Abstract
The ALCR/alcA (alc) two-component, ethanol-inducible gene expression system provides stringent control of transgene expression in genetically modified plants. ALCR is an ethanol-activated transcription factor that can drive expression from the ALCR-responsive promoter (alcA). However, the alc system has been shown to have constitutive expression when used in plant callus or cell suspension cultures, possibly resulting from endogenous inducer produced in response to lowered oxygen availability. To widen the use of the alc system in plant cell culture conditions, the receptor domain of the rat glucocorticoid receptor (GR) was translationally fused to the C terminus of ALCR to produce ALCR-GR, which forms the basis of a glucocorticoid-inducible system (alc-GR). The alc-GR switch system was tested in tobacco (Nicotiana tabacum) Bright Yellow-2 suspension cells using a constitutively expressed ALCR-GR with four alternative alcA promoter-driven reporter genes: β-glucuronidase, endoplasmic reticulum-targeted green fluorescent protein, haemagglutinin, and green fluorescent protein-tagged Arabidopsis (Arabidopsis thaliana) Arath;CDKA;1 cyclin-dependent kinase. Gene expression was shown to be stringently dependent on the synthetic glucocorticoid dexamethasone and, in cell suspensions, no longer required ethanol for induction. Thus, the alc-GR system allows tight control of alcA-driven genes in cell culture and complements the conventional ethanol switch used in whole plants.
The alc system is a two-component chemically inducible gene expression system, originally developed as a gene switch in Aspergillus nidulans (Waring et al., 1989). It consists of the alcR-encoded transcription factor (ALCR) that, in response to exogenous ethanol, drives gene expression from the alcA target promoter. This system has been used successfully in a wide range of plant species, including Arabidopsis (Arabidopsis thaliana), Brassica napus, tobacco (Nicotiana tabacum), and Solanum tuberosum (Salter et al., 1998; Roslan et al., 2001; Sweetman et al., 2002; Junker et al., 2003). The alc system has conferred conditional control over reporter, metabolic, and developmental genes (Caddick et al., 1998; Laufs et al., 2003) and can be induced with either ethanol or acetaldehyde (Junker et al., 2003). Furthermore, a range of tissue- and organ-specific alc systems have been characterized for use in Arabidopsis (Deveaux et al., 2003; Maizel and Weigel, 2004).
Plant-conditional gene expression systems (for review, see Gatz, 1997; Zuo and Chua, 2000; Padidam, 2003; Wang et al., 2003) are desirable because constitutively overexpressed transgenes, or RNA interference constructs, often display secondary phenotypes that are difficult to interpret or relate to the function of the gene or pathway in question. For example, plants constitutively expressing yeast invertase displayed an extreme stunted growth phenotype (Sonnewald et al., 1991). When expression of yeast invertase was temporally controlled in tobacco, using the alc system, it was possible to induce transgene expression and to assay carbon flux at different stages of development (Caddick et al., 1998). Therefore, conditional expression of genes can reveal a greater range of phenotypes.
We have previously used the alc system for the conditional restoration of gene function to the unusual floral organs (ufo) loss-of-function Arabidopsis mutant at different developmental stages (Laufs et al., 2003). UFO is required for the specification of organ identity in the second and third whorls and the proper primordium initiation pattern in the inner three whorls. Timed restoration of UFO gene function using the gene switch dissected the temporal requirements for UFO during floral development, revealing new roles of UFO in the outgrowth of petal primordia that previously were not apparent in loss-of-function or constitutive gain-of-function backgrounds (Ingram et al., 1995; Lee et al., 1997). Thus, complex phenotypes can be dissected by restoration of gene function with conditional gene expression. Use of the alc system as a switch for complementation experiments in plants appears to be generally applicable, as demonstrated by the conditional complementation of the Arabidopsis leafy-12 mutant (Maizel and Weigel, 2004).
Although the alc system has found widespread utility as a safe and effective expression system in whole plants, there are concerns about its efficacy under plant cell culture conditions. As will be demonstrated here, cell cultures often have significant, and sometimes high, levels of transgene expression in the absence of exogenous inducer. Although the reason for this is unclear, it is likely that plant cell culture conditions result in oxygen limitation leading to the production of inductive compounds. This limits the use of the alc system since it is often advantageous to test the effects of transgene expression in cell cultures. Furthermore, in those species in which transformation is reliant upon callus, transformation may be limited if expression of the transgene has deleterious effects during subculture or regeneration. To restore the requirement for an exogenously added chemical inducer, the rat glucocorticoid receptor (GR) domain has been fused to the ALCR transcription factor; conferring steroid-inducible control over alc-mediated gene expression. In animals, proteins containing a GR domain are sequestered outside of the nucleus with heat shock proteins and are released from these complexes upon binding of a steroid ligand (Picard, 1994). The rat GR domain, tagged to constitutively expressed transcription factors, has previously been used in plants to confer dexamethasone (Dex)-dependent control over transcription factor function (Sablowski and Meyerowitz, 1998).
In this article, we describe the leaky expression of alc in tomato (Lycopersicon esculentum L. var. Ailsa Craig Mill.) cell cultures derived from a characterized transgenic line and the specification of a Dex-inducible alc system (alc-GR system) for use in the tobacco Bright Yellow (BY)-2 cell culture (Nagata et al., 1992). We report that neither addition of ethanol or accumulation of endogenous inductive compounds activates the alc-GR system. Moreover, gene expression in the absence of inducer or on addition of ethanol is low or not detectable and responds quickly in a dose-dependant manner to Dex. Therefore, this system presents a useful gene switch for plant cell cultures, which is directly comparable and uses many of the same components as the alc system in whole plants.
RESULTS
The alc Gene Expression System Is Constitutively Active in Plant Cell Cultures
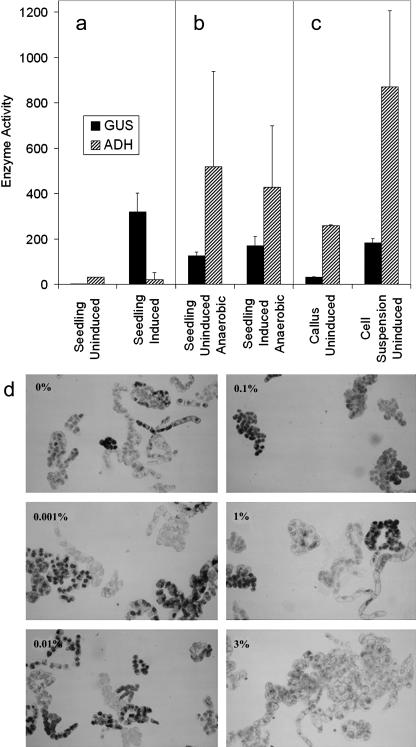
To test the efficacy of the alc system to regulate transgene expression, callus and cell suspension cultures were derived from previously characterized transgenic tomato plants (Garoosi, 1998; G.A. Garoosi, M.G. Salter, M.X. Caddick, and A.B. Tomsett, unpublished data). The highly inducible transgenic line (LeGUS20), carrying 35S∷ALCR/alcA∷GUS (Caddick et al., 1998; Roslan et al., 2001), had been selfed, and segregation was consistent with a 3:1 ratio indicating the presence of a single T-DNA insert (data not shown). Seedlings of the line grown in hydroponic solution consistently demonstrated tight regulation of alc-directed β-glucuronidase (GUS) expression, having a negligible background activity in the absence of inducer and a high GUS activity 96 h after induction with 0.1% (v/v) ethanol (Fig. 1a). Assays for the presence of alcohol dehydrogenase (ADH) activity in such lines demonstrated a low level of activity comparable with wild-type tomato seedlings grown under identical conditions. Under hypoxic/anoxic conditions, seedlings exhibited significant GUS activity whether ethanol was present or absent in the growth medium (Fig. 1b), and levels of ADH activity consistent with a switch to ethanolic fermentation, in which pyruvate is converted to acetaldehyde by pyruvate decarboxylase and the acetaldehyde is reduced to ethanol by ADH (Drew, 1997; Fukao and Bailey-Serres, 2004). As recently demonstrated, the alc system can be induced with either ethanol or acetaldehyde (Junker et al., 2003). Callus and cell suspension cultures derived from the same plant line were also tested for GUS and ADH activity. Such cells demonstrated significant levels of GUS activity in the absence of exogenously applied ethanol and ADH levels indicative of ethanolic fermentation (Fig. 1c). Similar data were obtained with tissue cultures derived from a second tightly regulated transgenic line in which chloramphenicol acetyltransferase was used as the reporter gene for the alc system. These cell suspensions were grown in both baffled (to increase aeration) and unbaffled conical flasks, but while expression from the alcA promoter declined with increased aeration, a significant amount of uninduced activity was retained (Supplemental Table I).
Figure 1.
alc expression in uninduced cell cultures. Enzyme Activity represents GUS (nmol 4MU h−1 mg−1 total protein) and ADH (μmol min−1 mg−1 fresh weight of tissue); the error bars represent se from at least four replicates for each value. a, Seedlings (20 d old) of LeGUS20, carrying cauliflower mosaic virus 35S∷ALCR/alcA∷GUS, were incubated in hydroponic solution without/with 0.1% (v/v) ethanol (Uninduced/Induced) for 96 h, before enzyme extraction to assay for GUS and ADH activity. b, Seedlings of LeGUS20 were incubated in hydroponic solution, purged with 100% N2 gas to decrease oxygen in the liquid medium, without/with 0.1% (v/v) ethanol (Uninduced/Induced), and sealed in a 3-L plastic box containing Anaerocult (Merck, Rahway, NJ) to induce anoxia for 96 h, before enzyme extraction. c, LeGUS20-derived callus was grown on solid Murashige and Skoog medium (without ethanol) in parafilm-sealed petri dishes for 20 d. Individual calli, 1.5 cm in diameter, were used for enzyme extraction. Cell suspension cultures (originally prepared from LeGUS20 callus) were grown for 2 weeks in the absence of ethanol in 50 mL of liquid medium in a 250-mL conical flask on an orbital shaker at 120 rpm, before enzyme extraction. d, Tobacco BY-2 cells transgenic for 35S∷ALCR/alcA∷GUS were induced with 0%, 0.001%, 0.01%, 0.1%, 1%, and 3% (v/v) ethanol overnight, stained for GUS activity with GUS-staining buffer, and visualized by light microscopy. GUS stained cells appear black.
Expression from the alc system in the absence of exogenously supplied inducer is not restricted to tomato. Tobacco BY-2 cells were transformed with 35S∷ALCR/alcA∷GUS using the pSRN/AGS plasmid (Roslan et al., 2001). A cell line derived from multiple transformant calli was treated overnight with various concentrations of ethanol and stained for GUS expression (Jefferson et al., 1987). It was clear that GUS expression occurred with or without treatment with ethanol (Fig. 1d). Cells treated with 0% to 1% (v/v) ethanol displayed distinct GUS staining, but 3% (v/v) ethanol was clearly toxic to cells as evidenced by weak GUS staining and distorted cell morphologies. Constitutive expression has also been observed in whole Arabidopsis plants when grown on agar or Phytagel-containing media (data not shown; Roslan et al., 2001).
Modification of the alc System and Selection of Transgenic Lines
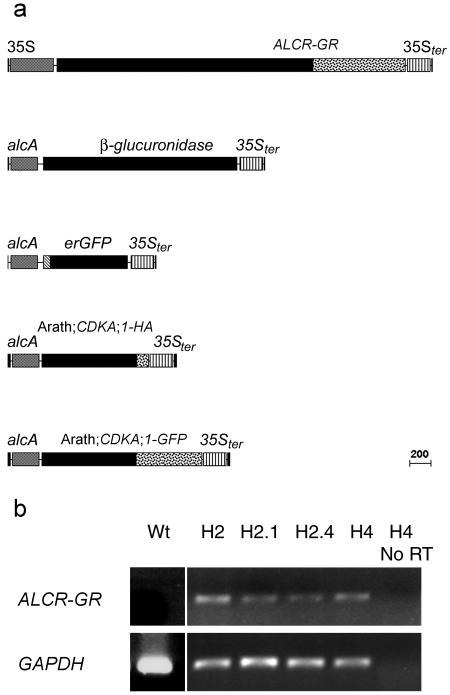
To confer inducible transgene expression to the alc system in BY-2 cells, a translational fusion was constructed that placed the rat GR domain gene fragment (GR) at the 3′-end of the ALCR gene. The ALCR-GR gene fusion was placed under the control of a 35S promoter in the pGreen 0129 plasmid (Fig. 2a; Hellens et al., 2000) and transformed into BY-2 cells. Eleven independently transformed 35S∷ALCR-GR lines (calli/clones) were generated, all of which expressed ALCR-GR when assayed by semiquantitative reverse transcription (RT)-PCR (Fig. 2b; data not shown). The H2, H2.1, H2.4, and H4 ALCR-GR background lines were chosen for further analysis as they were the first to become established in liquid culture. The ALCR-GR background lines were sequentially transformed with alcA∷GUS, and the resulting double transformants were assayed for Dex-dependent expression by staining for GUS activity. After induction, the proportion of alcA∷GUS sequentially transformed lines expressing GUS varied between the background lines. In the H2 background, five out of 12 were GUS positive, while nine out of 12 H2.1, four out of 12 H2.4, and 10 out of 12 of H4 secondary transformants also displayed GUS staining. Of the GUS positive lines a variable proportion in each background expressed GUS constitutively: three out of five of H2, one out of nine of H2.1, zero out of 12 of H2.4, and one out of 10 of H4, respectively (data not shown). We concluded from this that a screen for conditionally expressing ALCR-GR secondary transformants is appropriate prior to experimentation.
Figure 2.
Schematic representation of the constructs used and expression of ALCR-GR in primary alc-GR system transformants. a, 35S∷ALCR-GR in pGreen 0129, alcA∷GUS in pGreen 0029, alcA∷erGFP in pGreen 0029, alcA∷Arath;CDKA;1-HA in pGreen 0029, and alcA∷Arath;CDKA;1-GFP in pGreen 0029. b, ALCR-GR and the tobacco control glyceraldehyde-3-P-dehydrogenase (GAPDH) transcripts were assayed by RT-PCR in the H2, H2.1, H2.4, and H4 independent 35S∷ALCR-GR transformants. GAPDH transcript, but not ALCR-GR transcript, accumulated in untransformed (Wt) BY-2 cells and transcripts for neither are PCR amplified from RNA extracted from a 35S∷ALCR-GR line without reverse-transcriptase treatment (H4 No RT). All four 35S∷ALCR-GR transformants express both GAPDH and ALCR-GR transcripts.
The ALCR-GR Transactivator-Mediated Gene Switch Is Dex But Not Ethanol Dependent in BY-2 Cells and Responds to Dex in a Dose-Dependent Manner
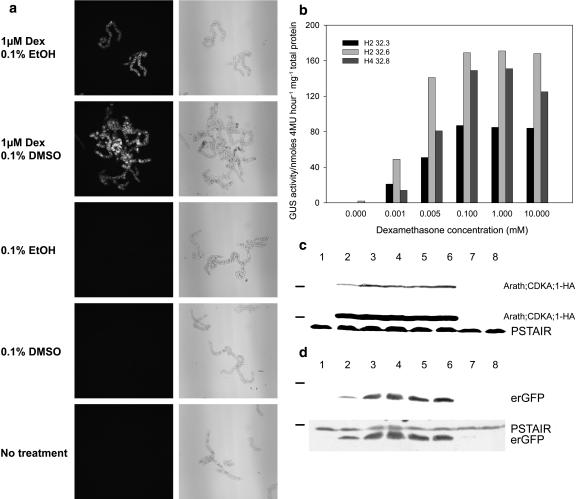
To test how the addition of the GR domain to the ALCR protein had altered its response to induction, ALCR-GR control over alcA∷erGFP expression was assayed following treatment of BY-2 cells with different combinations of Dex and/or solvents, in particular ethanol. We visually assayed the expression of endoplasmic reticulum-targeted green fluorescent protein (erGFP), in an H2.4 ALCR-GR background line sequentially transformed with alcA∷erGFP, by fluorescence microscopy, 24 h after treatment with 1 μm Dex dissolved in ethanol or dimethyl sulfoxide (DMSO). Both solvents were applied at a final concentration of 0.1% (v/v). The solvents were also applied independently to determine whether they alone could induce expression via ALCR-GR. erGFP expression was induced by Dex dissolved in ethanol and by Dex dissolved in DMSO (Fig. 3a). Dex was essential for the activation of the alc-GR system as neither solvent alone was able to induce reporter gene expression.
Figure 3.
Dex induces the alc-GR system in a dose-dependent manner. a, BY-2 cells, transgenic for both 35S∷ALCR-GR and alcA∷erGFP, were treated with Dex dissolved in either 0.1% (v/v) ethanol or DMSO, or either solvent without Dex, or finally without solvent or Dex. The erGFP expression was detected by fluorescence microscopy (left-hand column, GFP fluorescence appears white), and the cells are presented as brightfield images (right-hand column). b and c, One day after 1:10 subculture, ALCR-GR background lines and lines secondarily transformed with alcA driven reporter constructs were treated with 0 to 10 μm Dex. Twenty-four hours after inductions were applied, one sample of each line was harvested for transgene expression assays. b, Lines H2 32.3, H2 32.6, and H4 32.8, which are H2 and H4 ALCR-GR backgrounds secondarily transformed with alcA∷GUS. The values represent GUS activity (nmol 4MU−1 mg−1 total protein). c, Western blot using PSTAIR antibody to detect both Arath;CDKA;1-HA and endogenous tobacco CDK (PSTAIR) in protein extracts from an H2 ALCR-GR background secondarily transformed with alcA∷Arath;CDKA;1-HA. Lanes 1 to 6 contain protein extracts from an H2-derived BY-2 cell line carrying 35S∷ALCR-GR/alcA∷Arath;CDKA;1-HA, and lanes 7 to 8 contain protein extracts from the H2 ALCR-GR background line. Upper blot represents a shorter exposure time than the lower blot where endogenous CDK (PSTAIR) is also visible. d, Western blot first probed with GFP antibody (top) and later probed with PSTAIR antibody to detect erGFP and tobacco CDK, respectively, in protein extracts from an H2 ALCR-GR background line secondarily transformed with alcA∷erGFP. Lanes 1 to 6 contain protein extract from an H2-derived BY-2 line carrying 35S∷ALCR-GR/alcA∷erGFP, and lanes 7 to 8 contain protein extracts from the H2 ALCR-GR background line. Lane 1, 0 μm; lane 2, 0.001 μm; lane 3, 0.005 μm; lane 4, 0.1 μm; lane 5, 1 μm; lane 6, 10 μm; lane 7, 0 μm; and lane 8, 10 μm Dex. Molecular weight marker on the western-blot figure represents 36 kD.
To assess if ALCR-GR-regulated gene expression was dependent on the amount of Dex applied, the levels of alcA-driven GUS or Arath;CDKA;1-haemagglutinin (HA) were measured following treatment of cells with various concentrations of the inducer. GUS enzyme activity was assayed using a fluorometric assay following a 24-h induction with Dex (Fig. 3b). Generally, GUS activity was highest in a range of 0.1 to 10 μm Dex, with a slightly lower activity at 0.005 μm Dex. All three GUS reporter lines displayed a significantly lower GUS activity with 0.001 μm Dex. Two of the three GUS reporter lines displayed no background GUS activity on treatment with DMSO alone; however, line H4 32.8 displayed low GUS activity in the absence of Dex.
The Dex dose-dependent Arath;CDKA;1-HA expression profile was assayed by western blot using whole-cell extracts (Fig. 3c). Arath;CDKA;1-HA expression produces a protein that is 6 kD larger than the endogenous tobacco CDKA protein: both proteins are recognized by the monoclonal antibody raised against the cyclin-dependent kinase (CDK) PSTAIR motif. The alcA∷Arath;CDKA;1-HA in an H2 background displayed a similar Dex dose-dependent Arath;CDKA;1-HA expression profile as the GUS lines. Western blots of the Dex dose-dependent erGFP expression also displayed the same induction profile (Fig. 3d).
Time Course of Induction
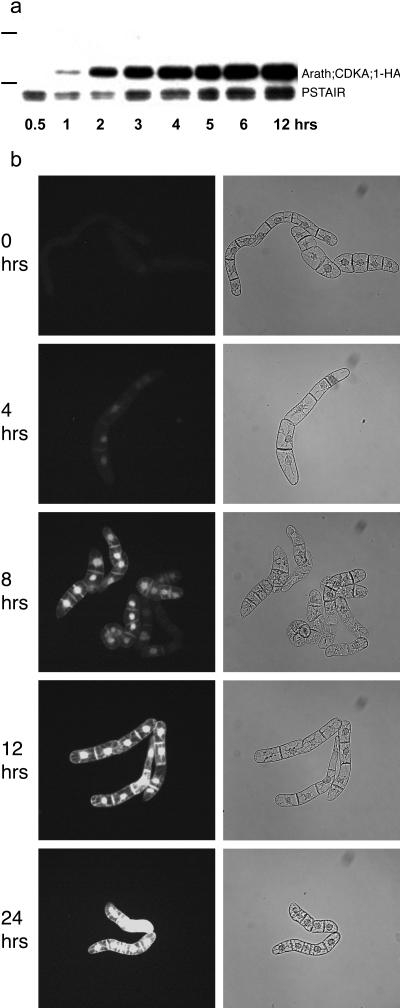
The dynamics of Dex induction of the ALC-GR system in tobacco BY-2 cells were defined by western blotting and fluorescence microscopy analyses of tagged Arath;CDKA;1. Following induction with 0.1 μm Dex, the accumulation of Arath;CDKA;1-HA, driven by the alc-GR system, was monitored by western blot using the PSTAIR antibody on whole-cell extracts. The Arath;CDKA;1-HA protein was detectable 1 h after induction, exceeded the level of endogenous CDKA proteins after 2 h, and maintained this high level for at least 12 h (Fig. 4a).
Figure 4.
Time course of Dex induction. a, Western blot using PSTAIR antibody to detect Arath;CDKA;1-HA and endogenous tobacco CDK (PSTAIR) in protein extracts from an H2 ALCR-GR background secondarily transformed with alcA∷Arath;CDKA;1-HA 0.5 to 12 h following induction with 0.1 μm Dex. Prior to induction, this line had been subcultured by dilution 1:10 from stationary phase cultures and grown for a further 24 h. Molecular weight markers represent 36 and 50 kD. b, Fluorescent microscopy of Arath;CDKA;1-GFP fluorescence in cells of an H2 ALCR-GR background line secondarily transformed with alcA∷Arath;CDKA;1-GFP, following treatment as in a. Left-hand column displays Arath;CDKA;1-GFP fluorescence (appears white) and right-hand brightfield view of cells.
Arath;CDKA;1-GFP, a GFP-tagged Arabidopsis CDK driven by the alc-GR system, was visually monitored by fluorescent microscopy, again following treatment with 0.1 μm Dex. The Arath;CDKA;1-GFP fluorescence was weakly fluorescent 4 h after induction and was strongest from 12 h onwards after induction (Fig. 4b). It is known that GFP takes up to 4 h to fold properly before it becomes active (Heim et al., 1994). Therefore, it is consistent that the Arath;CDKA;1-GFP fluorescence increases much slower than the Arath;CDKA;1-HA protein accumulates, following Dex induction.
DISCUSSION
Given the increasing use of the alc system (Ait-ali et al., 2003; Chen et al., 2003; Deveaux et al., 2003; Maizel and Weigel, 2004), there is a need to widen its applicability to plant culture growth conditions. In this article, we report the leaky expression of the alc system in plant tissue cultures and an adaptation of it for use in plant cell suspension cultures, in particular in tobacco BY-2 cells.
The alc gene switch has great potential for research and commercial application in part due to the nature of the inducer, ethanol, which is cheap and relatively nontoxic to plants and the environment. Ethanol is also highly penetrating and can induce gene expression deep within tissues such as meristems and even developing seeds (G.R. Roberts, L. Sakvarelidze, P. Laufs, and J.H. Doonan, unpublished data). However, plants can produce ethanol and acetaldehyde (the physiological inducer of the alc switch) during ethanolic fermentation, triggered by oxygen deficiency (see Drew, 1997). Typically, in whole plants, this can arise through excess water in the root environment of dry-land species. It has previously been reported that the alc system is activated by endogenous inducer in experiments designed to lower oxygen levels and when Arabidopsis was grown in agar plates, but this has not been observed for seedlings grown in hydroponic solution or for plants in soil flooded for 3 d (Salter et al., 1998; Roslan et al., 2001). It was possible however that plant cell suspension cultures would also experience oxygen limitation because of their submerged environment and hence induce alc in the absence of exogenously applied inducer. If affected, this would severely limit the widespread use of alc for those applications in which such cells are the material of choice, such as detailed cell biological studies (Nagata et al., 1992) or the production of bioactive molecules (James and Lee, 2001).
To address this, callus and cell suspension cultures were produced from transgenic plants already shown to contain a single T-DNA, which segregated normally, and that showed inducible regulation by alc but with negligible basal activity in the absence of exogenous inducer. This eliminated the possibility of constitutive expression in the cell cultures arising from the position of T-DNA integration, and further avoided the possibility of constitutive expression due to a series of different, or multiple, integration events within the selected transformed callus/cell line. Significant basal expression from alc was observed in callus cells, and high activity for both the alc-directed reporter gene activity and ADH were detected without exogenous inducer. The levels of activity in the cell suspension cultures were consistent with the artificial environment used to induce severe hypoxia in seedlings. Attempts to increase oxygen availability to such cells through the use of baffled flasks lowered but did not eliminate the leaky activity. This strongly suggested that ethanolic fermentation is activated in cell suspension cultures.
Since tobacco BY-2 cells are used widely, these were selected to test a modified alc system to ensure tight regulation of the transgene. Tests of the unmodified alc system showed expression in these cells in the absence of exogenous inducer, as in the tomato lines. We conferred chemical-inducible control over the alc system, in culture, by creating a translational fusion between the ALCR transcription factor and the GR domain of the GR to create the alc-GR Dex-inducible switch. A detailed characterization of the alc-GR switch in BY-2 cells demonstrated that this system mediates both tightly regulated and rapidly induced gene expression. The alc-GR system was activated by Dex whether dissolved in ethanol or DMSO, and it was tightly regulated as neither addition of ethanol or DMSO mediated gene expression in the absence of Dex. The switch was dose dependent with respect to Dex in a similar manner to other reports of glucocorticoid-inducible expression in BY-2 cells (Nara et al., 2000). Reporter protein was evident on western blots after only 1 h of induction and highly induced from 2 h onwards. GFP-tagged reporter fluorescence was visible at 4 h, which is consistent with the time it takes GFP to mature and fluoresce (Heim et al., 1994).
CONCLUSION
A modified alc switch (alc-GR) has been developed and characterized for use in cell culture that complements and extends the existing alc switch as used in whole plant systems. Although this has not yet been tested in whole plants, the modified switch should be resistant to endogenous activation by flooding and other stress conditions. Furthermore, constructs containing genes driven by the alcA promoter can now be conditionally expressed in plants by induction with ethanol (via ALCR) and in cell culture by induction with Dex (via ALCR-GR). The alc-GR system restores conditional control over gene expression for the alc system, enhancing the general utility of the alc gene switch.
MATERIALS AND METHODS
Plasmid Construction
DNA manipulations and cloning were carried out using standard procedures (Sambrook et al., 1989). The binary vector pSRN∷pAGS expressed the alcR gene from the cauliflower mosaic virus 35S promoter and the GUS reporter gene from the alcA promoter, as described previously (Caddick et al., 1998; Roslan et al., 2001). The 35S∷alcR-GR construct was introduced into the binary transformation vector pGreen0129 (Hellens et al., 2000). For this, a KpnI/EcoRV cassette from pJIT60 (Guerineau and Mullineaux, 1993), containing 2 × 35S promoter-35S terminator, was inserted into pGreen0129, generating pGR-2 × 35S. An XbaI (end polished with Klenow)/BamHI cassette from pDltaGRBX (gift from Dr. Robert Sablowski), containing GR, was inserted into pGR-2 × 35S using BamHI/SmaI, generating pGR-2 × 35S-GR. Finally, BclI and SalI restrictions sites were added to the alcR gene by PCR using AATGATCAAGAAAGCTGTCAACTTTCCCATTCAAACC and CCGTCGACGATATTCTCCTGCACACAGCATGG as primers and pSRN1 as template (Caddick et al., 1998). After BclI and SalI digestion, ALCR was inserted into pGR-2 × 35S-GR, generating pGR-35S∷alcR-GR.
The alcA∷GUS-35S terminator cassette was introduced into the pGreen0029 vector (Hellens et al., 2000) as a HindIII fragment from pSRN1 AGS/Bin19 (Roslan et al., 2001), generating pGR32. The HindIII alcA∷erGFP-35S terminator cassette from pMCB56 (Fernandez-Abalos et al., 1998) was introduced into pGreen0029, again by HindIII digestion/ligation, generating pGR33.
The alcA∷Arath;CDKA;1-HA cassette was introduced into pGreen0029. For this the EcoRI restriction site was added to a triple HA tag by PCR using primer set GCGGTAAATCTAGCAGTGCCTCAT and TAGCGAATTCACTGAGCAGCGTAATCTGGAA and pUC-HA as a template (gift from Dr. Laci Bogre), and PstI/NotI restriction sites were added to CDKA;1 by PCR using the primer set AAACTGCAGATGGATCAGTACGAGAAAGTTGAG (CDKA;1-PstI) and AAAGCGGCCGCCAGGCATGCCTCCAAGATCCTTG (CDKA;1-NotI) and pRS97 as template (gift from Dr. Robert Sablowski). The HA PCR product was digested with NotI/EcoRI and the Arath;CDKA;1 PCR product was digested with PstI/NotI, and both fragments were introduced into pL4 (gift from Syngenta, Norwich, UK), generating pGR40. A HindIII partially digested cassette from pGR40, containing alcA∷Arath;CDKA;1-HA-35S terminator, was inserted into pGreen0029, generating pGR42 (Supplemental Fig. 1).
The alcA∷ Arath;CDKA;1-GFP cassette was introduced into pGreen0029. For this, NotI/EcoRI restriction sites were added to a GFP tag by PCR using primer set CAGGGCGGCCGCGGGAGTAAAGGAGAAGAA and CTCGAATTCTTTATTTGTATAGTTCATCCATCGCA and pMCB5 as a template (Fernandez-Abalos et al., 1998), and PstI/NotI restriction sites were added to CDKA;1 by PCR using the CDKA;1-PstI and CDKA;1-NotI primer set as for amplification using pRS97 as template. The GFP PCR product was digested with NotI/EcoRI and the Arath;CDKA;1 PCR product was digested with PstI/NotI, and both fragments were introduced into pL4, generating pGR6. A HindIII partially digested cassette from pGR11, containing alcA∷CDKA;1-GFP -35S terminator, was inserted into pGreen0029, generating pGR14 (Supplemental Fig. 1).
Plant Material, Growth Conditions, Transformation, and Cell Culture Synchronization
The LeGUS20 transgenic tomato (Lycopersicon esculentum L. var. Ailsa Craig Mill.) line, transformed with pSRN∷pAGS∷kanR, has been described previously (Garoosi, 1998; G.A. Garoosi, M.G. Salter, M.X. Caddick, and A.B. Tomsett, unpublished data). For induction experiments, soil-grown seedlings (16 h light 300 μmol m−2 s−1, 25°C ± 2°C and 8 h dark, 16°C ± 2°C) were carefully removed, rinsed, and placed in 120 mL of 0.05% (w/v) Miracle-Gro (Marysville, OH) containing the required concentration of ethanol in a 400-mL Magenta pot. Callus was established from explants of young leaves of LeGUS20 on Murashige and Skoog medium (Murashige and Skoog, 1962), containing 0.8% agar, 2,4-dichlorophenoxyacetic acid (2 μg mL−1), 6-benzylaminopurine (0.5 μg mL−1), and kanamycin (80 μg mL−1) at 26°C under low light (150 μmol m−2 s−1) for 16 h light and an 8-h dark photoperiod, and subcultured at intervals of 2 to 3 weeks. Tomato cell suspension cultures were initiated from callus, as described by Patil et al. (1994), in the same medium (as callus) but lacking agar and kanamycin. It was maintained by dilution (1:5) every 2 weeks and incubated at 25°C and with shaking at 120 rpm in the dark. Note that ethanol was not used as a solvent for any components of the tissue culture media.
A rapidly growing suspension culture of tobacco BY-2 cells (Nicotiana tabacum cv BY-2) was maintained by weekly dilution (1:100) of culture into fresh medium (Nagata et al., 1992) and cultured at 27°C and 130 rpm in the dark. The pGreen plasmids described above were transformed into the Agrobacterium tumefaciens strain GV3101, which had previously been transformed with pSoup (Hellens et al., 2000) by electroporation.
Following cocultivation of BY-2 with pGR-35S∷alcR-GR harboring A. tumefaciens, stable transformants of BY-2 were selected on petri plates containing 0.4% Phytagel in fresh culture medium, supplemented with carbenicillin (500 μg/mL) and hygromycin (41.6 μg/mL). The resultant ALCR-GR transformants were sequentially transformed with pGR32, pGR33, pGR14, or pGR42, and secondary transformants were selected for on media containing plates supplemented with kanamycin (200 μg/mL; An, 1987). After 3 to 4 weeks, transgenic calli were recovered and replated on fresh Phytagel petri plates, as above. Individual calli were then cultured in liquid medium with antibiotics. Carbenicillin was used for three rounds of subculturing.
Chemicals and Induction
To induce the unmodified alc system, the appropriate concentration of ethanol (v/v) was added to the growth medium. Induction of the alc-GR system was achieved using Dex (Sigma-Aldrich, St. Louis), dissolved in either ethanol or DMSO. For selecting alc-GR lines with high inducible expression, portions of calli/clones were induced overnight in microfuge tubes or 96-well microtitre plates with 500 μL of fresh media containing Dex/DMSO or DMSO and assayed for gene expression as appropriate. Cultures were induced by addition of dilutions of Dex/DMSO or DMSO as described in the results.
RNA Extraction and RT-PCR
RNA was isolated using magnetic Oligo dT Dynabeads (Dynal, Bromborough, UK). Approximately 50 mg of cell pellet frozen in liquid nitrogen were ground in 100 μL of Lysis/Binding buffer (Dynal), centrifuged 1 min at 13,000 rpm in a benchtop centrifuge, then 50 μL of supernatant was added to 20 μL of Dynabeads prepared accordingly to manufacturer's instructions, incubated with gentle agitation for 5 min at room temperature to allow annealing of mRNA to the Oligo dT on the Dynabeads. Dynabeads were separated and washed accordingly to manufacturer's instructions, then resuspended in 20 μL of reaction mixture for RT (Omniscript, Qiagen USA, Valencia, CA) and incubated for 1 h at 37°C. PCR reactions were performed in a reaction volume of 25 μL (Taq Master Mix, Qiagen) with 4 μL of the cDNA reaction mixture containing suspended magnetic beads.
GAPDH amplification was used as loading control. The oligonucleotide primer pairs had the following sequences: alcR-forward CTCTAAATCCTTCGCAACCAGC and alcR-reverse GGACGTTTTGGAGAGCATCG for amplification of fragment 400 bp; and GAPDH -forward GGTTTGGCATTGTGGAGGGTC and GAPDH-reverse CCCTCCGATTCCTCCTTGATTGC for amplification of fragment 304 bp. After 15 to 20 rounds of amplification, with primer annealing temperature of 55°C, 10-μL samples of the PCR reaction mixture were separated on a 1% (w/v) agarose gel.
Protein Extraction and Western-Blotting Analysis
Liquid nitrogen-frozen BY-2 cell pellets were homogenized in microfuge tubes in extraction buffer (50 mm Tris-HCl, 5 mm EDTA, 5 mm NaF, 0.1% [v/v] Triton X-100, pH 7.5), at a volume of 1 μL/mg of cells; before quantification (Bradford, 1976), 20 μg of soluble extract was loaded per lane. CDKA and GFP proteins were detected on western blots using a mouse anti-PSTAIR monoclonal antibody (Sigma-Aldrich).
GUS Staining
BY-2 cells were suspended in GUS-staining buffer (Jefferson et al., 1987) and incubated overnight at 37°C. GUS-stained cells were washed and suspended in 70% (v/v) ethanol prior to photography. Cells were visualized by microscopy using a Nikon E600 microscope (Tokyo).
GUS and ADH Quantification
For GUS, protein was extracted from liquid nitrogen frozen plant cells/tissue in microfuge tubes by homogenization in GUS extraction buffer (50 mm NaH2PO4, 10 mm EDTA, 0.1% [v/v] Triton X-100, 1.0 g L−1 Sarcosyl). The GUS activity was determined with a fluorometric assay using 4-methylumbelliferyl β-d-glucuronidase (4MU; Sigma-Aldrich) as a substrate (Jefferson et al., 1987). GUS activity was quantified using either a Perkin-Elmer LS30 (tomato; Perkin-Elmer Applied Biosystems, Foster City, CA) or a Tecan Safire (BY-2) fluorimeter (Zurich) with 365 nm excitation and 455 nm emission wavelengths. ADH activity was determined as described by Rumpho and Kennedy (1981). Total soluble protein was determined as described by Bradford (1976).
Fluorescence Microscopy
The GFP fluorescence of Arath;CDKA;1-GFP and erGFP-expressing BY-2 cells was visualized using a Nikon E600 microscope with excitation with 465 to 495 nm and emission filter 515 to 555 nm.
Acknowledgments
We thank Wolfgang Schuch for initiating this project, and Andrew Hutchins, Grant Calder, and Max Bush for providing technical assistance and advice, and for critically reading the manuscript. In addition, we thank Jan Traas, Lieve Laurens, Hilde Nelissen, and Emma Pilling for critical reading.
This work was supported by a Biotechnology and Biological Sciences Research Council Industrial Case (Syngenta-sponsored) studentship and Marie Curie predoctoral fellowship (to G.R.R.), and a studentship from the Government of Iran (G.A.G.).
The online version of this article contains Web-only data.
References
- Ait-ali T, Rands C, Harberd NP (2003) Flexible control of plant architecture and yield via switchable expression of Arabidopsis gai. Plant Biotechnol J 1: 337–343 [DOI] [PubMed] [Google Scholar]
- An G (1987) Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol 153: 292–303 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Caddick MX, Greenland AJ, Jepson I, Krause KP, Qu N, Riddell KV, Salter MG, Schuch W, Sonnewald U, Tomsett AB (1998) An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nat Biotechnol 16: 177–180 [DOI] [PubMed] [Google Scholar]
- Chen S, Hofius D, Sonnewald U, Bornke F (2003) Temporal and spatial control of gene silencing in transgenic plants by inducible expression of double-stranded RNA. Plant J 36: 731–740 [DOI] [PubMed] [Google Scholar]
- Deveaux Y, Peaucelle A, Roberts GR, Coen E, Simon R, Mizukami Y, Traas J, Murray JAH, Doonan JH, Laufs P (2003) The ethanol switch: a tool for tissue-specific gene induction during plant development. Plant J 36: 918–930 [DOI] [PubMed] [Google Scholar]
- Drew MC (1997) Oxygen deficiency and root metabolism: injury and acclimation under hypoxia and anoxia. Annu Rev Plant Physiol Plant Mol Biol 48: 223–250 [DOI] [PubMed] [Google Scholar]
- Fernandez-Abalos JM, Fox H, Pitt C, Wells B, Doonan JH (1998) Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. Mol Microbiol 27: 121–130 [DOI] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J (2004) Plant responses to hypoxia: Is survival a balancing act? Trends Plant Sci 9: 449–456 [DOI] [PubMed] [Google Scholar]
- Garoosi GA (1998) A chemical gene switch for use in transgenic plants. PhD thesis. University of Liverpool, Liverpool, UK
- Gatz C (1997) Chemical control of gene expression. Annu Rev Plant Physiol Plant Mol Biol 48: 89–108 [DOI] [PubMed] [Google Scholar]
- Guerineau F, Mullineaux PM (1993) Plant transformation and expression vectors. In RD Croy, ed, Plant Molecular Biology Labfax. BIOS Scientific Publishers, Oxford, pp 121–148
- Heim R, Prasher DC, Tsien RY (1994) Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA 91: 12501–12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Ingram GC, Goodrich J, Wilkinson MD, Simon R, Haughn GW, Coen ES (1995) Parallels between UNUSUAL FLORAL ORGANS and FIMBRIATA, genes controlling flower development in Arabidopsis and Antirrhinum. Plant Cell 7: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James E, Lee JM (2001) The production of foreign proteins from genetically modified plant cells. Adv Biochem Eng Biotechnol 72: 127–156 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker BH, Chu C, Sonnewald U, Willmitzer L, Fernie AR (2003) In plants the alc gene expression system responds more rapidly following induction with acetaldehyde than with ethanol. FEBS Lett 535: 136–140 [DOI] [PubMed] [Google Scholar]
- Laufs P, Coen E, Kronenberger J, Traas J, Doonan J (2003) Separable roles of UFO during floral development revealed by conditional restoration of gene function. Development 130: 785–796 [DOI] [PubMed] [Google Scholar]
- Lee I, Wolfe DS, Nilsson O, Weigel D (1997) A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr Biol 7: 95–104 [DOI] [PubMed] [Google Scholar]
- Maizel A, Weigel D (2004) Temporally and spatially controlled induction of gene expression in Arabidopsis thaliana. Plant J 38: 164–171 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Science 245: 371–378 [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S (1992) Tobacco BY-2 cell line as the “Hela” cell in the cell biology of higher plants. Int Rev Cytol 132: 1–30 [Google Scholar]
- Nara Y, Kurata H, Seki M, Taira K (2000) Glucocorticoid-induced expression of a foreign gene by the GVG system in transformed tobacco BY-2 cells. Biochem Eng J 6: 185–191 [DOI] [PubMed] [Google Scholar]
- Padidam M (2003) Chemically regulated gene expression in plants. Curr Opin Plant Biol 6: 169–177 [DOI] [PubMed] [Google Scholar]
- Patil RS, Davey MR, Power JB (1994) Highly efficient plant regeneration from mesophyll protoplasts of Indian field cultivars of tomato (Lycopersicon esculentum). Plant Cell Tissue Organ Cult 36: 255–258 [Google Scholar]
- Picard D (1994) Regulation of protein function through expression of chimaeric proteins. Curr Opin Biotechnol 5: 511–515 [DOI] [PubMed] [Google Scholar]
- Roslan HA, Salter MG, Wood CD, White MR, Croft KP, Robson F, Coupland G, Doonan J, Laufs P, Tomsett AB, et al (2001) Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J 28: 225–235 [DOI] [PubMed] [Google Scholar]
- Rumpho ME, Kennedy RA (1981) Anaerobic mechanism in germinating seeds of Echinochloa crus-galli (barnyard grass): metabolic and enzyme studies. Plant Physiol 68: 165–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RW, Meyerowitz EM (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92: 93–103 [DOI] [PubMed] [Google Scholar]
- Salter MG, Paine JA, Riddell KV, Jepson I, Greenland AJ, Caddick MX, Tomsett AB (1998) Characterisation of the ethanol-inducible alc gene expression system for transgenic plants. Plant J 16: 127–132 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sonnewald U, Brauer M, von Schaewen A, Stitt M, Willmitzer L (1991) Transgenic tobacco plants expressing yeast-derived invertase in either the cytosol, vacuole or apoplast: a powerful tool for studying sucrose metabolism and sink/source interactions. Plant J 1: 95–106 [DOI] [PubMed] [Google Scholar]
- Sweetman JP, Chu C, Qu N, Greenland AJ, Sonnewald U, Jepson I (2002) Ethanol vapor is an efficient inducer of the alc gene expression system in model and crop plant species. Plant Physiol 129: 943–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zhou X, Wang X (2003) Chemically regulated expression systems and their applications in transgenic plants. Transgenic Res 12: 529–540 [DOI] [PubMed] [Google Scholar]
- Waring RB, May GS, Morris NR (1989) Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene 79: 119–130 [DOI] [PubMed] [Google Scholar]
- Zuo J, Chua NH (2000) Chemical-inducible systems for regulated expression of plant genes. Curr Opin Biotechnol 11: 146–151 [DOI] [PubMed] [Google Scholar]