 μg/ml aprotinin and incubation at 22°C for 1 h. The lysate was centrifuged at 15,000 rpm for 10 min, the supernatant mixed with 2.5 ml of Ni2+-agarose equilibrated with a buffer containing 8 M urea, 0.1 M sodium phosphate, and 0.01 M Tris-HCl pH 6.3, and the slurry rotated for 1 h at 22°C. The resin was washed three times with 10 volumes of equilibration buffer, and the His-tagged proteins were released by elution with 2 ml of buffer containing 100 mM EDTA, 8 M urea, 0.1 M sodium phosphate, and 0.01 M Tris-HCl pH 6.3. The eluted protein was ∼30 kDa and >95% pure, as judged by SDS-PAGE. Before being used for immunization, urea was removed by stepwise dialysis against phosphate-buffered saline. Fifty micrograms of protein was used to immunize rabbits. One of the immunized rabbits produced the polyclonal antiserum used in the present work (K7991).
Cell Culture and Transfections
COS-1 cells were maintained in Dulbecco's minimal essential medium containing penicillin and streptomycin (each 25 U/ml), and 10% (vol/vol) fetal bovine serum (FBS). Transfections for transactivation assays (∼3–6 × 104 cells) were performed with the FuGene reagent (Roche Applied Science, Indianapolis, IN) with 150 ng of an appropriate reporter vector and the amounts of plasmids depicted in the figure legends. pCMVβ was included to monitor transfection efficiency. At 18 h posttransfection, the medium was changed to one containing charcoal-stripped 2% (vol/vol) FBS and 100 nM testosterone or vehicle. Stably transfected PC-3 cells were maintained in F12 nutrient medium supplemented with charcoal-stripped 10% (vol/vol) FBS, 400 μg/ml G418 (Invitrogen), and penicillin and streptomycin (each 25 U/ml). For transactivation assays in PC-3 cells with stably integrated AR expression vector and probasin promoter, ∼1 × 106 cells were transfected using the FuGene reagent with 1 μg of pHOOK vector (Invitrogen), 0.1 μg of pCMVβ, and 0.5 μg of the appropriate expression vector as shown in the figure legend. Testosterone treatment was carried out in the same way as with COS-1 cells. Twenty-four hours after testosterone addition, the cells were isolated with Capture-Tec kit as recommended by the manufacturer and assayed for reporter gene activity. CV-1 cells transfected with the FuGene reagent were used in the mammalian two-hybrid experiments. For affinity-labeling with 8-azido-[γ-32P]ATP and ATPase assay, COS-1 cells were transfected by electroporation as described previously using 20 μg of the appropriate expression vectors (Moilanen et al., 1998b). Luciferase (LUC) and β-galactosidase activity measurements were carried out as described previously (Palvimo et al., 1996).
RNA Blotting, Immunocytochemistry, and Immunoblotting
Poly(A)-containing RNA was isolated from rat tissues, resolved by agarose gel electrophoresis under denaturing conditions, and transferred to Hybond membrane (Amersham Biosciences) as described previously (Moilanen et al., 1998b). In addition, mouse and rat multiple tissue RNA blots were purchased from OriGene Technologies (Rockville, MD). The blots were hybridized according to the manufacturer's instructions at 42°C in the presence of 50% formamide with a 32P-labeled ARIP4 cDNA fragment (2 × 106 cpm/ml) corresponding to the AR-ID (amino acids 91–230). Final washes were carried out at high stringency (0.1× SSC [1× SSC is 0.15 M NaCl, 0.015 M sodium citrate] and 0.1% SDS, 42°C), and the membranes subjected to autoradiography at −70°C.
ARIP4 antigen in transfected cells was detected by immunocytochemistry as described previously (Moilanen et al., 1998b). COS-1 cells seeded on coverslips were transfected using the FuGene reagent with 1 μg of pFLAG-ARIP4. Cells were fixed in 4% paraformaldehyde and permeabilized with Triton X-100. Ectopically expressed ARIP4 was detected either by anti-FLAG M2 monoclonal antiserum (1:50 dilution) or with anti-ARIP4 polyclonal rabbit antiserum (K7991, 1:1000 dilution) and fluorescein isothiocyanate-conjugated goat anti-mouse or anti-rabbit secondary antibody (1:200 dilution; Jackson Immunoresearch Laboratories, West Grove, PA), respectively. Immunoblotting was conducted as described previously (Poukka et al., 1999) except that ARIP4 was detected with anti-ARIP4 antiserum (K7991, 1:2000 dilution), and immunocomplexes were visualized with horseradish peroxidase-conjugated goat anti-(rabbit immunoglobulin G) antibody and ECL detection reagents (Amersham Biosciences).
Protein–Protein Interaction In Vitro and In Vivo
Affinity chromatography was carried out with bacterially expressed glutathione S-transferase (GST)-AR ZFR or GST alone bound to glutathione-Sepharose (Moilanen et al., 1998b; Poukka et al., 1999). Translation in vitro was performed using the TNT-coupled reticulocyte lysate system from Promega (Madison, WI). Ten microliters of [35S]methionine-labeled translation product was mixed with GST or GST-AR ZFR in a buffer containing 50 mM Tris-HCl pH 7.8, 50 mM KCl, 0.5 mM EDTA, 5 mM MgCl2, 0.05 mM ZnCl2, 10% (vol/vol) glycerol, 0.4% Nonidet P-40, 0.1% Triton X-100, and protein inhibitor cocktail (Sigma-Aldrich) in a total volume of 500 μl at 4°C overnight. The beads were washed three times with 1 ml of binding buffer, and bound proteins were resolved by SDS-PAGE and visualized by fluorography.
For coimmunoprecipitation experiments, COS-1 cells (∼3.5 × 105 cells) were transfected with 300 ng of pFLAG-ARIP4 and 50 ng of pSG5hAR. One day after transfection, the cells were exposed to 100 nM testosterone (or vehicle) for 2 h and then lysed in buffer containing 50 mM Tris-HCl pH 7.8, 150 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol, 0.3% Triton X-100, 0.5% Nonidet P-40, 0.3% Na-deoxycholate, 10 mM N-ethylmaleimide, 15 mM MgCl2, and protease inhibitor cocktail (Sigma-Aldrich). The lysates were clarified by centrifugation at 4°C for 20 min at 16,000 × g and precleared by incubation with 50 μl of GammaBind Sepharose (Amersham Biosciences) and 5 μl of mouse monoclonal anti-VP16 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 2 h at 4°C on a rotary shaker. After centrifugation, the precleared supernatants were adsorbed onto 30 μl of anti-FLAG M2 affinity matrix (Sigma-Aldrich) for 1 h at 4°C. The matrix was subsequently washed with 3 ml of lysis buffer, and bound proteins were eluted by boiling for 5 min in SDS-PAGE loading buffer and resolved by 7.5% SDS-PAGE. Immunoblotting was performed as described previously (Poukka et al., 1999) by using rabbit polyclonal anti-AR (K333, 1:7000 dilution) and anti-ARIP4 (K7991, 1:2500 dilution) antisera.
Immunoprecipitation and ATP-binding Assay
COS-1 cells electroporated with 10 μg of expression vectors encoding FLAG-tagged ARIP4, ARIP4K310A, ARIP4DE462AA, and AR were lysed in buffer containing 20 mM Tris-HCl pH 7.8, 140 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 0.5% Nonidet P-40, 1 mM dithiothreitol, 0.5 mM PMSF, and 10 μg/ml aprotinin. The cell lysates were clarified by centrifugation for 30 min at 16,000 × g at 4°C and adsorbed onto 50 μl of anti-FLAG M2 affinity matrix. After immunoadsorption, the matrix was washed with 4 ml of lysis buffer followed by 4 ml of binding buffer containing 20 mM HEPES pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 μM ZnCl2, 0.1% Tween 20, 0.5 mM PMSF, and 10 μg/ml aprotinin. The samples were kept as a 75% slurry. Ten microliters of antibody-immobilized proteins were incubated with 4 μCi of 8-azido-[γ-32P]ATP for 15 min at 22°C in 20 μl of binding buffer in the absence or presence of 5 mM ATP. The reaction mixtures were irradiated at 3 cm from 254-nm light bulbs of UV Stratalinker 2400 (Stratagene) by using the auto cross-link mode (120,000 μJ for 50 s). Cross-linked proteins were resolved by SDS-PAGE and visualized by autoradiography.
ATPase Assay
FLAG-tagged ARIP4, ARIP4K310A, ARIP4DE462AA, and AR were expressed in COS-1 cells and immunopurified as for the ATP-binding experiments except that proteins were eluted from the affinity matrix with buffer A (20 mM Tris-HCl pH 7.5, 2 mM MgCl2, 2 mM dithiothreitol, 5 mM KCl, and 150 mM NaCl) containing 0.2 mg/ml FLAG peptide. The assay mixture (15 μl) contained 11 μl of buffer B (buffer A supplemented with 100 μM ATP, 0.5 μCi of [γ-32P]ATP [3000 Ci/mmol], 3 μl of immunopurified protein sample) or 100 fmol of nonstructural protein 2 (nsP2) (Rikkonen et al., 1994) as the positive control, and 1 μl of double-stranded (ds)-DNA (1 mg/ml , pGL3-Basic vector; Promega) or water. After a 1-h incubation at 37°C, 0.5 μl of the reaction mixture was spotted onto a poly(ethyleneimine)-cellulose thin-layer plate that was developed in 1 M LiCl and 1 M formic acid to resolve 32Pi from [32P]ATP. The plates were subjected to autoradiography and scanned on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA) to quantify the amount of 32Pi released.
Production of Recombinant ARIP4 Proteins in Insect Cells
cDNA encoding FLAG-tagged wild-type ARIP4 or ARIP4K310A was cloned into XbaI/BglII-digested baculovirus transfer vector pVL1393 (BD PharMingen). Recombinant transfer plasmids were cotransfected into Spodoptera frugiperda (Sf9) cells with a modified linear baculovirus DNA by using the BaculoGold transfection kit (BD PharMingen). Sf9 cells were maintained and infected as monolayers (20 × 106 cells/150-cm2 flask at a multiplicity of infection of 10) in TNM-FH medium containing 10% FBS, 50 μg/ml gentamicin, and 2.5 μg/ml amphotericin B (Kallio et al., 1994). Infected cells were harvested 66–72 h postinfection and lysed in 1.5 ml of lysis buffer containing 20 mM Tris-HCl pH 7.8, 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% Triton X-100, 10% glycerol, 5 mM dithiothreitol, and protease inhibitor cocktail (Sigma-Aldrich). The lysates were clarified by centrifugation for 20 min at 16,000 × g and adsorbed onto 50 μl of anti-FLAG affinity gel. After immunoadsorption, the gel was washed with the lysis buffer containing 400 mM NaCl and the adsorbed protein was eluted with the lysis buffer containing 0.2 mg/ml FLAG peptide. The purity of the proteins was checked by SDS-PAGE and immunoblotting and was shown to be >95% in each case.
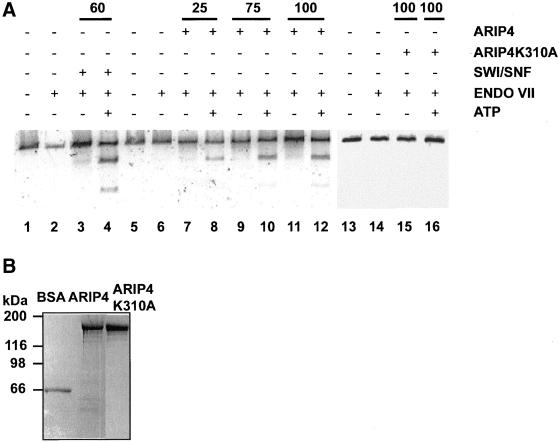
DNA Cruciform Formation Assay
The assay was performed as described previously (Havas et al., 2000). Briefly, the reactions (20 μl) contained 10 ng (0.4 nM) AvaI cut pGX540 (Greaves et al., 1985) and 25–100 ng of either wild-type or mutant ARIP4 as indicated in the legend to Figure 7. Reactions performed using SWI/SNF complex contained 1.5 nM SWI/SNF complex (60 ng) prepared from strain CY396. Standard reactions contained protein A-endonuclease VII (Giraud-Panis and Lilley, 1996) at 0.15 μg/ml. The reactions were incubated at 22°C for 1 h and terminated by the addition of stop buffer followed by an incubation for 30 min at 50°C.
Figure 7.
ARIP4 creates superhelical torsion within linear DNA fragments. (A) Cruciform formation assay was carried out essentially as described by Havas et al. (2000). Endonuclease VII fails to digest cruciform-free linear DNA (lanes 2, 6, and 14). In the presence of yeast SWI/SNF complex (lanes 3 and 4) or wild-type ARIP4 (lanes 7–12), cruciform formation occurs only in the presence of ATP in the reaction mixture (lanes 4, 8, 10, and 12). The mutant ARIP4K310A has no detectable activity both in the presence and absence of ATP (lanes 15 and 16). Indicated amounts (in ng) of ARIP4 or ARIP4K310A protein were used in the assay. (B) Purity of ARIP4 and ARIP4K310A proteins used in cruciform formation assay. The proteins were expressed in insect cells, and portions of the fractions eluted from the affinity matrix with the FLAG peptide (see MATERIALS AND METHODS) were subjected to SDS-PAGE. The gel was stained with GelCode Blue reagent (Pierce Chemical, Rockford, IL). For comparison, 500 ng of bovine serum albumin (BSA) was analyzed on the same gel.
RESULTS
Cloning of a New ATPase/Helicase-like Protein
Potential interaction partners for the ZFR of AR were identified by a yeast two-hybrid screen with hAR ZFR, encompassing the DNA-binding domain and 20 amino-terminal amino acids of the hinge region, as the bait (AR amino acids 544–644). AR ZFR was fused to LexA (LexA-AR ZFR), and the plasmid encoding the fusion construct was used to screen a size-selected 10.5-d-old mouse embryo cDNA library as described previously (Moilanen et al., 1998b). This screen yielded ∼30 positive clones, which corresponded to five unique cDNA sequences that were all represented multiple times among the isolates. Three of the encoded proteins have already been characterized (Moilanen et al., 1998a,b; 1999). To isolate a full-length cDNA encoding the fourth protein, mouse E11.5 λgt11 cDNA library was screened with two probes. The first sequence (Figure 1A, probe 1) representing the AR interaction domain (AR-ID) recognized in the yeast two-hybrid screen identified a 4.0-kb-long cDNA fragment containing a long open reading frame preceded by an in-frame stop codon. The second probe corresponded to the 3′ end of the 4.0-kb cDNA (Figure 1A, probe 2), and rescreening of the mouse embryo λgt11 cDNA library with this sequence led to the isolation of an additional clone that covered the complete 3′ end of the protein-coding sequence together with part of the 3′-untranslated region. The deduced sequence predicts a 1466-amino-acid-long protein that was termed ARIP4 (Figure 1B).
ARIP4 has a calculated molecular mass of 160 kDa, a net charge of −18.2 at pH 7.5, and an isoelectric point of 6.4. When ectopically expressed in COS-1 cells, the immunoreactive protein migrates on SDS-PAGE with an apparent molecular mass of ∼180 kDa (Figure 1C). Expression of pFLAG-ARIP4 in COS-1 cells results in nuclear localization of the ARIP4 antigen, as visualized by immunocytochemical analyses by using either anti-FLAG or anti-ARIP4 antibodies (Figure 1D). It is of note that ARIP4 is not evenly distributed in recipient cell nuclei but the immunoreactivity exhibits a speckled pattern. ARIP4 sequence includes at least three putative bipartite nuclear localization signals (amino acids 98–114, 412–428, and 1254–1271). There is a consensus SUMO modification site (ψKXE where ψ represents a large hydrophobic amino acid and X any amino acid; Yeh et al., 2000) starting at residue 663 and indeed, a sumoylated ARIP4 form is detectable in transfected cells (our unpublished data). There are three LXXLL motifs (Heery et al., 1997), also known as nuclear receptor boxes, in the ARIP4 sequence (amino acids 550–554, 724–728, and 1328–1332). The amino-terminal region (amino acids 21–260), including the AR interaction domain (AR-ID, amino acids 91–230), is very rich in negatively charged amino acids (Figure 1B), a feature typical of many proteins involved in transcriptional regulation.
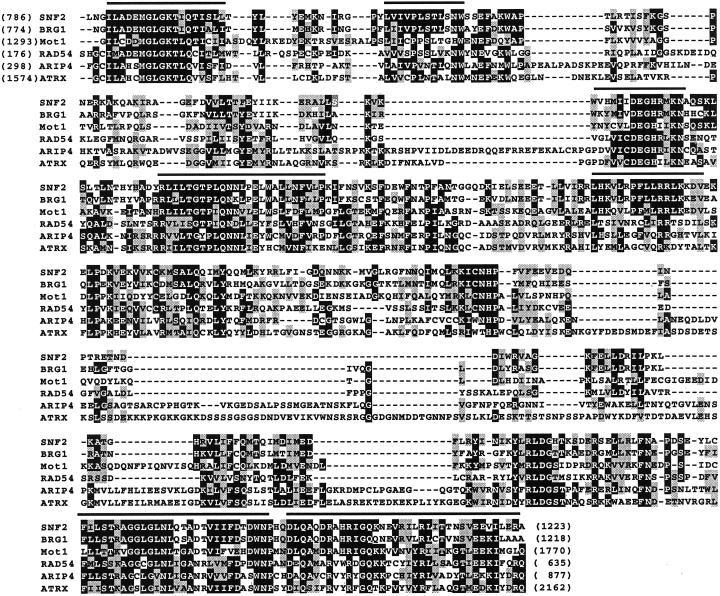
ARIP4 contains a region with a strong sequence homology to proteins in the SNF2-like family of ATPase subunits, and all the seven helicase motifs shared by the family members are included in the ARIP4 sequence (Figures 1B and 2), with the ATRX protein (Picketts et al., 1996) showing the highest homology in the helicase domains. The sequence similarity was restricted to the Snf2 domain, and no protein homologous to ARIP4 outside the Snf2 domain has thus far been isolated and characterized. However, a predicted protein sequence (KIAA0809 protein) of an unidentified human gene that exhibits >95% amino acid sequence identity with ARIP4 in their overlap, covering amino acids 225-1466 of ARIP4, has been deposited into GenBank (accession number BAA34529). Northern blot analysis of RNA samples from several mouse and rat tissues revealed that ARIP4 is encoded by an ∼10.0-kb mRNA that is expressed at a relatively low level, with the highest levels of ARIP4 mRNA accumulation occurring in testis, liver, and kidney (Figure 3). In addition, ARIP4 mRNA is present in rat prostate in levels comparable with those in the testis (our unpublished data).
Figure 2.
Homology of the ARIP4 SNF2 domain with that of some other members of the SNF2-like protein family. The numbers in parentheses depict the first and last amino acid taken for comparison in each case; the regions were as follows: SNF2, 786–1223; BGR1, 774–1218; Mot1, 1293–1770; RAD54, 176–635; ARIP4, 298–877; and ATRX, 1574–2162. The regions that include the seven-consensus helicase motifs (I, Ia, and II–VI) are depicted by a line above the sequence. Shaded amino acids are similar in at least three of the proteins compared. The following amino acid groups are considered to be similar: L, V, I, and M; F, Y, and W; S, T, A, P, and G; K, R, and H; and E, D, Q, and N. The sequence information is from Khavari et al. (1993), Laurent and Carlson (1992), Emery et al. (1991), Davis et al. (1992), and Picketts et al. (1996).
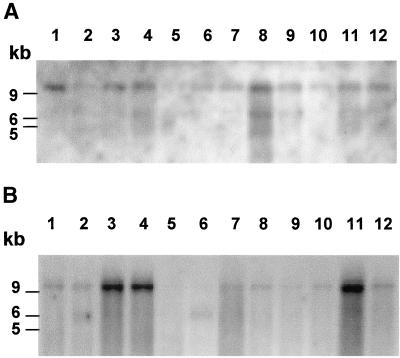
Figure 3.
Expression of ARIP4 mRNA in rat and mouse tissues. (A) Rat tissues. The blot contains 2 μg of poly(A)+ RNA per lane from the following tissues (the lane in parentheses): brain (1), thymus (2), lung (3), heart (4), muscle (5), stomach (6), small intestine (7), liver (8), kidney (9), spleen (10), testis (11), and skin (12). (B) Mouse tissues. The blot contains 2 μg of poly(a)+ RNA per lane from the following tissues (the lane in parentheses): brain (1), heart (2), kidney (3), liver (4), lung (5), muscle (6), skin (7), small intestine (8), spleen (9), stomach (10), testis (11), and thymus (12). The blots were from OriGene and were hybridized with 32P-labeled ARIP4 cDNA probe as described in MATERIALS AND METHODS. The integrity and equal loading of RNA were verified by the manufacturer with the human β-actin cDNA probe (our unpublished data).
ARIP4 Binds ATP and Possesses Intrinsic ATPase Activity
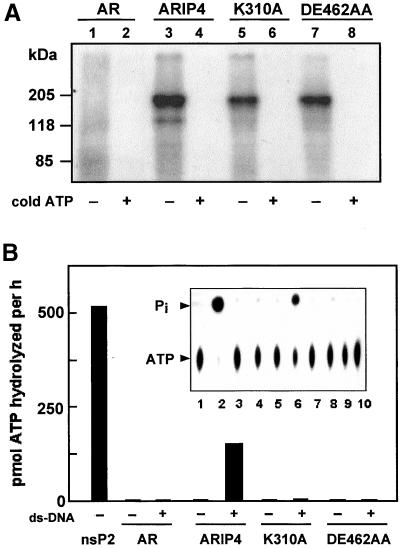
Several SNF2-like family members exhibit ATP-hydrolyzing activity (Laurent et al., 1993; Auble et al., 1994). To examine whether ARIP4 is capable of binding ATP, FLAG-tagged ARIP4 and two ARIP4 forms with mutations in the putative catalytic center (FLAG-ARIP4K310A and FLAGARIP4DE462AA) were immunopurified, and the isolated proteins were incubated with a photoreactive ATP analog, 8-azido-[γ-32P]ATP. UV irradiation of ARIP4 with 8-azido-[γ-32P]ATP resulted in cross-linking of [32P]ATP to the protein (Figure 4A, lane 3). ARIP4K310A and ARIP4DE462AA were also affinity labeled with 8-azido-[γ-32P]ATP, but somewhat less efficiently than the wild-type protein (Figure 4A, lanes 5 and 7). Immunopurified AR (a control protein) was not labeled by 8-azido-[γ-32P]ATP in a specific manner under the same conditions (Figure 4A, lane 1). Affinity labeling of ARIP4 proteins with 8-azido-[γ-32P]ATP was abolished by inclusion of a 1000-fold molar excess of nonradioactive ATP, attesting to the specificity of the cross-linking reaction.
Figure 4.
Affinity-labeling with 8-azido-[γ-32P]ATP and ATPase activity of ARIP4. (A) COS-1 cells were transfected with plasmids encoding the following FLAG-tagged proteins: AR (lanes 1 and 2); ARIP4 (lanes 3 and 4); ARIP4K310A (K310A, lanes 5 and 6); and ARIP4DE462AA (DE462AA, lanes 7 and 8). Proteins were purified by immunoadsorption onto anti-FLAG antibody matrix and affinity labeled with 8-azido-[γ-32P]ATP as described in MATERIALS AND METHODS. [32P]ATP-labeled proteins were resolved by SDS-PAGE and visualized by autoradiography. Labeling specificity was assessed by the inclusion of 1000-fold molar excess of nonradioactive ATP during affinity labeling (lanes 2, 4, 6, and 8). The amounts of AR as well as wild-type and mutant ARIP4 proteins in each lane were similar, as judged by immunoblot analysis (our unpublished data). (B) COS-1 cells were transfected with plasmids encoding FLAG-tagged AR, ARIP4, ARIP4K310A, and ARIP4DE462AA, and the cell extracts were immunopurified by anti-FLAG affinity matrix. The proteins eluted from the resin with FLAG peptide were subjected to ATPase assay with [γ-32P]ATP as the substrate. Semliki forest virus nsP2 was included as the positive control, and the amount of ATP hydrolyzed was calculated on the basis of its specific activity (91 pmol of 32Pi released/pmol protein × min−1). The assays were performed in the absence (−) and presence (+) of 1 μg of ds-DNA as depicted. The inset shows the autoradiogram of the thin-layer plate used to resolve 32Pi from [32P]ATP. The lanes contained the following samples: 1, blank; 2, nsP2; 3 and 4, AR ± ds-DNA; 5 and 6, ARIP4 ± ds-DNA; 7 and 8, ARIP4K310A ± ds-DNA; and 9 and 10, ARIP4DE462AA ± ds-DNA. The spots corresponding to 32Pi and [32P]ATP are depicted.
To measure ATPase activity of ARIP4, the FLAG-tagged wild-type protein together with the mutants ARIP4K310A and ARIP4DE462AA were purified by immunoadsorption. Semliki forest virus nsP2 (Rikkonen et al., 1994) and FLAG-tagged AR were used as positive and negative controls. ATPase activity of ARIP4 was strictly dependent on the presence of double-stranded DNA in the reaction mixture, and the two ARIP mutants along with AR were completely devoid of ATPase activity (Figure 4B). The amounts of FLAG-tagged ARIP4 proteins were not significantly different in the experiments presented in Figure 4 (our unpublished data). Under these conditions, ARIP4 hydrolyzed ∼50–100 ATP/min; a turnover number lower than that (∼1000 ATP/min) reported for other ATPases of the SNF2-like protein family (Peterson, 2000). However, the conditions used in the assay were not optimized for kinetic analysis of ARIP4 ATPase.
ARIP4 Interacts with AR In Vivo and In Vitro
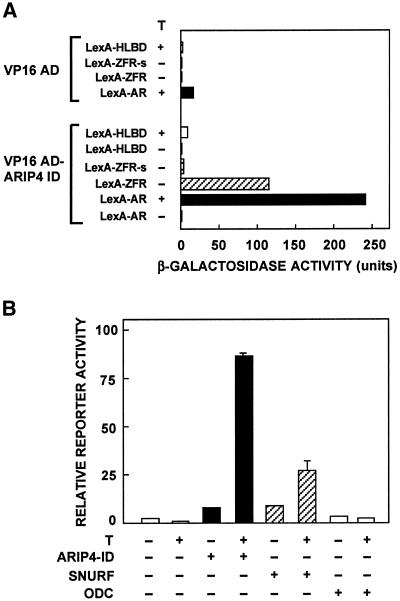
The interaction between AR and ARIP4 was assessed in yeast and in mammalian cells. In L40 yeast cells, different regions of AR fused to LexA, including the original bait construct (LexA-AR ZFR, AR amino acids 554–644), were coexpressed with the herpex simplex VP16 activation domain (VP16 AD) alone or with VP16 AD fused to amino acids 86–227 of ARIP4 (VP16-ARIP4 ID). Coexpression of VP16-ARIP4 ID and LexA-AR ZFR increased the reporter gene activity by ∼100-fold over that with Lex-AR ZFR and VP16 AD alone (Figure 5A). VP16-ARIP4 ID also interacted very strongly with full-length AR fused to LexA (LexA-AR); this interaction was strictly dependent on the presence of androgen (Figure 5A). Deletion of the 20 hinge region residues from the original bait construct (LexA ZFR-s, AR amino acids 554–623) weakened the interaction with VP16-ARIP4 ID markedly, suggesting that the amino-terminal hinge residues either participate in the interaction with ARIP4 or are essential for the AR ZFR to fold properly. A LexA fusion protein, including the hinge residues and the AR ligand-binding domain (LexA-HLBD, AR amino acids 624–919) exhibited rather weak interaction with VP16-ARIP4 ID in the presence of androgen; it was approximately fourfold higher that with VP16 AD alone (Figure 5A). When ZFRs of ER and progesterone receptors (PR) were fused to LexA, the fusion proteins possessed measurable interaction with VP16-ARIP4 ID, which was 20–30% of that with AR ZFR (our unpublished data).
Figure 5.
ARIP4 interacts with AR and in yeast and mammalian cells. (A) Yeast two-hybrid assay. Plasmids expressing different LexA fusion proteins (LexA-AR, LexA-ZFR [amino acids 554–644 of AR], LexA-ZFR-s [amino acids 554–623 of AR], LexA-HLBD [amino acids 624–919 of AR]) were used to cotransform into Saccharomyces cerevisiae L40 strain with plasmids expressing VP16 AD or VP16 AD fused to ARIP4 (amino acids 86–227) (VP16 AD-ARIP4 ID). Transformants were grown in the presence (+) or absence (−) of 50 nM testosterone (T) as depicted. β-Galactosidase activity in extracts of liquid culture is shown, and each bar gives the average of three independent yeast transformants. (B) Interaction of ARIP4 with AR in mammalian cells. CV-1 cells were transfected using the FuGene reagent with a reporter plasmid pG5LUC (150 ng), 30 ng of rat AR (amino acids 3–902) fused to the DBD of Gal4 (Gal4-AR), and 30 ng of the indicated VP16 AD fusion proteins depicted by the + signs. The proteins fused to VP16 AD were ARIP4-ID (ARIP4 amino acids 86–227), SNURF (SNURF residues 20–177), and human ODC. T (100 nM) was present in the culture medium as indicated. Each bar corresponds to mean ± SEM values of at least three independent experiments, and the values were calculated relative to that of Gal-AR in the presence of androgen and VP16 AD alone (= 1).
In mammalian cells, ARIP4 ID fused to VP16 AD interacts with full-length AR fused in-frame to Gal4 DBD, as judged by the activation of a reporter gene driven by Gal4 DNA-binding motifs (Figure 5B). There was some interaction with ARIP4 ID and Gal4-AR in the absence of androgen (approximately fivefold increase), but the interaction was much stronger in the presence than absence of 100 nM testosterone in culture medium (>10-fold increase). SNURF and human ODC fused to VP16 AD served in these experiments as positive and negative controls (Figure 5B). SNURF is an AR coregulator (Moilanen et al., 1998b), and it interacts with AR ZFR both in vitro and in vivo (Moilanen et al., 1998b). In contrast, ODC is a cytoplasmic protein that is not anticipated to interact with nuclear receptors. The interaction of ARIP4 ID with Gal4-AR was stronger than that of SNURF with the receptor, whereas ODC failed to recognize Gal4-AR (Figure 5B).
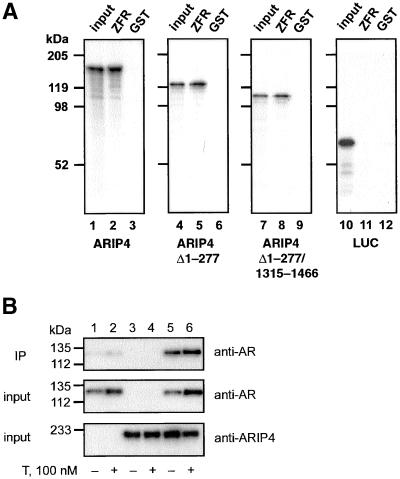
To assess whether ARIP4 interacts directly with AR under cell-free conditions, GST pull-down experiments were performed using full-length ARIP4 and ARIP4 with an amino-terminal deletion (ARIP4Δ1–277) or ARIP4 with both amino- and carboxyl-terminal deletions (ARIP4Δ1–277/1315–1466). The proteins were labeled with [35S]methionine by translation in vitro and adsorbed to glutathione-Sepharose beads containing AR ZFR (GST-ZFR). After the incubation of 35S-labeled protein with the GST-ZFR matrix, the beads were washed, and bound proteins were eluted and resolved by SDS-PAGE. Full-length ARIP4 interacted specifically with GST-ZFR (Figure 6A, lane 2), and no ARIP4 adhered to the control beads (lane 3). Neither an amino-terminal deletion (ARIP4Δ1–277; Figure 6A, lane 5) nor the combined amino- and carboxyl-terminal deletions (ARIP4Δ1–277/1315–1466; Figure 6A, lane 8) influenced markedly the in vitro interaction of ARIP4 with AR ZFR, indicating that there are other regions in ARIP4 capable of interacting with AR besides the AR-ID (residues 91–230) recognized in the yeast two-hybrid screen. [35S]Methionine-labeled LUC served as a negative control in these studies, and it did not exhibit any binding to GST-ZFR (Figure 6A, lane 11). The results of the GST pull-down experiments suggest that ARIP4 and AR have a complex interaction pattern, perhaps owing to the multiple LXXLL motifs, and that the region 91–230 of ARIP4 identified as the AR-ID in the yeast two-hybrid screen is not the only region of ARIP4 involved in the interaction with AR.
Figure 6.
Interaction of androgen receptor with ARIP4. (A) In vitro interaction of ARIP4 with the AR ZFR. Wild-type ARIP4, ARIP4Δ1–277, ARIP4Δ1–277/1315–1466 or LUC were synthesized by translation in vitro by using reticulocyte lysate in the presence of [35S]methionine, and the labeled proteins were incubated with the GST-AR ZFR matrix (ZFR) or with the GST matrix (GST) alone. After washing of the matrix, the bound proteins were released by boiling in the electrophoresis sample buffer, resolved by SDS-PAGE, and visualized by fluorography. Lanes 1, 4, 7, and 10 represent 5% of the input of the appropriate [35S]methionine-labeled protein. (B) Coimmunoprecipitation of ARIP4 and AR from COS-1 cell lysates. The cells were transiently transfected with 50 ng of pSG5hAR (lanes 1, 2, 5, and 6) and 300 ng of pFLAG-ARIP4 (lanes 3–6). Testosterone was included in culture medium as indicated for 2 h before harvesting the cells. The cells were lysed and lysates adsorbed onto anti-FLAG M2 affinity matrix, and the bound proteins resolved by SDS-PAGE as described in MATERIALS AND METHODS. The presence of AR in immunoprecipitates (IP) was detected by immunoblotting with polyclonal rabbit anti-AR antiserum (K333). Also shown are portions (2.5%) of the extract subjected to immunoprecipitation (input) and immunoblotted with polyclonal rabbit anti-AR (K333) and anti-ARIP4 (K7991) antisera.
ARIP4 and AR also interacted under the conditions, in which the two protein were ectopically expressed in COS-1 cells (Figure 6B). Unlike the interaction observed in a mammalian two-hybrid assay between full-length AR and ARIP4 ID, the interaction between full-length AR and ARIP4 proteins under the coimmunoprecipitation conditions was not markedly influenced by the presence of androgen (Figure 6B, cf. lanes 5 and 6), further supporting the notion that there are multiple potential interaction interfaces between the two proteins.
ARIP4 Generates Superhelical Torsion within Linear DNA Fragments
Recent results have shown that several members of the SNF2 superfamily of proteins can generate superhelical torsion within linear DNA fragments (Havas et al., 2000). To examine whether ARIP4 also possesses this activity, FLAG-tagged wild-type ARIP4 and the ATPase-deficient ARIP4K310A mutant were produced in insect cells by using the baculovirus expression system. The proteins were subsequently purified to >90% homogeneity by immunoaffinity chromatography (Figure 7B) and assayed for their ability to generate superhelical torsion on free DNA in the cruciform formation assay. The yeast SWI/SNF complex is active in this assay, and its activity is ATP dependent (Havas et al., 2000). It served as a positive control in our experiments (Figure 7, lanes 3 and 4), which indicated that ARIP4 is capable of generating a negative supercoiling within the linear DNA molecule and that this activity is dependent on the presence of catalytic ATPase activity (Figure 7, lanes 7–12). The ATPase-deficient ARIP4K310A mutant exhibited no detectable activity in the cruciform formation assay (Figure 7, lanes 15 and 16).
Effect of ARIP4 on AR-dependent Transcription
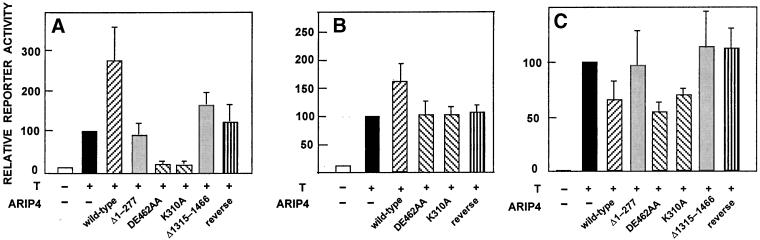
Other proteins in the SNF2-like family have been demonstrated to cooperate with the activity of some nuclear receptors (Yoshinaga et al., 1992; Muchardt and Yaniv, 1993; Chiba et al., 1994; Ichinose et al., 1997). To assess the influence of ARIP4 on the transactivation ability of AR, transient cotransfections were performed in COS-1 cells by using reporter constructs driven by different promoters. In addition to wild-type ARIP4, several mutated ARIP4 forms were transfected with AR. Coexpression of ARIP4 with AR increased the transcriptional activity of AR on a minimal promoter (ARE4-tk; Figure 8A) approximately threefold, whereas an ARIP4 form devoid of the amino terminus, including the AR interaction domain identified in the yeast two-hybrid screen (AR-ID; ARIP4Δ1–277), did not influence AR function. Truncation of ARIP4 at the carboxyl terminus (ARIP4Δ1315–1466) generated a protein that was significantly less active than wild-type ARIP4, suggesting that there are functionally important interaction surfaces for other proteins in the carboxyl-terminal region of ARIP4. It is of note that ARIP4 sequence contains three LXXLL motifs (amino acids 550–554, 724–728, and 1328–1332), and therefore, both ARIP4Δ1–277 and ARIP4Δ1315–1466 may still interact with AR through these nuclear receptor boxes. Interestingly, the ARIP4 mutants devoid of ATPase activity (ARIP4K310A and ARIP4DE462AA) were unable to activate AR function; rather, when coexpressed with AR, these two ARIP4 forms behaved as trans-dominant negative regulators of AR under the transient transfection conditions (Figure 8A). ARIP4 cDNA sequence expressed in reverse orientation did not modulate AR function significantly. ARIP4 also activated ARE4-tk promoter in PC-3 cells (our unpublished data). Similar to ARE4-tk, wild-type ARIP4 activated AR function and the ATPase-deficient mutants behaved as trans-dominant negative regulators of AR on another minimal promoter, ARE2-TATA (our unpublished data).
Figure 8.
Effect of ectopic ARIP4 expression on AR-dependent transactivation. (A) COS-1 cells were transiently transfected using the FuGene reagent with 150 ng of ARE4-tk-LUC, 20 ng of pSG-rAR and 30 ng of wild-type ARIP4 or ARIP4 mutants as indicated. Testosterone (T, 100 nM) was added 24 h after transfection as depicted by the + signs, and the cells were harvested 24 h later. The values are expressed relative to that of AR in the presence of androgen without ARIP4 expression plasmids (= 100), and each bar represents mean ± SEM values of at least three independent experiments. The amount of wild-type ARIP4 expression vector used (30 ng) produced a response that was 30–50% of maximal. (B) PC-3 cells with stably integrated pcDNA3.1-FLAG-hAR and pPB(−285/+32)-LUC were transfected using the FuGene reagent with 1 μg of pHOOK vector, 0.1 μg of pCMVβ, and 0.5 μg of the expression vector encoding wild-type ARIP4 or ARIP4 mutants as indicated. T (100 nM) was added 18 h after transfection as depicted by the + signs, and the cells were harvested 24 h later using Capture-Tec kit. The values are normalized to transfection efficiency and expressed relative to that of AR in the presence of androgen without ARIP4 expression plasmids (= 100). Each bar represents mean ± SEM values of at least four independent experiments. (C) COS-1 cells were transiently transfected as in A, except that 150 ng of pPB(−285/+32)-LUC was used as the reporter construct. The values (mean ± SEM, n = 3) are expressed as in A.
We next examined the influence of ectopic ARIP4 expression on AR function in PC-3 cells that were stably transfected with a human AR expression vector and the luciferase reporter gene driven by the probasin promoter (−285/+32). Under these conditions, the reporter gene was activated by AR ∼10-fold in the presence of androgen (Figure 8B). Wild-type ARIP4 elicited a modest increase (1.5–1.6-fold) in ligand-dependent transcription by AR from the stably integrated promoter, whereas the ATPase-deficient mutants ARIP4K310A and ARIP4DE462AA did not modify AR function.
In contrast to the stably integrated probasin promoter, ARIP4 failed to activate AR function on this promoter under transient transfection conditions in COS-1 (Figure 8C) or PC-3 cells (our unpublished data); rather, wild-type ARIP4 and the ATPase-deficient mutants attenuated AR-dependent transactivation to some extent. The mutants ARIP4Δ1–277 and ARIP4Δ1315–1466 did not alter AR function (Figure 8C). Immunoblot analyses indicated that wild-type and mutant ARIP4 proteins were expressed to comparable levels under the experimental conditions used (Figure 1C; our unpublished data).
To examine activator specificity of ARIP4, parallel series of cotransfection experiments were performed with AR, GR, PR, and ER by using minimal promoters in COS-1 cells; ARE4-tk for AR, GR, and PR, and ERE2-TATA for ER. Under these conditions, ARIP4 did not influence GR or PR function (1.0 ± 0.03- and 0.97 ± 0.06-fold vs. GR and PR alone, respectively), whereas AR function was activated in a manner similar to that depicted in Figure 8A (2.2 ± 0.2-fold). ARIP4 failed to activate ER function; rather, ER activity in the presence of ARIP4 was approximately one-half of that with ER alone.
DISCUSSION
Like other steroid receptors, AR contains two transactivation functions; AF1 in the amino-terminal region and AF2 in the hormone-binding domain (Quigley et al., 1995). In contrast to other members of this nuclear receptor subfamily, the activity of AR AF1 is much stronger than that of AF2 (Moilanen et al., 1997). The AF2 domain, however, can be activated through interaction with the amino-terminal region of AR and/or with multiple coregulatory proteins (Ikonen et al., 1997; He et al., 1999; Glass and Rosenfeld, 2000). The AR ZFR also presents an interaction interface for proteins, including those needed in trans-repression of activating protein 1 (AP1)- and nuclear factor-κB–activated genes, in part through a mechanism involving the cAMP response element-binding protein-binding protein-binding protein (Kallio et al., 1995; Palvimo et al., 1996; Aarnisalo et al., 1998). We have previously characterized three proteins interacting with AR ZFR and shown that, even though they do not belong to related protein families, each one behaves as an AR coregulator (Moilanen et al., 1998a,b; 1999). The fourth AR ZFR-interacting protein described herein, ARIP4, is a novel member of the SNF2-like family. ARIP4 binds ATP and possesses DNA-dependent ATPase activity and generates unconstrained negative superhelical torsion on free DNA, similar to several other proteins of this family, including the yeast SWI/SNF complex, human BGR1 as well as Drosophila ISWI and Xenopus Mi-2 complexes (Havas et al., 2000).
The interaction interface for AR ZFR recognized in the yeast two-hybrid screen resides in a nonconserved region located amino-terminal to the ATPase domain of ARIP4. The AR-ID region does not seem to be the only domain for the ARIP4–AR interaction, because the ARIP4 sequence contains three so-called nuclear receptor boxes, the LXXLL motifs (amino acids 550–554, 724–728, and 1328–1332) that form potential interaction interfaces for the ligand-binding domain of AR (Heery et al., 1997). Thus, the ATPase activity of ARIP4 may be recruited to AR-regulated chromatin regions through interaction with multiple regions of the receptor.
The proteins of the SNF2-like family comprise members that play multiple roles in the regulation of protein–DNA interactions, such as those involved in transcription, replication, and repair, and contain a large conserved Snf2 domain that confers ATPase activity on the proteins (Pazin and Kadonaga, 1997; Kingston and Narlikar, 1999). The initial connection of the SWI/SNF complex to steroid receptor-dependent transcription was the demonstration that GR activity, expressed in yeast, requires this complex and that reporter activation brought about by GR or ER is lost in yeast strains deficient for SWI1, SWI2, and SWI3 proteins of the SWI/SNF complex (Yoshinaga et al., 1992). Subsequent studies showed that ectopically expressed mammalian SWI2/SNF2 homologs hbrm and BGR1 cooperate with GR or ER in trans-activation of reporter genes (Muchardt and Yaniv, 1993; Chiba et al., 1994; Fryer and Archer, 1998), that GR increases the SWI/SNF nucleosome remodeling activity when bound to a nucleosomal glucocorticoid response element (Östlund Farrangs et al., 1997), and that glucocorticoids promote hormone-induced association of GR to the BRG1 complex in vivo (Fryer and Archer, 1998).
Diverse regions of steroid receptors are implicated in the interaction with SNF2-like proteins. An intact DNA-binding domain (zinc-finger region) of GR was shown to be needed for the receptor's cooperation with SWI2/SNF2 proteins and their mammalian homologs (Yoshinaga et al., 1992; Muchardt and Yaniv, 1993). The SWI/SNF complex may also potentiate GR action through the amino-terminal transactivation domain of the receptor (Wallenberg et al., 2000). In contrast, the ligand-binding domain of ER comprising the AF2 function interacted in a yeast two-hybrid assay with the amino-terminal region preceding the ATPase domain of hbrm or BRG1 (Ichinose et al., 1997). Another ATPase, p68 RNA helicase, was recently shown to be a coactivator of ERα function and to interact with the amino terminus containing AF1 of this receptor (Endoh et al., 1999). Our present data link AR function to a novel member of the SNF2-like family, ARIP4, the amino-terminal region of which was recognized in the yeast two-hybrid screen with AR ZFR. Deletion of this AR interaction interface (AR-ID) abolished the activity of ARIP4 on AR function, despite the fact that other regions of ARIP4, such as the LXXLL motifs may also interact with AR. Comparison of the AR-ID sequence with those in three other proteins interacting with AR ZFR (Moilanen et al., 1998a,b; 1999) fails to define a consensus sequence, although each surface contains a Ser-rich cluster of charged amino acids (Asp, Arg, Glu, and Lys) flanked by nonpolar residues (Ile, Leu, and Val).
The ATPase activity of ARIP4 was mandatory for its ability to activate AR function, a situation similar to that with GR activation by hbrm (Muchardt and Yaniv, 1993) and to the function of many other, but not all, SNF2-like family members (Khavari et al., 1993; Laurent et al., 1993; Auble et al., 1994; Kingston and Narlikar, 1999; Tyler and Kadonaga, 1999). Similar to the ATPase-deficient ARIP4 mutants ARIP4K310A and ARIP4DE462AA in mammalian cells under transient transfection conditions, MOT1 mutants devoid of ATPase activity behaved as trans-dominant negative alleles in yeast (Auble et al., 1994). Mechanisms for this feature are currently unknown, but perhaps ARIP4 without catalytic ATPase activity locks other partners interacting with endogenous ARIP4, or a related protein, in a nonfunctional complex. Determination of the nature of proteins complexed with ARIP4 in vivo would help to resolve this issue. The carboxyl-terminal region of ARIP4 does not exhibit sequence homology with any SNF2-like family members or contain recognizable functional domains, such a bromodomain or a SANT domain (Kingston and Narlikar, 1999; Vignali et al., 2000). The ATRX protein, a transcriptional regulator (McDowell et al., 1999), exhibits the highest homology with the ARIP4 Snf2 domain, but ARIP4 does not contain a plant homeodomain-like domain, such as that in ATRX, in the sequence flanking the helicase region. However, the deletion of the ∼150 carboxyl-terminal residues attenuated ARIP4 activity, implying that ATPase activity is mandatory but not sufficient for the ability of ARIP4 to modulate AR function.
ATP-driven chromatin remodeling factors facilitate not only transcriptional activation but also repression (Kingston and Narlikar, 1999; Tyler and Kadonaga, 1999). For example, genome-wide expression analysis in yeast revealed that, of the genes dependent on the SWI/SNF complex (∼6% of all yeast genes), a greater number was negatively rather than positively regulated by SWI/SNF (Holstege et al., 1998). Likewise, the c-fos promoter is repressed by BRG1, the mechanism of which requires the presence of the Rb protein (Murphy et al., 1999). Transcription activation by SWI/SNF is attributed to interaction of the SWI/SNF complex with acidic activators, whereas recruitment of histone deacetylase activity by chromatin remodeling complexes is involved in transcriptional repression (Tong et al., 1998; Neely et al., 1999; Tyler and Kadonaga, 1999). In view of this, it was not totally unexpected that AR-dependent minimal promoters ARE4-tk and ARE2-TATA were activated but the more complex probasin promoter was, if anything, attenuated by ectopic ARIP4 expression in transient transfection assays. However, ARIP4 was able to activate modestly the probasin promoter under the conditions, where this promoter is in a proper chromatin context, i.e., stably integrated in the PC-3 cell genome. We cannot exclude the possibility that, similar to another SNF2-like family member, MOT1 (Auble et al., 1997), ARIP4 influences DNA binding of the receptor itself in a promoter-specific manner.
Targeting of Snf2 domain-containing chromatin remodeling complexes in vivo to specific DNA sequences can be achieved 1) by recruitment of sequence-specific transcription factors, 2) through interaction with the RNA polymerase holoenzyme, or 3) by intrinsic DNA-binding ability (Björklund et al., 1999; Kingston and Narlikar, 1999; Lemon and Freedman, 1999). It is tempting to suggest that targeting of ARIP4-containing complexes to AR-dependent genes occurs through AR, a sequence-specific transcription factor. This notion is supported by the results showing that, at least under transient transfection conditions, ARIP4 exhibits receptor selectivity, because it failed to activate GR, PR, and ER function. However, it remains to be established whether ARIP4 possesses intrinsic DNA-binding activity that would, in turn, be able to target AR to requisite regulatory regions. In any event, ARIP4 is the first SNF2-like protein shown to interact with the AR, and it is also a novel member of this protein family. Better understanding of its function in steroid receptor signaling requires improved knowledge of the proteins that form complexes with ARIP4 in vivo and the ways by which ARIP4 facilitate nucleosome assembly and mobilization. It would be of particular interest to examine whether ARIP4 forms complexes in vivo with other AR ZFR-interacting proteins; SNURF (Moilanen et al., 1998b); the nuclear Ser/Thr kinase ANPK (Moilanen et al., 1998a); and ARIP3, a member of the PIAS family (Moilanen et al., 1999). And finally, it is intriguing that ARIP4 is a potential target for SUMO-1 modification, in a manner similar to AR itself (Poukka et al., 2000).
ACKNOWLEDGMENTS
We thank Leena Pietilä, Pirjo Kilpiö, Seija Mäki, and Kati Saastamoinen for skillful technical assistance; Hetti Poukka, Ulla Karvonen, and Marika Häkli for help with some experiments; Taneli Raivio for the PC-3 cell line; and Petri Auvinen for advice with the ATPase assay. This work was supported by grants from the Medical Research Council (Academy of Finland), the Finnish Foundation for Cancer Research, the Sigrid Jusélius Foundation, Biocentrum Helsinki, the Helsinki University Central Hospital, and Association for the Cure of Cancer of Prostate (CaP CURE).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–10–0484. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–10–0484.
REFERENCES
- Aarnisalo P, Palvimo JJ, Jänne OA. CREB-binding protein in androgen receptor-mediated signaling. Proc Natl Acad Sci USA. 1998;95:2122–2127. doi: 10.1073/pnas.95.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: John Wiley & Sons; 1997. [Google Scholar]
- Auble DT, Hansen KE, Mueller CGF, Lane WS, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- Auble DT, Wang D, Post KW, Hahn S. Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol Cell Biol. 1997;17:4842–4851. doi: 10.1128/mcb.17.8.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M, Herrlich P, Schütz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- Björklund S, Almouzni G, Davidson I, Nightingale KP, Weiss K. Global transcription regulators of eukaryotes. Cell. 1999;96:759–767. doi: 10.1016/s0092-8674(00)80586-3. [DOI] [PubMed] [Google Scholar]
- Blanco JCG, Wang I-M, Tsai SY, Tsai M-J, O'Malley BW, Jurutka PW, Haussler MR, Ozato K. Transcription factor TFIIB and the vitamin D receptor cooperatively activate ligand-dependent transcription. Proc Natl Acad Sci USA. 1995;92:1535–1539. doi: 10.1073/pnas.92.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophilabrahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Davis JL, Kunisawa R, Thorner J. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery HS, Schild D, Kellogg DE, Mortimer RK. Sequence of RAD54, a Saccharomyces cerevisiaegene involved in recombination and repair. Gene. 1991;104:103–106. doi: 10.1016/0378-1119(91)90473-o. [DOI] [PubMed] [Google Scholar]
- Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H, Yanagisawa J, Metzger D, Hashimoto S, Kato S. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor α. Mol Cell Biol. 1999;19:5363–5372. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Freedman LP. Increasing the complexity of coactivation in nuclear receptor signaling. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- Fryer CJ, Archer TJ. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Lilley DM. T4 endonuclease VII. Importance of a histidine-aspartate cluster within the zinc-binding domain. J Biol Chem. 1996;271:33148–33155. doi: 10.1074/jbc.271.51.33148. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Greaves DR, Patient RK, Lilley DM. Facile cruciform formation by an (A-T)34 sequence from a Xenopusglobin gene. J Mol Biol. 1985;185:461–478. doi: 10.1016/0022-2836(85)90064-6. [DOI] [PubMed] [Google Scholar]
- Hadzic E, Desai-Yajnik V, Helmer E, Guo S, Wu S, Koudinova N, Casanova J, Raaka BM, Samuels HH. A 10-amino-acid sequence in the N-terminal A/B domain of thyroid hormone receptor alpha is essential for transcriptional activation and interaction with the general transcription factor TFIIB. Mol Cell Biol. 1995;15:4507–4517. doi: 10.1128/mcb.15.8.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havas K, Flaus A, Phelan M, Kingston R, Wade PA, Lilley DM, Owen-Hughes T. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell. 2000;103:1133–1142. doi: 10.1016/s0092-8674(00)00215-4. [DOI] [PubMed] [Google Scholar]
- He B, Kemppainen JA, Voegel JJ, Gronemeyer H, Wilson EM. Activation function 2 in the human androgen receptor ligand binding domain mediates inter-domain communication with the NH2-terminal domain. J Biol Chem. 1999;274:37219–37225. doi: 10.1074/jbc.274.52.37219. [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Brown SA, Clark CD, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- Holstege FCP, Jennings EG, Wyrick JJ, Ihn Lee T, Hengartner CJ, Green MR, Golub TR, Lander EC, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Ichinose H, Garnier J-M, Chambon P, Losson R. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene. 1997;188:95–100. doi: 10.1016/s0378-1119(96)00785-8. [DOI] [PubMed] [Google Scholar]
- Ikonen T, Palvimo JJ, Jänne OA. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- Ing NH, Beekman JM, Tsai SY, Tsai M-J, O'Malley BW. Members of the steroid hormone receptor superfamily interact with TFIIB (S300-II) J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- Kallio PJ, Palvimo JJ, Mehto M, Jänne OA. Analysis of androgen receptor-DNA interactions with receptor proteins produced in insect cells. J Biol Chem. 1994;269:11514–11522. [PubMed] [Google Scholar]
- Kallio PJ, Poukka H, Moilanen A, Jänne OA, Palvimo JJ. Androgen receptor mediated transcriptional regulation in the absence of direct interaction with a specific DNA element. Mol Endocrinol. 1995;9:1017–1028. doi: 10.1210/mend.9.8.7476976. [DOI] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Bunker CA, Imbalzano AN. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Laurent BC, Carlson M. Yeast SNF2/SWI2, SNF5, and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and Biocoid. Genes Dev. 1992;6:1707–1715. doi: 10.1101/gad.6.9.1707. [DOI] [PubMed] [Google Scholar]
- Laurent BC, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- Lee DK, Duan HO, Chang C. From androgen receptor to general transcription factor TFIIH. Identification of cdk activating kinase (CAK) as an androgen receptor NH2-terminal associated coactivator. J Biol Chem. 2000;275:9308–9313. doi: 10.1074/jbc.275.13.9308. [DOI] [PubMed] [Google Scholar]
- Lemon BD, Freedman LP. Nuclear receptor cofactors as chromatin remodelers. Curr Opin Genet Dev. 1999;9:499–504. doi: 10.1016/s0959-437x(99)00010-6. [DOI] [PubMed] [Google Scholar]
- McEwan IJ, Gustafsson J-Å. Interaction of the human androgen receptor transactivation function with the general transcription factor TFIIF. Proc Natl Acad Sci USA. 1997;94:8485–8490. doi: 10.1073/pnas.94.16.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell TL, et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci USA. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocrin Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- Moilanen A, Rouleau N, Ikonen T, Palvimo JJ, Jänne OA. The presence of a transcription activation function in the hormone-binding domain of androgen receptor is revealed by studies in yeast cells. FEBS Lett. 1997;412:355–358. doi: 10.1016/s0014-5793(97)00791-6. [DOI] [PubMed] [Google Scholar]
- Moilanen A-M, Karvonen U, Poukka H, Jänne OA, Palvimo JJ. Activation of androgen receptor function by a novel nuclear protein kinase. Mol Biol Cell. 1998a;9:2527–2543. doi: 10.1091/mbc.9.9.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moilanen A-M, Karvonen U, Poukka H, Yan W, Toppari J, Jänne OA, Palvimo JJ. A testis-specific coregulator of androgen receptor that belongs to a novel family of nuclear proteins. J Biol Chem. 1999;274:3700–3704. doi: 10.1074/jbc.274.6.3700. [DOI] [PubMed] [Google Scholar]
- Moilanen A-M, Poukka H, Karvonen U, Häkli M, Jänne OA, Palvimo JJ. Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol Cell Biol. 1998b;18:5128–5139. doi: 10.1128/mcb.18.9.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophilabrm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Hardy S, Engel DA. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol Cell Biol. 1999;19:2724–2733. doi: 10.1128/mcb.19.4.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näär AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–5001. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Wallberg AE, Steger DJ, Cairns BR, Wright APH, Workman JL. Activation domain-mediated targeting of the SWI/SNF complex to promoter stimulates transcription from nucleosome arrays. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- Östlund Farrangs A-K, Blomquist P, Kwon H, Wrange Ö. Glucocorticoid receptor–glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol Cell Biol. 1997;17:895–905. doi: 10.1128/mcb.17.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palvimo JJ, Reinikainen P, Ikonen T, Kallio PJ, Moilanen A, Jänne OA. Mutual transcriptional interference between RelA and androgen receptor. J Biol Chem. 1996;271:24151–24156. doi: 10.1074/jbc.271.39.24151. [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein–DNA interactions? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- Perlmann T, Evans RM. Nuclear receptors in Sicily: all in the famiglia. Cell. 1997;90:391–397. doi: 10.1016/s0092-8674(00)80498-5. [DOI] [PubMed] [Google Scholar]
- Peterson CL. ATP-dependent chromatin remodeling. Going mobile. FEBS Lett. 2000;476:68–72. doi: 10.1016/s0014-5793(00)01673-2. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Workman JL. Promoter targeting, and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OWJ, Gibbons RJ. ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet. 1996;5:1899–1907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- Poukka H, Aarnisalo P, Karvonen U, Palvimo JJ, Jänne OA. Ubc9 interacts with the androgen receptor and activates receptor-dependent transcription. J Biol Chem. 1999;274:19441–19446. doi: 10.1074/jbc.274.27.19441. [DOI] [PubMed] [Google Scholar]
- Poukka H, Karvonen U, Jänne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc Natl Acad Sci USA. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley CA, de Bellis A, Marschke KB, El-Awady MK, Wilson EM, French FS. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr Rev. 1995;16:271–321. doi: 10.1210/edrv-16-3-271. [DOI] [PubMed] [Google Scholar]
- Rikkonen M, Peränen J, Kääriäinen L. ATPase and GTPase activities associated with Semliki forest virus nonstructural protein nsP2. J Virol. 1994;68:5804–5810. doi: 10.1128/jvi.68.9.5804-5810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. Brahma: a regulator of Drosophilahomeotic genes structurally related to the yeast transcriptional activator SWI2/SNF2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Torchia J, Glass C, Rosenfeld MG. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- Tsuchiya E, Hosotani T, Miyakawa T. A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiaecentromeres. Nucleic Acids Res. 1998;26:3286–3292. doi: 10.1093/nar/26.13.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Kadonaga JT. The “dark side” of chromatin remodeling: repressive effects on transcription. Cell. 1999;99:443–446. doi: 10.1016/s0092-8674(00)81530-5. [DOI] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallenberg AE, Neely KE, Hassan AH, Gustafsson J-Å, Workman JL, Wright APH. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor τ1 activation domain. Mol Cell Biol. 2000;20:2004–2013. doi: 10.1128/mcb.20.6.2004-2013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins. new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]