Abstract
The application of DNA array technology and chromatographic separation techniques coupled with mass spectrometry to transcriptomic and metabolomic analyses in plants has resulted in the generation of considerable quantitative data related to transcription and metabolism. The integration of “omic” data is one of the major concerns associated with research into identifying gene function. Thus, we developed a Web-based tool, KaPPA-View, for representing quantitative data for individual transcripts and/or metabolites on plant metabolic pathway maps. We prepared a set of comprehensive metabolic pathway maps for Arabidopsis (Arabidopsis thaliana) and depicted these graphically in Scalable Vector Graphics format. Individual transcripts assigned to a reaction are represented symbolically together with the symbols of the reaction and metabolites on metabolic pathway maps. Using quantitative values for transcripts and/or metabolites submitted by the user as Comma Separated Value-formatted text through the Internet, the KaPPA-View server inserts colored symbols corresponding to a defined metabolic process at that site on the maps and returns them to the user's browser. The server also provides information on transcripts and metabolites in pop-up windows. To demonstrate the process, we describe the dataset obtained for transgenic plants that overexpress the PAP1 gene encoding a MYB transcription factor on metabolic pathway maps. The presentation of data in this manner is useful for viewing metabolic data in a way that facilitates the discussion of gene function.
The total number of metabolites produced in the plant kingdom is estimated to exceed 200,000 (Dixon and Strack, 2003). Given the ever-increasing applications of plant metabolites, either directly or indirectly as foods, medicines, and industrial materials, the new research area referred to as “metabolomics,” in which metabolites are investigated holistically in conjunction with functional genomics, will have a strong influence on plant biotechnology and breeding. Comprehensive analyses of transcripts can be performed using DNA array technology (Aharoni and Vorst, 2002; Donson et al., 2002), especially for plant species such as Arabidopsis (Arabidopsis thaliana; Arabidopsis Genome Initiative, 2000) and rice (Oryza sativa) in which genome sequencing has been completed (Feng et al., 2002; Goff et al., 2002; Sasaki et al., 2002; Yu et al., 2002) or species for which expressed sequence tag sequences are available, such as soybean (Glycine max), wheat (Triticum aestivum), barley (Hordeum vulgare), and tomato (Solanum esculentum; http://www.ncbi.nlm.nih.gov/dbEST/). Recently, the application of compound separation techniques coupled to mass spectrometry (MS), such as gas chromatography-MS, liquid chromatography (LC)-MS, and capillary electrophoresis-MS, have been applied to the comprehensive analysis of plant metabolites (Sumner et al., 2003; Sato et al., 2004). These analyses have produced large amounts of transcriptomic and metabolomic (“omic”) quantitative data, the integration of which is one of the major concerns for researchers identifying genes and for “omic” approaches to systems biology.
Several metabolic pathway databases are available to facilitate our understanding of transcriptome and metabolome data. The Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.ad.jp/kegg/) has a pathway database (PATHWAY) that contains information of metabolites and genes, as well as graphical representations of metabolic pathways and complexes derived from various biological processes (Goto et al., 2002; Kanehisa et al., 2002). The metabolic pathways for 218 organisms, including Arabidopsis and rice, have been constructed to date. Organism-specific metabolic pathway maps can be generated according to assignment information on the KEGG/GENES database. The metabolic pathway reference database named MetaCyc (Krieger et al., 2004) contains pathways from 302 organisms (December 2004; http://metacyc.org/). The Arabidopsis pathway database AraCyc (http://www.arabidopsis.org/tools/aracyc/) was constructed by adding plant-specific pathways and reactions to basic pathway sets in the MetaCyc pathway collection (Mueller et al., 2003). While the comprehensive database has metabolic pathway data that is representative of the plant kingdom, the pathways and reactions involved in alkaloid and isoflavonoid biosyntheses are not well represented, as these are not found in Arabidopsis.
A relatively common characteristic of plants is that several homologous gene products are often assigned to a single enzymatic reaction. Multigene families are considerably more prevalent among plant genomes than among animal genomes (Arabidopsis Genome Initiative, 2000). Recent research has revealed that multiple genes are not simple repeats, but rather they exhibit diverse gene expression and, consequently, have a diverse array of functions. For example, the XTH gene family, a group of genes that encode xyloglucan endotransglucosylase/hydrolase involved in xyloglucan metabolism, is constituted by 33 and 29 member genes in Arabidopsis (Yokoyama and Nishitani, 2001) and rice (Yokoyama et al., 2004), respectively. The individual gene members exhibit tissue-specific and growth stage-dependent expression in dicot and monocot plants. In strawberry fruit (Fragaria × ananassa cv Elsanta), 5,884 masses are detected using Fourier-transform ion-cyclotron MS (Aharoni et al., 2002). Gene duplications might explain the extent of chemical diversity found in secondary metabolism of plants. Plants have several sets of high-copy genes, such as those involved in hydroxylation and glycosylation. For example, the number of genes responsible for various hydroxylation steps in the P450 gene family is 273 in Arabidopsis and more than 400 genes in rice, respectively (Nelson et al., 2004). In the Arabidopsis genome, more than 300 genes code for glycosyltransferase and 63 for acyltransferase, respectively. Given that the hydroxylation of carbon skeletons is the precursor of glycosylation and acylation, the high copy numbers of these genes are thought to facilitate the considerable diversity of plant metabolites. However, the contribution of individual gene behaviors and functions are not fully understood in plant metabolism.
The diagrammatic representation of individual transcript data together with enzymatic reaction information on a metabolic pathway map that also contains quantitative information of the substrate and product might aid in our interpretation of the gene function. One of the tools on the AraCyc database has the ability to paint data values of transcripts onto the metabolic overview diagram. However, when multigene families are thought to be involved in single reactions, only representative data are used for the painting. Individual transcript data are not shown on individual metabolic pathway maps. A recent version of AraCyc can also represent metabolite data but only onto the overview diagram. Recently, a user-driven tool, MAPMAN, was developed for representing transcript data on the pictorial diagrams, in which all Arabidopsis genes were categorized on the basis of biological function (Thimm et al., 2004). Metabolites were also categorized on pictorial diagrams for representation of quantitative values of each metabolite. However, as MAPMAN only provides several metabolic pathways, users must prepare their own diagrams using the user-driven tool. As a complementary approach to AraCyc and MAPMAN, we created a set of comprehensive metabolic pathway maps for Arabidopsis, in which 1,263 metabolic reactions were grouped together. We also developed a Web-based tool for the analysis of plant metabolic pathways, KaPPA-View, for displaying quantitative data for individual transcripts and metabolites on the same set of metabolic pathway maps. To facilitate dynamic document generation displaying rich graphical features and for the user's edition of the pathway maps, we adapted Scalable Vector Graphics (SVG) format to represent transcript/metabolite data. We demonstrated the usefulness of the KaPPA-View tool by displaying the dataset for transgenic plants that overexpress the PAP1 gene that encodes a MYB transcription factor (Tohge et al., 2005) on the metabolic pathway maps.
RESULTS
Overall Features of the KaPPA-View Tool
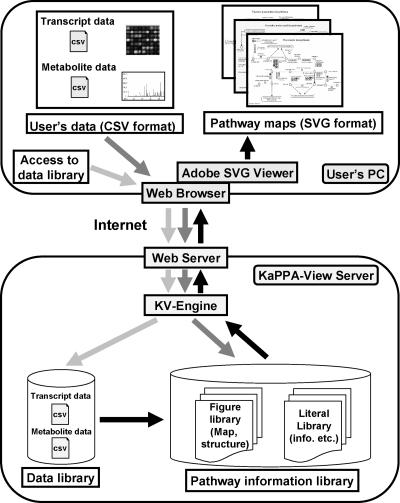
We designed a Web-based tool for the analysis of plant metabolic pathways, called KaPPA-View, with which users can display the changes of individual transcripts and metabolites on the comprehensive metabolic pathway maps that we prepared. In addition, each metabolic pathway map was designed to serve as a source of information for metabolites and genes involved in the pathway. To meet the computational requirements of such a task, we set up the program KV-Engine to manage the pathway information library that generates metabolic pathway maps in SVG format (see below), the data library, and the Web server on the KaPPA-View server (Fig. 1). Using Internet browsers, users can access and apply the tool to their own datasets formatted as Comma Separated Value (CSV) files. In addition, users can access the transcript and metabolite data libraries that are uploaded from the KaPPA-View administrator. Given that KaPPA-View is a JAVA application, it is platform independent and can be used on a variety of popular operation systems (OS), such as Windows 2000/XP (Microsoft, Redmond, WA) and Macintosh OS 9/X (Apple Computer, Cupertino, CA), all of which have the SVG plug-in SVG Viewer supplied by Adobe Systems (San Jose, CA). However, we recommend use of Windows 2000/XP and the Web browser Internet Explorer 6.0 or higher, which allows users to access the full functions of KaPPA-View. Macintosh users can use the basic functions but cannot access directly information windows from each SVG tag on maps, while information windows are accessible from the element list provided on the screen.
Figure 1.
General workflow for analysis of metabolic pathways using the KaPPA-View tool. A user sends a CSV-formatted dataset of transcripts and/or metabolites from their personal computer (PC) to the Web server through the Internet. The application engine (KV-Engine) processes the data and interacts with the pathway information library. The SVG-formatted files generated by the engine are returned to the user's PC through the Adobe SVG Viewer installed on the PC. Users also can access the transcript and metabolite data library located on the KaPPA-View server.
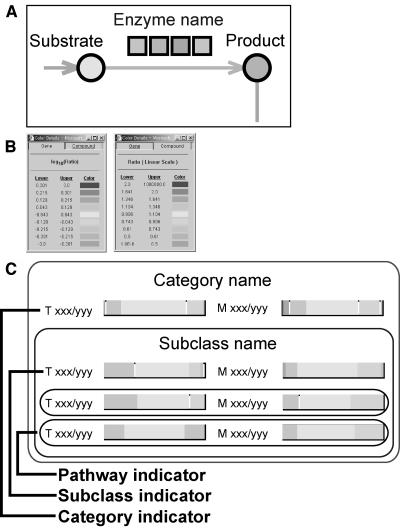
We used symbols to depict the various enzymatic reactions and transcripts involved in the metabolic pathway maps (Fig. 2A). In this way, circles were used to represent substrates and the reaction products, arrows for reactions, and squares for transcripts involved in, or putatively assigned to, the reaction. The changes in individual transcripts and metabolites were represented using squares and circles of different colors, respectively, that were defined by color charts (Fig. 2B). We also designed color pathway indicators for overall changes to transcripts and metabolites in individual pathways, subclasses, and categories (Fig. 2C).
Figure 2.
Symbolic representation of enzymatic reactions and transcripts on the metabolic pathway maps. A, Diagrammatic representation of an enzymatic reaction with substrate and the reaction product represented by circles, an arrow representing the reaction, and squares for the transcripts involved, or putatively assigned to be involved, in the reaction. Names for the substrate, product, and enzyme encoded by transcripts are also given. The colors of the symbols change according to quantitative data of either transcripts or metabolites. B, The color charts for defining quantitative data for transcripts (left) and metabolites (right). C, Three color levels for pathway maps, subclass, and metabolism category for transcripts (T) and metabolites (M). The numbers of either transcripts or metabolites analyzed by the dataset (xxx) and listed on the metabolic pathway map (yyy) are also indicated to the left of the pathway indicators.
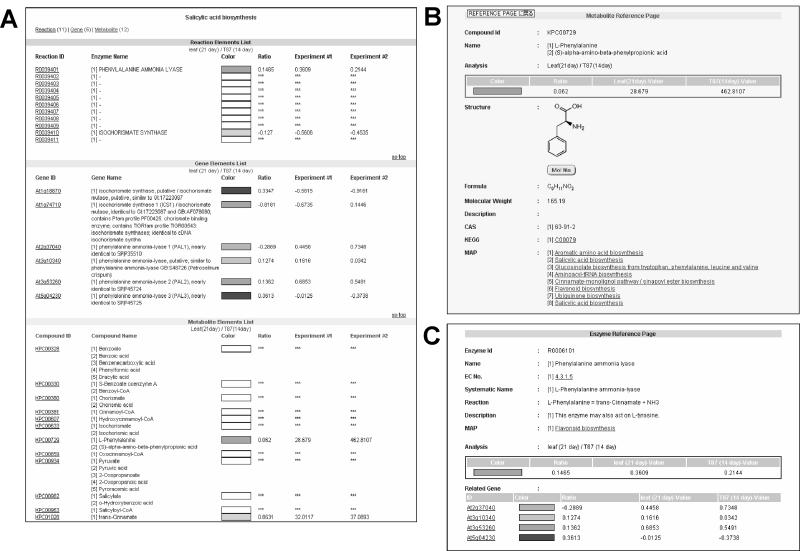
Information on the metabolites and genes in individual pathways can be retrieved for each map in pop-up windows (Fig. 3). An element list listing all of the information of the elements (the reaction, genes involved, and metabolites) on the map currently being displayed on a user's browser can be shown on the screen (Fig. 3A). This means that users are capable of knowing the quantitative values and other information for each transcript or metabolite listed. Using the element list, users can select and display the metabolite reference page (Fig. 3B); the gene reference page, in which each gene identifier is linked to the relevant gene information page of The Arabidopsis Information Resource database (http://www.arabidopsis.org/; data not shown); and the enzyme reference page (Fig. 3C). Users can extract all of the pathways that relate to the element currently being displayed by clicking on symbols of the elements.
Figure 3.
Pop-up windows for displaying information on the metabolic pathway maps. A to C, The element list containing all of the information available for the elements (the reaction, genes involved, and metabolites) on the map being displayed on the user's browser (A), the metabolite reference page (B), and the enzyme reference page (C). The gene reference page (not shown) can also be chosen from the element lists.
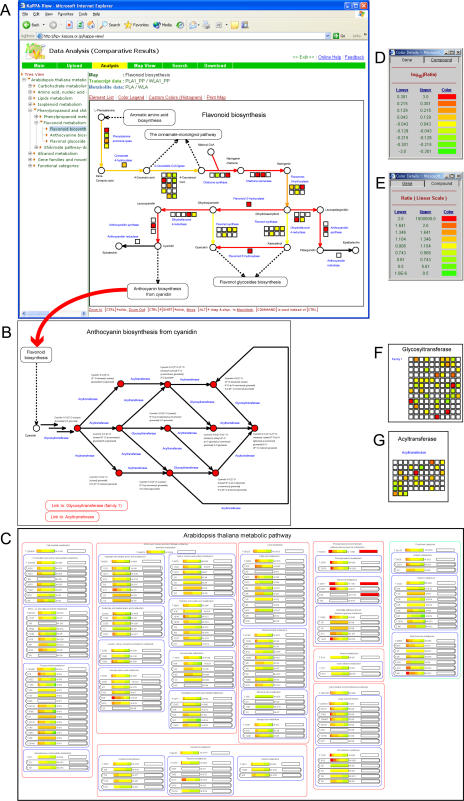
The manner in which diagrams are displayed gives users the opportunity to grasp the contents of the image at a glance, which is useful in plant research as multiple transcripts are often associated with a single reaction (Fig. 4A). Furthermore, the names of pathways immediately up or downstream are indicated on each map, and related pathway maps can be displayed in pop-up windows at the users request (Fig. 4B). The metabolic pathways thus generated were classified as being one of 25 subclasses that were further subdivided under seven major metabolic categories (described below). Using this classification, all of the pathway indicators are represented on the bird's-eye map (Fig. 4C), allowing users to appreciate the overall picture of the changes in transcripts and metabolites. Furthermore, users can access individual maps by clicking on the pathway indicators on the bird's-eye map.
Figure 4.
Representation of the transcript and metabolite dataset for wild type and PAP1-overexpressing plants. The dataset for wild-type and transgenic plants (Tohge et al., 2005) were displayed on the metabolic pathway maps for flavonoids (A) and anthocyanins (B) and the bird's-eye map (C). The changes in individual transcripts and metabolites on the map are shown as squares and circles with different colors matched using the color charts (D and E). The changes in the transcripts for glycosyltransferase (F) and acyltransferase (G) are shown.
To achieve a dynamic graphical representation of the quantitative changes in transcripts and/or metabolites, we adapted the SVG format for map drawing. We also ensured that the dimensions of the maps were compliant with standard computational viewing sizes and that printing A4 or letter-sized maps was possible. Due to the vector nature of SVG format, the sizes of maps generated using the program can be changed on screen without any loss of picture quality using a browser function.
Plant Metabolic Pathway Maps
We prepared a set of comprehensive metabolic pathway maps for Arabidopsis. These maps include 1,263 enzymatic reactions that could be classified into seven major metabolic categories with 25 subclasses (Table I). We classified the enzymatic reactions along with the carbon flows derived from the intake carbon dioxide by plants during photosynthesis. Therefore, we avoided functional categories such as “plant hormone” and “secondary metabolism” in our classification, but rather positioned such reactions as branches of the metabolic flows. For example, the biosynthesis pathway of brassinosteroids, which are known to be plant hormones, was classified as a subclass of isoprenoid metabolism. Isoprenoid metabolism and phenylpropanoid metabolism, which are often categorized in secondary metabolism, were classified in independent categories. Furthermore, to avoid the production of fragmentary maps and facilitate presentation of the image of transcript/metabolite changes in a particular metabolic pathway, we integrated related metabolic reactions into single maps. However, considerable care was taken to avoid too much integration, e.g. the mevalonate terpenoid pathway, the nonmevalonate terpenoid pathway, the isoprenoid pathway, the sterol pathway, the carotenoid and abscisic acid pathway, the tocopherol pathway, and the brassinosteroid pathway, which are included in a single map as in steroid biosynthesis in KEGG/PATHWAY, are all classified independently. Consequently, KaPPA-View has fewer maps (n = 130) than AraCyc (n = 220) but more than KEGG (n = 98 for Arabidopsis). To help users find a metabolic pathway based on functional category, however, we included link indicators to functional categories in a classification list. In cases where the same metabolites are localized in distinct subcellular compartments, such as lipid metabolism in the cytoplasm and plastids, both pathways were shown using distinct metabolic pathway maps. Otherwise, information of cellular location was not explicitly given on the maps.
Table I.
List of Arabidopsis metabolic pathways in KaPPA-View
We classified the 130 metabolic pathways in seven major categories with 25 subclasses.
| Category Name/Subclass Name/Pathway Name |
|---|
| Carbohydrate metabolism |
| CO2 fixation and central carbohydrate metabolism |
| Calvin cycle |
| Glycolate pathway |
| Glycolysis/gluconeogenesis |
| Phosphoenolpyruvate and pyruvate metabolism |
| TCA cycle |
| Glyoxylate cycle |
| Glycerol metabolism |
| Mono-, di-, and oligosaccharide metabolism |
| Hexose phosphate pool |
| Pentose phosphate cycle |
| Sucrose metabolism |
| Trehalose metabolism |
| UDP-sugar metabolism |
| GDP-sugar and ascorbate metabolism |
| dTDP-sugar biosynthesis |
| Inositol phosphate metabolism |
| Polysaccharide metabolism |
| Starch biosynthesis |
| Starch degradation |
| Cellulose biosynthesis |
| Cellulose degradation |
| Callose/glucan biosynthesis |
| Callose/glucan degradation |
| Xyloglucan biosynthesis and modification |
| Xyloglucan degradation |
| Homogalacturonan biosynthesis |
| Homogalacturonan degradation |
| Rhamnogalacturonan I biosynthesis |
| Rhamnogalacturonan I degradation |
| Rhamnogalacturonan II biosynthesis |
| Rhamnogalacturonan II degradation |
| Miscellaneous carbohydrate metabolism |
| Aminosugars metabolism |
| Pyridoxal 5-phosphate metabolism |
| Amino acid, nucleic acid, and nitrogen-containing derivative metabolism |
| Aspartate and related amino acid metabolism |
| Aspartate and asparagine metabolism |
| Lysine, threonine, and methionine biosynthesis |
| Lysine degradation |
| Methionine metabolism |
| Ethylene biosynthesis from methionine |
| Threonine and methylglyoxal metabolism |
| Pyridine nucleotide biosynthesis |
| Glutamate and related amino acid metabolism |
| Glutamate and glutamine metabolism/nitrate assimilation |
| Arginine and proline metabolism |
| Proline and 4-hydroxyproline metabolism |
| Biosynthesis of chlorophyll, proto, and siroheme |
| Leucine, valine, isoleucine, and alanine metabolism |
| Leucine, valine, isoleucine, and alanine biosynthesis |
| Leucine, valine, and isoleucine degradation |
| Aromatic amino acid metabolism |
| Aromatic amino acid biosynthesis |
| Tryptophan metabolism |
| Tyrosine metabolism |
| Salicylic acid biosynthesis |
| Auxin biosynthesis |
| Camalexin biosynthesis |
| Serine, glycine, and cysteine metabolism |
| Serine and glycine metabolism |
| Sulfur and cysteine metabolism |
| Glycine degradation |
| Homocysteine and cysteine interconversion |
| l-Cysteine degradation |
| Glutathione biosynthesis |
| Histidine and nucleic acid metabolism |
| Histidine metabolism |
| Purine biosynthesis |
| Purine metabolism |
| Ureide metabolism |
| Pyrimidine biosynthesis |
| Pyrimidine metabolism |
| Cytokinin metabolism |
| Glucosinolate metabolism |
| Methionine chain elongation pathway |
| Glucosinolate biosynthesis from chain elongated methionine |
| Glucosinolate biosynthesis from tryptophan, phenylalanine, leucine, and valine |
| Secondary modification of methylthioalkyl glucosinolate |
| Secondary modification of indole-3-methyl glucosinolate |
| Miscellaneous amino-acid-related metabolism |
| Aminoacyl-tRNA biosynthesis |
| Pantothenate and coenzyme A biosynthesis |
| Betaine biosynthesis |
| Folic acid biosynthesis |
| Formyl THF biosynthesis |
| Lipids metabolism |
| Fatty acid metabolism |
| Fatty acid biosynthesis |
| Fatty acid α-oxidation pathway |
| β-Oxidation of saturated fatty acid |
| β-Oxidation of unsaturated fatty acid |
| Linolenic acid metabolism (plastidial pathway) |
| Linolenic acid metabolism (cytosolic pathway) |
| Linoleic acid metabolism (plastidial pathway) |
| Linoleic acid metabolism (cytosolic pathway) |
| Jasmonic acid biosynthesis |
| Lipoic acid synthesis in mitochondria |
| Sphingolipids synthesis |
| Membrane lipid metabolism |
| Glycerolipid biosynthesis (prokaryotic pathway) |
| Phospholipid biosynthesis (prokaryotic pathway) |
| Glycolipid biosynthesis (prokaryotic pathway) |
| Glycerolipid biosynthesis (eukaryotic pathway) |
| Phospholipid biosynthesis (eukaryotic pathway) |
| Glycolipid biosynthesis (eukaryotic pathway) |
| Glycerolipid biosynthesis in mitochondria |
| Phosphatidyl inositol metabolism |
| Lipases pathway |
| Structural lipid metabolism |
| Cutin biosynthesis |
| Long-chain fatty acid and wax biosynthesis |
| Suberin biosynthesis |
| Storage lipid metabolism |
| Triacylglycerol biosynthesis |
| Triacylglycerol metabolism |
| Isoprenoid metabolism |
| Isoprenoid biosynthesis |
| Nonmevalonate terpenoid biosynthesis |
| Mevalonate pathway |
| Isoprenoid biosynthesis |
| Polyisoprenoid biosynthesis |
| Terpenoid metabolism |
| Monoterpenoid biosynthesis |
| Sesquiterpenoid biosynthesis |
| Gibberellin biosynthesis |
| Triterpenoid biosynthesis |
| Carotenoid and abscisic acid biosynthesis |
| Steroid metabolism |
| Sterol biosynthesis |
| Brassinosteroid biosynthesis |
| Phenylpropanoid and shikimate pathway-derived quinone metabolism |
| Phenylpropanoid metabolism |
| Cinnamate-monolignol pathway/sinapoyl ester biosynthesis |
| Flavonoid metabolism |
| Flavonoid biosynthesis |
| Anthocyanin biosynthesis from cyanidin |
| Flavonol glucoside biosynthesis |
| Shikimate pathway-derived terpenoid quinone metabolism |
| Tocopherol biosynthesis |
| Plastoquinone biosynthesis |
| Phylloquinone biosynthesis |
| Ubiquinone biosynthesis |
| Gene families and miscellaneous pathways |
| Large enzyme families |
| Cytochrome P450 |
| Acyltransferase |
| Glycosyltransferase |
| Glycoside hydrolase |
| Polysaccharide lyases |
| Carbohydrate esterases |
| Peroxidase, class III |
| Miscellaneous pathways |
| Catechol, protocatechuate, 1,4-dichlorobenzene and pentachlorophenol degradation pathway |
| Removal of superoxide radical |
| Carnitine metabolism |
| Thiamine biosynthesis |
| Biotin biosynthesis |
| Indole alkaloid biosynthesis |
In the initial version, we prepared the metabolic pathway maps for the model plant Arabidopsis. Although only a limited number of the metabolic reactions included in our maps have been proven experimentally in Arabidopsis, we included the reactions that are assigned with the Arabidopsis gene annotation. Thus, although alkaloid biosynthesis has not been reported in Arabidopsis, we included the indole alkaloid synthesis pathway under miscellaneous pathways because a putatively assigned gene for indole alkaloid synthesis has been found in Arabidopsis (see below).
The genes assigned to the metabolic reactions in AraCyc and the Arabidopsis metabolic reactions in KEGG/PATHWAY were used as queries to search for the Arabidopsis protein sequences. The genes that were not found in the gene sets were manually refined and incorporated in the metabolic pathway maps. Furthermore, the metabolic reactions that are not included in AraCyc and KEGG/PATHWAY were incorporated on the basis of recent knowledge of plant metabolism, as described in the following comments for each metabolic category.
Carbohydrate Metabolism
The genes involved in cell wall polysaccharide synthesis were cited from Yokoyama and Nishitani (2004). Cell wall polysaccharide biosynthesis, xyloglucan biosynthesis, and rhamnogalacturonan I and II biosyntheses were included in the metabolic pathway maps. Some gene families, such as glycoside hydrolase, glycosyltransferase, and carbohydrate esterase, are thought to be included partly in cell wall polysaccharide synthesis, but the functions of most of these genes have not been determined. Therefore, these genes were classified in the subclass “large enzyme families” in the miscellaneous pathway category.
Amino Acids, Nucleic Acids, and Nitrogen-Containing Derivative Metabolism
On the basis of the biosynthetic origins of carbon skeletons, amino acid metabolism was divided into eight subclasses (Table I). For simplicity, Ile biosynthesis was classified into a branched-chain amino acid pathway, which includes Leu and Val, although the origin of the carbon skeleton of Ile is distinct from that of the other amino acids. Nitrogen fixation and sulfur assimilation were also classified under the amino acid metabolism category because nitrogen and sulfur are incorporated into Gln/Asn and Cys, respectively. Amino acyl-tRNA synthesis was also classified in this category. It has been proposed that camalexin, which is present as a phytoalexin in Arabidopsis, is synthesized from Trp (Glawischnig et al., 2004). Consequently, we classified the pathway as being amino acid metabolism.
Biosynthesis of the nitrogen- and sulfur-containing glucosinolate metabolites, which are known to have physiological activity in the Brassicaceae (which includes Arabidopsis), was cited from the review article by Wittstock and Halkier (2002). In the process of side-chain elongation of amino acids, glucosinolate skeleton formation, and their modification, Phe, Val, Leu and Ile, Met, and Trp were all reported as the precursors in Arabidopsis.
Lipid Metabolism
Lipid metabolism was classified into five subclasses according to the Arabidopsis Lipid Gene Database (Beisson et al., 2003; http://www.plantbiology.msu.edu/lipids/genesurvey/index.htm). Lipoxygenation reactions were classified into four metabolic pathways on the basis of the subcellular localization of metabolites.
Isoprenoid Metabolism
The isoprenoid synthetic pathways in Arabidopsis and the genes involved in the pathways were cited from the review article by Lange and Ghassemian (2003). Intermediate reactions in brassinosteroid biosynthesis and the stigmasterol biosynthesis pathway were included in the metabolic pathway maps on the basis of recent findings regarding their biosynthesis. Given that three monoterpenes and 11 sesquiterpenes were identified in Arabidopsis (Chen et al., 2003), the biosynthesis pathways were included.
Phenylpropanoid Metabolism
The biosynthesis of monolignols was cited by Raes et al. (2003) and Goujon et al. (2003). The biosynthetic pathways for lignans, neolignans, and norlignans were not included in our metabolic pathway maps, as these metabolites and the genes assigned to the pathways have not yet been identified in Arabidopsis.
As several flavonol glucosides (Graham, 1998; Veit and Pauli, 1999) were reported in Arabidopsis, the biosynthesis reactions were included in the map for the flavonol glucoside biosynthesis pathway. We prepared a pathway map for anthocyanin biosynthesis on the basis of recent reports (Bloor and Abrahams, 2002; Tohge et al., 2005).
We included the biosynthesis of tocopherols, plastoquinones, phylloquinones, and ubiquinones in the terpenoid quinone pathway (Lange and Ghassemian, 2003).
Miscellaneous Pathways
Genes whose products are not assigned to individual enzymatic reactions but are thought to be involved in metabolism were classified as being “miscellaneous pathways.” They include the gene families for P450 (http://www.p450.kvl.dk/), glycosyltransferase, glucosidase, and polysaccharide lyase (Carbohydrate-Active enZymes server; http://afmb.cnrs-mrs.fr/CAZY/; Yokoyama and Nishitani, 2004).
While there are no reports of alkaloids in Arabidopsis, the genome contains the genes (At1g74000, At1g74010, At1g74020, At2g41290, At2g41300, At3g57010, At3g57020, At3g57030, and At3g59530) that share homology with the genes for strictosidine synthase, which is involved in the synthesis of indole alkaloids in Eschscholtzia californica. The gene products are also referred to as FAD-binding domain-containing proteins. Therefore, we included the indole alkaloid pathway in miscellaneous pathways.
Presentation of Transcript and Metabolite Data Using Metabolic Pathway Maps
To demonstrate the usefulness of our KaPPA-View tool, we represent quantitative data of transcripts and metabolites of wild-type and the PAP1-overexpressing plants on a bird's-eye map and on individual metabolic pathway maps (Fig. 4). Overexpression of the PAP1 gene encoding a MYB transcription factor in Arabidopsis was recently reported to cause the accumulation of high levels of anthocyanins and to induce the transcription of a set of genes, including those of the flavonoid biosynthesis pathway (Tohge et al., 2005). Tohge and colleagues analyzed transcription by wild-type and PAP1-overexpressing plants with the Arabidopsis Genome ATH1 GeneChip array, which contains 22,810 genes. They also used HPLC-MS (LC-MS) to analyze the levels of flavonoid-related compounds, including anthocyanins, and performed Fourier-transform ion-cyclotron MS for analysis of nontargeted metabolites. These microarray and LC-MS data were then examined by KaPPA-View, and we show here only the dataset for leaves of wild-type and a PAP1-overexpressing line as an example of the usefulness of the KaPPA-View analysis.
The pathway indicators in the bird's-eye map (Fig. 4A) are colored according to a key (Fig. 4, section D for transcripts and section E for metabolites). The bird's-eye map reveals activation of the flavonoid biosynthesis pathway as was reported by Tohge et al. (2005). Moreover, the map highlights specific activation of the pathway among other metabolic pathways, which was previously suggested from the induction of 38 genes by ectopic PAP1 overexpression. This demonstrates the usefulness of the bird's-eye map for reviewing the differences in the transcriptome and metabolome between two plant samples.
The flavonoid synthesis pathway map shows the overall activation of the pathway at the transcriptional level (Fig. 4A), which is consistent with the accumulation of anthocyanins (Fig. 4B) as reported by Tohge et al. (2005). Tohge and his colleagues indicated that, in addition to well-known genes involved in anthocyanin production, several genes with unidentified function or annotated only with putative functions (a putative glycosyltransferase, acyltransferase, glutathione S-transferase, sugar transporters, and transcription factors) were induced by PAP1. The biosynthesis of anthocyanin from cyanidins requires several glycosylation and acylation steps (Fig. 4B). The analysis highlighted that the transcripts for glycosyltransferase and acyltransferase are up-regulated among the gene families analyzed (Fig. 4, F and G), implying that these genes are involved in these enzymatic processes. However, users should not conclude that the functions of these genes are necessarily for anthocyanin biosynthesis based only on the KaPPA-View presentation and without further experimentation. For example, Tohge and colleagues confirmed that the two putative glycosyltransferase genes, At5g17050 and At4g14090, induced by PAP1 expression encoded flavonoid 3-O-glucosyltransferase and anthocyanin 5-O-glucosyltransferase, respectively, based on in vitro enzymatic assays using the recombinant proteins and analysis of anthocyanins in the respective T-DNA-inserted mutants.
DISCUSSION
Given the wealth of transcriptomic data and the increasing amounts of metabolomic data, we designed a tool for the analysis of plant metabolic pathways, KaPPA-View, to display individual quantitative changes in transcripts and/or metabolites on a set of comprehensive metabolic pathway maps. As exemplified by comparative analysis between wild type and transgenic plants that overexpress the PAP1 gene that encodes a MYB transcription factor (Tohge et al., 2005; Fig. 4), simultaneous presentation of both transcripts and metabolites on the bird's-eye and metabolic pathway maps allows users to quickly grasp the differences in the samples being examined. Although the visualization tool is user friendly, intuitive, and simplifies the review of complex results of transcription analyses, the data presented on the metabolic maps should be interpreted with caution; simple conclusions should not be made in the absence of a statistical treatment of the data and/or biochemical experimentation. In this regard, we are currently preparing various transgenic Arabidopsis plants carrying various genes under the control of a strong promoter, and we are comparing these with the wild-type host plants using the KaPPA-View tool. These comparisons will provide an understanding of the effects of the introduced genes on the transcriptome and metabolome, leading to hypotheses on the function of specific genes. In this context, the KaPPA-View tool works well as a generator of hypotheses rather than for making definitive conclusions about the role of specific genes. Comparison of transcriptome and/or metabolome of distinct tissues, such as leaves and roots, under various growth conditions might also be useful for finding trends in transcription and metabolism.
The KaPPA-View tool is complementary to other metabolic pathway tools and databases. The user-driven tool MAPMAN is designed to present quantitative data of all Arabidopsis transcripts that are categorized on the basis of functionality of their products on pictorial graphs of various biological processes (Thimm et al., 2004). In the “metabolism category” of MAPMAN, several metabolic pathway graphs are provided for independent representation of transcripts. The manner in which this data is presented differs markedly from that of the KaPPA-View tool that simultaneously displays transcripts and metabolites with their respective enzymatic reactions on the same maps. The KaPPA-View tool also provides a way to access all of the information contained on the map that the user is viewing. To appreciate the full extent of the reactions involving the various transcripts and metabolites, we tried to present closely related reactions on single maps, taking care to avoiding too much integration. This approach is what differentiates the KaPPA-View tool from those of pathway databases such as KEGG/PATHWAY and AraCyc. Furthermore, the size of the metabolic pathway maps has been adjusted such that they can be printed at A4 or at letter size, which makes them suitable for electronic retrieval and presentation in articles or talks.
The KaPPA-View tool differs conceptually from other “omics” data tools such as MetNet (Wurtele et al., 2003), PathMAPA (Pan et al., 2003), Pathway Processor (Grosu et al., 2002), and GiGA (Breitling et al., 2004). MetNet is designed to enable visualization, statistical analyses, and modeling of metabolic and regulatory networks in Arabidopsis. PathMAPA can be used to examine Arabidopsis gene expression patterns associated with metabolic pathways. It generates pathway diagrams without building image files manually; visualizes gene expression for each pathway; and performs statistical tests at pathway, enzyme, and gene levels. Pathway Processor features a graphical output that displays differences in expression on metabolic charts of the biochemical pathways for which open reading frames are assigned. GiGA is used to enhance interpretation of microarray data with a statistically rigorous identification of the subgraphs of interest, such as maps of metabolic and signaling pathways or protein interaction. Our presentation of individual transcripts assigned to each reaction on metabolic pathway maps is unique among these tools. However, our tool does not provide statistical treatment of transcriptomic or metabolomic data and does not generate networking images. Because the 130 metabolic pathway maps were designed for KaPPA-View on the basis of major routes of carbon flow in plant metabolism, they could be modules for networking and statistical treatments for clarifying the function of each metabolic pathway in complex metabolic systems. Future versions of KaPPA-View will incorporate such functions.
We adapted the SVG format to be able to display and change the colors of different symbols used to represent quantitative data of transcripts and metabolites on an Internet browser. The SVG technology proved suitable not only for the generation of figures, but also for submission of new or edited pathway maps from researchers to the KaPPA-View administrator using the SVG editor SVG Map Drawer we provide. Consequently, the present versions of our maps will change along with new knowledge of metabolic pathways. The method of submitting new maps will facilitate quick updates and maintenance of pathway information. In future versions of KaPPA-View, we will incorporate appropriate suggestions from users regarding the addition of genes, compounds, and reactions. If users wish to alter information, such as metabolite names and the names of genes presented on our maps, for their presentations, and we believe such a demand is likely given the variety of chemical and genetic nomenclature in use, they may edit the SVG source text files using a text file editor.
The SVG-formatted maps for Arabidopsis can be used as universal plant metabolic pathway maps for analysis of other plant species, although some pathways, such as those for isoflavonoids, have not been included. In the SVG Arabidopsis maps, the Arabidopsis Genome Initiative (AGI) gene numbers given to all Arabidopsis genes on the basis of their genome sequences were used to assign SVG tags (square symbols). Therefore, if users prepare a correspondence table between AGI numbers and gene identification numbers for the plant species of interest, the DNA array data assigned to the identification numbers can be represented using maps as well as metabolite data. Such a correspondence table could be generated using BLAST match of one's own gene sequence dataset to that of the Arabidopsis genome sequence. However, in cases where the number of genes assigned to a reaction number more than that assigned to the reaction in the Arabidopsis map, the user must choose the genes according to the numbers of assigned Arabidopsis genes. Therefore, to increase the applicability of the tool, the use of immovable square symbols on the maps will be changed such that they will be flexible and ordered according to the numbers assigned to the reaction in future versions of KaPPA-View. Other plant metabolic pathways not found in Arabidopsis also will be included. Because the BLAST match strategy could produce a significant number of incorrect functional annotations, however, users must interpret the results with caution. We are currently preparing a comprehensive set of metabolic pathways for the legume Lotus japonicus.
Recently, a new tool for integrating the Arabidopsis transcriptomic and metabolomic data, BioPathAt, which operates as a visual interface in a commercial software package, GeneSpring, was reported (Lange and Ghassemian, 2005).
MATERIALS AND METHODS
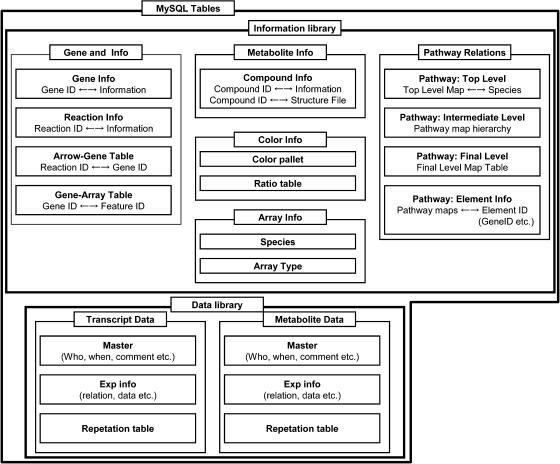
Architecture of the KaPPA-View Tool
The application engine (KV-Engine) was designed to control the pathway information library that contained the pathway map files and the metabolite structure files, the data library with its experimental data files, and the information library that contained information such as the correlations between maps and genes (Fig. 5). The pathway information library and the data library were managed using a relational database management system (MySQL server; http://www.mysql.com/). The script for the KV-Engine was written using JAVA 1.4.2 (Sun Microsystems, Santa Clara, CA) and run on a Debian GNU/Linux 3.0 operating system (http://www.debian.org). The KV-Engine and MySQL (4.0.20) were connected using Java Database Connectivity Application Program Interface technology. The Web server was constructed using Tomcat 4_4.0.3_3 woody3 application server and Apache 1.3.26_0 woody3 WWW server controller. JavaServer Pages technology was used to generate the dynamic Web contents. The KaPPA-View tool can be accessed at http://kpv.kazusa.or.jp/kappa-view/.
Figure 5.
Data components of the MySQL-KaPPA-View tool.
To generate SVG-formatted files for the metabolic pathway maps and pathway indicators, we drew the maps using a drawing tool, SVG Map Drawer, available for download from the KaPPA-View Web site. The symbols on the metabolic pathway maps were assigned using SVG identification tags to receive the values from the information library. The AGI code numbers for Arabidopsis (Arabidopsis thaliana) genes were given SVG identification tags for transcripts on the SVG-formatted maps as the tag value. The EC numbers for enzymes were named according to International Union of Biochemistry and Molecular Biology Enzyme Nomenclature (http://www.chem.qmw.ac.uk/iubmb/enzyme).
All compound names appearing on the KaPPA-View maps were proofread using ontology data files of the Chemical Entities of Biological Interest site of EMBL-European Bioinformatics Institute (http://www.ebi.ac.uk/chebi/) and the GENE ONTOLOGY site (http://www.geneontology.org/GO.downloads.shtml).
The chemical structures of plant metabolites were drawn using ChemDrawUltra 7.0 (CambridgeSoft, Cambridge, UK) and stored as gif-formatted figures and mol-formatted files in the metabolite structure files.
The data library was designed to store quantitative data of transcripts and metabolites. As the file format of the transcript data of the initial version of the KaPPA-View tool does not correspond to the Minimum Information About a Microarray Experiment (MIAME) format (http://www.mged.org/Workgroups/MIAME/miame.html), the data included in the data library are imported from other transcript databases such as the ArrayExpress held at European Bioinformatics Institute (http://www.ebi.ac.uk/arrayexpress/) and Gene Expression Omnibus held at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/), where MIAME-formatted descriptions of experimental details are accessible. Information of the imported data, such as experimental identification numbers in the original database, is kept in the comment field of the data library. In future versions of KaPPA-View, the data library will be formatted using MIAME as the standard.
User Setup
To display SVG-formatted pathway maps on client computers, users need to install the Adobe SVG Viewer Web-browser plug-in, which is freely available from the Adobe SVG Viewer download site (http://www.adobe.com/svg/viewer/install/main.html). The KaPPA-View tool can be used with any personal computers using Windows OS or Macintosh OS X. The user's transcriptome/metabolome data should be prepared as CSV-formatted text files (see the KaPPA-View manual).
Acknowledgments
We thank Dr. Kentaro Yano (Kazusa DNA Research Institute) for his help in analyzing Arabidopsis genes and Dr. Takayuki Tohge (Chiba University) for providing transcript and metabolite data for PAP1-overexpressing plants. We thank Dr. Youji Takeuchi for his help preparing pathway maps, Kanami Moriya and Mayumi Hasegawa for their advice on the KaPPA-View manual, and Yuuko Tazawa for drawing the SVG-formatted metabolic pathway maps.
This work was supported by New Energy and Industrial Technology Development (as part of the project called Development of Fundamental Technologies for Controlling the Process of Material Production of Plants).
The online version of this article contains Web-only data.
References
- Aharoni A, Ric de Vos CH, Verhoeven HA, Maliepaard CA, Kruppa G, Bino R, Goddenowe DB (2002) Nontargeted metabolome analysis by use of Fourier Transform Ion Cyclotron Mass Spectrometry. OMICS 6: 217–234 [DOI] [PubMed] [Google Scholar]
- Aharoni A, Vorst O (2002) DNA microarrays for functional plant genomics. Plant Mol Biol 48: 99–118 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Beisson F, Koo AJ, Ruuska S, Schwender J, Pollard M, Thelen JJ, Paddock T, Salas JJ, Savage L, Milcamps A, et al (2003) Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a Web-based database. Plant Physiol 132: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloor SJ, Abrahams S (2002) The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry 59: 343–346 [DOI] [PubMed] [Google Scholar]
- Breitling R, Amtmann A, Herzyk P (2004) Graph-based iterative Group Analysis enhances microarray interpretation. BMC Bioinformatics 5: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tholl D, D'Auria JC, Farooq A, Pichersky E, Gershenzon J (2003) Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15: 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Strack D (2003) Phytochemistry meets genome analysis, and beyond. Phytochemistry 62: 815–816 [DOI] [PubMed] [Google Scholar]
- Donson J, Fang Y, Espiritu-Santo G, Xing W, Salazar A, Miyamoto S, Armendarez V, Volkmuth W (2002) Comprehensive gene expression analysis by transcript profiling. Plant Mol Biol 48: 75–97 [PubMed] [Google Scholar]
- Feng Q, Zhang Y, Hao P, Wang S, Fu G, Huang Y, Li Y, Zhu J, Liu Y, Hu X, et al (2002) Sequence and analysis of rice chromosome 4. Nature 420: 316–320 [DOI] [PubMed] [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, Halkier BA (2004) Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc Natl Acad Sci USA 101: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Goto S, Okuno Y, Hattori M, Nishioka T, Kanehisa M (2002) LIGAND: database of chemical compounds and reactions in biological pathways. Nucleic Acids Res 30: 402–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon T, Sibout R, Eudes A, MacKay J, Jouanin L (2003) Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana. Plant Physiol Biochem 41: 677–687 [Google Scholar]
- Graham TL (1998) Flavonoid and flavonol glycoside metabolism in Arabidopsis. Plant Physiol Biochem 36: 135–144 [Google Scholar]
- Grosu P, Townsend JP, Hartl DL, Cavalieri D (2002) Pathway Processor: a tool for integrating whole-genome expression results into metabolic networks. Genome Res 12: 1121–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Nakaya A (2002) The KEGG databases at GenomeNet. Nucleic Acids Res 30: 42–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger CJ, Zhang P, Müller LA, Wang A, Paley S, Arnaud M, Pick J, Rhee SY, Karp PD (2004) MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res 32: D438–D442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Ghassemian M (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 51: 925–948 [DOI] [PubMed] [Google Scholar]
- Lange BM, Ghassemian M (2005) Comprehensive post-genomic data analysis approaches integrating biochemical pathway maps. Phytochemistry 66: 413–451 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Zhang P, Rhee SY (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S (2004) Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol 135: 756–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Sun N, Cheung KH, Guan Z, Ma L, Holford M, Deng X, Zhao H (2003) PathMAPA: a tool for displaying gene expression and performing statistical tests on metabolic pathways at multiple levels for Arabidopsis. BMC Bioinformatics 4: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Matsumoto T, Yamamoto K, Sakata K, Baba T, Katayose Y, Wu J, Niimura Y, Cheng Z, Nagamura Y, et al (2002) The genome sequence and structure of rice chromosome 1. Nature 420: 312–316 [DOI] [PubMed] [Google Scholar]
- Sato S, Soga T, Nishioka T, Tomita M (2004) Simultaneous determination of the main metabolites in rice leaves using capillary electrophoresis mass spectrometry and capillary electrophoresis diode array detection. Plant J 40: 151–163 [DOI] [PubMed] [Google Scholar]
- Sumner LW, Mendes P, Dixon RA (2003) Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62: 817–836 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Veit M, Pauli GF (1999) Major flavonoids from Arabidopsis thaliana leaves. J Nat Prod 62: 1301–1303 [DOI] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7: 263–270 [DOI] [PubMed] [Google Scholar]
- Wurtele ES, Li J, Diao L, Zhang H, Foster CM, Fatland B, Dickerson J, Brown A, Cox Z, Cook D, et al (2003) MetNet: software to build and model the biogenetic lattice of Arabidopsis. Comp Funct Genomics 4: 239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K (2001) A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol 42: 1025–1033 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K (2004) Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis. Plant Cell Physiol 45: 1111–1121 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Rose JK, Nishitani K (2004) A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol 134: 1088–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]