Abstract
The SOL Genomics Network (SGN; http://sgn.cornell.edu) is a rapidly evolving comparative resource for the plants of the Solanaceae family, which includes important crop and model plants such as potato (Solanum tuberosum), eggplant (Solanum melongena), pepper (Capsicum annuum), and tomato (Solanum lycopersicum). The aim of SGN is to relate these species to one another using a comparative genomics approach and to tie them to the other dicots through the fully sequenced genome of Arabidopsis (Arabidopsis thaliana). SGN currently houses map and marker data for Solanaceae species, a large expressed sequence tag collection with computationally derived unigene sets, an extensive database of phenotypic information for a mutagenized tomato population, and associated tools such as real-time quantitative trait loci. Recently, the International Solanaceae Project (SOL) was formed as an umbrella organization for Solanaceae research in over 30 countries to address important questions in plant biology. The first cornerstone of the SOL project is the sequencing of the entire euchromatic portion of the tomato genome. SGN is collaborating with other bioinformatics centers in building the bioinformatics infrastructure for the tomato sequencing project and implementing the bioinformatics strategy of the larger SOL project. The overarching goal of SGN is to make information available in an intuitive comparative format, thereby facilitating a systems approach to investigations into the basis of adaptation and phenotypic diversity in the Solanaceae family, other species in the Asterid clade such as coffee (Coffea arabica), Rubiaciae, and beyond.
The SOL Genomics Network (SGN; http://sgn.cornell.edu) is a genomics information resource for the Solanaceae family and related families in the Asterid clade, with the aim of building a comparative bioinformatics platform for answering questions about adaptation, evolution, development, defense, biochemistry, and other facets of this clade. To date, SGN's efforts have focused primarily on four areas: (1) cataloging and maintaining genetic maps and markers of the Solanaceae species; (2) disseminating sequence information for the different species of Solanaceae, mostly in the form of expressed sequence tags (ESTs), for which SGN generates and publishes unigene builds; (3) cataloging and publishing phenotypic information; and (4) assembling, analyzing, and publishing data from the recently commenced sequencing of the tomato (Solanum lycopersicum) genome. Unlike many other plant resources on the Web, which often focus on a single plant species, such as the Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org) on Arabidopsis (Arabidopsis thaliana; Rhee et al., 2003) or maizeGDB (http://www.maizeGDB.org) on Zea mays (Lawrence et al., 2004), SGN has always had a strong comparative bias because of its wider scope. With the advent of the tomato genome sequence, this framework will be leveraged to place the tomato genome in the center of comparisons, thus tying together all the other Solanaceae species and relating these sequences to the other known plant genomes, such as Arabidopsis, Medicago, and rice (Oryza sativa).
The Solanaceae have held great interest for many researchers, breeders, and consumers for a long time. Indeed, the Solanaceae family is composed of more than 3,000 species, including the tuber-bearing potato (Solanum tuberosum), a number of fruit-bearing vegetables (tomato, eggplant [Solanum melongena], and peppers [Capsicum annuum]), ornamental plants (petunias [Petunia hybrida], Nicotiana), plants with edible leaves (Solanum aethiopicum, Solanum macrocarpon), and medicinal plants (e.g. Datura, Capsicum; Knapp, 2002). The Solanaceae are the third most important plant taxon economically, the most valuable in terms of vegetable crops, and the most variable of crop species in terms of agricultural utility. In addition to their role as important food sources, many solanaceous species have a role as scientific model plants, such as tomato and pepper, for the study of fruit development (Gray et al., 1992; Fray and Grierson, 1993; Hamilton et al., 1995; Brummell and Harpster, 2001; Alexander and Grierson, 2002; Adams-Phillips et al., 2004; Giovannoni, 2004; Tanksley, 2004), potato for tuber development (Prat et al., 1990; Fernie and Willmitzer, 2001), petunia for the analysis of anthocyanin pigments, and tomato and tobacco (Nicotiana tabacum) for plant defense (Bogdanove and Martin, 2000; Gebhardt and Valkonen, 2001; Li et al., 2001; Pedley and Martin, 2003). The Solanaceae genomes have undergone relatively few genome rearrangements and duplications and therefore have very similar gene content and order. This exceptionally high level of conservation of genome organization at the macro and micro levels makes this family a model to explore the basis of phenotypic diversity and adaptation to natural and agricultural environments. Recognizing these unique features of the Solanaceae, the International Solanaceae Project (SOL) was launched, setting research goals for the next 10 years. The SOL project addresses two key questions: (1) How can a common set of genes/proteins give rise to the wide range of morphologically and ecologically distinct organisms that occupy our planet? (2) How can a deeper understanding of the genetic basis of plant diversity be harnessed to better meet the needs of society in an environmentally friendly and sustainable manner?
To meaningfully analyze the gene-to-phenotype relationships, a large amount of sequencing information is necessary. The most cost-effective way to get sufficient sequence information to address the SOL questions is to sequence a high-quality reference genome and then map sequences from other genomes onto the reference sequence. Hence, the first cornerstone of the SOL project is the sequencing of the full euchromatic portion of the tomato genome by an international consortium of 10 countries. Concomitantly, SOL will build a bioinformatics platform that allows intuitive and unrestricted access for researchers, and integrates information from all Solanaceae research into a one-stop shop on the Web that will ultimately allow approaching Solanaceae biology from a systems biology perspective. In collaboration with other bioinformatics centers involved in SOL, SGN is actively building this infrastructure, which will be distributed in nature. It will rely on bioMOBY (Wilkinson and Links, 2002) and other technologies to implement a virtual online center of information for the Solanaceae.
OVERVIEW OF THE DATABASE AND WEB SITE
Like most other plant databases, SGN can be accessed through an easy-to-use Web interface. The SGN homepage was recently revamped to improve the usability of the site. It now contains an intuitively organized Getting Started section that provides links to the major features and Web pages, such as data overview pages, search pages, map and markers, resources, etc. In the lower part of the screen is a section providing links to related sites of interest, such as the Tomato Expression Database (TED; Fei et al., 2004) and the Tomato Genomics Resource Center (TGRC) germplasm collection at the University of California, Davis (http://tgrc.ucdavis.edu). To make the site more interesting and accessible to casual or new users, the entry page also contains a News section that lists new features on the Web site and news from the community, an image of the week, a link to a recent article of interest, and a profile of a lab involved in Solanaceae research.
All pages on SGN contain a toolbar at the top with links to the most frequently used sections of the site for easy navigation. The toolbar consists of the SGN logo, which is also a link to the SGN homepage, a quick search function that searches the SGN databases and Web pages, and a menu bar with pull-down menus providing quick links to specific pages grouped by menu topic. Search pages for several types of data are available, such as searches for markers, unigenes, expressed sequence tags (ESTs), EST libraries, bacterial artificial chromosomes (BACs), and profiles of registered SGN users. Using the marker search, markers can be queried by name, map, organism, map position, and whether a marker has associated information such as an overgo probe. Using the BAC search, BACs can be searched by name, including wild cards, presence of end sequence, and matches to overgo probes. Currently, about 75,000 BACs have been end sequenced from a HindIII library (Budiman et al., 2000) and 10,000 BACs have been end sequenced from a newly generated MboI library. These numbers will grow to approximately 200,000 total BACs for a nominal 400,000 BAC end reads over the next few months. The Maps and Markers section links to the different maps available on SGN, which are rendered using the newly developed Comparative Viewer that allows visualizing the maps in an interactive comparative format (see below). The tools available on SGN include the SGN Web BLAST, an identifier conversion tool, the intron finder tool (recently developed at SGN), the Genes That Make Tomatoes database (Menda et al., 2004), the real-time quantitative trait loci tool (Gur et al., 2004), and the bulk download tool pages, which provide utilities to download partial and complete datasets for further analysis by the user. For the download of partial datasets, lists of identifiers (for unigenes, microarray spots, or ESTs) can be entered to download associated information. Entire datasets can be downloaded from the SGN FTP server (ftp://ftp.sgn.cornell.edu), including tomato genome data such as BAC end sequences and BLAST databases, full BAC sequences, and supporting data.
On SGN, all information is freely accessible to all users. A login system exists only for the purpose of a user-managed database of Solanaceae researchers and for submission of EST sequences, and is required for sequencing centers that participate in the tomato sequencing program to update the BAC status information in the SGN database. In the near future, login will also allow users to comment on data objects, such as markers and unigenes, and to make user-contributed annotations to genes. In summary, SGN strives to operate under these guiding principles: (1) all data should be accessible without restrictions; (2) original data should be stored wherever possible (chromatograms, assembly files, gel images, etc.) to ensure complete reproducibility; (3) all data should be attributed to the submitters and data generators; (4) all annotations should be carried out using standard vocabularies and annotation guidelines; (5) free and open-source software is used where possible and SGN-developed software is made available to all as open source; and (6) all the data are loaded into interconnected SGN relational databases such that, ultimately, a systems approach to Solanaceae biology becomes possible.
SGN SITE ARCHITECTURE AND IMPLEMENTATION
The SGN database consists of a number of interrelated relational databases implemented in MySQL (http://www.mysql.com). Most software is written in Perl. The Web site uses the Apache (http://www.apache.org) Web server with the mod_perl integrated Perl interpreter. In keeping with the philosophy of open systems and open-source software, all servers and most development machines run the Debian distribution of the GNU/Linux operating system. More information on the database schemas, software, and setup at SGN can be found on the SGN Web site (http://sgn.cornell.edu).
SGN DATA AND TOOLS
SGN Solanaceae Unigene Builds
As no full genome sequence of a representative Solanaceae species is yet available, much of the existing sequence data on SGN consists of EST datasets for Solanaceae species. However, as the tomato sequencing project begins to bear fruit, SGN's focus will change more to genomic sequence data. From these EST datasets and other known transcript sequences, unigenes are assembled in an effort to approximate the transcriptome set of each organism. SGN currently produces unigene builds for Solanaceae species that have EST sequences available with associated chromatograms. Unigene builds are available for tomato, potato, pepper, eggplant, and petunia (see Table I). A Web interface is also available for submitting new sequence datasets.
Table I.
A snapshot of the SGN unigene builds
Currently, SGN has unigene builds for tomato, potato, pepper, petunia, and eggplant.
| Build | Species Included | No. ESTs | No. Unigenes | No. Singletons | No. Contigs |
|---|---|---|---|---|---|
| Tomato | Solanum lycopersicum, Solanum pennellii, Solanum hirsutum | 184,860 | 30,576 | 11,160 | 19,416 |
| Potato | Solanum tuberosum | 97,425 | 24,931 | 9,117 | 15,814 |
| Pepper | Capsicum annuum | 20,738 | 9,554 | 6,618 | 2,936 |
| Petunia | Petunia hybrida | 11,479 | 5,135 | 3,476 | 1,659 |
| Eggplant | Solanum melongena | 3,181 | 1,841 | 1,224 | 617 |
In contrast to many other unigene assembly methods, the SGN custom unigene assembly pipeline starts at the level of the raw chromatogram in order to apply the same quality standards to all data, thereby increasing the consistency and overall quality of the builds. The assembly pipeline, which is tightly integrated with the SGN database, works as follows. First, the chromatograms are base called with phred and the raw sequences are loaded into our database. Next, the sequences are processed to determine a high-quality region excluding low-quality or cloning vector sequences. Then, Escherichia coli or lambda phage contamination is detected with an automated National Center for Biotechnology Information (NCBI) BLAST search, and contaminated sequences are flagged in the database. Inserts that contain sequences matching the multiple cloning site of the vector are flagged as chimeric. A second chimera screen is also applied that attempts to align the ends of a read with any Arabidopsis coding sequences. If the two ends match unrelated Arabidopsis genes, the sequence is flagged as chimeric. Flagged sequences are not used in subsequent unigene builds. The unigene assembly proceeds through a custom preclustering program, which generates clusters that are fed into the cap3 program (Huang and Madan, 1999). cap3 is run with the following parameter settings: -e 5000 -p 90 -d 10000 -b 60. The overlap identity is set to a stringent 90% (default 75%, option -p), and the effects of some options are minimized by setting them to the maximum allowed values. These parameters have been fine tuned over a period of years and comparisons to tomato mRNAs (that were not part of the input sequences) indicate that the unigenes are of high quality. Table I summarizes the unigene builds currently available from SGN. These unigene builds also serve as the basis for a number of analyses, including prediction of coding regions, Interpro domains, Markov clustering into gene families, and development of simple sequence repeat and conserved ortholog set (COS) markers. Some of these results are available from the Web interface and some can be downloaded from the FTP site.
Markers, Maps, and the SGN Comparative Viewer
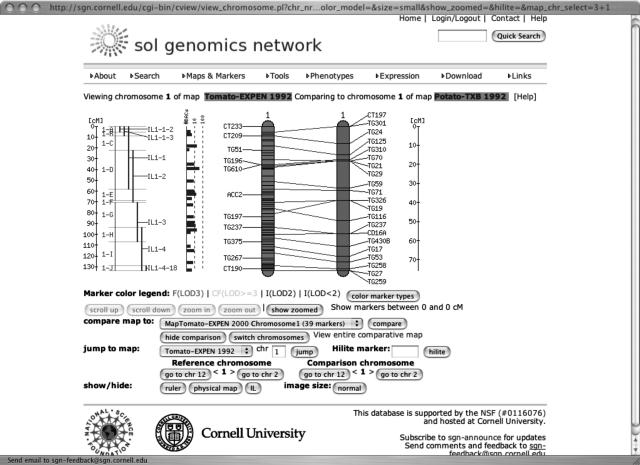
The SGN database currently houses six maps, the Tomato E × PEN 2000 (Fulton et al., 2002), the Tomato E × PEN 1992 (Tanksley et al., 1992), the Tomato E × HIR, the Tomato E × PIMP, the Potato T × B (Tanksley et al., 1992), and the Eggplant L × M (Doganlar et al., 2002; Table II). A classic and an integrated pepper map will soon be added to the database, and some maps, such the pepper SNU2 map (Lee et al., 2004), are available as static images only. For the nonstatic maps, all marker and mapping data are stored, searchable, and browsable in the SGN database. The maps and markers are a good example of how SGN attempts to integrate and present information intuitively in a comparative format. The SGN Comparative Viewer accesses the SGN database information and displays the maps as shown in Figure 1. By default, the Comparative Viewer shows a reference chromosome on the left-hand side of the screen with the option of showing additional information tracks, such as a physical map, inbred lines (currently, the Zamir lines can be viewed), or a centiMorgan ruler. Because there are usually many more markers than can be displayed on a full-chromosome map representation, only a selection of approximately a dozen markers is shown by name; the other markers are shown as tick marks only until the view is zoomed in. Clicking a point on the chromosome will show an enlarged view of the selected region on the right, giving all markers by name within the enlarged interval. A zoom level button adjusts the size of the interval so that a convenient number of markers can be displayed in the zoomed-in section. All marker labels are conveniently linked to their corresponding marker detail pages. When markers appear on more than one map, comparative views of two maps can be displayed. A pull-down menu shows all other chromosomes from other maps in the database that have markers in common with this one. Choosing a chromosome from this menu adds that chromosome to the display alongside the reference chromosome and draws connecting lines between markers that are present on both maps, facilitating comparison between different mapping populations or different species. Many of these common markers in the database are COS markers, which were developed for the purpose of comparative mapping (Fulton et al., 2002) and which are currently expanded with a new set of markers called COSII (F. Wu and S.D. Tanksley, unpublished data). To get a bird's eye view of how two different maps relate to each other, the View Entire Comparative Map link provides a view of the reference map chromosomes vertically displayed on the left and the comparison map on the right, with all chromosomal connections shown between the two maps. A powerful and user-friendly marker search is also available, allowing searching for markers by type, chromosome, position, map, species, and other criteria. Used together, SGN's Comparative Viewer and marker search capabilities give the biological community a powerful resource for comparing genetic maps of species in our database.
Table II.
SGN maps and markers
Maps for pepper and coffee will be added to the site in the near future.
| Species | Parents | Map Name | No. Markers |
|---|---|---|---|
| Tomato | S. lycopersicum × S. pennellii | F2-2000 | 1575 |
| S. lycopersicum × S. pennellii | E × PEN1992 | 553 | |
| S. lycopersicum × S. habrochaites | E × HIR1997 | 135 | |
| S. lycopersicum × S. pimpenellifolium | E × PIMP2001 | 139 | |
| Potato | S. tuberosum × S. berthaultii | T × B1992 | 178 |
| Eggplant | S. linnaeanum × S. melongena | L × M2002 | 220 |
Figure 1.
The SGN Comparative Viewer. The viewer shows the relationships between different genetic maps stored in the SGN database and allows users to visualize other information associated with the maps, such as rulers, inbred lines, and the tomato physical map. A zooming function lets the user examine the maps in detail. A second map can be displayed on the right-hand side of the screen; the relationships between markers on the two maps are shown with lines. The interactive Comparative Viewer is controlled with the toolbar visible at the bottom of the screen.
Phenotypic Information
SGN houses a collection of phenotypic information in the Genes That Make Tomatoes database describing a mutant population of 13,000 Solanum lycopersicum M2 families of the M82 variety. This comprehensive mutant population is a useful basic resource for exploring gene function. These mutants were generated using ethyl methanesulfonate and fast-neutron mutagenesis, and the plants were visually phenotyped in the field and then categorized into a morphological catalog encompassing 15 primary and 48 secondary categories. Currently 3,417 mutations have been cataloged; among them are most of the previously described phenotypes from the monogenic mutant collection of the TGRC, plus over 1,000 new mutants with multiple alleles per locus. The phenotypic database indicates that most mutations fall into more than a single category (they are pleiotropic), with some organs (e.g. leaves) more prone to alterations than others. All data and images can be searched and accessed through SGN. The mutants were generated and phenotyped and the database is administered by the Zamir lab at the Hebrew University in Jerusalem, Israel.
Another tool is real-time quantitative trait loci, which presents the results of eight independent phenotyping experiments on an isogenic inbred line population and allows viewing correlations online in real time. A tighter integration of phenotypic data with the molecular and mapping data on SGN is planned for the future.
TOMATO SEQUENCING PROJECT
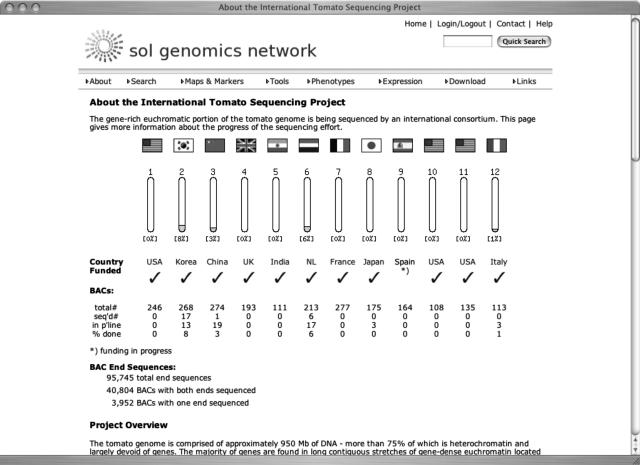
The tomato sequencing project was initiated in 2004 by a consortium of 10 countries, with each of the following countries sequencing one chromosome: Korea (chromosome 2), China (chromosome 3), Great Britain (chromosome 4), India (chromosome 5), The Netherlands (chromosome 6), France (chromosome 7), Japan (chromosome 8), Spain (chromosome 9), and Italy (chromosome 12). The United States sequenced three chromosomes (chromosomes 1, 10, and 11; see Fig. 2). The tomato genome is composed of approximately 950 Mb of DNA, more than 75% of which is heterochromatin and largely devoid of genes. The majority of genes are found in long contiguous stretches of gene-dense euchromatin located on the distal portions of each chromosome arm. The sequencing strategy is to sequence a minimal tiling path of BAC clones through the approximately 220 Mb of euchromatin. The starting points for sequencing the genome will be 1,500 anchor points, where the physical map has been linked to the genetic map using overgo probes. The results of the overgo analysis are available on SGN (http://sgn.cornell.edu). In addition, SGN is involved in setting up part of the infrastructure for the tomato sequencing project, such as a BAC registry, so that the status of each BAC in the sequencing pipeline can be tracked by users and sequencers alike. The finished BAC sequences will be deposited in both GenBank and SGN. On SGN, annotations will be included that will be viewable online, based on Gbrowse (Stein et al., 2002). The tomato sequence will provide the resource for mapping sequences of other species onto it and, in conjunction with the comparative maps, generate virtual sequences for other Solanaceae species, and to compare the features of the Solanaceae genomes with other sequenced species, such as Arabidopsis, rice, and Medicago. SGN will produce a unified interface for viewing this information. More details on the SOL project and the tomato sequencing project, including the SOL white paper and the tomato sequencing standards document, can be found on SGN at http://sgn.cornell.edu/solanaceae-project.
Figure 2.
The overview page for the tomato sequencing project. The chromosomes are shown as small bar graphs, the size corresponding to the chromosome size in centiMorgans, which will fill in as sequencing progresses. The page is available at http://sgn.cornell.edu/help/about/tomato_sequencing.html.
CURATIONAL ACTIVITIES AT SGN
To date, the curational activities have focused on maps and markers, and the functional annotation, primarily by automatic means, of the unigene sets for Solanaceae species generated at SGN. However, with the start of tomato sequencing, SGN will be involved in the annotation of the tomato genome. A pipeline is currently being developed by the tomato sequencing consortium that allows the automatic annotation of gene structures, repeats, RNAs, and other features on BAC sequences. The pipeline will be based on a distributed system such that, for each analysis, the best tool available for a given task can be pulled into the pipeline as a Web service, such as a bioMOBY service (Wilkinson and Links, 2002), from a number of tools made available by any consortium member. Standard, open-source tools, such as those made available by the GMOD project (http://www.gmod.org), will be used to review the results of these structural annotations, in particular the Apollo gene editor (Lewis et al., 2002). Functional annotation will be based on homology searches such as BLAST and identification of Interpro (Zdobnov and Apweiler, 2001; Mulder et al., 2003) domains. The latter will allow gene ontology (Ashburner et al., 2000) terms to be assigned automatically. In parallel, a list of Solanaceae genes that are experimentally described in the literature will be compiled and loaded into a Web-accessible, searchable database, forming the basis for a set that will be manually annotated with the gene ontology and plant ontology systems. The use of these shared, common vocabularies will be important for allowing easy comparisons to the other genomes that were annotated using these same vocabularies. Manual annotation based on literature is an expensive and time-consuming process. Community-based annotation systems have been developed, but circumstantial evidence indicates that community participation is usually not high. As SGN attempts to develop into a more community-driven resource, a higher degree of user participation in annotation will be a major goal, although user submission motivation is still an unsolved problem. In addition, user-based annotations have to be verified by curators, which suggests that such a distributed model cannot fully replace in-house curators.
OTHER SOLANACEAE RESOURCES ON THE WEB
There are myriad other Solanaceae resources on the Web besides SGN. The most important ones are listed on the Solanaceae Resources links page on SGN, some of which are summarized in Table III.
Table III.
Other Solanaceae resources available on the Web
More links can be found on SGN at http://sgn.cornell.edu (click on Solanaceae Resources).
| Name | Description | URL |
|---|---|---|
| Solanaceae in General | ||
| Solanum source | The official Web site of the Solanum PBI project, containing species descriptions, specimen data from herbaria, nomenclature data about names and their places of publication, and a bibliography of scientific articles about members of the family | http://www.nhm.ac.uk/botany/databases/solanum |
| Nijmegen Botanical and Experimental Garden Web site | A botanical garden with one of the major collections of Solanaceae plants; The Netherlands | http://www.bgard.science.ru.nl |
| TIGR plant gene indices | TIGR ESTs and consensus sequences for plants including the Solanaceae; U.S. | http://www.tigr.org/tdb/tgi/plant.shtml |
| Tomato | ||
| Kazusa Microtom Site | EST and unigene dataset from the Microtom tomato variety, including GO annotations and Pathway Viewer; Japan | http://www.kazusa.or.jp/microtom |
| The Tomato Genetics Cooperative | Homepage of the Tomato Genetics Cooperative with access to publications online | http://gcrec.ifas.ufl.edu/tgc |
| TGRC | Large collection of Solanaceae germplasms, with a special focus on tomato; University of California, Davis, CA | http://tgrc.ucdavis.edu |
| Genes that make tomatoes | Web site of a large-scale tomato mutagenesis and phenotyping project; Israel | http://zamir.sgn.cornell.edu/mutants |
| TED | Site dedicated to tomato microarrays, BTI; U.S. | http://ted.bti.cornell.edu |
| Potato | ||
| PoMaMo database | Potato genome and map data, including single-nucleotide polymorphism data, marker sequences, references, and more from the Max-Planck Institute for Plant Breeding Research; Cologne, Germany | https://gabi.rzpd.de/PoMaMo.html |
| Centro Internacional de la Papa (International Potato Center) | The International Potato Center with potato-specific databases and resources such as the CIP Potato GenBank and the World Potato Atlas; near Lima, Peru | http://www.cipotato.org |
| Intergenebank for potato | A database containing accession data from different gene banks around the world, including an simple sequence repeat database; Peru | http://www.potgenebank.org |
| Potatogenome.org | The official Web site of the NSF potato genome project; U.S. | http://www.potatogenome.org |
| Pepper | ||
| KRIBB pepper resources | A pepper EST database at KRIBB; Korea | http://plant.pdrc.re.kr/ks200201/pepper.html |
| Embrapa Capsicum site | A site dedicated to pepper; Brazil | http://www.cnph.embrapa.br/capsicum |
| Petunia | ||
| Petuniaplatform.net | A Web site dedicated to Petunia, with contacts, techniques, bibliography and other information; The Netherlands | http://www.petuniaplatform.net |
| Coffee | ||
| CoffeeDNA | A site about coffee and coffee genomics. Login required for certain functions; Italy | http://www.coffeedna.net |
CONCLUSION AND OUTLOOK
SGN is a relatively new database that is rapidly developing into a comprehensive resource for comparative biology between members of the Solanaceae family, closely related plants, such as coffee (Coffea arabica), and beyond. The tomato genome sequence will provide the foundation for a new sequenced-based, comparative resource that will aid in linking the Solanaceae to each other and outward to other species and families where enough sequence information is available. As time progresses, the hypothesis-driven research enabled by large repositories of biological data like SGN will continue to gain importance. Instead of only answering simple queries, data repositories and plant Web sites should strive to provide the enabling context that leads to further questions, allowing a researcher to posit new hypotheses that can be verified in silico or in vitro. The ultimate goal of SGN is to make information available in an intuitive comparative format that will make this type of science possible, thus facilitating a systems approach to investigating the basis of adaptation and phenotypic diversity in the Solanaceae.
This work was supported by the National Science Foundation (grant nos. 0116076, 9872617, 975866, and 0421634) for the SGN and the tomato sequencing project.
References
- Adams-Phillips L, Barry C, Giovannoni J (2004) Signal transduction systems regulating fruit ripening. Trends Plant Sci 9: 331–338 [DOI] [PubMed] [Google Scholar]
- Alexander L, Grierson D (2002) Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot 53: 2039–2055 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Martin GB (2000) AvrPto-dependent Pto-interacting proteins and AvrPto-interacting proteins in tomato. Proc Natl Acad Sci USA 97: 8836–8840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH (2001) Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol Biol 47: 311–340 [PubMed] [Google Scholar]
- Budiman MA, Mao L, Wood TC, Wing RA (2000) A deep-coverage tomato BAC library and prospects toward development of an STC framework for genome sequencing. Genome Res 10: 129–136 [PMC free article] [PubMed] [Google Scholar]
- Doganlar S, Frary A, Daunay MC, Lester RN, Tanksley SD (2002) A comparative genetic linkage map of eggplant (Solanum melongena) and its implications for genome evolution in the Solanaceae. Genetics 161: 1697–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Z, Tang X, Alba RM, White JA, Ronning CM, Martin GB, Tanksley SD, Giovannoni JJ (2004) Comprehensive EST analysis of tomato and comparative genomics of fruit ripening. Plant J 40: 47–59 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Willmitzer L (2001) Molecular and biochemical triggers of potato tuber development. Plant Physiol 127: 1459–1465 [PMC free article] [PubMed] [Google Scholar]
- Fray RG, Grierson D (1993) Molecular genetics of tomato fruit ripening. Trends Genet 9: 438–443 [DOI] [PubMed] [Google Scholar]
- Fulton TM, Van der Hoeven R, Eannetta NT, Tanksley SD (2002) Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell 14: 1457–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt C, Valkonen JP (2001) Organization of genes controlling disease resistance in the potato genome. Annu Rev Phytopathol 39: 79–102 [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ (2004) Genetic regulation of fruit development and ripening. Plant Cell (Suppl) 16: S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J, Picton S, Shabbeer J, Schuch W, Grierson D (1992) Molecular biology of fruit ripening and its manipulation with antisense genes. Plant Mol Biol 19: 69–87 [DOI] [PubMed] [Google Scholar]
- Gur A, Semel Y, Cahaner A, Zamir D (2004) Real time QTL of complex phenotypes in tomato interspecific introgression lines. Trends Plant Sci 9: 107–109 [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Fray RG, Grierson D (1995) Sense and antisense inactivation of fruit ripening genes in tomato. Curr Top Microbiol Immunol 197: 77–89 [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S (2002) Tobacco to tomatoes: a phylogenetic perspective on fruit diversity in the Solanaceae. J Exp Bot 53: 2001–2022 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dong Q, Polacco ML, Seigfried TE, Brendel V (2004) MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Res 32: D393–D397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Nahm SH, Kim YM, Kim BD (2004) Characterization and molecular genetic mapping of microsatellite loci in pepper. Theor Appl Genet 108: 619–627 [DOI] [PubMed] [Google Scholar]
- Lewis SE, Searle SM, Harris N, Gibson M, Lyer V, Richter J, Wiel C, Bayraktaroglir L, Birney E, Crosby MA, et al (2002) Apollo: a sequence annotation editor. Genome Biol 3: RESEARCH0082 [DOI] [PMC free article] [PubMed]
- Li L, Li C, Howe GA (2001) Genetic analysis of wound signaling in tomato. Evidence for a dual role of jasmonic acid in defense and female fertility. Plant Physiol 127: 1414–1417 [PMC free article] [PubMed] [Google Scholar]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D (2004) In silico screening of a saturated mutation library of tomato. Plant J 38: 861–872 [DOI] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood RK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P, et al (2003) The Interpro database, 2003 brings increased coverage and new features. Nucleic Acids Res 31: 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2003) Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu Rev Phytopathol 41: 215–243 [DOI] [PubMed] [Google Scholar]
- Prat S, Frommer WB, Hofgen R, Keil M, Kossmann J, Koster-Topfer M, Liu XJ, Muller B, Pena-Cortes H, Rocha-Sosa M, et al (1990) Gene expression during tuber development in potato plants. FEBS Lett 268: 334–338 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al (2002) The generic genome browser: a building block for a model organism system database. Genome Res 12: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD (2004) The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell (Suppl) 16: S181–S189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB, et al (1992) High density molecular linkage maps of the tomato and potato genomes. Genetics 132: 1141–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Links M (2002) BioMOBY: an open source biological web services proposal. Brief Bioinform 3: 331–341 [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R (2001) InterProScan—an intergration for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848 [DOI] [PubMed] [Google Scholar]