To survive and develop normally, plants must constantly perceive changes in their environment and respond properly through a variety of molecular mechanisms. What are the processes by which plant cells sense extracellular modifications, specifically transmit the signal from the outside to the inside of the cell, and ultimately to the nucleus where changes in gene expression may occur? These questions, which are crucial for plant survival and consequently for animal life, are becoming well documented in some aspects. Current knowledge of plant signaling networks was first based on the identification of some receptors that perceive the signal as well as many transcription factors and target genes that mediate the responses. However, the list of the identified regulatory components linking receptors to cellular responses is still largely incomplete and the understanding of molecular mechanisms regulating signal transduction pathways is really poor.
One of the most important abiotic stresses for crop productivity concerns plant dehydration. Plants suffer from dehydration under high salinity and drought, as well as low-temperature conditions, all of which cause hyperosmotic stress characterized by a decreased turgor pressure and water loss. Dehydration triggers the biosynthesis of the abscisic acid (ABA) hormone and it has been known for a long time that a significant set of genes, induced by drought, salt, and cold stresses, are also activated by ABA. The use of common components and pathways in plant response to related stresses allows plants to acclimate partially to a range of adverse conditions after exposure to only one specific stress. In addition to these common signaling elements, highly specific signaling mechanisms occur, allowing precise plant adaptation. For example, several genes induced by salt, drought, and cold stress are not responsive to exogenous ABA treatment, indicating the existence of ABA-independent signal transduction cascades in addition to the ABA-mediated pathways.
This article briefly describes actual knowledge of the signaling pathways induced by conditions of drought, cold, and high salinity. Little information is available concerning the receptors of osmolarity, whereas much is known about the numerous transcription factors acting in the regulation of gene expression. This abundant knowledge, including recent advances, is summarized here. But the main focus of this article concerns the growing number of identified molecules that catalyze the phosphorylation and dephosphorylation of water stress-signaling proteins. Limited information is available regarding the different groups of plant protein phosphatases and their involvement in osmotic signaling. On the contrary, a significant amount of data concerns the role of plant mitogen-activated protein kinases (MAPKs) in osmoregulation. More generally, MAPK cascades are now recognized as major signal transduction mechanisms in plants, as in yeast (Saccharomyces cerevisiae) and mammals. Interestingly, recent studies have identified novel plant-specific families of protein kinases that play important roles in osmotic signaling pathways. Their phosphorylation activity can be modulated by calcium, either directly (calcium-dependent protein kinase [CDPK]) or via a calcium-binding protein (calcineurin B-like [CBL] sensor and CBL-interacting protein kinase [CIPK]), but can be independent of calcium as well (Suc nonfermenting-related kinase [SnRK] 2). We have particularly emphasized these three emerging families of protein kinases that are presumed to function in plants as mediators of osmotic adaptation. A glossary of different names used for the same signaling component is included to facilitate reading.
GLOSSARY OF DIFFERENT NAMES ATTRIBUTED TO THE SAME SIGNALING COMPONENT
AtCPK10: AtCDPK1; AtCPK30: AtCDPK1a; McCPK1: McCDPK1; SOS3: AtCBL4; AtCBL1: SCaBP5; AtCBL2: SCaBP1; AtCBL3: SCaBP3; SOS2: CIPK24, SnRK3.11; CIPK3: PKS12, SnRK3.17; AtMKK1: AtMEK1; SnRK2.4: ASK1, OSKL7; SnRK2.6: OST1, SRKE; SnRK2.8: SRKC, OSKL4; CBF1: DREB1B; CBF2: DREB1C; CBF3: DREB1A; CBF4: DREB1D.
OSMOSENSORS AND SECOND MESSENGERS
Searching for Osmosensors in Plants
In yeast, hyperosmolarity can be sensed by a two-component system composed of the SLN1 His kinase, the YPD1 phosphorelay intermediate, and the SSK1 response regulator, leading to the activation of the HOG1 MAPK pathway. The Arabidopsis (Arabidopsis thaliana) SLN1 homolog, AtHK1, is able to suppress the salt-sensitive phenotype of the yeast double-mutant sln1Δ sho1Δ, which lacks both yeast osmosensors (Urao et al., 1999). However, direct evidence for a role of AtHK1 as an osmosensor in plants is still lacking. Although it could interact with the phosphorelay AtHP1 in the yeast two-hybrid system, no interaction was observed between AtHP1 and the response regulators (Urao et al., 2000). Another His kinase, CRE1, which was identified as a cytokinin receptor, is also able to complement the yeast sln1Δ mutant in the presence of cytokinin (Inoue et al., 2001). Interestingly, a recent work reported that SLN1 and CRE1 perceive the osmotic signal by turgor sensing in yeast (Reiser et al., 2003). It was shown that the integrity of the periplasmic region of SLN1 is essential for its sensor function. This suggests that osmotic stress may trigger a conformational change of SLN1 due to a stress-induced modification of the cell wall-plasma membrane interaction. It is tempting to speculate that a similar turgor-sensing mechanism might regulate hyperosmotic signaling in plants. On the other hand, the involvement of receptor-like kinases (RLK) in osmosensing has been suggested by the increased osmotic stress tolerance induced by overexpression of the tobacco (Nicotiana tabacum) NtC7 (Tamura et al., 2003). This membrane-located receptor-like protein is proposed to form dimers whose conformation may be sensitive to changes in membrane architecture to sense hyperosmolarity. In addition, NtC7 displays the special feature of lacking the kinase catalytic domain. This suggests that NtC7 transduces the osmotic signal to cytoplasmic components by interacting with protein partners through its C-terminal tail region. Thus, several osmosensors seem to be involved in plant stress signal perception, although definitive evidence is still scarce. As salinity, drought, and cold can induce specific stress responses, it is probable that distinct osmosensors are involved in different stress signaling pathways.
Calcium and Newly Recognized Second Messengers
Signal perception at the plasma membrane leads to the production of second messengers that initiate cascades of signaling events. Among them, calcium has been extensively studied and its involvement in osmotic signaling has been recently reviewed (Chinnusamy et al., 2004). It was shown that osmotic stresses induced calcium fluxes characterized by different kinetics and magnitudes, resulting in calcium signatures specific to a particular stress and cell type. The calcium signal results from extracellular calcium influx and/or calcium release from intracellular stores. It has been shown that hyperosmotic stresses induce an increase in inositol 1,4,5-trisphosphate (IP3), which is blocked by phospholipase C inhibitors (Takahashi et al., 2001). As IP3 is known to activate vacuolar calcium channels, it was proposed that calcium is released from intracellular stores in response to hyperosmotic stresses as a result of the activation of the IP3-dependent calcium channels.
Other phospholipids, in particular phosphatidic acid (PA), seem to have important roles in osmotic signaling (Munnik and Meijer, 2001). PA was reported to accumulate in response to cold treatment (Ruelland et al., 2002) and water deficit (Frank et al., 2000). It can be formed directly by phospholipase D or indirectly by phospholipase C after phosphorylation of diacylglycerol. It is worth noting that both pathways are involved in PA production in response to osmotic stresses, allowing a subtle regulation in PA concentration (Ruelland et al., 2002). Moreover, the stress-activated alfalfa MAPK (SAMK), was shown to be activated by PA in a dose-dependent manner (Lee et al., 2001), and a recent study using PA affinity chromatography identified several transduction proteins such as kinases, phosphatases, and 14-3-3 proteins as potential PA targets (Testerink et al., 2004). All these data strongly support the view that PA plays an important role in osmotic transduction pathways.
It is well known that osmotic stresses induce oxidative damage, which can be reduced by the activation of antioxidant enzymes and the biosynthesis of osmolytes acting as reactive oxygen species (ROS) scavengers. However, an increasing number of studies suggest that ROS could also play a signaling role, as in response to biotic stresses in which ROS production is partly due to a plasma membrane NADPH oxidase. Although major ROS production induced by hyperosmotic stress occurs at intracellular sites, it was also shown that a cell wall diamine oxidase (Lin and Kao, 2002) and a plasma membrane NADPH oxidase (Jiang and Zhang, 2002) were activated by hyperosmolarity and drought, respectively. Interestingly, the hyperosmotic induction of the catalase gene CAT1 was shown to be mediated by H2O2 (Guan et al., 2000). Moreover, H2O2 is able to activate the osmotic-responsive MAPK AtMPK6 in Arabidopsis cell suspensions (Yuasa et al., 2001). Further work is needed to better understand the role of ROS in osmotic signaling, but the data presented confirmed that ROS also act as second messengers despite their toxicity at high concentration.
PROTEIN KINASES AND PHOSPHATASES
Calcium Sensing: Involvement of CDPKs and CBLs-CIPKs
Changes in calcium concentration can be sensed by two kinds of proteins: the sensor relays, including CBL proteins, and the sensor responders, including CDPKs. For sensor relays, the conformational changes induced by calcium binding are relayed to an interacting partner whose structure or activity is then modified. On the contrary, sensor responders display their own enzyme activity, which is directly affected by calcium binding.
The involvement of CDPKs in osmotic signaling has been suggested by the transcriptional induction of CPK genes in response to salinity, cold, or drought (Saijo et al., 2000; Chehab et al., 2004). In the common ice plant (Mesembryanthemum crystallinum), a potential substrate of McCPK1, CSP1, was isolated by the two-hybrid method (Patharkar and Cushman, 2000). The CSP1 gene encodes a pseudoresponse regulator that is constitutively localized in the nucleus. It was shown that McCPK1 could phosphorylate CSP1, in vitro, in a calcium-dependent manner. Moreover, McCPK1 undergoes a change in subcellular localization from the plasma membrane to the nucleus in response to salt stress and dehydration (Patharkar and Cushman, 2000; Chehab et al., 2004). These results suggest that McCPK1 may regulate the function of CSP1 by phosphorylation in response to hyperosmotic stresses. In rice (Oryza sativa), overexpression of OsCDPK7 conferred cold, drought, and salt tolerance by inducing the expression of stress-responsive genes (Saijo et al., 2000). In protoplast transient expression assays, constitutively active Arabidopsis AtCPK10 and AtCPK30 were shown to activate the HVA1 promoter, which is responsive to ABA and abiotic stresses (Sheen, 1996). This effect was not observed with other CDPKs, indicating a specific role of these two kinases in osmotic signaling.
The nonenzymatic CBLs interact with kinase partners, the CIPKs, also called salt overly sensitive (SOS) 2-like protein kinases (PKSs) or SnRK3s. A recent review of these two families indicated a complex network of interactions between members of the CBL and CIPK families, which was observed in two-hybrid and in vitro experiments (Gong et al., 2004). This suggests the existence of cross-talk between different transduction pathways as well as highly regulated signaling events based on competition for the appropriate CBL-CIPK interaction. The most-studied members of these families, SOS3 and SOS2, interact together in the SOS pathway, leading to sodium ion homeostasis in response to salt stress. This model (for review, see Chinnusamy et al., 2004), which was supported by a transgenic approach in Arabidopsis (Guo et al., 2004), is based on the release of SOS2 autoinhibition by interaction with SOS3, which targets the kinase to the plasma membrane. The SOS2/SOS3 complex activates the SOS1 Na+/H+ antiporter by phosphorylation. On the other hand, SOS2 also regulates vacuolar transporters like the CAX1 Ca2+/H+ antiporter (Cheng et al., 2004) and the AtNHX1 Na+/H+ antiporter (Qiu et al., 2004), but the kinase activity does not seem to be required. Moreover, SOS3 is not involved in this mechanism, which is consistent with its role of targeting SOS2 to the plasma membrane. However, other members of the CBL family could also target SOS2 to the tonoplast.
Concerning the other proteins of the families, the transcripts of the CIPK3 gene accumulated in response to ABA, cold, and salt stress, and the corresponding protein was proposed to regulate the response pathways to these treatments (Kim et al., 2003). Although the cipk3 mutant did not display any modification in stress tolerance, the induction of stress-responsive genes was altered. AtCBL1 could also be involved in abiotic stress responses. However, some divergent results were observed. The first report on AtCBL1 function suggested that the protein mediated ABA signaling, but was not involved in abiotic responses (Guo et al., 2002). This study was based on the analysis of an RNAi cbl1 mutant in which the expression of other CBLs (AtCBL2 and AtCBL3) was not affected. As the expression of the most closely related gene, AtCBL9, was not analyzed, it was proposed that the observed phenotype in the mutant was due to cosuppression of both AtCBL1 and AtCBL9 (Pandey et al., 2004). This hypothesis is supported by the analysis of the T-DNA insertional cbl9 mutant, which is also affected in ABA responses (Pandey et al., 2004). On the contrary, overexpression of AtCBL1 was shown to trigger an increase in drought and salt induction of stress genes correlated with an enhanced tolerance to both stresses, whereas T-DNA knockout mutants displayed the opposite phenotype (Albrecht et al., 2003; Cheong et al., 2003). Based on cold induction of marker genes, a positive (Cheong et al., 2003) or a negative role (Albrecht et al., 2003) of AtCBL1 on cold responses was proposed. However, it should be noted that different sets of marker genes were used in each study, except RD29A, which displayed the same absolute level in the two analyses despite distinct kinetics. These results highlight the complexity of CBL-CIPK signaling and demonstrate the importance of these proteins in osmotic transduction pathways.
MAPK Proteins: At the Crossroads of Signaling Pathways
Much interest was focused on MAPKs because of the well-known HOG1 MAPK cascade involved in hyperosmotic signaling in yeast. So far, several MAPKs were shown to be activated by abiotic stresses in different plant species. In Arabidopsis detached leaves, AtMPK4 and AtMPK6 were activated by cold, drought, saline stress, and sorbitol, indicating an important role of these two kinases in a general response to hyperosmotic stresses (Ichimura et al., 2000). AtMPK6 was also activated by hyperosmolarity in Arabidopsis cell suspensions (Droillard et al., 2002). Although AtMPK4 activation was neither observed in cell suspensions nor in seedlings exposed to hyperosmotic and saline stresses, the mpk4 knockout mutant displayed an enhanced tolerance to hyperosmolarity and an increased expression of RAB18, a drought-inducible gene, suggesting a negative role for AtMPK4 in hyperosmolarity tolerance (Droillard et al., 2004). In addition, the MAPKK AtMKK1, which was also activated by cold, drought, and high salinity (Matsuoka et al., 2002), phosphorylated and activated AtMPK4 in vitro (Huang et al., 2000). These results suggest the existence of a MAPK cascade in response to hyperosmotic stresses involving AtMPK4 and AtMKK1. The upstream kinase could be AtMEKK1, which is able to interact in the two-hybrid system with the MAPKK. In addition, a recent work based on protoplast transient expression assays and mutant lines identified a complete MAPK cascade in response to cold and salt stress involving the MAPKKK, AtMEKK1, the MAPKK, AtMKK2, and the two MAPKs, AtMPK4 and AtMPK6 (Teige et al., 2004). This questions the specificity of signal transduction because AtMEKK1 is also involved in the AtMPK3 and AtMPK6 activation in response to the biotic stress induced by flagellin, mediated by the MAPKKs, AtMKK4 and AtMKK5 (Asai et al., 2002). One possibility to maintain specificity in signal transduction pathways is the involvement of scaffold proteins, which associate the different elements of a same-signaling cascade in specific modules. Little is known about these proteins in plants and it will become an important biological issue in the following years.
In comparison with Arabidopsis, less data are available in other plant species. In tobacco, a rapid and transient activation of the MAPK salicylic acid-induced protein kinase (SIPK) was demonstrated in response to hyperosmolarity induced by several osmolytes (Droillard et al., 2000; Hoyos and Zhang, 2000; Mikolajczyk et al., 2000). The upstream MAPKK could be NtMEK2, which was shown to phosphorylate and activate SIPK in vitro, as well as in vivo, in a tobacco leaf transient expression assay (Yang et al., 2001). In alfalfa, the salt-induced activation of SIMK is mediated by the SIMKK (Kiegerl et al., 2000; Cardinale et al., 2002). The cold and drought activation of SAMK may occur through a phosphorylation cascade involving the MAPKK PRKK and the MAPKKK NPK1 because SAMK activity is further induced when the three kinases are coexpressed in protoplasts (Cardinale et al., 2002). Both MAPKs are also activated in response to elicitors, providing other evidence for the key position of the MAPKs at the crossroads of signaling pathways.
SnRK2 Proteins: An Emerging Osmotic Signaling Family
Using in-gel kinase assays, several proteins were shown to be activated by hyperosmotic stresses with different features than MAPKs. They are activated in a sustained manner throughout the stress kinetics and they can phosphorylate both myelin basic protein and histone. In tobacco cells, Mikolajczyk et al. (2000) first identified one of these kinases, which is homologous to Arabidopsis SnRK2.4. Similarly, it was shown that two soybean SnRK2s (SPK1 and SPK2) were activated by high hyperosmolarity when overexpressed in yeast (Monks et al., 2001). A specific role of SnRK2.6 in stomatal closure was demonstrated with the withering phenotype of the corresponding mutant exposed to dehydration (Mustilli et al., 2002; Yoshida et al., 2002). It has been shown that SnRK2.6 is activated by drought and ABA, and that the drought hypersensitivity of the mutant is due to its inability to trigger ABA-induced stomatal closure. ABA-induced ROS production is disrupted in the mutant guard cells whereas exogenous H2O2 can restore stomatal closure, positioning SnRK2.6 between ABA perception and ROS production (Mustilli et al., 2002). In addition, ABA-induced gene expression of two drought-inducible genes, RD22 and RD29B, is suppressed in the mutant, further indicating an important role of SnRK2.6 in ABA signaling in response to water stress (Yoshida et al., 2002). Similarly, the Vicia faba ABA-activated protein kinase, which is specifically expressed in guard cells, was shown to mediate stomatal closure by activating plasma membrane anion channels (Li et al., 2000).
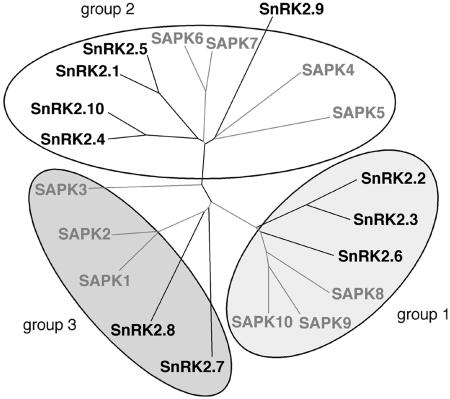
Very recently, other members of the Arabidopsis SnRK2 family were also demonstrated to be involved in osmotic signaling. Using a family-specific antibody able to recognize the 10 Arabidopsis SnRK2s, it was shown that hyperosmolarity activated at least four members of the family in vivo (Boudsocq et al., 2004). In fact, protoplast transient expression assays allowed us to demonstrate that hyperosmotic and saline stresses activated all SnRK2 proteins, except SnRK2.9 (Boudsocq et al., 2004). Using the same approach, the 10 rice SnRK2s were shown to be activated by salt stress in protoplasts (Kobayashi et al., 2004). It is interesting to note that the only Arabidopsis kinase that is not activated by hyperosmotic stresses is the most divergent of the Arabidopsis family (Fig. 1), suggesting that SnRK2.9 may be involved in a different signaling pathway. In addition, the two rice SAPK1 and SAPK2 proteins were shown to be hyperosmotically activated by phosphorylation (Kobayashi et al., 2004) as it was observed for the nine Arabidopsis SnRK2s (M. Boudsocq and C. Laurière, unpublished data). On the other hand, only a part of the osmotically activated SnRK2 proteins was also activated by ABA both in Arabidopsis and rice, suggesting the involvement of ABA-dependent and ABA-independent signaling pathways. Thus, SnRK2 proteins can be divided into three groups (Fig. 1). Group 1 corresponds to proteins activated both by hyperosmolarity and ABA, and their osmotic activation can be mediated by ABA. Group 2 is composed of SnRK2s only activated by hyperosmotic stresses. Group 3 includes kinases activated by hyperosmolarity but poorly or not activated by ABA. Interestingly, these three groups, based on functional analyses by transient expression assays, reflect the three branches of the relation tree (Fig. 1), suggesting that distinct evolutions of the SnRK2 proteins have led to specific responses. Although in planta analysis is required to confirm this classification, it is supported by the in vitro study of chimeric constructs showing that the relatively divergent SnRK2 C-terminal domain is mainly involved in ABA responsiveness (Kobayashi et al., 2004). The fact that the complete families from Arabidopsis and rice are activated in a similar manner by hyperosmotic stresses suggests a high conservation of plant mechanisms involved in osmotic responses and highlights a crucial role for SnRK2 proteins in hyperosmotic signaling. However, it also raises the question of functional redundancy. Why do plants need to activate so many kinases? One explanation could be that each kinase possesses specific targets and that the swift regulation of all these target proteins will lead to a rapid induction of stress responses. Expression profiles of the 10 rice SAPK genes indicate that they are all present in leaf blades, sheaths, and roots (Kobayashi et al., 2004). However, it is possible that the proteins display more specific localization at the tissue or subcellular level. For example, it was shown in Arabidopsis that SnRK2.8 was mainly expressed in root tips, which could explain the specific drought-induced root hypersensitivity of the corresponding knockout mutant (Umezawa et al., 2004). On the contrary, overexpression of SnRK2.8 enhanced drought tolerance, which was correlated with increased expression of stress-responsive genes like RD29A, COR15A, and CBF3. Thus, the activation of the complete family could allow a concerted action to induce the same response in different organs and subcellular compartments. On the other hand, it has been shown that each SnRK2 displays distinct activation levels (Boudsocq et al., 2004; Kobayashi et al., 2004). It is thus possible that each member of the family displays distinct activation profiles according to kinetics, intensities, and threshold, which will lead to subtle and specific responses for each stress condition.
Figure 1.
Relation tree of the SnRK2 family from Arabidopsis and rice. Arabidopsis SnRK2s are drawn with black lines and rice SAPKs are drawn with gray lines. Based on transient expression assays, SnRK2 proteins activated by hyperosmotic conditions can be divided into three groups, depending on additional activation by ABA. Group 1 corresponds to kinases strongly activated by ABA, whereas in group 2, SnRK2s are not responsive to ABA. Group 3 includes kinases poorly or not activated by ABA. The Arabidopsis SnRK2.9 does not fit into any of the groups due to the lack of activation by hyperosmolarity.
Figure 2 summarizes the different kinase families that have been identified so far as components of hyperosmotic signaling. In addition to these kinases, some plant homologs of shaggy-like protein kinases are likely to play a role, since overexpression of one AtSK induces NaCl stress responses and results in enhanced NaCl tolerance in Arabidopsis (Piao et al., 2001). The large diversity of involved kinases reflects the need of rapid and elaborate transduction mechanisms for plants to adapt to and survive constantly changing environmental conditions. Cross-talks between signaling pathways, for example at the calcium level, contribute to the swiftness and the efficiency of transduction mechanisms. On the other hand, some pathways appear to be specific to one stress condition, such as the SOS pathway in response to saline stress or the drought-induced stomata closure. This can be explained by the specificity of each environmental condition, which only partially corresponds to osmotic stress. In this regard, the multiple kinases could play crucial roles for specific regulation of plant stress responses.
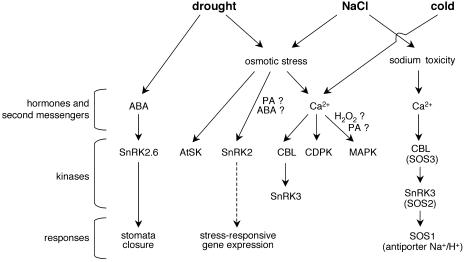
Figure 2.
Regulatory network of stress responses to drought, salt, and cold: specificity and cross-talk. The common osmotic component of drought, salt, and cold leads to common signaling pathways mediated by several kinases like AtSK, SnRK2, SnRK3, MAPK, and CDPK proteins. Calcium was shown to be an upstream element in some SnRK3, CDPK, and MAPK activation, whereas PA, ABA, and H2O2 are presumed to be involved in SnRK2 and/or MAPK regulation. In addition to these common signaling events, some pathways are specific to one stress condition like the SOS pathway regulating sodium homeostasis in response to NaCl, and the SnRK2.6-mediated stomatal closure induced by ABA in response to drought. In major cases, downstream responses regulated by protein kinases are still unknown.
Protein Phosphatases: The Other Side of the Mirror
Studies have been focused mainly on phosphatases involved in the regulation of MAPKs, which are transiently activated by phosphorylation. Several phosphatases, including protein Tyr phosphatase (PTP), dual-specificity protein Tyr phosphatase (DsPTP), and protein Ser/Thr phosphatase 2C (PP2C), were shown to down-regulate MAPKs that are involved in osmotic signaling. AtPTP1 dephosphorylates AtMPK4 and AtMPK6 in vitro on Tyr, leading to inactivation of the kinases (Huang et al., 2000; Gupta and Luan, 2003). Similarly, AtDsPTP1 dephosphorylates and inactivates AtMPK4 in vitro (Gupta et al., 1998). In addition, a mutant of the DsPTP MKP1 exhibits an enhanced salt resistance in comparison to wild-type plants, indicating that this phosphatase is a negative regulator of salt stress tolerance (Ulm et al., 2002). On the other hand, MKP1 interacts with both AtMPK6 and AtMPK4 in the yeast two-hybrid system, suggesting that MKP1 may regulate these two MAPKs. However, the saline stress-induced activation of AtMPK6 and AtMPK4 is not altered in the mkp1 mutant, and the mpk4 knockout mutant exhibits an increased hyperosmotic tolerance (Droillard et al., 2004). These results suggest that MKP1 could be involved in an osmotic pathway independent of the two MAPKs. Interestingly, the alfalfa MP2C, a PP2C, is able to suppress the lethal phenotype induced by overexpression of a constitutively active STE11, which is the MAPKKK involved in the yeast hyperosmotic HOG1 MAPK cascade (Meskiene et al., 1998). This suggests that MP2C can also inactivate the plant MAPK cascade involved in osmotic signaling. In this regard, it was shown that MP2C dephosphorylates and inactivates the stress-activated SIMK when coexpressed in parsley protoplasts (Meskiene et al., 2003). Coimmunoprecipitation experiments from transfected protoplasts demonstrated that SIMK and MP2C could directly interact in vivo, providing additional clues to assume MP2C as a negative regulator of the SIMK signaling pathway. The role of two Arabidopsis PP2Cs, ABI1 and ABI2, in the negative regulation of ABA signaling involved in drought-induced stomata closure has been recently reviewed (Schweighofer et al., 2004). Together, these results suggest two kinds of roles for the protein phosphatases in osmoregulation. A regulatory role may be to bring the stress-activated MAPKs back to their basal activity level, allowing a new activation of the signaling pathway after a second stimulation. On the other hand, the different mutants affected in protein phosphatases obtained to date display an increased stress tolerance, indicating a negative role of the phosphatases. It is probable that future research will also reveal positive roles for phosphatases, as dephosphorylation processes have been described to be positive regulation mechanisms in other signaling contexts.
TRANSCRIPTION FACTORS
Environmental stresses induce the expression of many genes that can be classified into two groups. The first group corresponds to proteins involved in transduction pathways, such as transcription factors, whereas the second group includes effector proteins like the enzymes of osmolyte biosynthesis. Many studies have been focused on transcription factors involved in gene expression regulation and they have been extensively reviewed (Shinozaki et al., 2003). This article does not aim to discuss in detail all these stress-responsive transcription factors. The purpose is to summarize briefly the main osmotic signaling pathways, including recent works in cold signaling (Fig. 3). For each signal (salt, drought, and cold), several pathways can be distinguished depending on ABA involvement.
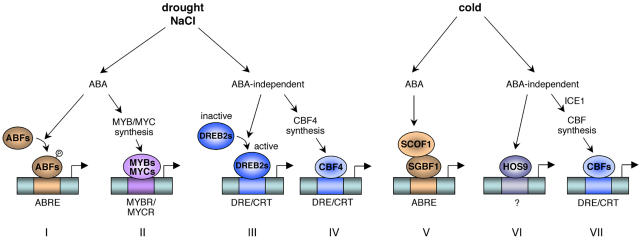
Figure 3.
Regulation of gene expression in response to drought, salt, and cold: involvement of ABA-dependent and ABA-independent pathways. Cis-acting elements are shown in boxes and transcription factors are in ovals. Pathways I, II, and V are ABA dependent and based on ABRE or MYBR/MYCR cis-acting elements. Pathways III, IV, VI, and VII are ABA independent and based on DRE/CRT or still unidentified cis-acting sequences. Both types of pathways are implemented in drought, salt, and cold signaling. Some transcription factors like ABFs and DREB2s are stress activated to induce gene expression, whereas others, like CBFs, require biosynthesis first.
Concerning ABA-dependent pathways, two kinds of cis-acting sequences are involved in ABA-mediated gene expression. The first one is the ABA-responsive element (ABRE). It has been shown that the bZIP transcription factors ABRE-binding factor (ABF)/ABRE binding protein (AREB) can activate the stress-responsive RD29A promoter through binding to the ABRE motifs (Fig. 3, pathway I). This activation is increased by an ABA-induced post-translational modification, e.g. phosphorylation (Uno et al., 2000). In response to cold, the soybean zinc finger protein SCOF-1 increases the DNA-binding activity of the bZIP SGBF-1 to ABRE sequence (Fig. 3, pathway V), suggesting a cooperative role of the two proteins to induce ABA-mediated cold-responsive gene expression (Kim et al., 2001). The second promoter sequences are the MYB and MYC recognition sites (MYBR and MYCR, respectively). It has been shown that MYB and MYC transcription factors are involved in an ABA-dependent pathway leading to the expression of drought-responsive genes like RD22 (Abe et al., 2003).
On the other hand, ABA-independent expression of stress-responsive genes can occur through dehydration-responsive element (DRE)/C-repeat (CRT) cis-acting elements. The binding factors CBF/DREB1 (CRT-binding factor/DRE-binding factor 1) and DREB2 mediate gene expression in response to cold and drought/salinity, respectively (Fig. 3, pathways VII and III). Interestingly, the CBF4 protein seems to mediate drought response unlike the other CBFs (Haake et al., 2002). A particular feature of CBF proteins is their early and transient cold induction, which precedes the expression of cold-responsive genes. This requires the involvement of a constitutively expressed CBF-transcriptional inducer, which would be activated by cold treatment. The identification of such a regulator, the MYC-like protein inducer of CBF expression 1 (ICE1), has been reported to specifically induce the expression of CBF3 (Chinnusamy et al., 2003). On the other hand, the increased stress tolerance of the cbf2 mutant supported the view of CBF2 acting as a negative regulator of CBF1 and CBF3 expression to ensure their transient expression (Novillo et al., 2004). This is in contradiction to the enhanced freezing tolerance induced by overexpression of each CBF, suggesting redundant functional activities of the three proteins (Gilmour et al., 2004). However, it can be explained by a double role of CBF2 as a positive regulator of cold-induced genes and a negative regulator for CBF genes. Indeed, in the cbf2 knockout mutant, the release of transcriptional repression of CBF1 and CBF3 led to enhanced expression of these two CBF genes followed by enhanced expression of cold-responsive genes and an increased freezing tolerance (Novillo et al., 2004). On the contrary, overexpression of CBF2 is sufficient to increase cold-induced gene expression and subsequently freezing tolerance because of the common gene targets of the three CBF proteins (Gilmour et al., 2004). The role of CBFs in cold-induced gene expression has been extensively studied. However, microarray analyses have indicated that multiple regulatory pathways are activated in response to cold, involving several other transcription factors (Fowler and Thomashow, 2002). For example, the Arabidopsis hos9 mutant displays hypersensitivity to freezing and impairment in cold acclimation ability without any alteration in cold-induced CBF expression (Zhu et al., 2004). Thus, the homeodomain transcription factor encoded by HOS9 is required for freezing tolerance in a CBF-independent pathway (Fig. 3, pathway VI).
This classification in seven distinct pathways suggests that each signal transduction is independent. However, transient expression assays have indicated that CBF and DREB2 proteins could cooperate with ABFs to better activate the RD29A promoter, indicating the existence of cross-talk between ABA-dependent and ABA-independent pathways (Narusaka et al., 2003). In addition, overexpression of transcription factors like CBFs leads to enhanced tolerance to freezing as well as drought and salinity, further indicating that transduction pathways are interconnected. The next challenge for future research will be to establish links between transduction proteins like kinases and transcription factors.
CONCLUSIONS AND PERSPECTIVES
Signal transduction cascades, from sensing dehydration to the expression of various stress-responsive genes, have been studied in plants in some aspects, leading to significant knowledge. First of all, the model system of stomata closure in guard cells has opened many interesting studies mainly focused on the involvement of the ABA hormone. Besides this model, information is also available on transcription factors responding to drought, salt, and cold via ABA-dependent or ABA-independent pathways. Concerning the kinase-mediated signal transduction, the SOS pathway, which is specific to salt stress, is probably the most documented and represents a major calcium-mediated pathway for the regulation of plant salt tolerance and ion homeostasis. On the contrary, the other kinase-mediated pathways are still poorly understood, although some components have been identified at the molecular level. For example, the involvement of two Arabidopsis MAPKs, AtMPK4 and AtMPK6, in response to salt or hyperosmolarity, has been well characterized, but studies on upstream protein kinases and phosphatases, which both regulate the MAPKs, are still scarce.
Major research domains have been opened recently, with emerging osmotic signaling compounds, like the osmosensors, the second messengers PA and ROS, and the three families of kinases more precisely discussed here. It is worth noting that CDPKs and CIPKs are regulated by calcium, whereas SnRK2 kinases are completely independent of calcium for both their activity and upstream signaling steps. A focus on this novel component is required, notably to get functional evidence in planta on the putative osmosensors or to discover the roles of the numerous members of the CDPK and CIPK families. It is interesting that SnRK2s seem to play an important function in osmotic signaling, with the complete family appearing specifically activated by drought and salt, in contrast to the other involved kinase families, like CDPKs or CIPKs.
Many other components remain to be discovered and some clues can already be mentioned. In particular, information on protein kinase substrates, which may be transcription factors, as well as cytoplasmic or membrane proteins, is very limited. In this regard, the first plant MAPK substrate has been recently identified and corresponds to the rate-limiting enzyme of ethylene biosynthesis. The MAPK acting on this substrate is AtMPK6, which is already known as an important mediator in stress signaling (Liu and Zhang, 2004). It is interesting to note that this result allows us to link osmotic and ethylene signaling pathways. This assumption is reinforced by the discovery of a novel ethylene-responsive factor in tomato (Lycopersicon esculentum), which also binds to DRE. In addition, this transcription factor enhances osmotic stress tolerance by activating expression of downstream genes (Huang et al., 2004). Further studies, combining genetic and molecular approaches, will increase the knowledge of the complex networks of osmotic signaling in plants. How the signals interact to confer cross-tolerance and developmental traits, and how this interaction influences crop productivity, are surely important questions in the research area aimed at improving plant stress tolerance.
Acknowledgments
We thank Dr. H. Barbier-Brygoo for a critical reading of the manuscript and Dr. R. Oomen for helpful comments.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht V, Weinl S, Blazevic D, D'Angelo C, Batistic O, Kolukisaoglu U, Bock R, Schulz B, Harter K, Kudla J (2003) The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J 36: 457–470 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Barbier-Brygoo H, Laurière C (2004) Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem 279: 41758–41766 [DOI] [PubMed] [Google Scholar]
- Cardinale F, Meskiene I, Ouaked F, Hirt H (2002) Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell 14: 703–711 [PMC free article] [PubMed] [Google Scholar]
- Chehab EW, Patharkar OR, Hegeman AD, Taybi T, Cushman JC (2004) Autophosphorylation and subcellular localization dynamics of a salt- and water deficit-induced calcium-dependent protein kinase from ice plant. Plant Physiol 135: 1430–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH, Pittman JK, Zhu JK, Hirschi KD (2004) The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J Biol Chem 279: 2922–2926 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Kim KN, Pandey GK, Gupta R, Grant JJ, Luan S (2003) CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15: 1833–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Schumaker K, Zhu JK (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55: 225–236 [DOI] [PubMed] [Google Scholar]
- Droillard MJ, Boudsocq M, Barbier-Brygoo H, Laurière C (2002) Different protein kinase families are activated by osmotic stresses in Arabidopsis thaliana cell suspensions. Involvement of the MAP kinases AtMPK3 and AtMPK6. FEBS Lett 527: 43–50 [DOI] [PubMed] [Google Scholar]
- Droillard MJ, Boudsocq M, Barbier-Brygoo H, Laurière C (2004) Involvement of MPK4 in osmotic stress response pathways in cell suspensions and plantlets of Arabidopsis thaliana: activation by hypoosmolarity and negative role in hyperosmolarity tolerance. FEBS Lett 574: 42–48 [DOI] [PubMed] [Google Scholar]
- Droillard MJ, Thibivilliers S, Cazalé AC, Barbier-Brygoo H, Laurière C (2000) Protein kinases induced by osmotic stresses and elicitor molecules in tobacco cell suspensions: two crossroad MAP kinases and one osmoregulation-specific protein kinase. FEBS Lett 474: 217–222 [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank W, Munnik T, Kerkmann K, Salamini F, Bartels D (2000) Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. Plant Cell 12: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54: 767–781 [DOI] [PubMed] [Google Scholar]
- Gong D, Guo Y, Schumaker KS, Zhu JK (2004) The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol 134: 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan LM, Zhao J, Scandalios JG (2000) Cis-elements and trans-factors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2 is the likely intermediary signaling molecule for the response. Plant J 22: 87–95 [DOI] [PubMed] [Google Scholar]
- Guo Y, Qiu QS, Quintero FJ, Pardo JM, Ohta M, Zhang C, Schumaker KS, Zhu JK (2004) Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 16: 435–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu JK (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3: 233–244 [DOI] [PubMed] [Google Scholar]
- Gupta R, Huang Y, Kieber J, Luan S (1998) Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from Arabidopsis. Plant J 16: 581–589 [DOI] [PubMed] [Google Scholar]
- Gupta R, Luan S (2003) Redox control of protein tyrosine phosphatases and mitogen-activated protein kinases in plants. Plant Physiol 132: 1149–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyos ME, Zhang SQ (2000) Calcium-independent activation of salicylic acid-induced protein kinase and a 40-kilodalton protein kinase by hyperosmotic stress. Plant Physiol 122: 1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YF, Li H, Gupta R, Morris PC, Luan S, Kieber JJ (2000) ATMPK4, an Arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol 122: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Zhang Z, Zhang X, Zhang H, Huang D, Huang F (2004) Tomato TERF1 modulates ethylene response and enhances osmotic stress tolerance by activating expression of downstream genes. FEBS Lett 573: 110–116 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000) Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24: 655–665 [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Jiang M, Zhang J (2002) Involvement of plasma-membrane NADPH oxidase in abscisic acid- and water stress-induced antioxidant defense in leaves of maize seedlings. Planta 215: 1022–1030 [DOI] [PubMed] [Google Scholar]
- Kiegerl S, Cardinale F, Siligan C, Gross A, Baudouin E, Liwosz A, Eklof S, Till S, Bogre L, Hirt H, et al (2000) SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12: 2247–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Lee SH, Cheong YH, Yoo CM, Lee SI, Chun HJ, Yun DJ, Hong JC, Lee SY, Lim CO, et al (2001) A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J 25: 247–259 [DOI] [PubMed] [Google Scholar]
- Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S (2003) CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15: 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T (2004) Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16: 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Hirt H, Lee Y (2001) Phosphatidic acid activates a wound-activated MAPK in Glycine max. Plant J 26: 479–486 [DOI] [PubMed] [Google Scholar]
- Li J, Wang XQ, Watson MB, Assmann SM (2000) Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287: 300–303 [DOI] [PubMed] [Google Scholar]
- Lin CC, Kao CH (2002) Osmotic stress-induced changes in cell wall peroxidase activity and hydrogen peroxide level in roots of rice seedlings. Plant Growth Regul 37: 177–183 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka D, Nanmori T, Sato K, Fukami Y, Kikkawa U, Yasuda T (2002) Activation of AtMEK1, an Arabidopsis mitogen-activated protein kinase kinase, in vitro and in vivo: analysis of active mutants expressed in E. coli and generation of the active form in stress response in seedlings. Plant J 29: 637–647 [DOI] [PubMed] [Google Scholar]
- Meskiene I, Baudouin E, Schweighofer A, Liwosz A, Jonak C, Rodriguez PL, Jelinek H, Hirt H (2003) Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. J Biol Chem 278: 18945–18952 [DOI] [PubMed] [Google Scholar]
- Meskiene I, Bogre L, Glaser W, Balog J, Brandstotter M, Zwerger K, Ammerer G, Hirt H (1998) MP2C, a plant protein phosphatase 2C, functions as a negative regulator of mitogen-activated protein kinase pathways in yeast and plants. Proc Natl Acad Sci USA 95: 1938–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajczyk M, Awotunde OS, Muszynska G, Klessig DF, Dobrowolska G (2000) Osmotic stress induces rapid activation of a salicylic acid-induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell 12: 165–178 [PMC free article] [PubMed] [Google Scholar]
- Monks DE, Aghoram K, Courtney PD, DeWald DB, Dewey RE (2001) Hyperosmotic stress induces the rapid phosphorylation of a soybean phosphatidylinositol transfer protein homolog through activation of the protein kinases SPK1 and SPK2. Plant Cell 13: 1205–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Meijer HJ (2001) Osmotic stress activates distinct lipid and MAPK signalling pathways in plants. FEBS Lett 498: 172–178 [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34: 137–148 [DOI] [PubMed] [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J (2004) CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 101: 3985–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Cheong YH, Kim KN, Grant JJ, Li L, Hung W, D'Angelo C, Weinl S, Kudla J, Luan S (2004) The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16: 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patharkar OR, Cushman JC (2000) A stress-induced calcium-dependent protein kinase from Mesembryanthemum crystallinum phosphorylates a two-component pseudo-response regulator. Plant J 24: 679–691 [DOI] [PubMed] [Google Scholar]
- Piao HL, Lim JH, Kim SJ, Cheong GW, Hwang I (2001) Constitutive overexpression of AtGSK1 induces NaCl stress responses in the absence of NaCl stress and results in enhanced NaCl tolerance in Arabidopsis. Plant J 27: 305–314 [DOI] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, Zhu JK (2004) Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt overly sensitive (SOS) pathway. J Biol Chem 279: 207–215 [DOI] [PubMed] [Google Scholar]
- Reiser V, Raitt DC, Saito H (2003) Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J Cell Biol 161: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelland E, Cantrel C, Gawer M, Kader JC, Zachowski A (2002) Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol 130: 999–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Overexpression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23: 319–327 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9: 236–243 [DOI] [PubMed] [Google Scholar]
- Sheen J (1996) Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274: 1900–1902 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6: 410–417 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Katagiri T, Hirayama T, Yamaguchi-Shinozaki K, Shinozaki K (2001) Hyperosmotic stress induces a rapid and transient increase in inositol 1,4,5-trisphosphate independent of abscisic acid in Arabidopsis cell culture. Plant Cell Physiol 42: 214–222 [DOI] [PubMed] [Google Scholar]
- Tamura T, Hara K, Yamaguchi Y, Koizumi N, Sano H (2003) Osmotic stress tolerance of transgenic tobacco expressing a gene encoding a membrane-located receptor-like protein from tobacco plants. Plant Physiol 131: 454–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, Doczi F, Ichimura K, Shinozaki K, Dangl JL, Hirt H (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15: 141–152 [DOI] [PubMed] [Google Scholar]
- Testerink C, Dekker HL, Lim ZY, Johns MK, Holmes AB, Koster CG, Ktistakis NT, Munnik T (2004) Isolation and identification of phosphatidic acid targets from plants. Plant J 39: 527–536 [DOI] [PubMed] [Google Scholar]
- Ulm R, Ichimura K, Mizoguchi T, Peck SC, Zhu T, Wang X, Shinozaki K, Paszkowski J (2002) Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J 21: 6483–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K (2004) SRK2C, a SNF1-related protein kinase 2,improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 17306–17311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Miyata S, Yamaguchi-Shinozaki K, Shinozaki K (2000) Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett 478: 227–232 [DOI] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K (1999) A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11: 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KY, Liu YD, Zhang SQ (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K (2002) ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Yuasa Y, Ichimura K, Mizoguchi T, Shinozaki K (2001) Oxidative stress activates ATMPK6, an Arabidopsis homologue of MAP kinase. Plant Cell Physiol 42: 1012–1016 [DOI] [PubMed] [Google Scholar]
- Zhu J, Shi H, Lee BH, Damsz B, Cheng S, Stirm V, Zhu JK, Hasegawa PM, Bressan RA (2004) An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc Natl Acad Sci USA 101: 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]