Introduction
Periodontitis is caused by irreversible breakdown of periodontal epithelial and connective tissue attachment followed by alveolar bone loss, which may be due to local pathogenic bacterial infection and the host's protective inflammatory reactions.1 Epidemiological and animal model studies have disclosed that various diseases and pathological conditions, such as vascular disorders, diabetes, premature birth and low birth weight in children, aspiration pneumonia, rheumatoid arthritis, chronic kidney diseases, Alzheimer's disease, and non-alcoholic fatty liver disease (NAFLD-including non-alcoholic fatty liver [NAFL] and non-alcoholic steatohepatitis [NASH]) were exacerbated by periodontitis.2, 3, 4, 5
The liver has various roles for maintaining whole body homeostasis, including lipid, carbohydrate, iron, vitamin, copper, and drug metabolisms.6,7 Gut barrier destruction increases gut permeability, thereby inducing the translocation of microbiota from the gut to the liver via the portal vein; this process activates natural immunity and thus contributes to the progression of NASH.8 NASH poses high risks for cirrhosis and hepatocellular carcinoma; therefore, its onset and progression is considered a life-threatening event.9 Similarly, oral pathogenic bacteria, such as Porphyromonas gingivalis (P.g.), can enter the systemic circulation and reach the liver, thereby facilitating NASH progression.10
The pre-symptomatic state is defined as the state of health prior to the clinical appearance of the signs and symptoms of a disease; it is biologically regarded as “the breakdown of homeostasis”.11,12 Therefore, appropriate interventions during the pre-symptomatic disease state may provide greater advantages in increasing healthy life expectancy and reducing medical costs. From this point of view, periodontal disease is one of the most common adult inflammatory diseases.13 Periodontal disease establishes a pre-symptomatic disease state in various tissues and organs by circulating periodontal bacteria and toxins, periodontal tissue-originated inflammatory cytokines, and immunological cells, thereby causing dysbiosis.5,14 However, previous studies mainly focused on the mechanistic link between liver and periodontal diseases and were based on systemic disease/condition models such as high-fat feeding and diabetes mellitus.
In this study, the transcriptome profiles of the periodontal and liver tissues of ligature-induced periodontitis and healthy mice were generated. The key molecules and pathways of the liver and their roles in the onset of periodontitis-induced, pre-symptomatic liver disease were clarified.
Materials and methods
Reagents
Eicosapentaenoic (EPA) (HY–B0660) and docosahexaenoic acids (DHA) (HY–B2167) were purchased from MedChem Express (Monmouth Junction, NJ, USA). The 17,18-epoxy eicosatetraenoic (EpETE) and 17,18-dihydroxyeicosatetraenoic acids (diHETE) were purchased from Cayman Chemical Co. (Ann Arbor, MI, USA). Lipopolysaccharides (LPS) (127–05141) was purchased from FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan). Recombinant tumor necrosis factor (TNF)-α (210-TA-005) was obtained from R&D Systems (Minneapolis, MN, USA).
Experimental animals
All experimental procedures conformed to the Regulations for Animal Experiments and Related Activities at Tohoku University, were reviewed by the Institutional Laboratory Animal Care and Use Committee of Tohoku University, and were approved by the President of the University (Permit No. 2023DnA-008). Eleven-week-old male C57BL6/J mice (specific pathogen-free grade) were purchased from CLEA Japan, Inc. (Tokyo, Japan). The mice were randomly allocated; five mice were housed per cage. The mice were fed with standard rodent chow and water ad libitum under a 12:12-h, controlled light/dark photoperiod cycle (lights on at 8:00 A.M.); mice were given 1 week to adapt to the new environment before the experiments.
The mice were randomly allocated to experimental and control groups. Each group comprised two mice for RNA-seq and four mice for lipidomic analysis. The mice were anesthetized; silk ligatures (Elp Sterile Blade Silk, Black, 5–0, Akiyama Medical MFG, Tokyo, Japan) were then tied around their second maxillary molars and maintained for 14 days to induce severe periodontal tissue breakdown, as described previously.15, 16, 17 After the 14-day periodontal inflammation period, the mice were anesthetized; the periodontal and liver tissues were subsequently dissected and immediately immersed in RNAiso Plus (Takara Bio Inc., Otsu, Japan) and sonicated using a homogenizer (Tomy, Tokyo, Japan) for RNA purification or immediately immersed in nitrogen and used for lipidomic analysis.
Poly-A selected and strand-specific RNA-seq and data analyses
Total RNA was purified from the dissected periodontal and liver tissues, DNase-treated, and used in RNA-seq analyses, as described previously.18,19 For the processing of the RNA-seq data, adapter trimming was conducted using Trim Galore version 0.6.5 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with default settings, and then aligned to a reference genome (mm10) using HISAT2 version 2.2.1.20 Transcript expression at the exons was quantified using the “analyzeRepeast.pl” command in HOMER21 with “-strand both” and “-count exons,” considering periodontal and liver tissues collected from periodontitis-affected mice as the target and those from healthy mice as the background, respectively. The original raw data of the RNA-seq analyses and the processed gene expression profiles were deposited in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) database22 under the accession number, GSE264546.
Quantitative PCR analysis
Total RNA purification, complimentary DNA (cDNA) preparation, and quantitative polymerase chain reaction (qPCR) were conducted, as described previously.23, 24, 25 Human hypoxanthine phosphoribosyltransferase 1 (HPRT) was used as an internal reference control. The PCR primer sequences for the target genes are listed in Table 1.
Table 1.
Primer pairs used in this study.
| Primer name | Species | Direction | Sequence |
|---|---|---|---|
| TNF-α | Human | forward | AGCCCATGTTGTAGCAAACC |
| reverse | ATGAGGTACAGGCCCTCTGA | ||
| HPRT | Human | forward | TGGCGTCGTGATTAGTGATG |
| reverse | CGAGCAAGACGTTCAGTCCT |
TNF-ɑ = tumor necrosis factor-alpha; HPRT = hypoxanthine phosphoribosyltransferase 1.
Histology
Liver samples were removed and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) at 4 °C for 24 h. The sections were then dehydrated using a graded ethanol series, placed in xylene, and embedded in paraffin. Hematoxylin and eosin staining was performed on 5-μm-thick paraffin sections, as described previously.26
Liquid chromatography coupled with tandem mass spectrometry analysis of fatty acid metabolites
Liver fatty acid metabolites were identified by Lipidome Lab Co. Ltd. (Akita, Japan). Fatty acid metabolites were purified from the lipid fractions by solid-phase extraction using Oasis HLB columns (Waters Corporation, Milford, MA, USA). The purification and liquid chromatography methods were the same as described previously.27 Briefly, fatty acid metabolites were extracted from 50-mg frozen liver samples using internal standards (2 ng each of 12,13-dihydroxy-octadecenoic acid-d4, 11,12-epoxy-eicosatrienoic acid-d11, and 20 ng of arachidonic acid-d8; Cayman Chemicals). They were separated using a high-performance liquid chromatography system (Nexera LC-30AD, Shimadzu Corporation, Kyoto, Japan) equipped with an XBridge C18 column (particle size 3.5 μm, length 150 mm; inner diameter 1.0 mm; Waters) and analyzed on a triple quadrupole mass spectrometer (LCMS-8040; Shimadzu Corporation). The metabolites were then identified and quantified by multiple reaction monitoring, as previously reported.27 For quantification, calibration curves were prepared for each compound; recoveries were monitored using deuterated internal standards. Data analysis was performed using LabSolutions software (Shimadzu Corporation).
Lipopolysaccharide or tumor necrosis factor-α (TNF-α) challenge in HepG2 cells
Human hepatic HepG2 cells were obtained from the RIKEN Cell Bank (Tsukuba, Japan). Cells were maintained in low-glucose Dulbecco's Modified Eagle Medium (DMEM; Thermo Fisher Scientific, Carlsbad, CA, USA) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and a heat-inactivated 10% fetal bovine serum. They were then cultivated at 37 °C under humidified atmospheric conditions (5% CO2 and 95% air). For analyzing the roles of omega-3 fatty acids and their metabolites in the inflammatory response of HepG2 cells, cells were cultured with the medium containing a heat-inactivated 2.5% fetal bovine serum and treated with or without LPS or TNF-α in the presence or absence of EPA, DHA, 17,18-EpETE, or 17,18-diHETE for 24 h. Total RNAs were collected for qPCR analysis.
Statistical analysis
Statistical analyses were performed using a two-tailed, unpaired Student's t-test (Fig. 3) and one-way analysis of variance, followed by Tukey's test (Fig. 4). Statistical significance was considered at P ≤ 0.05.
Figure 3.
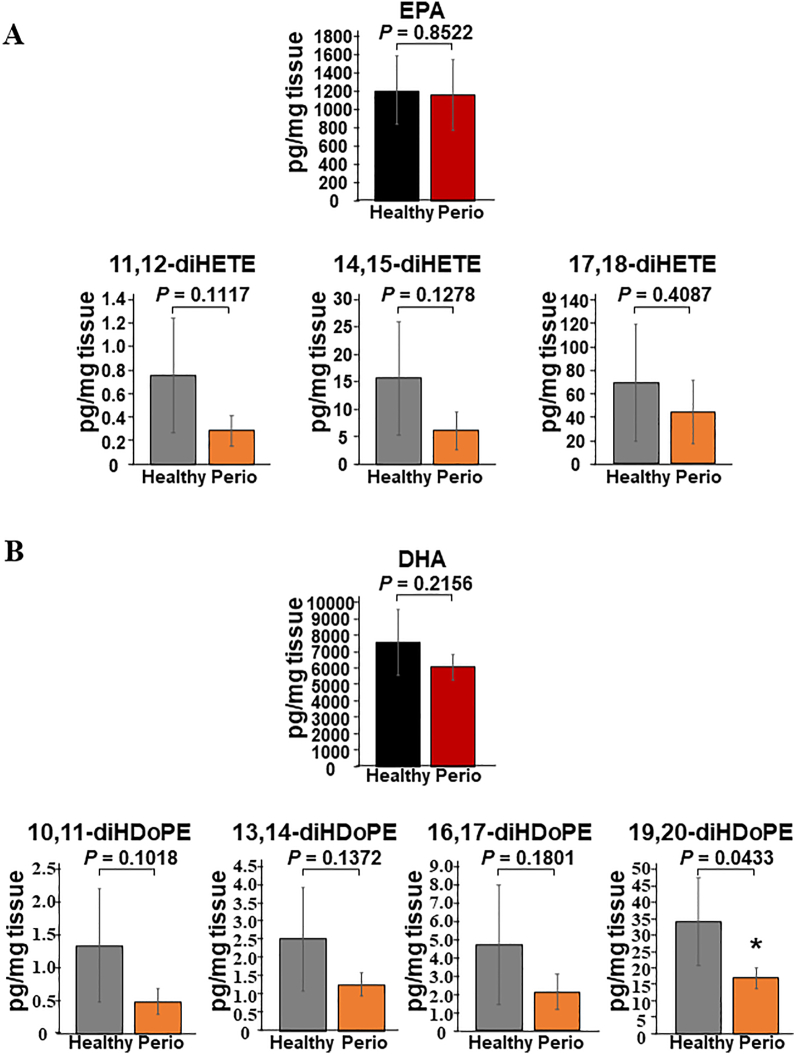
Comparative lipidomic analysis of the liver of periodontitis-affected and healthy control mice. (A) Hepatic EPA and EPA-metabolites, 11,12-diHETE, 14,15-diHETE, and 17,18-diHETE, of periodontitis-affected (periodontitis) and healthy control mice (healthy). (B) Hepatic DHA and DHA-metabolites, 10,11-diHDoPE, 13,14-diHDoPE, 16,17-diHDoPE, and 19,20-diHDoPE, of periodontitis-affected (periodontitis) and healthy control mice (healthy). EPA metabolites, 5,6-EpETE, 8,9-EpETE, 11,12-EpETE, 14,15-EpETE, 17,18-EpETE, 5,6-diHETE, and 8,9-diHETE, and DHA metabolites, 4,5-EpDPA, 7,8-EpDPA, 10,11-EpDPA, 13,14-EpDPA, 16,17-EpDPA, and 19,20-EpDPA, and 7,8-diHDoPE, were below the detection limit. P < 0.05 significantly lower in periodontitis-affected mice (n = 4) than in healthy control mice (n = 4). EPA = eicosapentaenoic acid, diHETE = dihydroxyeicosatetraenoic acid, DHA = docosahexaenoic acid, diHDoPE = dihydroxydocosapentaenoic acid.
Figure 4.
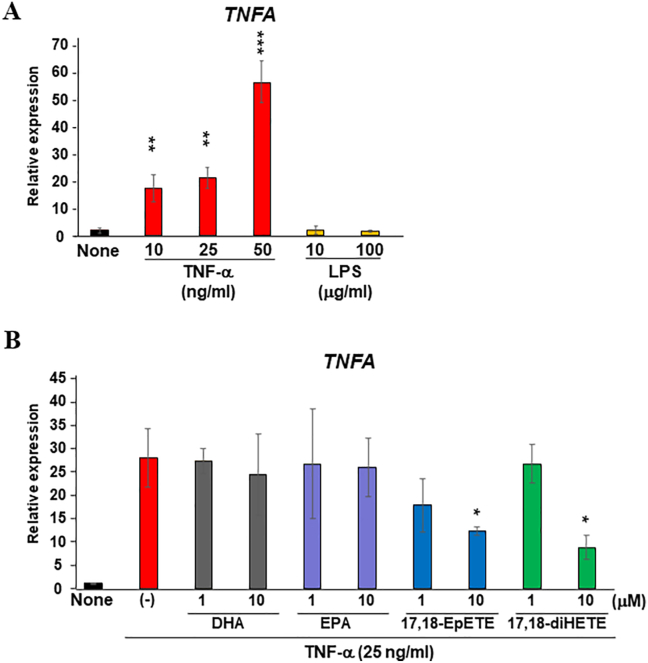
EPA metabolites inhibit inflammatory responses of HepG2 cells. (A) HepG2 cells were stimulated with LPS (10 and 100 μg/ml) or recombinant TNF-α (10, 25, 50 ng/ml) for 24 h; total RNA was collected to analyze the expression changes of TNF-α. (B) HepG2 cells were stimulated with recombinant TNF-α (25 ng/ml) for 24 h in the presence or absence of EPA (1 and 10 μM), DHA (1 and 10 μM), 17,18-EpETE (1 and 10 μM), and 17,18-diHETE (1 and 10 μM); total RNA was collected to analyze the expression changes of TNF-α. Each column represents the mean ± SD; n = 3 for each group. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 significantly higher than non-stimulated cells (A) and lower than the cells stimulated with recombinant TNF-α alone (B). LPS = lipopolysaccharide, TNF-α = tumor necrosis factor-α, EPA = eicosapentaenoic acid, diHETE = dihydroxyeicosatetraenoic acid, DHA = docosahexaenoic acid, diHDoPE = dihydroxydocosapentaenoic acid, SD = standard deviation.
Results
Genes associated with extracellular matrix proteins and chemokines are upregulated in the ligature-induced, inflamed periodontal tissue
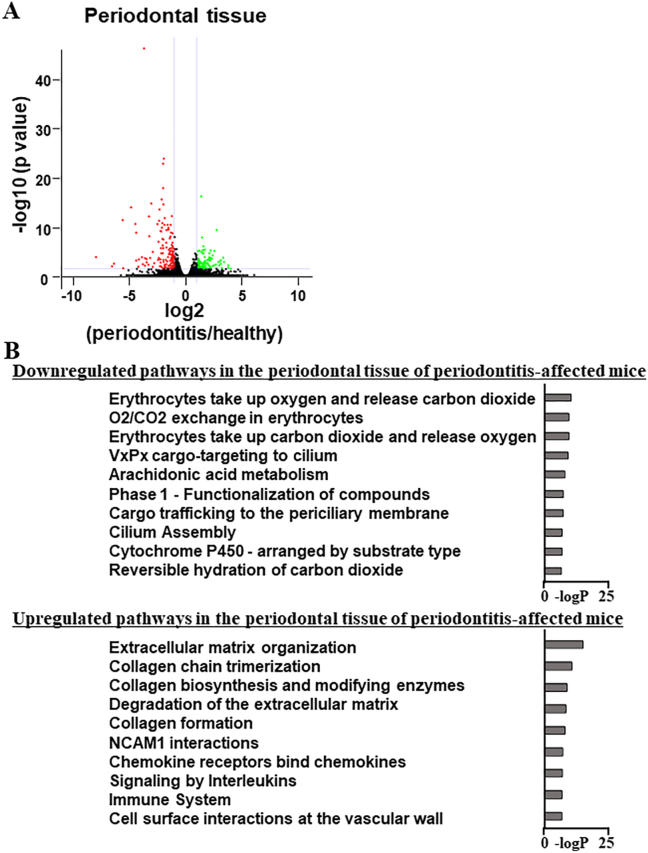
Total RNA was collected from the periodontal tissues of periodontitis-affected and healthy mice; whole-transcriptome analyses were then conducted using RNA-seq. In the inflamed periodontal tissues, 107 and 133 genes were significantly downregulated and upregulated, respectively, compared with healthy periodontal tissue (Fig. 1A). The processed whole-transcriptome profiles obtained from RNA-seq analyses were deposited in the NCBI GEO database. Gene ontology analysis of the downregulated genes in the inflamed periodontal tissues identified terms associated with the erythrocytes; however, their -logP scores were low. Conversely, gene ontology analysis of upregulated genes in the inflamed periodontal tissues identified enriched terms associated with extracellular matrix (ECM) formation and degradation, such as “extracellular matrix organization” and “collagen chain trimerization,” and with inflammatory responses, such as “chemokine receptors bind chemokines” and “immune system“ (Fig. 1B).
Figure 1.
Enhanced expression of the genes coding extracellular matrix proteins and chemokines in inflamed periodontal tissue. (A, B) Total RNAs were collected from ligature-induced inflamed and healthy periodontal tissues; whole-genomic transcriptional changes were assessed using RNA-seq. The volcano plot shows a comparison of inflamed periodontal (periodontitis) and healthy periodontal tissues (healthy). The genes whose expression level is significantly higher in healthy and periodontitis tissues are shown in green and red, respectively. The genes whose expression level was equivalent between healthy and periodontitis tissues are shown in black (A). The top ten pathways of downregulated (107 genes) and upregulated genes (133 genes) in the periodontal tissue of periodontitis-affected mice (periodontitis) compared with those of healthy mice (healthy) (B). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Terms associated with cytochrome P450 are enriched in the downregulated genes of the liver of periodontitis-affected mice
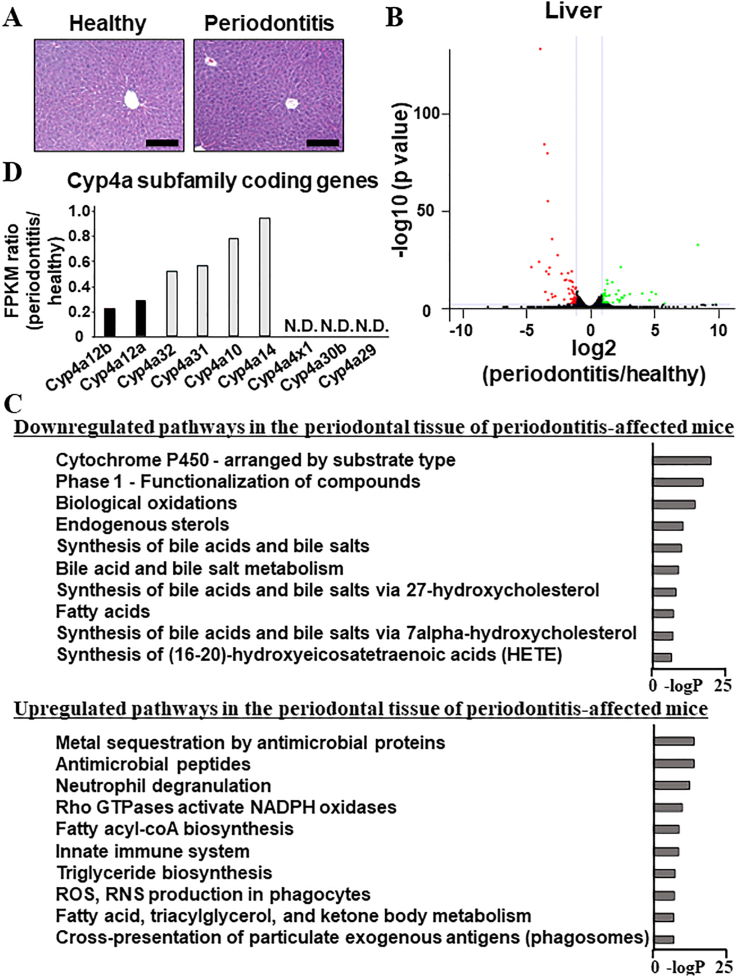
Induced periodontal breakdown did not result in obvious inflammatory responses, i.e., immunological cell infiltration in the liver (Fig. 2A). However, 61 and 71 genes were identified in the livers of the periodontitis-affected mice as significantly downregulated and upregulated, respectively, compared with those of healthy mice (Fig. 2B). Gene ontology analysis of downregulated genes indicated that all ten top-ranked terms, such as “cytochrome P450-arranged by substrate type” and “phase 1-functionalization of compounds,” were associated with cytochrome P450 (CYP) enzymes (Fig. 2C). The processed whole-transcriptome profiles obtained from RNA-seq analyses were deposited in the NCBI GEO database. The ratio of fragments per kilobase of exon per million mapped reads (FPKM) obtained using RNA-seq analyses revealed that among the CYP-coding genes, which were consisted of approximately 110 kinds in mice, Cyp4a12a and Cyp4a12b were the most enriched; among the Cyp4a subfamily, only Cyp4a12a (adjusted P value = 1.21E-14) and Cyp4a12b (adjusted P value = 9.06E-18) were significantly downregulated (threshold = 0.5) in the livers of periodontitis-affected mice (Fig. 2D).
Figure 2.
Suppressed expression of the genes coding cytochrome P450 enzymes in the liver of periodontitis-affected mice. (A) Hematoxylin and eosin staining of the livers of healthy and periodontitis-affected mice. (B) Total RNAs were collected from the liver of ligature-induced periodontitis-affected (periodontitis) and healthy mice (healthy); whole-genomic transcriptional changes were assessed using RNA-seq. The volcano plot shows a comparison of the liver tissues of periodontitis-affected mice (periodontitis) and healthy mice (healthy). The genes whose expression level was significantly higher in healthy and periodontitis tissues are shown in green and red, respectively. The genes whose expression level was equivalent between healthy and periodontitis tissues are shown in black. (C) The top ten pathways of downregulated (61 genes) and upregulated (72 genes) genes in the liver tissues of periodontitis-affected mice (periodontitis) as compared with those of healthy mice (healthy). (D) FPKM values of the gene coding for the CYP enzymes belonging to the Cyp4a subfamily in the liver tissues of periodontitis-affected (periodontitis) and healthy mice (healthy) were extracted from the processed whole transcriptome profiles deposited in the NCBI GEO database (accession number, GSE264546). FPKM ratios are shown as the quotient of the FPKM values in the liver tissues of periodontitis-affected mice (periodontitis) by the FPKM values in the liver tissues of healthy mice (healthy). Scale bars correspond to 200 μm. NCBI GEO = National Center for Biotechnology Information Gene Expression Omnibus, FPKM = fragments per kilobase of exon per million mapped reads, N.D. = not detected. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Suppression of omega-3 fatty acids metabolism in the livers of periodontitis-affected mice
CYP enzymes are required for the epoxidation of EPA and DHA to generate epoxy eicosatetraenoic (EpETE) (5,6-EpETE, 8,9-EpETE, 11,12-EpETE, 14,15-EpETE, and 17,18-EpETE) and epoxy docosapentaenoic acids (EpDPA) (4,5-EpDPA, 7,8-EpDPA, 10,11-EpDPA, 13,14-EpDPA, 16,17-EpDPA, and 19,20-EpDPA). Epoxide hydrolases hydrolyze EpETE and EpDPA to generate dihydroxyeicosatetraenoic (diHETE) (5,6-diHETE, 8,9-diHETE, 11,12-diHETE, 14,15-diHETE, and 17,18-diHETE) and dihydroxydocosapentaenoic acids (diHDoPE) (7,8-diHDoPE, 10,11-diHDoPE, 13,14-diHDoPE, 16,17-diHDoPE, and 19,20-diHDoPE). Quantification of EPA, EpETEs, and diHETEs revealed that the uptake of EPA in the liver tissues of periodontitis-affected mice was equivalent to that in healthy control mice (Fig. 3A). However, 11,12-diHETE, 14,15-diHETE, and 17,18-diHETE levels tended to decrease in the periodontitis-affected mice. The levels of 5,6-EpETE, 8,9-EpETE, 11,12-EpETE, 14,15-EpETE, 17,18-EpETE, 5,6-diHETE, and 8,9-diHETE were below the detection limits. Quantification of DHA, EpDPAs, and diHDoPEs revealed that DHA uptake in the liver tissues of periodontitis-affected mice was equivalent to that in healthy control mice. However, 19,20-diHDoPE, which was the most abundant among five kinds of diHDoPEs, was significantly decreased (P = 0.0433) (Fig. 3B). The levels of 10,11-diHDoPE, 13,14-diHDoPE, and 16,17-diHDoPE decreased in the periodontitis-affected mice. The levels of 4,5-EpDPA, 7,8-EpDPA, 10,11-EpDPA, 13,14-EpDPA, 16,17-EpDPA, 19,20-EpDPA, and 7,8-diHDoPE were below the detection limits.
Eicosapentaenoic acid metabolites inhibits inflammatory responses of HepG2 cells
Hepatic HepG2 cells were stimulated with recombinant TNF-α (10, 25, 50 ng/ml) or LPS (10 and 100 μg/ml) for 24 h. Inflammatory responses was then assessed by TNF-α expression. Recombinant TNF-α at 10, 25, and 50 ng/ml concentrations significantly induced TNF-α expression; in contrast, LPS did not induce TNF-α expression. To examine the biological functions of omega-3 fatty acids and their metabolites, HepG2 cells were stimulated with recombinant TNF-α at 25 ng/ml concentration in the presence or absence of EPA (1 and 10 μM), DHA (1 and 10 μM), 17,18-EpETE (1 and 10 μM) as the representatives of epoxidized metabolites of omega-3 fatty acids, or 17,18-diHETE (1 and 10 μM) as the representative of hydrolyzed metabolites of epoxidized omega-3 fatty acids. The 17,18-EpETE and 17,18-diHETE at 10-μM concentration significantly suppressed recombinant TNF-α-induced TNF-α expression; however, using EPA or DHA did not result in suppression.
Discussion
Ligature-induced, periodontal tissue breakdown increased the expression of genes associated with ECM organization, such as ECM formation and collagen trimerization, in addition to genes associated with inflammatory responses and immunological cell infiltration (Fig. 1). This indicates that cells in the periodontal tissue strive to maintain tissue homeostasis in an inflammatory environment.
In the liver, the top-ranked enriched terms associated with downregulated genes in periodontitis-affected mice were limited to CYP enzymes (Fig. 2). Humans and mice have more than 50 and 100 kinds of CYP enzymes, respectively.28 CYP enzymes mainly localize in the liver to facilitate drug metabolism.29 Particularly, Cyp4a12a and Cyp4a12b participate in the metabolism of EPA and DHA.30,31 Consistent with the suppressed expression of Cyp4a12a and Cyp4a12b, the liver of periodontitis-affected mice contained relatively lower amounts of diHETE and diHDoPE in spite of equivalent EPA and DHA amounts (Fig. 3). Lipidomic analysis successfully detected diHETE and diHDoPE, which were generated by the hydrolysis of EpETE and EpDPA (Fig. 3). However, EpETE and EpDPA were not detected, presumably due to the metabolic instability of the epoxidized metabolites in vivo.32 The epoxidation pathway of omega-3 fatty acids is key for anti-inflammatory effects.33,34 These results indicate that periodontal tissue inflammation remains indispensable for developing a CYP-dysregulated, pre-symptomatic state, thus triggering the onset of inflammatory liver diseases and exacerbating NAFLD by inhibiting the production of anti-inflammatory metabolites, such as EpETE, EpDPA, diHETE, and diHDoPE.
The oral administration of P.g. suppressed the expression of hepatic anti-inflammatory factors such as peroxisome proliferator activated receptor gamma (Pparg), enhanced the expression of hepatic various genes coding for pro-inflammatory cytokines, increased serum endotoxin, and altered gut microbiota in mice.35 Moreover, previous studies have shown that P.g. administered from the pulp chamber was detectable in the liver.10,36 A limitation of the present study involved the exploration of how periodontal inflammation evokes the downregulation of hepatic CYP enzymes in a ligature-induce periodontitis mouse model. HepG2 cells induced TNF-α by recombinant TNF-α stimulation, which supposed hematogenous arrival of periodontal tissue-derived cytokines at the liver, rather than LPS, which supposed hematogenous arrival of periodontal bacteria toxin and gut pathobionts at the liver. Therefore, inflammatory cytokines secreted from periodontal tissue may be the key for downregulating CYP enzymes in vivo (Fig. 4A).
Various signaling pathways, such as nuclear factor κB (NF-κB), mitogen-activated protein kinases (MAPKs), and CCAAT/enhancer binding protein (C/EBP), contribute in the suppressed expression of the genes coding for CYP enzymes in an inflammatory environment by regulating the transcriptional activity of the CYP response element.37 Moreover, individual CYP exhibits distinct sensitivity to inflammation, which implies that distinct mechanisms are involved in downregulating individual CYP enzyme expression and activity.37 Future studies may be warranted to investigate whether downregulating specific CYPs deeply involved in omega-3 fatty acid epoxidation, such as Cyp4a12a and Cyp4a12b, is unique to a periodontitis-affected inflammatory environment.
In conclusion, by analyzing hepatic gene expression profiles and conducting lipidomic analyses of EPA and DHA metabolites, omega-3 fatty acid metabolism was suppressed by periodontitis, despite invisible hepatic phenotypic changes. The relationship between periodontitis and the development of liver disease has been the focus of previous studies. Further studies are warranted to identify the key periodontal tissue-delivered pathogenic factors which directly link to the downregulation of hepatic CYP enzymes. However, the murine model used in this study may have demonstrated, for the first time, that periodontitis is sufficient to establish a hepatic pre-symptomatic state, in which omega-3 fatty acid metabolism-induced anti-inflammatory functions are eliminated. Therefore, early interventions for periodontitis may contribute to the prevention of inflammatory hepatic diseases.
Declaration of competing interest
The authors have no conflicts of interest relevant to this article.
Acknowledgments
This study was financially supported by JSPS KAKENHI Grant Numbers 22H03266 and 22K19611 to Shigeki Suzuki, and 23H03077 and 23K18350 to Satoru Yamada. We would like to thank Editage (www.editage.jp) for the English language editing.
References
- 1.Usui M., Onizuka S., Sato T., Kokabu S., Ariyoshi W., Nakashima K. Mechanism of alveolar bone destruction in periodontitis-periodontal bacteria and inflammation. Jpn Dent Sci Rev. 2021;57:201–208. doi: 10.1016/j.jdsr.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen T.P., Yu H.C., Lin W.Y., Chang Y.C. The role of microbiome in the pathogenesis of oral-gut-liver axis between periodontitis and nonalcoholic fatty liver disease. J Dent Sci. 2023;18:972–975. doi: 10.1016/j.jds.2023.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mao S., Huang C.P., Lan H., Lau H.G., Chiang C.P., Chen Y.W. Association of periodontitis and oral microbiomes with Alzheimer's disease: a narrative systematic review. J Dent Sci. 2022;17:1762–1779. doi: 10.1016/j.jds.2022.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung Y.L., Lee J.J., Chien H.H., Chang M.C., Jeng J.H. Interplay between diabetes mellitus and periodontal/pulpal-periapical diseases. J Dent Sci. 2024;19:1338–1347. doi: 10.1016/j.jds.2024.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez-García M., Hernández-Lemus E. Periodontal inflammation and systemic diseases: an overview. Front Physiol. 2021;12 doi: 10.3389/fphys.2021.709438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strnad P., Tacke F., Koch A., Trautwein C. Liver - guardian, modifier and target of sepsis. Nat Rev Gastroenterol Hepatol. 2017;14:55–66. doi: 10.1038/nrgastro.2016.168. [DOI] [PubMed] [Google Scholar]
- 7.Trefts E., Gannon M., Wasserman D.H. The liver. Curr Biol. 2017;27:1147–1151. doi: 10.1016/j.cub.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolodziejczyk A.A., Zheng D., Shibolet O., Elinav E. The role of the microbiome in NAFLD and NASH. EMBO Mol Med. 2019;11 doi: 10.15252/emmm.201809302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musso G., Cassader M., Cohney S., et al. Fatty liver and chronic kidney disease: novel mechanistic insights and therapeutic opportunities. Diabetes Care. 2016;39:1830–1845. doi: 10.2337/dc15-1182. [DOI] [PubMed] [Google Scholar]
- 10.Nagasaki A., Sakamoto S., Chea C., et al. Odontogenic infection by Porphyromonas gingivalis exacerbates fibrosis in NASH via hepatic stellate cell activation. Sci Rep. 2020;10:4134. doi: 10.1038/s41598-020-60904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu R., Li M., Liu Z.P., Wu J., Chen L., Aihara K. Identifying critical transitions and their leading biomolecular networks in complex diseases. Sci Rep. 2012;2:813. doi: 10.1038/srep00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L., Liu R., Liu Z.P., Li M., Aihara K. Detecting early-warning signals for sudden deterioration of complex diseases by dynamical network biomarkers. Sci Rep. 2012;2:342. doi: 10.1038/srep00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards D. Oral diseases affect some 3.9 billion people. Evid Base Dent. 2013;14:35. doi: 10.1038/sj.ebd.6400925. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y.C.G., Lan S.J., Hirano H., et al. Update and review of the gerodontology prospective for 2020's: linking the interactions of oral (hypo)-functions to health vs. systemic diseases. J Dent Sci. 2021;16:757–773. doi: 10.1016/j.jds.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato A., Suzuki S., Yuan H., et al. Pharmacological activation of YAP/TAZ by targeting LATS1/2 enhances periodontal tissue regeneration in a murine model. Int J Mol Sci. 2023;24:970. doi: 10.3390/ijms24020970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto T, Yuan H, Suzuki S, Nemoto E, Saito M, Yamada S. Procyanidin B2 enhances anti-inflammatory responses of periodontal ligament cells by inhibiting the dominant negative pro-inflammatory isoforms of peroxisome proliferator-activated receptor. J Dent Sci 2024;19:1801–1810. [DOI] [PMC free article] [PubMed]
- 17.Wang X, Suzuki S, Yuan H, et al. Periodontal ligament fibroblasts utilize isoprenoid intermediate farnesyl diphosphate for maintaining osteo/cementogenic differentiation abilities. J Dent Sci 2025;20:560–568. [DOI] [PMC free article] [PubMed]
- 18.Yuan H., Suzuki S., Hirata-Tsuchiya S., et al. PPARγ-induced global H3K27 acetylation maintains osteo/cementogenic abilities of periodontal ligament fibroblasts. Int J Mol Sci. 2021;22:8646. doi: 10.3390/ijms22168646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida K., Suzuki S., Kawada-Matsuo M., et al. Heparin-LL37 complexes are less cytotoxic for human dental pulp cells and have undiminished antimicrobial and LPS-neutralizing abilities. Int Endod J. 2019;52:1327–1343. doi: 10.1111/iej.13130. [DOI] [PubMed] [Google Scholar]
- 20.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinz S., Benner C., Spann N., et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett T., Wilhite S.E., Ledoux P., et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki S., Fukuda T., Nagayasu S., et al. Dental pulp cell-derived powerful inducer of TNF-α comprises PKR containing stress granule rich microvesicles. Sci Rep. 2019;9:3825. doi: 10.1038/s41598-019-40046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki K., Suzuki S., Fahreza R.R., Nemoto E., Yamada S. Dynamic changes in chromatin accessibility during the differentiation of dental pulp stem cells reveal that induction of odontogenic gene expression is linked with specific enhancer construction. J Dent Sci. 2024;19:1705–1713. doi: 10.1016/j.jds.2023.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki S., Sasaki K., Fahreza R.R., Nemoto E., Yamada S. The histone deacetylase inhibitor MS-275 enhances the matrix mineralization of dental pulp stem cells by inducing fibronectin expression. J Dent Sci. 2024;19:1680–1690. doi: 10.1016/j.jds.2023.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaha H., Husein D., Ohyama Y., et al. N-terminal dentin sialoprotein fragment induces type I collagen production and upregulates dentinogenesis marker expression in osteoblasts. Biochem Biophys Rep. 2016;6:190–196. doi: 10.1016/j.bbrep.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umeda N., Hirai T., Ohto-Nakanishi T., Tsuchiya K.J., Matsuzaki H. Linoleic acid and linoleate diols in neonatal cord blood influence birth weight. Front Endocrinol. 2022;13 doi: 10.3389/fendo.2022.986650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nebert D.W., Wikvall K., Miller W.L. Human cytochromes P450 in health and disease. Philos Trans R Soc Lond B Biol Sci. 2013;368 doi: 10.1098/rstb.2012.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanger U.M., Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Isobe Y., Itagaki M., Ito Y., et al. Comprehensive analysis of the mouse cytochrome P450 family responsible for omega-3 epoxidation of eicosapentaenoic acid. Sci Rep. 2018;8:7954. doi: 10.1038/s41598-018-26325-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller D.N., Schmidt C., Barbosa-Sicard E., et al. Mouse Cyp4a isoforms: enzymatic properties, gender- and strain-specific expression, and role in renal 20-hydroxyeicosatetraenoic acid formation. Biochem J. 2007;403:109–118. doi: 10.1042/BJ20061328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucas D., Goulitquer S., Marienhagen J., et al. Stereoselective epoxidation of the last double bond of polyunsaturated fatty acids by human cytochromes P450. J Lipid Res. 2010;51:1125–1133. doi: 10.1194/jlr.M003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang G., Kodani S., Hammock B.D. Stabilized epoxygenated fatty acids regulate inflammation, pain, angiogenesis and cancer. Prog Lipid Res. 2014;53:108–123. doi: 10.1016/j.plipres.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamabata T., Nakamura T., Tachibana Y., Horikami D., Murata T. 5,6-DiHETE attenuates vascular hyperpermeability by inhibiting Ca2+ elevation in endothelial cells. J Lipid Res. 2018;59:1864–1870. doi: 10.1194/jlr.M085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arimatsu K., Yamada H., Miyazawa H., et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci Rep. 2014;4:4828. doi: 10.1038/srep04828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furusho H., Miyauchi M., Hyogo H., et al. Dental infection of Porphyromonas gingivalis exacerbates high fat diet-induced steatohepatitis in mice. J Gastroenterol. 2013;48:1259–1270. doi: 10.1007/s00535-012-0738-1. [DOI] [PubMed] [Google Scholar]
- 37.de Jong L.M., Jiskoot W., Swen J.J., Manson M.L. Distinct effects of inflammation on cytochrome P450 regulation and drug metabolism: lessons from experimental models and a potential role for pharmacogenetics. Genes. 2020;11:1509. doi: 10.3390/genes11121509. [DOI] [PMC free article] [PubMed] [Google Scholar]