Abstract
Background/purpose
Bone reconstruction in the maxillofacial region typically relies on autologous bone grafting, which presents challenges, including donor site complications and graft limitations. Recent advances in tissue engineering have identified highly pure and proliferative dedifferentiated fat cells (DFATs) as promising alternatives. Herein, we explored the capacity for osteoblast differentiation and the osteoinductive characteristics of extracellular vesicles derived from DFATs (DFAT-EVs).
Materials and methods
DFATs were isolated from human buccal fat pads, cultured to confluency, and placed in either a standard or osteogenic induction medium. After culturing for 3 days, the conditioned medium was used to generate EVs using the size-exclusion chromatography and concentration filter method.
Results
Characterization of DFAT-EVs revealed typical EV morphology and positive markers (CD9 and CD63), with no differences between the two groups. In vitro assays demonstrated that EVs derived from the osteogenic induction medium (OI-EVs) significantly increased alkaline phosphatase activity and osteogenesis-related genes (Runx2 and collagen type I) compared to control EVs. Next-generation sequencing identified differentially expressed miRNAs, and gene ontology analysis suggested pathways involved in osteoblast differentiation.
Conclusion
Isolating DFATs from buccal fat pads under osteogenic induction conditions offers a procedure confined to the oral cavity, eliminating the need for harvesting from other sites. Thus, DFAT-EVs hold promise for promoting bone regeneration in maxillofacial applications.
Keywords: Buccal fat pad, Dedifferentiated fat cells, DFAT-Derived extracellular vesicles, Extracellular vesicles, miRNA, Osteoblast differentiation
Introduction
Dedifferentiated fat cells (DFATs) can be harvested with minimal invasiveness from all age groups and are produced using the ceiling culture method.1 As they are derived from the mature adipose layer of fat tissues, DFATs represent a highly pure cell population with a high potential for proliferation and multipotent differentiation into bone, cartilage, and fat cells. These characteristics make DFATs suitable donor cells for regenerative medicine.1,2
The buccal fat pad (BFP) offers a convenient and accessible location for oral surgeons and oral dentists, allowing for minimally invasive surgical removal. BFP can be incidentally collected during surgeries in the oral and maxillofacial regions without expanding the surgical field. Kishimoto et al. reported that cell surface antigens of DFATs derived from BFP were essentially equivalent to those established from subcutaneous adipose tissues; further, DFATs originating from the buccal mucosa demonstrated enhanced osteoblastic differentiation capabilities compared to adipose-derived stem cells (ADSCs) from the same source.2,3 However, Kou et al. reported that DFATs possess weak osteogenic capacity,4 prompting further investigations to enhance their bone-forming potential.
Research on extracellular vesicles (EVs) aims to elucidate their extensive biological functions and therapeutic potential, offering insights into their role in cellular communication and potential applications in medical science. Extensive studies have demonstrated that EVs derived from stem cells enhance osteogenesis, angiogenesis, and bone mineralization. These results clearly suggest that therapies based on stem cell-derived EVs, which do not involve cells, hold potential for bone repair.5 Through paracrine effects, EVs can influence cell phenotype, mobilization, proliferation, and differentiation, offering potential applications in regenerative medicine. Recent exploration of stem cell-derived EVs for applications in tissue regeneration,6 particularly those from mesenchymal stem cells (MSCs), has revealed their impact on osteoinductive differentiation.5,7, 8, 9 Changes in culture conditions can affect the properties of isolated EVs. Liu et al. reported that EVs cultured in osteoblast differentiation induction medium promote bone differentiation in MSCs.10
To date, there have been no reports on the isolation of DFAT-derived EVs (DFAT-EVs). This study compared the osteoinductive potentials of DFAT-EVs obtained under different conditions for optimized osteogenic DFAT-EVs. Additionally, the component miRNA differences among DFAT-EVs were analyzed using next-generation sequencing (NGS) to understand key signaling pathways and the mechanisms for optimized osteogenic DFAT-EVs.
Materials and methods
Isolation and culture of human dedifferentiated fat cells
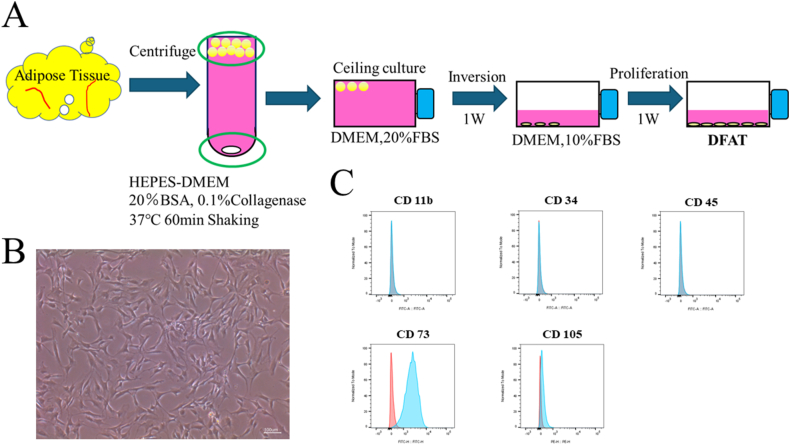
To isolate DFATs, human BFP samples were collected from healthy female and male donors treated at Osaka Dental University Hospital. Informed consent was obtained from all donors, and the research protocol was endorsed by the Ethics Committee of Osaka Dental University (approval number: 111242). DFATs were isolated using the ceiling culture technique, as outlined in a previous study (Fig. 1A).2 Surface antigens of cells were analyzed using flow cytometry (FACSVerse; BD Biosciences, San Jose, CA, USA). FlowJo software (BD Biosciences) was used to acquire the data and count the number of positive cells. We evaluated the following surface antigens with antibodies: CD11b (Cat#301329, BioLegend, San Diego, CA, USA), CD34 (Cat#343503, BioLegend), CD45 (Cat#304005, BioLegend), CD73 (Cat#11073942, Thermo Fisher Scientific Inc., Waltham, MA, USA), CD105 (Cat#323206, BioLegend).
Figure 1.
“Ceiling culture” method to isolate DFAT. (A) Adipose tissue acquired from the rat inguinal region was degraded using 0.1% collagenase. After centrifugation, monocular adipocytes were isolated from the floating fat layer. The isolated cells were cultured for 7 days in culture flasks filled with Dulbecco's modified Eagle's medium containing 20% fetal bovine serum. During the culture, cells adhering to the upper surface of the flasks changed to fibroblast-like cells. After a certain amount of culture fluid had been removed, the flasks were inverted and cultured in the usual way. (B) Morphology of adipocytes on day 7 of culture. Cells indicated by arrowheads proliferated, formed colonies, and were found to be dedifferentiated. Scale bar: 100 μm. (C) Flow cytometry results of human dedifferentiated adipocytes, expressing cluster of differentiation CD11b (−), CD34 (−), CD45 (−), CD73 (+), CD105 (+). DFAT, dedifferentiated fat cells.
Isolation and purification of dedifferentiated fat cell-derived extracellular vesicles
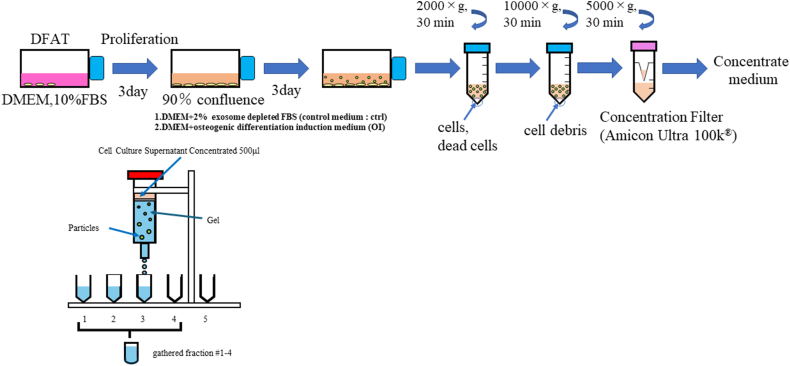
In this study, five passages of DFATs were used for EV production. Upon reaching 90% confluency, the culturemedium was changed to fresh Dulbecco's Modified Eagle Medium (Fujifilm Wako, Osaka, Japan) containing 2% fetal bovine serum (FBS) with exosomes depletion (Biosera, Cholet, France) or bone induction medium (PT-3002, Human Mesenchymal Stem Cell (hMSC) Osteogenic Differentiation Medium BulletKit™, without MCGS) plus 10% exosome-depleted FBS. After cultivation for 72 h, the supernatant was collected. The collected supernatant was then centrifuged at 2000 × g for 30 min at 4 °C to remove detached cells, followed by centrifugation at 10,000 × g for 30 min at 4 °C to remove cell debris. Size-exclusion chromatography (SEC) was used to isolate EVs.11 Before SEC, the volume of the centrifuged supernatant was concentrated up to 500 μL with an ultra-filtration filter with a cutoff molecular weight of 100 kD (Amicon® Ultra-15 Centrifugal Filter Unit; Merck Millipore, Burlington, MA, USA) at 5000 × g and 4 °C. A SEC column (qEV original/70 nm; Izon, Christchurch, New Zealand) was flushed with phosphate-buffered saline (PBS) before sample loading. The concentrate (500 μL) was loaded into a column, followed by flushing with PBS. The automated fraction collector (Izon) was set to collect 1 mL of elute fraction, and elutions from fraction numbers 2−5 were used as EVs (Fig. 2). Two types of EVs were obtained from DFATs before and after osteogenic induction (OI): DFAT-ctrl-EV and DFAT-OI-EV, respectively.
Figure 2.
Isolation of DFAT-EVs using size-exclusion chromatography. DFAT-EVs, dedifferentiated fat cells-derived extracellular vesicles.
Characterization of extracellular vesicles
The morphology of EVs was observed using a 30 kV Scanning Transmission Electron Microscope (STEM, HITACHI SU 9000, Japan) after negative staining with 2% uranyl acetate. Size distribution and concentration of EVs were analyzed using NanoSight LM 10 (MalvernPanalytical, Worcestershire, UK) equipped with nanoparticle tracking analysis (NTA) software (version 2.2, NanoSight). PS Capture™ Exosome Flow Cytometry Kit (297–79701; Fujifilm Wako) and PE-conjugated primary anti-human antibodies for cluster of differentiation CD9 (Cat#312103, Bio Legend) and CD63 (Cat#353003, Bio Legend) were used to evaluate the surface of EVs. Flow cytometry was used to analyze the surface of EVs.
Internalization of extracellular vesicles
EVs were fluorescently labeled using the ExoSparkler Exosome Membrane Labeling Kit-Red (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer's manual. DFATs were cultured with the labeled EVs for 4 h, followed by fixation with paraformaldehyde and nucleus staining with Hoechst33342 (Nacalai Tesque Inc., Kyoto, Japan). The fluorescent image was acquired using a fluorescence microscope (BZ-X800, KEYENCE, Osaka, Japan).
Osteogenic induction
After 48 h post-seeding confluences, the DFATs (3 × 104 cells/well) were seeded in 24-well plates and cultured in an osteogenic induction medium (PT-3002, hMSC-Osteogenic Differentiation Medium BulletKit™). The culture media of the experimental groups were:1 Osteogenic induction (OI),2 OI + DFAT-ctrl-EVs, and3 OI + DFAT-OI-EVs. The medium was replaced every 2 days. The assay was conducted after 7 and 14 days for alkaline phosphatase (ALP) activity and after 3 and 7 days for RNA analysis.
Alkaline phosphatase activity
The cells were stained using an ALP staining kit (AK20; Cosmo Bio Co., Ltd., Tokyo, Japan) according to the manufacturer's protocol. The absorbance at 405 nm measured by a SpectraMax iD 3 spectrophotometer (Molecular Devices, San Jose, CA) was divided by the DNA content obtained from a Pico Green dsDNA Assay Kit (Thermo Fisher Scientific Inc.).
Ribonucleic acid analysis
Extraction of total RNA from cells after 3 and 7 days of culturing and subsequent cDNA synthesis were performed using QIAcube PREMIUM (Qiagen, Hilden, Germany) and SuperScript™ IV VILO™ Master Mix (Thermo Fisher Scientific Inc.), respectively. Real-time quantitative polymerase chain reaction (RT-qPCR) was conducted to measure the expression of Runt-related transcription factor (Runx2) (Hs1047973_m1 PN4453320) and Collagen Type I (Hs00164004_m1 PN 4453320) on a QuantStudio3 (Thermo Fisher Scientific Inc.) using TaqMan™ Fast Advanced Master Mix (Thermo Fisher Scientific Inc.). PCR was performed at 50 °C for 2 min and 95 °C for 20 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 20 s. Glyceraldehyde-3-phosphate dehydrogenase was selected as an internal reference (4310884E-1709071).
Detection of small ribonucleic acids
Total RNAs in EVs were obtained using a microRNA Extractor kit for purified EVs (Fujifilm Wako). Libraries were prepared through ligation of 3′ and 5′ adaptors, reverse transcription, and PCR amplification using a NEB Next Multiplex Small RNA Library Prep Set for Illumina (Illumina Inc., San Diego, CA) according to the manufacturer's protocols. NGS was carried out under a 100-bp single read condition (Illumina Inc.). All procedures for EV-derived small RNA preparation and NGS analysis were performed by Rhelixa Company (Tokyo, Japan). Normalization of raw read counts and differential expression analysis were performed using the trimmed mean of M values and edgeR (version 3.26.8), respectively.
Gene ontology enrichment and pathway analysis
GOATOOLS (version 1.1.6) was used to perform gene ontology (GO) enrichment analysis of differentially expressed miRNAs target genes.
Statistical analysis
ALP activity and RNA analysis data are presented as mean ± standard deviation. Parametric data from the three groups were analyzed using a one-way analysis of variance with Tukey's Honest Significant Difference post-hoc test. Statistical significance was set at P < 0.05. Graphs were created using GraphPad 10 (Dotmatics, Boston, MA, USA).
The P-value of the GO enrichment analysis was corrected using the Benjamini−Hochberg method for multiple testing calibrations. Statistical significance was set at P < 0.05.
Results
Expression of surface antibodies of dedifferentiated fat cells
The flow cytometry results revealed that DFATs positively expressed markers CD73 and CD105 while they negatively expressed markers CD11b, CD34 and CD45 (Fig. 1C). These results are consistent with previous reports on DFATs.2
Extraction and characterization of extracellular vesicles from dedifferentiated fat cells
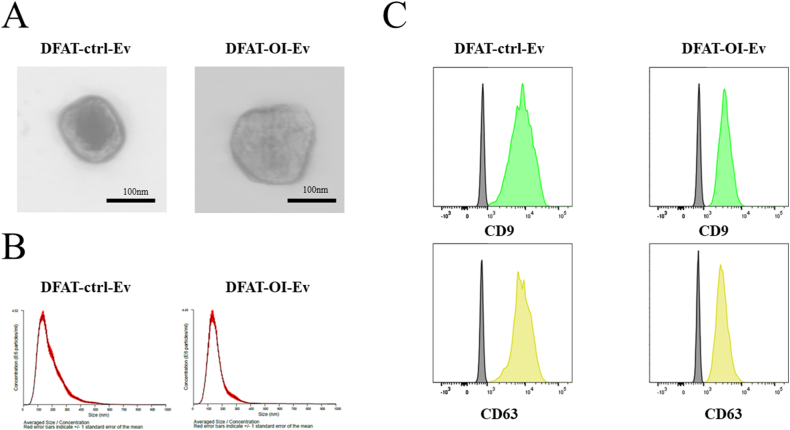
This finding aligned with the TEM observations, which indicated that the EV sizes varied from 50–200 nm and were round-shaped (Fig. 3A). The NTA results showed that the size and concentration of extracted EVs were approximately 140 nm and 7 × 109 particles/mL, respectively, irrespective of the extent of osteogenic differentiation of DFAT and extraction method (Table 1, Fig. 3B).
Figure 3.
Characterization of DFAT-EVs (A) Morphology observed by TEM. Scale bar: 200 μm. (B) The size distribution and particle concentration of DFAT-EVs were measured using nanoparticle tracking analysis. (C) Flow cytometry results for DFAT-EVs, expressing cluster of differentiation CD9 (+), CD63 (+). DFAT-EVs, dedifferentiated fat cells-derived extracellular vesicles.
Table 1.
Particle sizes and concentrations of DFAT-ctrl-EV and DFAT-OI-EV measured by nanoparticle tracking analysis (mean ± SD).
| Average particle size (nm) | Concentration ( × 109 particle/mL) | |
|---|---|---|
| DFAT-ctrl-EVs | 132−/+3.9 | 6.18−/+0.20 |
| DFAT-OI-EVs | 136−/+4.8 | 8.78−/+0.50 |
DFAT-ctrl-EVs, dedifferentiated fat cells-derived control extracellular vesicles; DFAT-OI-EVs, dedifferentiated fat cells-derived osteoinductive extracellular vesicles.
Flow cytometry positively detected EV surface protein markers, CD9 and CD63, in the prepared DFAT-ctrl-EVs and DFAT-OI-EVs samples (Fig. 3C).
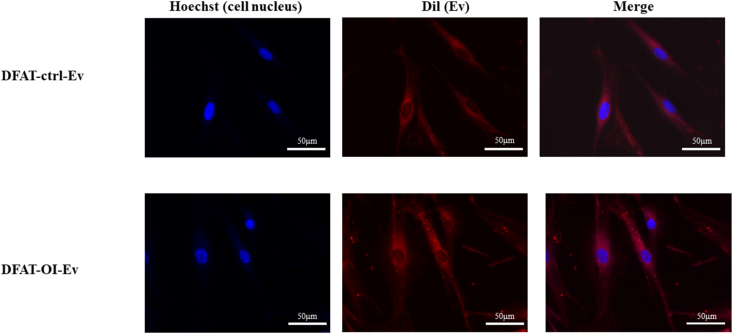
On fluorescent microscopy, EVs with red fluorescence were observed within the cytoplasm (Fig. 4), suggesting successful EV internalization into the target stem cells. This successful delivery was essential for their subsequent application.
Figure 4.
Fluorescent images of DFATs 4 h after culturing with DFAT-EVs. DFATs, dedifferentiated fat cells; DFAT-EVs, dedifferentiated fat cells; -derived extracellular vesicles.
Effect of extracellular vesicles on the osteoblast differentiation of dedifferentiated fat cells
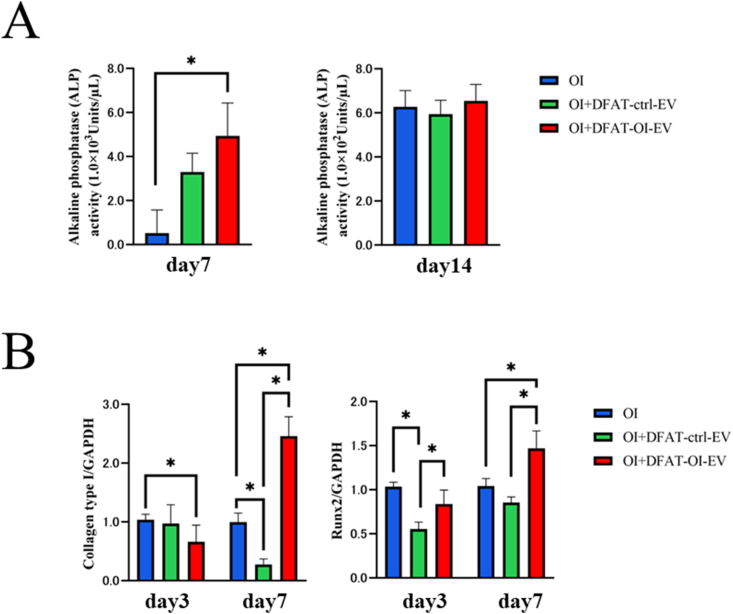
The ALP activity of cells cultured for 7 days in the OI + DFAT-OI-EVs group was significantly higher than that in the OI group. However, after 14 days, no significant difference was observed in the three groups tested (Fig. 5A).
Figure 5.
Evaluation of the osteoinductivity of DFAT-EVs (A) ALP activity of DFAT-induced EVs at different concentrations on days 7 and 14. (B) Expression levels of Runx2 and collagen type I 7 and 14 days after osteoblast induction (Statistical comparisons with blank ctrl: P < 0.05). DFAT-EVs, dedifferentiated fat cells-derived extracellular vesicles; ALP, alkaline phosphatase.
The RT-PCR analysis showed elevated expression levels of Runx2, an early marker of osteoblast differentiation in the DFAT-OI-EVs group on day 7. Collagen type I had an overall low expression level on day 3 but significantly increased by day 7, reaching levels 2.5 times higher than the control group (Fig. 5B).
Profiling of miRNA expression in dedifferentiated fat cells-derived osteoinductive extracellular vesicles vs. dedifferentiated fat cells-derived control extracellular vesicles
The difference between samples [log2 fold change > |2|] was calculated. Fold-up or downregulated miRNA were considered differentially expressed. As a result of miRNA profiling, 162 differential miRNAs were detected in DFAT-EVs, with 92 downregulated and 70 upregulated in DFAT-OI-EVs compared to DFAT-ctrl-EVs. As shown in Table 2, DFAT-OI-EVs had nine downregulated miRNAs (miR-20a-5p, miR-98-5p, miR-543, miR-30c-2-3p, miR-133a-3p, miR-485-5p, miR-30c-2-3p, miR-877-5p, and miR-221-3p) and five upregulated miRNAs (miR-130b, miR-16-5p, miR-143-5p, miR-142-5p, and miR-204-5p), and many of these miRNAs have been documented as participating in osteogenic differentiation and associated mediating mechanisms.12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23 However, some miRNAs showed results differing from those reported in previous literature.
Table 2.
Differentially expressed miRNAs in DFAT cultured in OI medium, including DFAT-ctrl-EV or DFAT-OI-EV and their predicted target genes.
| miRNA | Up/Down | Fold change | Previously reported mechanisms |
|---|---|---|---|
| miR-485-5p | Down | 8.69 | Upregulation inhibits osteogenic differentiation by BMP/SMAD pathway |
| miR-30c-2-3p | Down | 7.41 | Upregulation inhibits osteogenic differentiation |
| miR-133a-3p | Down | 6.06 | Runx2 decreases the differentiation of osteoblasts |
| miR-20a-5p | Down | 5.84 | Regulates BAMBI to activate the phosphorylation of SMAD5 and P38 |
| miR-877-5p | Down | 4.73 | Promote osteoblast differentiation by targeting EIF4G2 expression |
| miR-98-5p | Down | 4.3 | Inhibits preosteoblast proliferation and differentiation via BMP2 targeting in the PI3K/AKT/GSK3β pathway. |
| miR-221-3p | Down | 4 | Downregulation promotes osteogenic differentiation by the IGF-1/ERK pathway |
| miR-130b | Up | 8.07 | Upregulation promotes osteogenic differentiation |
| miR-143-5p | Up | 7.56 | Odontoblasts target Runx1 via the OPG/RANKL signaling pathway |
| miR-142-5p | Up | 6.91 | Promotes osteoblast differentiation by targeting nuclear factor IA |
| miR-16-5p | Up | 6.68 | Inhibits osteogenesis by targeting SMAD5 and ACVR2A |
| miR-204-5p | Up | 2.01 | Regulates Runx2 and mesenchymal progenitor cell differentiation |
DFAT-ctrl-EVs, dedifferentiated fat cells-derived control extracellular vesicles; DFAT-OI-EVs, dedifferentiated fat cells-derived osteoinductive extracellular vesicles.
Gene ontology enrichment analysis
Several signaling pathways are related to osteoblast differentiation, including β-catenin, bone morphogenetic protein (BMP), transforming growth factor β (TGF-β), Hedgehog, and mitogen-activated protein kinase (MAPK). We identified multiple pathways associated with osteoblast differentiation, highlighting the number of genes (gene count) involved in each pathway and their statistical significance (P-value), providing valuable insights into the mechanisms underlying osteoblast differentiation. These pathways play crucial roles in osteoblast differentiation, as widely reported. The results of the GO analysis related to these signaling pathways are shown in Table 3.
Table 3.
Gene ontology terms for the predicted target genes.
| GO term | Description | Gene count | P-Value |
|---|---|---|---|
| GO:0016055 | Wnt signaling pathway | 162 | 0.000748∗ |
| GO:0030509 | BMP signaling pathway | 47 | 0.033484∗ |
| GO:0007179 | TGF-β signaling pathway | 61 | 0.00838 |
| GO:0007224 | Hedgehog signaling pathway | 60 | 0.175039 |
| GO:0000165 | MAPK signaling pathway | 40 | 0.273356 |
∗P < 0.05.
GO, gene ontology; BMP, bone morphogenetic protein; TGF-β, transforming growth factor β; MAPK, mitogen-activated protein kinase.
Discussion
Several methods exist for EV isolation. SEC use has increased rapidly, indicating a shift towards this method.24 Ultracentrifugation (UC) exposes EVs to substantial gravitational force, leading to degradation, aggregation, and fusion of EVs, phenomena not observed with SEC. The absence of applied force in SEC makes it the most gentle method, enabling the isolation of EVs in a state that closely resembles their original condition in the starting material.25 However, SEC typically yields fewer isolated EVs compared to UC.26
In this study, we isolated EVs from the DFATs culture medium using the SEC and concentration filter method. Characterization using TEM combined with NTA revealed EVs with disc-like morphology and diameters of 100–150 nm. Flow cytometry data indicated that DFATs positively expressed EV surface markers, including CD9 and CD63. An in vitro osteogenic differentiation assay showed that DFAT-OI-EVs significantly upregulated ALP activities and significantly upregulated Runx2 and collagen type I for 7 days. Liu et al. reported that EVs purified from the culture supernatant of MSCs and ADSCs cultured in osteogenic differentiation media affected osteogenesis more significantly than EVs from standard culture,10 which aligns with our findings. Additionally, DFAT-ctrl-EVs notably reduced the expression of Runx2 and collagen type I, indicating potential suppression of osteoblastic differentiation. Liu et al. found that exosomes from control ADSCs (ADSCs-ctrl-exosomes) did not positively affect osteogenesis-related genes and even significantly inhibited BMP-2 and BMPR1A expressions. This inhibition may stem from the intrinsic adipogenic properties of ADSCs, which adversely affect osteogenic differentiation.10 This finding corroborates our results. As suggested by Liu et al., comparing DFAT-EVs with EVs from other stem cell sources could further underscore the distinct advantages of DFAT-EVs in specific applications.
Runx2 promotes the differentiation of multipotent mesenchymal cells into immature osteoblasts, guiding the development of nascent bone structures. This effect is facilitated by its DNA-binding domain (Runt domain), which targets specific gene promoter regions to initiate transcription. Additionally, Runx2 regulates its activity by interacting with other transcription factors and co-activators.27 In the present study, we report on miRNAs that directly act on Runx2. These include miR-30c-2-3p, miR-133a-3p, and miR-204-5p, which directly bind to the 3’ UTR of Runx2 mRNA, suppressing its translation, reducing the expression of Runx2 and decreasing the differentiation of osteoblasts.13,14 Furthermore, these miRNAs may decrease the interaction between Runx2 and β-catenin, inhibiting the Wnt cascade and the activation of genes that require the β-catenin-Runx2 complex, resulting in reduced bone formation.27 The Wnt signaling pathway is primarily divided into the classical Wnt/β-catenin signaling pathway and the non-classical Wnt signaling pathway. Osteogenic differentiation is mainly due to the activation of the classical Wnt/β-catenin signaling pathway.28 In this study, the expression levels of miR-30c-2-3p, miR-133a-3p, and miR-485-5p in DFAT-ctrl-EVs were higher than those in DFAT-OI-EVs. miR-485-5p is associated with the Wnt/β-catenin signaling pathway, although this has not been in the context of bone differentiation.29 Our future research will focus on investigating whether miR-485-5p influences osteogenic differentiation in DFATs. Additionally, GO analysis revealed that the gene count for the Wnt signaling pathway was 162, with a P -value of 0.000748. This suggests that the Wnt signaling pathway may be related to the results of this study.
In conclusion, we successfully isolated EVs from BFP-derived DFATs for the first time. DFAT-derived osteoinductive EVs promoted the induction of osteoblast differentiation in DFATs. Furthermore, NGS analysis of miRNAs suggested that DFAT-derived osteoinductive EVs might enhance osteogenic differentiation by activating the Wnt signaling pathway. For bone regeneration, an in vivo study will require DFATs, DFAT-derived osteoinductive EVs, and the appropriate scaffold for bone tissue regeneration.
Declaration of competing interest
The authors have no conflicts of interest relevant to this article.
Acknowledgments
The authors thank Yurie Taniguchi (Department of Anesthesiology, Osaka Dental University) for her assistance during the study. This work was supported by JSPS KAKENHI Grant Number JP23K09366.
References
- 1.Matsumoto T., Kano K., Kondo D., et al. Mature adipocyte-derived dedifferentiated fat cells exhibit multilineage potential. J Cell Physiol. 2008;215:210–222. doi: 10.1002/jcp.21304. [DOI] [PubMed] [Google Scholar]
- 2.Kishimoto N., Momota Y., Hashimoto Y., et al. The osteoblastic differentiation ability of human dedifferentiated fat cells is higher than that of adipose stem cells from the buccal fat pad. Clin Oral Invest. 2014;18:1893–1901. doi: 10.1007/s00784-013-1166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kishimoto N., Honda Y., Momota Y., Tran S.D. Dedifferentiated Fat (DFAT) cells: a cell source for oral and maxillofacial tissue engineering. Oral Dis. 2018;24:1161–1167. doi: 10.1111/odi.12832. [DOI] [PubMed] [Google Scholar]
- 4.Kou L., Lu X.W., Wu M.K., et al. The phenotype and tissue-specific nature of multipotent cells derived from human mature adipocytes. Biochem Biophys Res Commun. 2014;444:543–548. doi: 10.1016/j.bbrc.2014.01.077. [DOI] [PubMed] [Google Scholar]
- 5.Lu Z., Chen Y., Dunstan C., Roohani-Esfahani S., Zreiqat H. Priming adipose stem cells with tumor necrosis factor-alpha preconditioning potentiates their exosome efficacy for bone regeneration. Tissue Eng. 2017;23:1212–1220. doi: 10.1089/ten.tea.2016.0548. [DOI] [PubMed] [Google Scholar]
- 6.Stoorvogel W., Kleijmeer M.J., Geuze H.J., Raposo G. The biogenesis and functions of exosomes. Traffic. 2002;3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 7.Lopatina T., Bruno S., Tetta C., Kalinina N., Porta M., Camussi G. Platelet-derived growth factor regulates the secretion of extracellular vesicles by adipose mesenchymal stem cells and enhances their angiogenic potential. Cell Commun Signal. 2014;12:26. doi: 10.1186/1478-811X-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin Y., Wang L., Gao Z., Chen G., Zhang C. Bone marrow stromal/stem cell-derived extracellular vesicles regulate osteoblast activity and differentiation in vitro and promote bone regeneration in vivo. Sci Rep. 2016;6 doi: 10.1038/srep21961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martins M., Ribeiro D., Martins A., Reis R.L., Neves N.M. Extracellular vesicles derived from osteogenically induced human bone marrow mesenchymal stem cells can modulate lineage commitment. Stem Cell Rep. 2016;6:284–291. doi: 10.1016/j.stemcr.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu A., Lin D., Zhao H., et al. Optimized BMSC-derived osteoinductive exosomes immobilized in hierarchical scaffold via lyophilization for bone repair through Bmpr2/Acvr2b competitive receptor-activated Smad pathway. Biomaterials. 2021;272 doi: 10.1016/j.biomaterials.2021.120718. [DOI] [PubMed] [Google Scholar]
- 11.Lu Y., Eguchi T., Sogawa C., et al. Exosome-based molecular transfer activity of macrophage-like cells involves viability of oral carcinoma cells: size exclusion chromatography and concentration filter method. Cells. 2021;10:1328. doi: 10.3390/cells10061328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H.O., Zhang L., Tang Z.Y., Gong Z.M. MiR-485-5p promotes the development of osteoarthritis by inhibiting cartilage differentiation in BMSCs. Eur Rev Med Pharmacol Sci. 2018;22:3294–3302. doi: 10.26355/eurrev_201806_15148. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y., Xie R.L., Croce C.M., et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011;108:9863–9868. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu T., Zhou H., Hong Y., Li J., Jiang X., Huang H. miR-30 family members negatively regulate osteoblast differentiation. J Biol Chem. 2012;287:7503–7511. doi: 10.1074/jbc.M111.292722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asila A., Yang X., Kaisaer Y., Ma L. SNHG16/miR-485-5p/BMP7 axis modulates osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. J Gene Med. 2021;23 doi: 10.1002/jgm.3296. [DOI] [PubMed] [Google Scholar]
- 16.Laxman N., Rubin C.J., Mallmin H., et al. Global miRNA expression and correlation with mRNA levels in primary human bone cells. RNA. 2015;21:1433–1443. doi: 10.1261/rna.049148.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cen X., Pan X., Zhang B., et al. miR-20a-5p contributes to osteogenic differentiation of human dental pulp stem cells by regulating BAMBI and activating the phosphorylation of Smad5 and p38. Stem Cell Res Ther. 2021;12:421. doi: 10.1186/s13287-021-02501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.You Y., Ma W., Wang F., et al. Ortho-silicic acid enhances osteogenesis of osteoblasts through the upregulation of miR-130b which directly targets PTEN. Life Sci. 2021;264 doi: 10.1016/j.lfs.2020.118680. [DOI] [PubMed] [Google Scholar]
- 19.Gan K., Dong G.H., Wang N., Zhu J.F. miR-221-3p and miR-222-3p downregulation promoted osteogenic differentiation of bone marrow mesenchyme stem cells through IGF-1/ERK pathway under high glucose condition. Diabetes Res Clin Pract. 2020;167 doi: 10.1016/j.diabres.2020.108121. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y., Zhang Y., Wang Q., Jiang B., Jiang X., Luo B. MicroRNA-877-5p promotes osteoblast differentiation by targeting EIF4G2 expression. J Orthop Surg Res. 2024;19:134. doi: 10.1186/s13018-023-04396-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhan F.L., Liu X.Y., Wang X.B. The role of microRNA-143-5p in the differentiation of dental pulp stem cells into odontoblasts by targeting Runx2 via the OPG/RANKL signaling pathway. J Cell Biochem. 2018;119:536–546. doi: 10.1002/jcb.26212. [DOI] [PubMed] [Google Scholar]
- 22.Garcia J., Delany A.M. MicroRNAs regulating TGFβ and BMP signaling in the osteoblast lineage. Bone. 2021;143 doi: 10.1016/j.bone.2020.115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan H., Li M., Feng X., Zhu E., Wang B. miR-142a-5p promoted osteoblast differentiation via targeting nuclear factor IA. J Cell Physiol. 2021;236:1810–1821. doi: 10.1002/jcp.29963. [DOI] [PubMed] [Google Scholar]
- 24.Liangsupree T., Multia E., Riekkola M.L. Modern isolation and separation techniques for extracellular vesicles. J Chromatogr A. 2021;1636 doi: 10.1016/j.chroma.2020.461773. [DOI] [PubMed] [Google Scholar]
- 25.Sidhom K., Obi P.O., Saleem A. A review of exosomal isolation methods: is size exclusion chromatography the best option. Int J Mol Sci. 2020;21:6466. doi: 10.3390/ijms21186466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardiner C., Di Vizio D., Sahoo S., et al. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles. 2016;5 doi: 10.3402/jev.v5.32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaur T., Lengner C.J., Hovhannisyan H., et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- 28.Han X., Fan Z. MicroRNAs regulation in osteogenic differentiation of mesenchymal stem cells. Front Dent Med. 2021;2 [Google Scholar]
- 29.Wang Y., Sun L., Wang L., et al. Correction: Long non-coding RNA DSCR8 acts as a molecular sponge for miR-485-5p to activate Wnt/β-catenin signal pathway in hepatocellular carcinoma. Cell Death Dis. 2022;13:679. doi: 10.1038/s41419-022-05141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]