Abstract
The apical wall of growing pollen tubes must be strong enough to withstand the internal turgor pressure, but plastic enough to allow the incorporation of new membrane and cell wall material to support polarized tip growth. These essential rheological properties appear to be controlled by pectins, which constitute the principal component of the apical cell wall. Pectins are secreted as methylesters and subsequently deesterified by the enzyme pectin methylesterase (PME) in a process that exposes acidic residues. These carboxyls can be cross-linked by calcium, which structurally rigidifies the cell wall. Here, we examine the role of PME in cell elongation and the regulation of its secretion and enzymatic activity. Application of an exogenous PME induces thickening of the apical cell wall and inhibits pollen tube growth. Screening a Nicotiana tabacum pollen cDNA library yielded a pollen-specific PME, NtPPME1, containing a pre-region and a pro-region. Expression studies with green fluorescent protein fusion proteins show that the pro-region participates in the correct targeting of the mature PME. Results from in vitro growth analysis and immunolocalization studies using antipectin antibodies (JIM5 and JIM7) provide support for the idea that the pro-region acts as an intracellular inhibitor of PME activity, thereby preventing premature deesterification of pectins. In addition to providing experimental data that help resolve the significance and function of the pro-region, our results give insight into the mechanism by which PME and its pro-region regulate the cell wall dynamics of growing pollen tubes.
Pollen tube growth, which delivers the sperm cells to the female gametophyte in the ovule, is essential for plant reproduction. The elongation process is driven by the secretion of Golgi-derived vesicles that dock and fuse with the plasma membrane at the extreme apex of the tube, providing new plasma membrane and cell wall components necessary for polarized pollen tube growth. Their fast growth and relative ease of culture in vitro make pollen tubes a well-established model system for studying cell elongation in plants. In the search for cellular components that regulate pollen tube growth, most of the attention has been drawn to secretory membrane traffic, intracellular motility, ion activities, and turgor pressure, while the contribution of the cell wall has been somewhat neglected (Holdaway-Clarke and Hepler, 2003). Yet, the apical cell wall certainly is a key component since it has to be strong enough to withstand the internal turgor pressure, but, at the same time, provide enough plasticity to allow the incorporation of new membrane and cell wall material to support tip growth (Steer and Steer, 1989).
The wall in the tip region of the pollen tube is composed of a single pectin layer, where neither cellulose nor callose has been detected (Ferguson et al., 1998). Homogalacturonan, a major component of pectins, is a linear polymer composed of (1,4)-α-d-galacturonic acid (GalUA) residues. Current evidence indicates that these pectins are synthesized and methylesterified in the Golgi and thereafter secreted into the wall in a highly methylesterified state (Li et al., 1995; Staehelin and Moore, 1995; Sterling et al., 2001). Subsequently, they are deesterified by wall-associated pectin methylesterases (PMEs) converting the methoxyl groups into carboxyl groups on the polygalacturonic acid chain and releasing both methanol and protons. It appears that most plant PMEs deesterify pectins in a blockwise fashion, e.g. linearly along the chain, which favors pectin gelation and concomitant cell wall stiffening due to the formation of cooperative Ca2+ cross-bridges between free carboxyl groups of adjacent pectin chains (Catoire et al., 1998). Alternatively, it has been proposed that the localized reduction in pH, due to the deesterification process, could promote cell wall extension or growth by stimulating the activity of several cell wall-loosening hydrolases, such as polygalacturonases and pectate lyases (Wen et al., 1999; Ren and Kermode, 2000). It is suggested that these opposing effects, cell wall stiffening versus cell wall loosening, are associated with specific PME isoforms or are regulated depending on the microenvironment, in particular apoplastic pH and the availability of divalent cations (Moustacas et al., 1991; Charnay et al., 1992; Bordenave, 1996; Catoire et al., 1998).
Immunolocalizations have shown that in both Nicotiana tabacum and Lilium longiflorum, esterified pectins are almost exclusively present in the apical region of the wall while relatively unesterified pectins are present along the entire length of the pollen tubes (Li et al., 1994; Geitmann et al., 1995). However, Parre and Geitmann (2005) have recently shown that unesterified pectins are absent from the apical cell wall of Solanum chacoense. Since PMEs affect the rheological properties of the cell wall, the regulated deesterification of methylesterified pectins by these PMEs may control the plasticity of the apical cell wall in pollen tubes. Although a few pollen and pollen tube-expressed PMEs and PME-like genes have been identified (Albani et al., 1991; Mu et al., 1994; Wakeley et al., 1998), the first functional analysis of a pollen tube-expressed PME has only been described very recently (Jiang et al., 2005).
It is known that many plant PME genes encode so-called pre-pro-proteins. The pre-region is required for protein targeting to the endoplasmic reticulum (ER), while only the mature part of the PME, without the pro-region, is extracted from the cell wall. Although several functions for the pro-region have been suggested (Micheli, 2001), including targeting of PME to the cell wall, correct folding of PME, and inhibition of PME enzyme activity, none has been conclusively established. In this study, we show that application of an exogenous PME from orange (Citrus sinensis) peel induces thickening of the cell wall at the tip, with the concomitant inhibition of pollen tube growth, confirming that PME can play an important role in the regulation of the wall dynamics. Screening of a N. tabacum pollen cDNA library resulted in the identification of a pre-pro-PME. Transient expression studies with green fluorescent protein (GFP) fusion proteins suggest that the pro-region is important for the correct targeting of the mature PME to the apoplasm. Results from in vitro growth analysis further support the idea that the pro-region also acts as an intramolecular inhibitor of PME activity. We also show that, using the JIM5 and JIM7 antipectin antibodies, in the absence of the pro-region, PME expression causes a dramatic change of the pectin methylesterification profile in the apical pollen tube wall. Our results confirm a functional contribution of PME in pollen tube growth, but, in addition, provide insight into the regulatory role that its pro-region may play in controlling PME activity and thus the wall dynamics of growing pollen tubes.
RESULTS
Effect of Exogenous PME on Pollen Tube Growth and Morphology
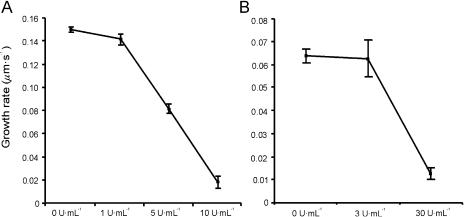
To determine whether PMEs affect the average pollen tube growth rate, the culture medium was supplemented with PME from orange peel (Fig. 1). When applied before germination of the pollen grains, PME caused a dramatic decrease of the germination rate for both Lilium formosanum and N. tabacum (data not shown). To avoid this effect from influencing the growth rate calculations, pollen grains were first allowed to germinate in standard growth medium and only afterward cultured in PME-supplemented medium (Holdaway-Clarke et al., 2003). The growth rate of L. formosanum pollen tubes decreased with increasing concentrations of PME, until almost no growth could be observed at a concentration of 10 units mL−1 (Fig. 1A). Also, N. tabacum pollen tubes decreased their growth when exposed to orange peel PME, although a higher PME concentration was needed for the inhibitory effect to be revealed, with nearly complete cessation of growth at 30 units mL−1 (Fig. 1B). These results are similar to those reported recently by Parre and Geitmann (2005) in Solanum pollen tubes.
Figure 1.
The effect of orange peel PME on L. formosanum (A) and N. tabacum (B) pollen tube growth. Data represent average ± sd of three independent experiments with n ≥ 100 each.
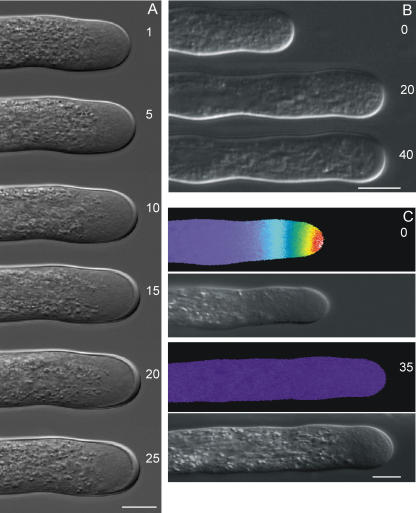
A closer look at the morphology of PME-treated tubes from both species showed a significant increase in the cell wall thickness over time, which is most prominent at the apex (Fig. 2, A and B). Application of boiled orange peel PME did not affect the pollen tube growth rate or morphology (data not shown). Pollen tubes possess an intracellular tip-focused Ca2+ gradient that has been related to cell elongation (Pierson et al., 1994, 1996). Ratiometric Ca2+ imaging of L. formosanum tubes revealed that PME treatment triggers a dissipation of this gradient, most probably caused by the apical cell wall thickening and subsequent inhibition of cell elongation (Fig. 2C).
Figure 2.
Morphology of L. formosanum pollen tubes (A) and pseudocolor ratio images of intracellular Ca2+ distribution in L. formosanum pollen tubes (C) exposed to 10 units mL−1 orange peel PME. B, Morphology of N. tabacum pollen tubes exposed to 30 units mL−1. Numbers represent minutes after addition of PME to the growth medium. The [Ca2+]i gradient shown extends from 1 to 3 μm at the apex of the growing tube to basal values of around 0.15 μm within 20 μm from the apex. Scale bar = 10 μm.
Characterization of a N. tabacum Pollen Tube PME
Knowing that exogenous PME induces apical cell wall thickening and growth inhibition of pollen tubes, we screened a N. tabacum pollen cDNA library for PMEs using the Petunia inflata pollen cDNA clone PPE1 (Mu et al., 1994) as a probe. Several independent cDNA clones were obtained that all encoded the same PME. Nucleotide sequencing revealed an open reading frame of 1,665 bp, which would encode a 60.3-kD protein. Analysis of the deduced amino acid sequence, shown in Figure 3A, indicates the presence of a signal peptide (SP) with the most likely cleavage site between the Ala residues at positions 22 and 23 (Nielsen et al., 1997). In addition to the PME domain, the National Center for Biotechnology (NCBI) Conserved Domain Search (Marchler-Bauer and Bryant, 2004) revealed a putative plant PME inhibitor domain, which is predicted to inhibit PMEs through formation of a noncovalent 1:1 complex (D'Avino et al., 2003). These data show that the isolated PME clone encodes what has been referred to as a pre-pro-PME, in which the pre-region is required for protein targeting to the ER, and only the mature part of the PME (without the pro-region) is found in the cell wall (Micheli, 2001).
Figure 3.
A, Deduced amino acid sequence of the NtPPME1 cDNA clone. Arrow indicates the predicted SP cleavage site. The Cys residues that are conserved between the pro-region and the PMEI from kiwi fruit are in bold. The catalytically important PME residues (Asp-365, Asp-386, and Arg-454) are underlined. B, Schematic representation indicates the pre-pro-PME that is encoded by NtPPME1. The SP used in the various experiments comprised amino acids 1 to 40 of the deduced NtPPME1 protein sequence.
Although several roles for the pro-region have been suggested, including being an autoinhibitory domain, a targeting factor, and/or a chaperone (Micheli, 2001), the biological function of this region remains to be established. Figure 3B shows a schematic representation of the deduced pre-pro-PME protein. At this stage, we can only speculate about the exact borders of the pro-region and the PME domain. Based on alignments with fungal PMEs, which do not possess a pro-region, and the predictions from the Conserved Domain Search, it seems that the PME domain starts between amino acids 175 and 200, resulting in a mature protein of 39 to 41.5 kD with a predicted pI of 8.6. The pro-region is expected to run from amino acids 23 to 175 to 200, resulting in a protein domain that translates into 17 to 19.5 kD and an pI of 4.5.
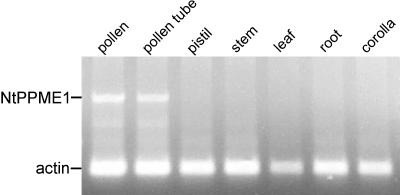
Although our PME was obtained from a N. tabacum pollen cDNA library, this does not necessarily imply that expression is confined to pollen or pollen tubes. The spatial expression pattern of the pre-pro-PME was examined using reverse transcription (RT)-PCR. Total RNA was isolated from all major N. tabacum tissues, and PCR was performed using cDNA from the first-strand reaction with primers specific for the PME clone. Primers specific for N. tabacum actin were included as an internal control. Expression of this pre-pro-PME was restricted to pollen and pollen tubes (Fig. 4), and, therefore, the isolated clone was named NtPPME1 (Nicotiana tabacum pollen tube PME1).
Figure 4.
RT-PCR for NtPPME1 gene expression in various N. tabacum tissues. Expression of actin was used as an internal control.
What Can We Learn from the Arabidopsis Transcriptome?
Several Arabidopsis (Arabidopsis thaliana) pollen transcriptome studies have shown that genes encoding for proteins related to cell wall biosynthesis and regulation are highly expressed in Arabidopsis pollen (Becker et al., 2003; Honys and Twell, 2003, 2004; Pina et al., 2005). The gene expression data used in Tables I and II were obtained from a comparative Arabidopsis transcriptome study using the Affymetrix Arabidopsis ATH1 genome array, which covers more than 80% of the Arabidopsis genome (Pina et al., 2005). Out of the 68 open reading frames that have been annotated as PMEs, 60 are represented on the ATH1 chip. Expression analysis shows that at least 12 different PME isoforms are expressed in Arabidopsis pollen (Table I), which is particularly significant considering that the pollen transcripts originate from single cells, while the vegetative tissues contain many different cell types. Furthermore, these transcriptome data emphasize that pollen has extremely high PME expression levels compared to the other tissues analyzed. Most of the PME isoforms highly expressed in Arabidopsis pollen contain a pro-region (Table II). However, the fact that some isoforms lack this region might signify the need for separate PME inhibitor (PMEI) proteins.
Table I.
PME and invertase/PME inhibitor expression levels in different Arabidopsis tissues
The number of genes whose expression falls into a given expression level is shown for several Arabidopsis tissues. The actual expression data of the genes encoding PME and invertase/PME inhibitor family proteins can be found in Pina et al. (2005).
| Expression Level | Pollen | Leaf | Flower | Seedling | Silique | |
|---|---|---|---|---|---|---|
| PME | >15,000 | 6 | 0 | 0 | 0 | 0 |
| 10,000–15,000 | 0 | 0 | 0 | 0 | 0 | |
| 5,000–10,000 | 1 | 0 | 2 | 0 | 3 | |
| 1,000–5,000 | 4 | 5 | 15 | 6 | 8 | |
| <1,000a | 1 | 8 | 13 | 12 | 10 | |
| Invertase/PME inhibitor | >15,000 | 5 | 0 | 0 | 0 | 0 |
| 10,000–15,000 | 0 | 0 | 0 | 0 | 0 | |
| 5,000–10,000 | 3 | 0 | 1 | 0 | 1 | |
| 1,000–5,000 | 2 | 2 | 11 | 2 | 3 | |
| <1,000a | 0 | 6 | 11 | 10 | 12 |
Only present calls with expression levels >200 are included.
Table II.
Analysis of PME isoforms expressed in Arabidopsis pollen
The deduced amino acid sequences of pollen-expressed PME isoforms are analyzed for the presence of a pro-region. The pI and molecular mass were calculated based on a prediction of the mature PME protein. The expression data of the PME genes are taken from Pina et al. (2005).
| Accession No. | Relative Expression Levela | Pro-Region Present | Estimated pI PME Domain | Estimated Molecular Mass PME Domain |
|---|---|---|---|---|
| kD | ||||
| At2g26450 | 15,650 | Yes | 9.0 | 37.9 |
| At2g47040 | 19,351 | Yes | 9.9 | 39.3 |
| At3g05610 | 15,779 | Yes | 6.4 | 47.5 |
| At3g06830 | 2,524 | Yes | 9.2 | 37.7 |
| At3g17060 | 15,145 | No | 8.7 | 36.2 |
| At3g62170 | 17,681 | Yes | 9.4 | 36.7 |
| At4g15980 | 1,600 | Yes | 10.0 | 38.8 |
| At4g33230 | 1,727 | Yes | 9.4 | 37.9 |
| At5g07420 | 4,283 | No | 9.1 | 37.4 |
| At5g07430 | 18,042 | No | 8.9 | 37.6 |
| At5g27870 | 5,515 | Yes | 6.7b | 50.5b |
| At5g64640 | 922 | Yes | 5.1 | 37.8 |
Only present calls with expression levels >200 are included.
C terminus containing PT-repeat (accession no. PF04886) with no homology to PME included in calculation.
The expression of genes encoding for invertase/PME inhibitors is very high in pollen compared to the other tissues (Table I). Here we note that, due to the sequence similarity between invertase inhibitors and PMEIs, it is impossible to separate the two from each other (Rausch and Greiner, 2004). Invertases, and thus also the regulation of their activity by invertase inhibitors, are likely to play an important role in pollen tube growth since these enzymes hydrolyze Suc to Glc and Fru, which are important metabolites for respiration and synthetic processes (Ylstra et al., 1998; Rausch and Greiner, 2004). It is known, however, that at least some of the Arabidopsis pollen-expressed inhibitor family members represent functional PMEIs (Wolf et al., 2003). Together, the transcriptome data underline the importance of PMEs and the regulation of their activity for pollen germination and/or tube growth.
PME Isoforms Present in N. tabacum Pollen Tubes
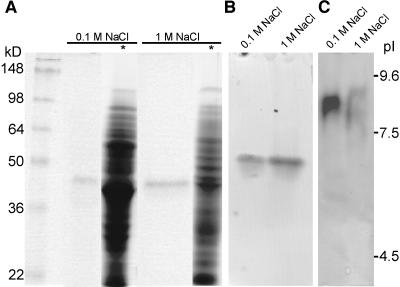
We have seen that in Arabidopsis pollen many different PME isoforms are highly expressed. To establish how many PME isoforms are present in N. tabacum pollen tubes, proteins were extracted with a low- and high-salt buffer and stained for PME activity after electrophoretic separation. A single band with an apparent molecular mass of approximately 40 kD appeared upon activity staining after SDS-PAGE, which corresponds to the predicted molecular mass of the mature PME encoded by NtPPME1 (Fig. 5A). Also, after native acidic continuous PAGE, only one active PME band could be detected (Fig. 5B). The pollen tube proteins extracted with a low-salt buffer showed one major band, corresponding to a pI of about 8.5, after separation by isoelectric focusing (IEF) followed by activity staining. Upon reextraction of the remaining pellet with the high-salt buffer, two bands corresponding to pIs of approximately 8 and 9 appear (Fig. 5C), which may indicate the presence of more isoforms or may reflect posttranslational modifications of the same isoform. These data suggest that the isolated NtPPME1 clone encodes the principal PME expressed in N. tabacum pollen tubes since both the apparent molecular mass of approximately 40 kD and pI of 8.5 match closely with the values predicted for the mature NtPPME1 (respectively, 39–41.5 kD and pI 8.6; Figs. 3B and 4). However, further analysis of the PME isoforms expressed in Arabidopsis pollen (Table II) reveals that the predicted pI (8.7–10.0) and molecular mass (36.2–39.3 kD) of most deduced PME domains are very similar to those obtained upon PME activity staining of N. tabacum pollen tube protein extracts. This indicates that, as in Arabidopsis pollen, multiple PME isoforms might be expressed in N. tabacum pollen tubes.
Figure 5.
PME activity staining after electrophoretic separation of N. tabacum pollen tube protein extracts. Activity staining after SDS-PAGE (A), after native acidic continuous PAGE (B), and IEF (C). *, Coomassie Brilliant Blue staining; 0.1 m NaCl, proteins extracted with low-salt extraction buffer; 1 m NaCl, remaining pellet after low-salt extraction reextracted with high-salt buffer; 25 μL protein extract was loaded in each lane.
The presence of multiple PME isoforms in N. tabacum pollen tubes (four acidic, one neutral, and two basic) is reported by Li et al. (2002). The fact that we are unable to detect the acidic and neutral isoforms might be related to the different PME activity-staining methods that have been used. However, it is a concern that the heterologous anti-flax PME antibody used by Li et al. (2002) recognized two isoforms (pI 5.5 and 7.3) with molecular masses of about 160 kD after SDS-PAGE. By contrast, the molecular masses of PMEs purified from other plant species range between 22 and 62 kD, with most of them being in the 30- to 40-kD range (Gaffe et al., 1997; Ren and Kermode, 2000; Table II). We have been unable to confirm the presence of these high molecular mass (160 kD) PME isoforms upon activity staining after SDS-PAGE (Fig. 5A) with the sensitive assay described by Hou and Lin (1998).
Localization of GFP-Tagged NtPPME1 Proteins
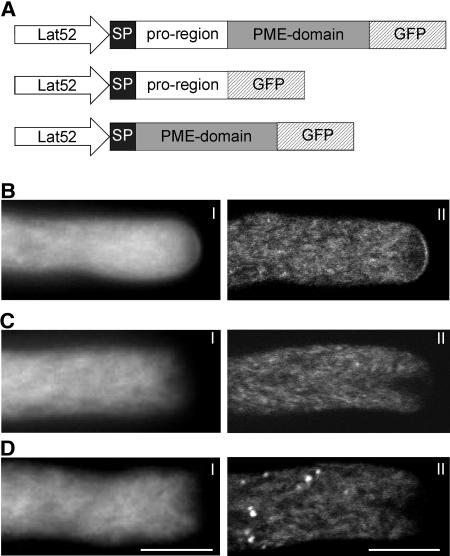
N. tabacum pollen tubes were transformed by microprojectile bombardment with various chimeric DNA constructs containing NtPPME1 cDNA, or its deletion variants, and a C-terminal GFP under the control of the pollen-specific Lat52 promoter (Twell et al., 1989; Fig. 6A).
Figure 6.
Localization of GFP-tagged NtPPME1 proteins. A, Chimeric gene constructs used for expression in N. tabacum pollen tubes, all under the control of the Lat52 promoter. Expression profiles of NtPPME1-GFP (B), SP-pro-region-GFP (C), and SP-PME-domain-GFP (D) are shown. For each expression profile, a wide-field (I) and a median plane confocal (II) image are shown. Scale bar = 10 μm.
In pollen tubes transformed with the complete NtPPME1 gene (pre-pro-PME-GFP), fluorescence labeling appeared to be associated with the ER network and Golgi dictyosomes, both of which are known components of the secretory pathway (Fig. 6B). Curiously, labeling is markedly reduced in the apical inverted cone, a domain previously thought to be an accumulation of secretory vesicles (Derksen et al., 1995). Importantly, a distinct fluorescent signal occurs at the pollen tube apical cell surface, which indicates that NtPPME1 has been correctly targeted via the secretory pathway to the apical cell wall. Note here that the fluorescence is brightest at the extreme apex of the tube, and that the intensity falls off within a few microns from the polar axis, such that no staining is observed along the shank of the tube.
In marked contrast to the results above, the expression of NtPPME1 in which the PME domain was deleted (pre-pro-GFP) shows no label in the pollen tube wall (Fig. 6C). However, within the cytoplasm, there is a marked similarity between this construct and the full-length pre-pro-PME-GFP. Thus, the ER network and Golgi dictyosomes are strongly labeled, while the region of the inverted cone is relatively unlabeled. A similar fluorescence pattern has been obtained in pollen tubes transformed with NtPPME1 in which the pro-region was deleted (pre-PME-GFP; Fig. 6D; Supplemental Movie 1). These results show that exocytosis to the cell wall requires the presence of both the pro-region and the PME domain since deletion of either one disrupts the targeting process.
The Effect of Overexpressed NtPPME1 Proteins on Pollen Tube Growth
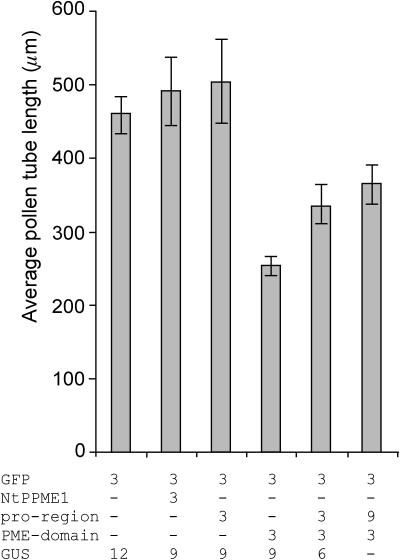
To determine whether overexpression of the whole protein as well as its separate domains has an effect on the pollen tube growth, pollen tubes were cotransformed with the gene of interest and Lat52-GFP, which served as a marker for transformation. Overexpression of NtPPME1, as well as the separate pro-region, did not significantly affect pollen tube growth (Fig. 7). However, overexpression of the PME domain dramatically reduced the pollen tube growth rate as the average pollen tube lengths reached only about one-half the size of the control tubes (Fig. 7). Interestingly, partial rescue of tube growth can be achieved by coexpression of the separate pro-region and PME domain (Fig. 7). These results suggest that the PME domain alone is sufficient for its hydrolytic activity and that the pro-region, directly or indirectly, can rescue the observed inhibitory effect caused by the PME domain.
Figure 7.
The effect of the various NtPPME1 protein domains on pollen tube growth. Average lengths of N. tabacum pollen tubes are shown 6 h after transformation by microprojectile bombardment. Pollen was transformed with the indicated amounts of DNA all under the control of the Lat52 promoter. All pollen grains were cobombarded with 3 μg of Lat52-GFP as a marker for transformation. Lat52-GUS was used as mock to ensure equal amounts (15 μg) of DNA used for each transformation. Bars represent the mean values from at least four independent experiments with n ≥ 50 each. Error bars indicate the se obtained for the mean values.
Expression of the PME Domain Alters the Methylesterification Profile of Pectins
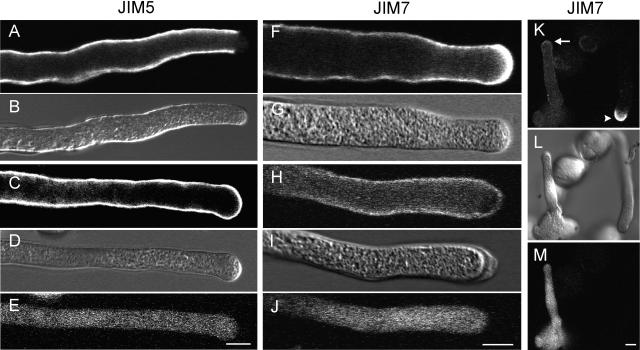
The distribution of methylesterified and deesterified pectins can be studied using the monoclonal antibodies JIM5 and JIM7. JIM5 has been shown to bind preferably to at least four contiguous unesterified GalUA residues with adjacent or flanking methylesterified residues. By contrast, JIM7 binds to a relatively highly methylesterified pectin epitope (Clausen et al., 2003).
To determine whether expression of the PME domain fused to the SP of NtPPME1 affects the degree of pectin methylesterification, pollen tubes were cotransformed with Lat52-pre-PME and Lat52-GFP. Pollen tubes were grown for 6 h, chemically fixed, and labeled with the antipectin antibodies. In nontransformed pollen tubes, very few JIM5 epitopes are present in the apical cell wall (Fig. 8A), indicating that unesterified pectins are scarce in this region. The accumulation of unesterified pectins only starts abruptly in the subapical cell wall region and continues evenly along the more distal parts of the pollen tube (Fig. 8A). By contrast, in pollen tubes that express the PME domain (lacking the pro-region), JIM5 epitopes are present along the entire pollen tube wall including the apical cell wall (Fig. 8C).
Figure 8.
Comparison of the pectin distribution between transformed pollen tubes expressing the SP-PME-domain (as well as a GFP marker) and nontransformed pollen tubes. DIC images are included as a reference (B, D, G, I, and L), while a GFP signal identifies transformed pollen tubes (E, J, and M). In nontransformed pollen tubes, labeling for JIM5, which detects preferably deesterified pectins, is evenly distributed along the cell wall but absent in the apical region (A). On the contrary, in transformed pollen tubes expressing the PME domain, the JIM5 signal is also present in the apical cell wall region (C). Labeling for JIM7, which detects esterified pectins, is very high in the apical region of nontransformed pollen tubes (F and K arrowhead), while in transformed tubes JIM7 labeling is much weaker at the apical cell wall (H and K arrow). Scale bar = 10 μm.
The methylesterified epitopes recognized by JIM7 are abundantly present at the very apex of nontransformed pollen tubes, while labeling is significantly lower in the subapical and distal wall regions (Fig. 8F). The apical cell wall labeling by JIM7 is dramatically reduced in pollen tubes expressing the PME domain (Fig. 8H). Figure 8K, in which the arrow indicates the tip of a transformed tube while the arrowhead points to the tip of an untransformed tube, shows another example for this change in the apical accumulation of methylesterified pectins caused by the expression of the PME domain. These immunolocalization data show that expression of the PME domain, lacking the pro-region, results in the accumulation of deesterified pectins in the apical pollen tube wall and thus support our hypothesis that the PME domain alone is sufficient for exerting its enzymatic activity.
DISCUSSION
PME Affects Pollen Tube Growth
PMEs have been implicated in a number of processes, including cell wall extension, fruit maturation and senescence, pathogenesis, systemic movement of tobacco mosaic virus, cambial cell differentiation, and border cell separation from root caps (Moustacas et al., 1991; Tieman and Handa, 1994; Bordenave, 1996; Wen et al., 1999; Micheli et al., 2000; Chen and Citovsky, 2003). Although it has been suggested that PMEs also participate in pollen tube growth (Mu et al., 1994; Wakeley et al., 1998), evidence for this has only recently been presented in a study in which the functional interruption of an Arabidopsis pollen PME, called VANGUARD1 (VGD1), reduced the total PME activity in pollen to 82% of the wild-type pollen. This leads to pollen tubes that burst when grown in vitro and display retarded growth in vivo (Jiang et al., 2005). Notably, At2g47040, which encodes VGD1, represents the Arabidopsis pollen PME isoform that showed the highest expression level in the transcriptome study by Pina et al. (2005; Table II).
The demethylesterification of pectins by PME can have opposite consequences for the plasticity of the cell wall. On one hand, PMEs produce acidic pectins, which can bind Ca2+ and reinforce the wall structure. On the other hand, the protons produced by the hydrolysis decrease the local cell wall pH and, consequently, can promote the activity of cell wall hydrolases and result in cell extension and growth (Gaffe et al., 1994; Bordenave, 1996). Which of these two effects prevails might depend on the PME isoform itself, the local pH, the availability of cations, the presence of cell wall-loosening hydrolases, or even the degree and distribution of esterification on the secreted pectins (Moustacas et al., 1991; Charnay et al., 1992; Bordenave, 1996; Catoire et al., 1998).
Experiments in which L. formosanum and N. tabacum pollen tubes were exposed to exogenous orange peel PME resulted in a significant thickening of the apical cell wall (Fig. 2, A and B) and inhibition of tube growth (Fig. 1), indicating that PME can alter the apical cell wall dynamics. This is in agreement with recent data from Parre and Geitmann (2005), which show that Solanum pollen tubes treated with orange peel PME exhibit a dramatic increase in stiffness and reduction of viscosity at the apex. We also found that the expression of the PME domain of the endogenous NtPPME1 inhibits N. tabacum pollen tube growth (Fig. 7). Often the tubes expressing the PME domain showed apical thickening of the cell wall similar to the tubes exposed to the exogenous PME (e.g. see Fig. 8I). Different mechanisms need to be considered for the inhibition of pollen tube growth caused by exogenous PME treatment versus endogenous PME expression. These are discussed below.
Exogenous PME
The simplest explanation for the induced inhibition of growth by exogenous PME is that the enhanced extracellular enzyme activity generates extra carboxyl groups on the pectin residues in the cell wall, which are then cross-linked by calcium (Goldberg et al., 1986; Holdaway-Clarke and Hepler, 2003). Although secretion continues, the calcium-stiffened cell wall at the tip inhibits turgor-induced cell expansion, and instead leads to cell wall thickening. It is known that inhibition of pollen tube elongation by various means results in a concomitant dissipation of the intracellular tip-focused Ca2+ gradient (Rathore et al., 1991; Pierson et al., 1994). This also occurs when L. formosanum pollen tube growth is inhibited by the application of exogenous PME (Fig. 2C). The dissipation of the Ca2+ gradient might result from the cell wall stiffening and thickening, which prevents the influx of extracellular Ca2+ across the plasma membrane (Malho et al., 1995; Holdaway-Clarke et al., 1997). It has been proposed that stretch-activated channels control Ca2+ influx at the apex (Pierson et al., 1994; Feijo et al., 1995; Malho et al., 1995), an idea that receives support from the recent identification of such channels in pollen tube tip protoplasts from L. longiflorum (Dutta and Robinson, 2004).
Endogenous PME
We know from our localization studies with pre-PME-GFP that the expressed PME domain is not targeted to the apical cell wall (Fig. 6D; Supplemental Movie 1) and therefore cannot exert its enzymatic activity on the pectins in the wall. How, then, can we explain the inhibition of pollen tube growth by endogenous expression of the PME domain (Fig. 7)? It is known that the homogalacturonan fraction of pectins is synthesized in the Golgi, where 70% to 80% of its GalUA residues are methylesterified (Staehelin and Moore, 1995; Willats et al., 2001). Upon secretion, these esterified pectins accumulate in the apical pollen tube wall, confirmed by our JIM7 labeling of nontransformed pollen tubes (Fig. 8F). Deesterified pectins, labeled with JIM5, are not detected at the very apex (Fig. 8A), indicating that there is little to no secretion of these pectins. Expression of pre-PME dramatically alters the methylesterification profile of pectins; deesterified pectins accumulate in the apical pollen tube wall while methylesterified pectins become far less abundant in this region (Fig. 8, C, H, and K). These results support the idea that the absence of the pro-region, and the resulting accumulation of PME enzyme activity in the Golgi, leads to premature, intravesicular, demethylesterification of pectins. In the presence of Ca2+, these deesterified pectins might already form pectate gels in the secretory vesicles or immediately upon secretion into the cell wall, thereby altering the rheological properties of the apical wall with the concomitant cessation of growth. The inhibitory effect of the PME domain, together with the observation that overexpression of the whole NtPPME1 protein, including the pro-region, does not significantly affect pollen tube growth (Fig. 7), indicates the tight spatial regulation that must exist for the control of PME enzyme activity.
Identification of a Pre-Pro-PME in N. tabacum Pollen Tubes
Screening of a N. tabacum pollen cDNA library resulted in the isolation of NtPPME1, which is specifically expressed in pollen and pollen tubes (Fig. 4). Based on sequence analysis of the Arabidopsis genome, it has been proposed that PME genes may be divided into two classes. Type I genes contain only two or three introns and encode a long pro-region, whereas type II genes contain five or six introns and encode either a short or no pro-region (Micheli, 2001). Although the genomic structure of the NtPPME1 gene is unknown, database analysis clearly indicates that this gene belongs to the type I genes, since it encodes a long pro-region (Fig. 3B). The positions of the catalytically important residues Asp-365, Asp-386, and Arg-454, which are predicted to be present at the bottom of the active site cleft of PMEs from both plants and phytopathogenic microorganisms (Jenkins et al., 2001; Johansson et al., 2002; D'Avino et al., 2003), are conserved in the PME domain of NtPPME1.
The Pro-Region Regulates PME Targeting and Enzyme Activity
The pro-region has been suggested to play a role in the targeting of PMEs toward the cell wall, to function as a chaperone in the correct conformational folding of the mature PME, and/or to act as an inhibitor of the enzyme activity carried out by the mature PME (Micheli, 2001). Our results provide direct support for a role of the pro-region in PME targeting. We also promote the concept of the pro-region being able to inhibit PME activity. However, we reject the idea that the pro-region functions as an intramolecular chaperone. These conclusions are discussed below.
Targeting of PME
Plant PMEs are generally secreted via the exocytotic pathway into the cell wall to which they are believed to be ionically bound based on their alkaline pIs. The labeling observed in our GFP expression studies for NtPPME1 is similar to the pattern typically found using fluorescence markers for the ER and Golgi (Parton et al., 2003). Thus, the labeling of these components of the secretory pathway, in combination with the clear labeling of the apical cell wall, show that NtPPME1 is also secreted via the exocytotic pathway. The inverted cone at the tube apex has traditionally been associated with the region to which secretory vesicles are transported prior to exocytosis at the apical plasma membrane (Derksen et al., 1995). Interestingly, we do not find significant labeling in this region, suggesting that the route followed by NtPPME1-loaded secretory vesicles is different from the one commonly assumed. In nonapical cell wall regions, fluorescence is low or absent, which might be related to quenching of the signal due to the acidic pH of the pollen tube cell wall (Brandizzi et al., 2004). Apparently, physiochemical conditions of the cell wall are such that fluorescence of GFP-tagged NtPPME1 proteins can only be detected at the moment of secretion into the apical cell wall or shortly thereafter. Deletion of either the pro-region or the PME domain from NtPPME1 disrupts the transport from the Golgi into the secretory vesicles and, consequently, also targeting to the cell wall (Fig. 6, C and D). Together, these results indicate that the presence of the pro-region is necessary for correct targeting of the mature protein to the cell wall.
Inhibitor of PME Activity
Conserved Domain Search predicts that the pro-region of NtPPME1 belongs to that group of proteins referred to as plant PMEIs (Marchler-Bauer and Bryant, 2004). This is a protein family predominantly derived from homology with the first functionally and structurally characterized PMEI isolated from kiwi fruit (Balestrieri et al., 1990; Giovane et al., 1995; Camardella et al., 2000). The kiwi (Actinidia chinensis) PMEI with a molecular mass of 16.3 kD and predicted pI of 4.45 was found to inhibit various plant PMEs through the formation of a 1:1 complex, whereas it was inactive against fungal PMEs (Balestrieri et al., 1990; Giovane et al., 1995; Camardella et al., 2000). Recently, two genes from Arabidopsis, AtPMEI1 and AtPMEI2, have been shown to encode functional PMEIs (Wolf et al., 2003). The formation of disulfide bridges is essential for the maintenance of the secondary structure and therefore important for PMEI activity (Camardella et al., 2000; Wolf et al., 2003; Giovane et al., 2004); these Cys residues are conserved in the pro-region of NtPPME1.
Provisional support for the inhibitory activity of the pro-region derives from our studies showing that the blockage of tube growth by expression of the PME domain can be partially rescued by coexpression of the pro-region (Fig. 7). That the recovery of growth is not complete might be explained by the fact that intramolecular inhibition is more effective than the intermolecular inhibition seen in our experiment. At this stage, we cannot resolve whether this recovery of growth is the result of direct enzymatic inhibition, due to the interaction of the pro-region with the active site of the PME domain, as has been shown for the interaction between the kiwi PMEI and a PME from tomato (Lycopersicon esculentum; D'Avino et al., 2003), or an indirect event, due to the fact that interaction between the pro-region and the PME domain somehow reestablishes correct targeting of the mature PME to the cell wall. Given the known structural similarity between the pro-region and the kiwi PMEI, a direct inhibition mechanism seems to be the more likely option.
Chaperone Function
Since our localization data indicate that the GFP-tagged PME domain appears to be retained in the ER and the Golgi (Fig. 5), there is a possibility that it might be folded incorrectly and that the pro-region thus functions in part as a chaperone. However, the fact that overexpression of the PME domain dramatically alters the apical methylesterification profile and significantly reduces the pollen tube growth indicates that this domain itself, without the presence of the pro-region, is enzymatically active and thus able to fold correctly. Therefore, we reject the idea that the pro-region functions as an intramolecular chaperone.
CONCLUSIONS
In this study, we confirm and extend the idea that PMEs contribute to the regulation of pollen tube growth. Both endogenous expression and exogenous application of PME inhibit pollen tube growth, probably through changes in the rheological properties of the apical pectinaceous tube wall. We also show that our pollen-specific NtPPME1 is encoded as a pre-pro-PME. However, we provide experimental data revealing that the pro-region participates in the correct targeting of PME and give support to the idea that this region can serve as an inhibitor of PME activity, possibly preventing the premature deesterification of pectins prior to secretion. Future studies need to address in more detail the mechanism by which correct targeting of PME is achieved, and how the inhibition as well as stimulation of PME activity is regulated.
MATERIALS AND METHODS
Pollen Tube Growth Conditions
Lilium formosanum pollen was germinated and cultured in growth medium consisting of 7% (w/v) Suc, 0.1 m KCl, 1.6 mm H3BO3, and 15 mm MES buffer adjusted to pH 5.7 with KOH. Nicotiana tabacum (cv Petit Havana SR1) pollen was germinated and cultured in medium consisting of 2% (w/v) Suc, 15% PEG-3350, 0.01% (w/v) H3BO3, 0.01% (w/v) HNO3, 0.02% (w/v) MgSO4, 0.07% (w/v) CaCl2, and 20 mm MES buffer, pH 6. Pollen tubes were germinated and grown on a rotor at room temperature. For microscopic observations, a pollen suspension was plated and immobilized with a growth medium solution containing a final concentration of 0.7% (w/v) low-melting agarose. The N. tabacum low-melting agarose solution contained 7% (w/v) Suc instead of PEG-3350. The immobilized pollen tubes were covered with 1 mL of liquid growth medium and placed in a moist chamber until observation.
In Vitro Growth Rate Experiments with Orange Peel PME
Pollen tubes were grown as described before. After at least 1 h of germination, a pollen tube sample was taken and stored at 4°C to stop growth. The remaining pollen tube culture was then divided into smaller aliquots, the pollen tubes were allowed to settle, and the growth medium was replaced with media containing the experimental concentrations of orange (Citrus sinensis) peel PME (Sigma, St. Louis). Orange peel PME stock solutions (0.1 unit mL−1 or 1 unit mL−1) were prepared in 25 mm sodium phosphate, pH 7.5. Boiled preparations (10 min) of orange peel PME did not show any PME activity staining (Hou and Lin, 1998) and were used as a control. The pollen tubes were cultured for at least 1 more hour, after which the cultures were stored at 4°C. Slides were prepared with pollen tube samples and low-magnification pictures were acquired with a digital camera coupled to the dissecting microscope. Scion image software (Scion, Frederick, MD) was used to measure the length of at least 100 pollen tubes in each sample, and the average pollen tube growth rate was calculated for each different sample. Data shown represent average ± sd of three independent experiments with n ≥ 100 each.
Screening of a N. tabacum Pollen cDNA Library
A N. tabacum (cv Petit Havana SR1) pollen cDNA library (Chen et al., 2002) was screened for PMEs according to standard protocols using the 32P-labeled Petunia inflata PPE1 cDNA (Mu et al., 1994) as a probe. After three rounds of screening, phagemids were excised from pure phages, according to the manufacturer's protocol (Stratagene, La Jolla, CA), and several of the cDNA inserts were sequenced leading to the isolation of the full-length NtPPME1 cDNA clone.
RT-PCR Expression Analysis
Total RNA from the different N. tabacum tissues was isolated using the RNeasy plant mini kit (Qiagen, Chatsworth, CA). First-strand cDNA synthesis was performed using AMV reverse transcriptase (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions, using 2.5 μg RNA and oligo(dT) primers. Two microliters of cDNA from the first-strand reaction were used as template for PCR with NtPPME1 specific primers: forward, 5′-GAAATGCAATCATGAACACTGATTTG-3′; reverse, 5′-CATTTGTGGTTTTGCTGCTAGGAAC-3′, giving a 1,081-bp product. As an internal control, N. tabacum actin was amplified in the same PCR reaction using the following primers: forward, 5′-GATGCCTATGTTGGTGATGAAGCTC-3′; reverse, 5′-CACCATCACCAGAGTCCAACACAAT-3′, giving a 400-bp product.
Arabidopsis Transcriptome Data
The relative expression levels used for the Arabidopsis (Arabidopsis thaliana) transcriptome data in Tables I and II were provided by Pina et al. (2005). For the analysis presented in Table I, expression levels were categorized as indicated, and the number of genes falling into each category counted. For the data presented in Table II, the deduced amino acid sequence of PME genes that have a present call with expression levels higher than 200 were analyzed using the NCBI Conserved Domain Search (Marchler-Bauer and Bryant, 2004). In the situation in which a PME gene encoded a pro-region, the putative start of the PME domain was positioned halfway between the end of the predicted pro-region (pfam04043; plant invertase/PME inhibitor) and the beginning of the predicted PME domain (pfam01095; pectinesterase). The amino acid sequence from the putative start of the PME domain until the carboxy terminus was used to estimate the pI and molecular mass. The pI and molecular mass from PMEs that do not contain a pro-region were calculated based on the deduced amino acid sequence from which the predicted SP (Nielsen et al., 1997) was excluded.
Extraction of PMEs from Pollen Tubes
N. tabacum pollen (100 mg) was germinated in 10-mL germination medium and cultured for 7 h at 25°C. After a mild centrifugation, the culture medium was removed and 0.5 mL low-salt extraction buffer (10 mm Tris buffer, pH 7.7, 0.1 m NaCl) was added to the pollen tube pellet and tubes were disrupted with a small Potter homogenizer. The homogenate was transferred into a new tube, and proteins were extracted by repeated vortexing (20 s) and incubation on ice (20 min). The extract was centrifuged (14,000g, 10 min), and the supernatant was collected (low-salt protein extract). The remaining proteins in the pellet were reextracted with 0.5-mL high-salt extraction buffer (10 mm Tris buffer, pH 7.7, 1 m NaCl) using the same procedure. The protein extracts were desalted using Microcon YM-10 centrifugal filter units (Millipore, Billerica, MA) into 10 mm Tris buffer, pH 7.7.
PME Activity Staining following SDS-PAGE
Protein samples were incubated overnight at room temperature with sample buffer in a 3:1 ratio (sample buffer: 60 mm Tris buffer [pH 6.8], 2% [w/v] SDS, 2% [v/v] β-mercaptoethanol, 25% [v/v] glycerol, and 0.05% [w/v] bromphenol blue). Samples were separated with SDS-PAGE, according to the discontinuous buffer system of Laemmli (1970), using 10% SDS-polyacrylamide gels (Mini-Protean II apparatus; Bio-Rad Laboratories, Hercules, CA). Proteins were visualized by Coomassie Brilliant Blue R-250 or subjected to PME activity staining, according to Hou and Lin (1998). Gels were incubated (twice for 10 min) in 10 mm Tris buffer (pH 7.9) containing 25% (v/v) isopropanol, followed by two washes in 10 mm Tris buffer, pH 7.9. The substrate dye solution was freshly prepared and consisted of 40 mg β-naphthyl acetate in 16 mL of N,N-dimethylformamide that was brought to 160 mL with 144 mL of 10 mm Tris buffer, pH 7.9, in which 80 mg of tetrazotized o-dianisidine was dissolved. The gels were incubated in this solution for 20 min in the dark at 37°C and destained with 10% acetic acid.
PME Activity Staining Following Acidic Continuous Native PAGE
Protein samples were separated by acidic continuous native PAGE, as described by Ren and Kermode (2000). The protein gel contained 10% (w/v) acrylamide, 24 mm KOH, 0.86% (v/v) glacial acetic acid, 0.075% (w/v) ammonium persulfate, and 0.5% (v/v) N,N,N′,N′-tetramethylethylenediamine, pH 4.3. The running buffer contained 24 mm KOH, 0.86% (v/v) glacial acetic acid, pH 4.3. Protein samples were mixed with an equal volume of sample buffer (24 mm KOH, 0.86% [v/v] glacial acetic acid, 10% [w/v] glycerol, and 0.5 μL of methyl green dye, pH 4.3). Electrophoresis took place at 100 V, 4°C for 3.5 h, with the polarity reversed (Mini-Protean II apparatus; Bio-Rad), after which the gel was equilibrated for 5 min in a solution containing 6.25 mL of 0.1 m citrate and 12.5 mL of 0.2 m Na2HPO4, pH 6.3, followed by a 90-min incubation in the same solution plus 0.5% (w/v) 90% esterified pectin as substrate at 37°C. Following a brief rinse with water, gels were stained with 0.02% (w/v) ruthenium red and destained with water.
PME Activity Staining following IEF Gel Electrophoresis
IEF gel electrophoresis was performed using IEF Ready Gels (pH 3–pH 10) together with the Mini-Protean II apparatus, according to the manufacturer's instruction (Bio-Rad). Following electrophoresis, the staining procedure with ruthenium red was the same as described for the acidic continuous native gel, except that the incubation time with the esterified pectin substrate was only 30 min.
Chimeric Gene Constructs and Microprojectile Bombardment
Standard recombinant DNA methodology was used in constructing all of the chimeric genes used in this study. All fusion genes were cloned behind the pollen-specific promoter Lat52 (Twell et al., 1990), and GFP was always expressed as a C-terminal fusion protein. For expression of the pro-region, including the SP (encoded by the pre-region), amino acids 1 to 190 of NtPPME1 were expressed, while, for the expression of the PME domain, including the SP, the nucleotide sequence corresponding to amino acids 175 to 555 was cloned behind the nucleotides that give rise to amino acids 1 to 40 to express the SP. Although the predicted cleavage site for the SP is between amino acids 22 and 23, the length of the N-terminal sequence was extended since the expression of amino acids 1 to 31 did not always result in correct targeting of the proteins to the secretory pathway. Similar strategies were followed and the same domain annotations were used for the chimeric constructs without GFP fusion.
Microprojectile bombardment was performed using the helium-driven PDS-1000/He biolistic system (Bio-Rad). Tungsten particles (1.1 μm) were coated with plasmid DNA, according to the manufacturer's recommendation (Bio-Rad). For the GFP localization studies, typically 1 to 3 μg DNA and, for the pollen tube growth experiments, 15 μg DNA, were used to coat 3 mg of tungsten particles, which were used to bombard an individual pollen sample (10 mg). For the growth experiments, Lat52-GUS was included in the DNA samples used for bombardment to ensure that the pollen grains were transformed with equivalent amounts of DNA. For immunofluorescence, individual pollen samples (10 mg) were bombarded with tungsten particles (3 mg) coated with a DNA mixture of 3 μg Lat52-pre-PME and 3 μg Lat52-GFP. The parameters for bombardment were as described by Chen et al. (2002).
Immunofluorescence
Pollen was cobombarded with Lat52-pre-PME and Lat52-GFP, and pollen tubes were germinated and grown for 6 h in liquid growth medium under the conditions described. After mild centrifugation with a hand centrifuge, the growth medium was removed and the pollen tubes were fixed in 3% (w/v) formaldehyde in 50 mm PIPES buffer, pH 6.7, containing 1 mm EGTA and 0.5 mm MgCl2 for 15 min. Following three brief washes in phosphate-buffered saline (PBS), the samples were incubated with the monoclonal antibodies JIM5 and JIM7, diluted 1:50 in PBS containing 1% (w/v) bovine serum albumin overnight at 4°C. Subsequently, the samples were washed three times in PBS and incubated with Alexa Fluor 555 goat anti-rat IgG (Molecular Probes, Eugene, OR), diluted 1:400 in PBS containing 1% (w/v) bovine serum albumin for 1 h at 37°C. After three brief washes in PBS, the pollen tubes were transferred to a microscope slide, mounted in an antifade solution (10% [v/v] PBS, 90% [v/v] glycerol, and 4% [w/v] n-propyl gallate), and examined on a fluorescence microscope. Transformed and nontransformed pollen tubes could be distinguished based on the expression of GFP in the transformed tubes.
Microscopy and Imaging
Differential interference contrast (DIC) and wide-field epi-fluorescence images were acquired using a Nikon Eclipse TE300 inverted microscope, 40×oil immersion lens, N.A. 1.3 (Nikon, Melville, NY), and a Cool Snap CCD camera (Photometrics, Tucson, AZ) driven by MetaMorph software (Universal Imaging, West Chester, PA). To test the effect of orange peel PME on pollen tube morphology, the growth medium in the microscope slide chamber was replaced by growth medium supplemented with the orange peel PME, and DIC images were taken at regular time intervals. Boiled preparations of orange peel PME were used as a control. Ratiometric imaging of [Ca2+]i was performed as previously described (Roy et al., 1999) by pressure injection of L. formosanum pollen tubes with fura-2-dextran (Molecular Probes), after which the growth medium was supplemented with 10 units mL−1 orange peel PME. Confocal GFP images were obtained using the Zeiss LSM 510 Meta confocal system (Zeiss, Jena, Germany) at the University of Massachusetts Central Microscopy Facility using a 60× oil immersion lens, N.A. 1.4, and enhanced GFP filter sets. For immunostaining with the JIM antibodies, dual GFP and immunofluorescence were imaged in a multichannel setting (excitation 488 and 543 nm, emission 505- to 530-nm bandpass filter and 560-nm cutoff filter, respectively, for GFP and immunofluorescence), using a 25× multi-immersion lens.
For length measurements of GFP-expressing tubes, wide-field epi-fluorescence images were taken using a Nikon Diaphot 300 inverted microscope with a 10× lens (Nikon). Pollen tube lengths were measured using MetaMorph software (Universal Imaging).
Sequence data of NtPPME1 have been deposited with the NCBI/GenBank data libraries under accession number AY772945.
Supplementary Material
Acknowledgments
We gratefully thank Teh-hui Kao (Department of Biochemistry and Molecular Biology, Penn State University), who generously provided the Petunia inflata PPE1 clone. José Feijó and Jörg Becker (Centro de Biologia do Desenvolvimento, Instituto Gulbenkian de Ciência, Oeiras, Portugal) are acknowledged for sharing their Arabidopsis ATH1 genome array data with us. Paul Knox (Centre for Plant Sciences, University of Leeds, UK) is thanked for his generous gift of the JIM5 and JIM7 antibodies. We also thank Hen-ming Wu for helpful suggestions and for sharing molecular tools and expertise, and Maura Cannon for the use of her biolistic microprojectile apparatus. Luis Cardenas is thanked for help with the ratiometric imaging. We thank Sheila McCormick for the Lat52 promoter. Confocal microscopy was performed at the Central Microscopy Facility of the University of Massachusetts.
This work was supported by the National Science Foundation (grant no. MCB–0077599 to P.K.H. and grant no. BBS–8714235 to the University of Massachusetts Central Microscope Facility), and by the U.S. Department of Agriculture (grant no. 0101936 to A.Y.C.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.059865.
References
- Albani D, Altosaar I, Arnison PG, Fabijanski SF (1991) A gene showing sequence similarity to pectin esterase is specifically expressed in developing pollen of Brassica napus. Sequences in its 5′ flanking region are conserved in other pollen-specific promoters. Plant Mol Biol 16: 501–513 [DOI] [PubMed] [Google Scholar]
- Balestrieri C, Castaldo D, Giovane A, Quagliuolo L, Servillo L (1990) A glycoprotein inhibitor of pectin methylesterase in kiwi fruit (Actinidia chinensis). Eur J Biochem 193: 183–187 [DOI] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijo JA (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenave M (1996) Analysis of pectin methyl esterases. In HF Linskens, JF Jackson, eds, Plant Cell Wall Analysis, Vol 17. Springer-Verlag, Berlin, pp 165–180
- Brandizzi F, Irons SL, Johansen J, Kotzer A, Neumann U (2004) GFP is the way to glow: bioimaging of the plant endomembrane system. J Microsc 214: 138–158 [DOI] [PubMed] [Google Scholar]
- Camardella L, Carratore V, Ciardiello MA, Servillo L, Balestrieri C, Giovane A (2000) Kiwi protein inhibitor of pectin methylesterase—amino-acid sequence and structural importance of two disulfide bridges. Eur J Biochem 267: 4561–4565 [DOI] [PubMed] [Google Scholar]
- Catoire L, Pierron M, Morvan C, du Penhoat CH, Goldberg R (1998) Investigation of the action patterns of pectinmethylesterase isoforms through kinetic analyses and NMR spectroscopy—implications in cell wall expansion. J Biol Chem 273: 33150–33156 [DOI] [PubMed] [Google Scholar]
- Charnay D, Nari J, Noat G (1992) Regulation of plant cell-wall pectin methyl esterase by polyamines—interactions with the effects of metal-ions. Eur J Biochem 205: 711–714 [DOI] [PubMed] [Google Scholar]
- Chen CY, Wong EI, Vidali L, Estavillo A, Hepler PK, Wu HM, Cheung AY (2002) The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 14: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Citovsky V (2003) Systemic movement of a tobamovirus requires host cell pectin methylesterase. Plant J 35: 386–392 [DOI] [PubMed] [Google Scholar]
- Clausen MH, Willats WGT, Knox JP (2003) Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr Res 338: 1797–1800 [DOI] [PubMed] [Google Scholar]
- D'Avino R, Camardella L, Christensen T, Giovane A, Servillo L (2003) Tomato pectin methylesterase: modeling, fluorescence, and inhibitor interaction studies—comparison with the bacterial (Erwinia chrysanthemi) enzyme. Proteins 53: 830–839 [DOI] [PubMed] [Google Scholar]
- Derksen J, Rutten T, Lichtscheidl IK, de Win AHN, Pierson ES, Rongen G (1995) Quantitative analysis of the distribution of organelles in tobacco pollen tubes: implications for exocytosis and endocytosis. Protoplasma 188: 267–276 [Google Scholar]
- Dutta R, Robinson KR (2004) Identification and characterization of stretch-activated ion channels in pollen protoplasts. Plant Physiol 135: 1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijo JA, Malho R, Obermeyer G (1995) Ion dynamics and its possible role during in-vitro pollen germination and tube growth. Protoplasma 187: 155–167 [Google Scholar]
- Ferguson C, Teeri TT, Siika AM, Read SM, Bacic A (1998) Location of cellulose and callose in pollen tubes and grains of Nicotiana tabacum. Planta 206: 452–460 [Google Scholar]
- Gaffe J, Tieman DM, Handa AK (1994) Pectin methylesterase isoforms in tomato (Lycopersicon esculentum) tissues—effects of expression of a pectin methylesterase antisense gene. Plant Physiol 105: 199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffe J, Tiznado ME, Handa AK (1997) Characterization and functional expression of a ubiquitously expressed tomato pectin methylesterase. Plant Physiol 114: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitmann A, Li YQ, Cresti M (1995) Ultrastructural immunolocalization of periodic pectin depositions in the cell wall of Nicotiana tabacum pollen tubes. Protoplasma 187: 168–171 [Google Scholar]
- Giovane A, Balestrieri C, Quagliuolo L, Castaldo D, Servillo L (1995) A glycoprotein inhibitor of pectin methylesterase in kiwi fruit—purification by affinity-chromatography and evidence of a ripening-related precursor. Eur J Biochem 233: 926–929 [DOI] [PubMed] [Google Scholar]
- Giovane A, Servillo L, Balestrieri C, Raiola A, D'Avino R, Tamburrini M, Clardiello MA, Camardella L (2004) Pectin methylesterase inhibitor. BBA-Proteins Proteom 1696: 245–252 [DOI] [PubMed] [Google Scholar]
- Goldberg R, Morvan C, Roland JC (1986) Composition, properties and localization of pectins in young and mature cells of the mung bean hypocotyl. Plant Cell Physiol 27: 417–429 [Google Scholar]
- Holdaway-Clarke TL, Feijo JA, Hackett GR, Kunkel JG, Hepler PK (1997) Pollen tube growth and the intracellular cytosolic calcium gradient oscillate in phase while extracellular calcium influx is delayed. Plant Cell 9: 1999–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdaway-Clarke TL, Hepler PK (2003) Control of pollen tube growth: role of ion gradients and fluxes. New Phytol 159: 539–563 [DOI] [PubMed] [Google Scholar]
- Holdaway-Clarke TL, Weddle NM, Kim S, Robi A, Parris C, Kunkel JG, Hepler PK (2003) Effect of extracellular calcium, pH and borate on growth oscillations in Lilium formosanum pollen tubes. J Exp Bot 54: 65–72 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou WC, Lin YH (1998) Activity staining of pectinesterase on polyacrylamide gels after acidic or sodium dodecyl sulfate electrophoresis. Electrophoresis 19: 692–694 [DOI] [PubMed] [Google Scholar]
- Jenkins J, Mayans O, Smith D, Worboys K, Pickersgill RW (2001) Three-dimensional structure of Erwinia chrysanthemi pectin methylesterase reveals a novel esterase active site. J Mol Biol 305: 951–960 [DOI] [PubMed] [Google Scholar]
- Jiang L, Yang S-L, Xie L-F, Puah CS, Zhang X-Q, Yang W-C, Sundaresan V, Ye D (2005) VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17: 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson K, El-Ahmad M, Friemanna R, Jornvall H, Markovic O, Eklund H (2002) Crystal structure of plant pectin methylesterase. FEBS Lett 514: 243–249 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during assembly of the heads of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Li YQ, Chen F, Linskens HF, Cresti M (1994) Distribution of unesterified and esterified pectins in cell walls of pollen tubes of flowering plants. Sex Plant Reprod 7: 145–152 [Google Scholar]
- Li YQ, Faleri C, Geitmann A, Zhang HQ, Cresti M (1995) Immunogold localization of arabinogalactan proteins, unesterified and esterified pectins in pollen grains and pollen tubes of Nicotiana tabacum L. Protoplasma 189: 26–36 [Google Scholar]
- Li YQ, Mareck A, Faleri C, Moscatelli A, Liu Q, Cresti M (2002) Detection and localization of pectin methylesterase isoforms in pollen tubes of Nicotiana tabacum L. Planta 214: 734–740 [DOI] [PubMed] [Google Scholar]
- Malho R, Read ND, Trewavas AJ, Pais MS (1995) Calcium-channel activity during pollen-tube growth and reorientation. Plant Cell 7: 1173–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32: W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli F (2001) Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci 6: 414–419 [DOI] [PubMed] [Google Scholar]
- Micheli F, Sundberg B, Goldberg R, Richard L (2000) Radial distribution pattern of pectin methylesterases across the cambial region of hybrid aspen at activity and dormancy. Plant Physiol 124: 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustacas AM, Nari J, Borel M, Noat G, Ricard J (1991) Pectin methylesterase, metal-ions and plant cell-wall extension—the role of metal-ions in plant cell-wall extension. Biochem J 279: 351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu JH, Stains JP, Kao TH (1994) Characterization of a pollen-expressed gene encoding a putative pectin esterase of Petunia inflata. Plant Mol Biol 25: 539–544 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, Von-Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Parre E, Geitmann A (2005) Pectin and the role of the physical properties of the cell wall in pollen tube growth of Solanum chacoense. Planta 220: 582–592 [DOI] [PubMed] [Google Scholar]
- Parton RM, Fischer-Parton S, Trewavas AJ, Watahiki MK (2003) Pollen tubes exhibit regular periodic membrane trafficking events in the absence of apical extension. J Cell Sci 116: 2707–2719 [DOI] [PubMed] [Google Scholar]
- Pierson ES, Miller DD, Callaham DA, Shipley AM, Rivers BA, Cresti M, Hepler P (1994) Pollen tube growth is coupled to the extracellular calcium ion flux and the intracellular calcium gradient: effect of BAPTA-type buffers and hypertonic media. Plant Cell 6: 1815–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson ES, Miller DD, Callaham DA, Van Aken J, Hackett G, Hepler P (1996) Tip-localized calcium entry fluctuates during pollen tube growth. Dev Biol 174: 160–173 [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijo JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138: 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore KS, Cork RJ, Robinson KR (1991) A cytoplasmic gradient of Ca2+ is correlated with the growth of lily pollen tubes. Dev Biol 148: 612–619 [DOI] [PubMed] [Google Scholar]
- Rausch T, Greiner S (2004) Plant protein inhibitors of invertases. BBA-Proteins Proteom 1696: 253–261 [DOI] [PubMed] [Google Scholar]
- Ren CW, Kermode AR (2000) An increase in pectin methyl esterase activity accompanies dormancy breakage and germination of yellow cedar seeds. Plant Physiol 124: 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SJ, Holdaway CT, Hackett GR, Kunkel JG, Lord EM, Hepler PK (1999) Uncoupling secretion and tip growth in lily pollen tubes: evidence for the role of calcium in exocytosis. Plant J 19: 379–386 [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Moore I (1995) The plant Golgi-apparatus—structure, functional organization and trafficking mechanisms. Annu Rev Plant Physiol 46: 261–288 [Google Scholar]
- Steer MW, Steer JM (1989) Tansley review no.16: pollen tube tip growth. New Phytol 111: 323–358 [DOI] [PubMed] [Google Scholar]
- Sterling JD, Quigley HF, Orellana A, Mohnen D (2001) The catalytic site of the pectin biosynthetic enzyme alpha-1,4-galacturonosyltransferase is located in the lumen of the Golgi. Plant Physiol 127: 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Handa AK (1994) Reduction in pectin methylesterase activity modifies tissue integrity and cation levels in ripening tomato (Lycopersicon esculentum Mill.) fruits. Plant Physiol 106: 429–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D, Wing R, Yamaguchi J, McCormick S (1989) Isolation and expression of an anther-specific gene from tomato. Mol Gen Genet 217: 240–245 [DOI] [PubMed] [Google Scholar]
- Twell D, Yamaguchi J, Mccormick S (1990) Pollen-specific gene expression in transgenic plants—coordinate regulation of 2 different tomato gene promoters during microsporogenesis. Development 109: 705–713 [DOI] [PubMed] [Google Scholar]
- Wakeley PR, Rogers HJ, Rozycka M, Greenland AJ, Hussey PJ (1998) A maize pectin methylesterase-like gene, ZmC5, specifically expressed in pollen. Plant Mol Biol 37: 187–192 [DOI] [PubMed] [Google Scholar]
- Wen FS, Zhu YM, Hawes MC (1999) Effect of pectin methylesterase gene expression on pea root development. Plant Cell 11: 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WGT, McCartney L, Mackie W, Knox JP (2001) Pectin: cell biology and prospects for functional analysis. Plant Mol Biol 47: 9–27 [PubMed] [Google Scholar]
- Wolf S, Grsic-Rausch S, Rausch T, Greiner S (2003) Identification of pollen-expressed pectin methylesterase inhibitors in Arabidopsis. FEBS Lett 555: 551–555 [DOI] [PubMed] [Google Scholar]
- Ylstra B, Garrido D, Busscher J, vanTunen AJ (1998) Hexose transport in growing petunia pollen tubes and characterization of a pollen-specific, putative monosaccharide transporter. Plant Physiol 118: 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.