Abstract
Many developmental and environmental signals are transduced through changes in intracellular calcium concentrations, yet only a few calcium-binding proteins have been identified in plants. Calcineurin B-like (CBL) proteins are calcium-binding proteins that are thought to function as plant signal transduction elements. RNA profiling using a rice (Oryza sativa cv Nipponbare) oligonucleotide microarray was used to monitor gene expression in de-embryonated rice grains. This analysis showed that a putative rice CBL gene responded to gibberellic acid, but not abscisic acid, treatment. The CBL gene family in rice contains at least 10 genes and these have extensive similarity to the CBLs of Arabidopsis (Arabidopsis thaliana). In yeast (Saccharomyces cerevisiae) two-hybrid assays, rice CBLs interact with the kinase partners of Arabidopsis CBLs. Only one rice CBL gene, OsCBL2, is up-regulated by GA in the aleurone layer. A homolog with 91% sequence identity to OsCBL2 was cloned from barley (Hordeum vulgare cv Himalaya), and designated HvCBL2. We examined the localization and function of OsCBL2 and HvCBL2 in rice and barley aleurone because changes in cytosolic calcium have been implicated in the response of the aleurone cell to GA. Green fluorescent protein translational fusions of OsCBL2 and OsCBL3 were localized to the tonoplast of aleurone cell protein storage vacuoles and OsCBL4-green fluorescent protein was localized to the plasma membrane. Data from experiments using antisense expression of OsCBL2 and HvCBL2 are consistent with a role for OsCBL2 in promoting vacuolation of barley aleurone cells following treatment with GA.
Environmental and developmental signals are perceived by plants and communicated to downstream effector molecules by second messengers. The concentration of cytosolic free calcium [Ca2+]cyt is one of the most commonly utilized second messengers and roles for [Ca2+]cyt in plant responses to GAs, abscisic acid (ABA), drought, salinity, and cold have been reported (for review, see Sanders et al., 1999, 2002). Despite the importance of the calcium signal to plants, only a few calcium-binding proteins have been identified and shown to have a role in signaling (Liu and Zhu, 1998; Zielinski, 1998; Harmon et al., 2001; Snedden and Fromm, 2001; Luan et al., 2002; Zhang and Lu, 2003). Among the Ca2+-binding proteins that feature prominently in plant signal transduction are calmodulin (CaM) and the CaM domain protein kinases that may be unique to plants (Harmon et al., 2001).
Calcium-binding proteins with similarity to calcineurin B have been cloned recently from plants (Liu and Zhu, 1998; Shi et al., 1999; Luan et al., 2002). These calcineurin B-like proteins (CBLs) contain calcium-binding EF hands and are similar to the regulatory B-subunit of calcineurin and to the neuronal calcium sensor (Liu and Zhu, 1998). CBLs, therefore, have the potential to transduce [Ca2+]cyt signals and are thought to play roles in stress and hormone signaling in plants (Luan et al., 2002). The first CBL gene to be cloned was a salt overly sensitive (SOS) gene from Arabidopsis (Arabidopsis thaliana) that was designated SOS3 (Liu and Zhu, 1998). SOS3 is identical to AtCLB4, a salt-responsive CBL gene cloned independently from Arabidopsis (Kudla et al., 1999; Kim et al., 2000). At least 10 expressed CBL genes and proteins from Arabidopsis have now been identified (Luan et al., 2002), and many CBL genes are present in the sequenced rice (Oryza sativa) genome (Albrecht et al., 2001; Kolukisaoglu et al., 2004).
AtCBLs interact with novel protein kinases, designated calcineurin B-like interacting protein kinases (CIPKs; Shi et al., 1999), some of which also respond to stress and ABA (Kim et al., 2003). The yeast (Saccharomyces cerevisiae) two-hybrid system has been used to show that AtCIPK1 interacts with AtCBL1, and that SOS3 interacts with SOS2, a salt-sensitive protein kinase (Halfter et al., 2000) also referred to as CIPK24 (Albrecht et al., 2001). The Arabidopsis CIPKs contain the three-amino acid motif Asn-Ala-Phe, and this so-called NAF domain is required for interaction with CBLs (Albrecht et al., 2001). The NAF domain is also found in CIPKs from maize (Zea mays), rice, and sorghum (Sorghum vulgare), and the NAF domain of the sorghum kinase SNFL3 interacts with AtCBL2 (Albrecht et al., 2001).
In this article, we describe the CBL genes of rice. Throughout we adhere to the CBL/CIPK nomenclature to describe calcineurin B-like proteins and their interacting kinases because these have been described in detail by others (Kolukisaoglu et al., 2004). Many of the experiments described below were done with cereal aleurone cells because calcium signaling is central to the hormonal responses of aleurone cells in small grains such as rice, barley (Hordeum vulgare), and wild oat (Avena fatua; Bethke et al., 1995, 1997; Chen et al., 1997; Lovegrove and Hooley, 2000; Ritchie et al., 2002). In aleurone cells, GA stimulates the synthesis and secretion of hydrolytic enzymes including α-amylase, promotes the vacuolation of the aleurone protoplast, and initiates programmed cell death. All of these processes require an increase in [Ca2+]cyt. The plant hormone ABA antagonizes the effects of GA on hydrolase synthesis, vacuolation, and programmed cell death, and ABA prevents the GA-induced increase in [Ca2+]cyt. Other GA-stimulated processes, such as the transcription of α-amylase genes, are Ca2+ independent (Gilroy, 1996). Ca2+-binding proteins have been identified in aleurone, including several isoforms of CaM (W. Lin and R. Jones, unpublished data). GA perception results in increased synthesis of CaM protein (Schuurink et al., 1996), and a Ca2+-dependent protein kinase has been shown to be required for the response of aleurone cells to GA (Ritchie and Gilroy, 1998).
Here we show that the expression of one gene in the rice CBL family is up-regulated in aleurone by GA, but not by ABA. We show that other rice CBLs are not differentially expressed by GA and ABA in aleurone or in vegetative tissues of the shoot or root. We present data showing that OsCBL2 is localized to the aleurone tonoplast (TN), and transient expression assays with rice and barley CBLs in barley aleurone cells indicate that they are likely to be involved in a GA-signaling pathway that leads to the vacuolation of the aleurone cell.
RESULTS
RNA Profiling Analysis of a Rice Aleurone CBL Gene
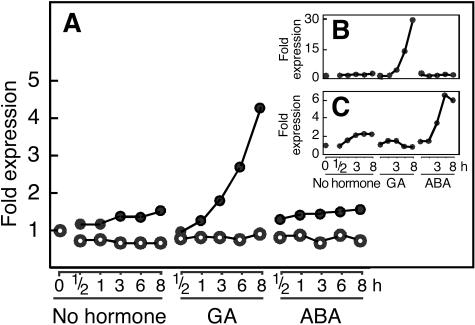
In order to identify novel components in GA or ABA signaling, we used a custom-designed GeneChip microarray (noNOVA002a; Zhu et al., 2003) with probe sets for 21,000 rice genes to monitor gene expression in rice aleurone treated with ABA, GA, or no added hormone. Because the aleurone layer of rice grains cannot be removed easily from the dead starchy endosperm, all experiments with rice reported in this article were carried out with de-embryonated half-grains. Since the aleurone layer contains the only living cells in the rice half-grain, the changes in transcription that we measured reflect those that occur in the aleurone layer. As seen in Figure 1A, a putative CBL gene was up-regulated rapidly by GA in rice aleurone. Three hours after treating tissue with GA, expression of the CBL gene was twice that at time zero, and by 8 h after GA treatment, expression of the CBL gene was almost 4.5 times that at time zero. The rice CBL gene showed little change in expression in response to ABA or no hormone, and the expression profile of the CBL gene in response to ABA or no hormone was similar to that of the constitutively expressed actin gene (Fig. 1A).
Figure 1.
The rice calcineurin B-like gene OsCBL2 is up-regulated by GA treatment of rice aleurone layers. Transcript abundance of OsCBL2 (black circles) and actin (white circles) as measured by hybridization to a rice oligonucleotide chip (A). Total RNA was extracted from embryoless rice half-grains treated with GA, ABA, or no hormone for the indicated time. Expression of GA-induced α-amylase, RAmy1A (B), and ABA-induced dehydrin (C) genes in the same chip experiment are shown for comparison.
α-Amylase and dehydrin genes were used as positive controls in this experiment. Microarray analysis of the expression of these two genes in rice aleurone demonstrates that hormonally regulated changes in transcription were occurring as expected. Figure 1B shows that α-amylase gene expression was strongly up-regulated by GA, but not ABA, treatment of rice half-grains, and Figure 1C shows that dehydrin gene expression was strongly up-regulated by ABA, but not by GA. For both α-amylase and dehydrin, gene expression 3 h after hormone treatment was greater than twice that at time zero, and expression of both genes was high by 8 h.
Molecular Cloning and Identification of Rice CBL Genes
To better understand rice CBLs, we identified the predicted CBL genes in the rice genome. Rice genome databases, including the Japanese rice genome annotation database (http://ricegaas.dna.affrc.go.jp) and The Institute for Genomic Research (TIGR) whole rice genome automated annotation database (http://www.tigr.org/tdb/e2k1/osa1/irgsp.shtml), were searched for homologs of CBL genes, and this search identified more than 20 predicted genes. A search of the TIGR expressed sequence tag (EST) database (rice gene index: http://www.tigr.org/tdb/tgi/ogi), the rice genome annotation database (http://ricegaas.dna.affrc.go.jp), and the National Center for Biotechnology Information (NCBI) dbEST (http://www.ncbi.nlm.nih.gov/dbEST) identified a total of eight different EST clones, and these are listed in Table I. When possible, EST sequences were used to eliminate duplicated and incorrectly predicted genes from the list of 20 genes. Gene structure has been shown to be highly conserved between the CBL genes of rice and Arabidopsis, and we used these conserved features to confirm gene predictions (Kolukisaoglu et al., 2004). As shown in Table II, the sequences of rice CBLs have high similarity and identity to Arabidopsis CBLs. There was 67% or more amino acid sequence similarity (52% identity) between AtCBL1 and OsCBL1 to 10 (Table II). Although these data make it clear that rice CBLs are very similar to Arabidopsis CBLs, they also make it clear that sequence information alone is insufficient to identify CBL orthologs in rice and Arabidopsis. To minimize confusion in CBL nomenclature, we selected gene designations for the rice CBLs so that genes with the same numerical suffix in rice and Arabidopsis would have very high homology, often the highest of all pairwise comparisons.
Table I.
Rice calcineurin B-like sequences are present in genomic sequence databases and in EST databases
Accession numbers in GenBank, BAC clone designations and location, and EST clone designations are cross-referenced for OsCBL1-10. BAC, Bacterial artificial chromosome.
| Gene Name | GenBank Accession No. | BAC Clone | Location | EST Clone |
|---|---|---|---|---|
| OsCBL1 | AC084763 | OSJNBa0027P10 | 94244–103391 | TC139060a |
| OsCBL2 | AL713904 | OJ1306_H03 | 34126–30072 | TC138681 |
| AL928754 | OSJNBa0056D07 | 9844–5790 | ||
| OsCBL3 | AC092390 | OSJNBb0013K08 | 28690–34606 | TC149447 |
| AC091246 | OSJNBa0002I03 | 144719–150635 | ||
| OsCBL4 | AC121365 | OSJNBa0053E05 | 182887–184367 | C27482a |
| AC097111 | OJ1014_C08 | 26564–28044 | ||
| OsCBL5 | AP003504 | P0025A05 | 59100–61578 | TC156374 |
| OsCBL6 | AL713906 | OJ1587_D05 | 69628–73277 | TC138902 |
| AL731748 | OSJNBa0095K18 | 6807–10456 | ||
| OsCBL7 | AP004212 | OJ1086_G08 | 16316–18296 | |
| OsCBL8 | AP004229 | OJ1124_E11 | 23843–25922 | |
| OsCBL9 | AP002908 | P0013G02 | 121268–123745 | TC136805a |
| AP003546 | P0672C09 | 48136–50613 | ||
| OsCBL10 | AP002901 | P0456F08 | 128202–131015 | TC140870a |
| AP003410 | B1142C05 | 31748–34561 |
Indicates a partial clone.
Table II.
Amino acid similarity and identity of rice CBLs (OsCBL1–10) and Arabidopsis CBLs (AtCBL1–10)
For each pairwise comparison, similarity values are followed by identity values in parentheses.
| AtCBL1 | AtCBL2 | AtCBL3 | AtCBL4 | AtCBL5 | AtCBL6 | AtCBL7 | AtCBL8 | AtCBL9 | AtCBL10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| OsCBL1 | 87(78) | 73(63) | 74(66) | 73(63) | 60(51) | 64(54) | 60(50) | 70(58) | 87(79) | 70(59) |
| OsCBL2 | 74(64) | 91(85) | 93(87) | 69(57) | 63(50) | 77(70) | 77(68) | 69(56) | 72(63) | 68(58) |
| OsCBL3 | 75(64) | 93(89) | 94(88) | 69(58) | 63(48) | 79(74) | 75(67) | 67(55) | 73(64) | 67(59) |
| OsCBL4 | 69(55) | 64(52) | 64(53) | 74(66) | 61(47) | 61(50) | 54(43) | 73(64) | 68(55) | 65(55) |
| OsCBL5 | 67(52) | 68(55) | 68(56) | 71(61) | 61(47) | 62(50) | 59(47) | 69(59) | 67(56) | 66(55) |
| OsCBL6 | 73(61) | 86(81) | 87(81) | 72(58) | 62(47) | 75(69) | 75(67) | 68(56) | 73(61) | 71(61) |
| OsCBL7 | 71(62) | 68(56) | 68(56) | 77(68) | 64(50) | 62(50) | 56(44) | 74(62) | 71(63) | 69(60) |
| OsCBL8 | 70(62) | 67(54) | 68(55) | 75(67) | 61(52) | 61(48) | 55(44) | 72(62) | 70(62) | 67(59) |
| OsCBL9 | 73(61) | 70(60) | 70(59) | 70(58) | 62(47) | 61(50) | 62(49) | 68(55) | 74(62) | 74(67) |
| OsCBL10 | 68(54) | 68(58) | 70(61) | 65(53) | 58(42) | 61(52) | 60(49) | 65(51) | 70(58) | 74(67) |
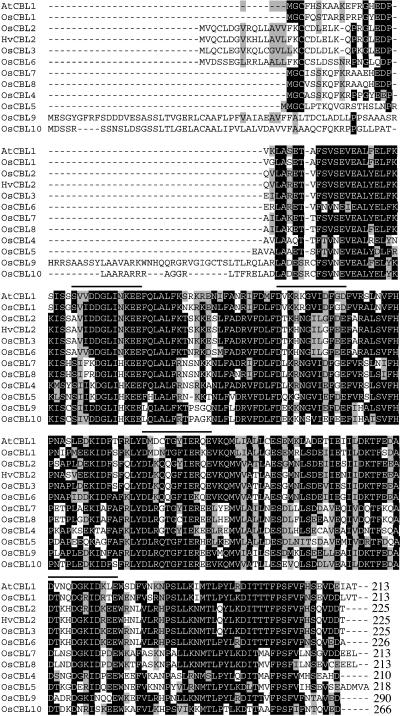
In addition to the sequences listed in Tables I and II, several additional putative CBL genes were retrieved from GenBank. A cDNA from a rice root EST library (GenBank accession no.: CA758757) has been reported to be a CBL homolog, but we could not confirm the genomic sequence for this clone in published rice genome databases. Interestingly, this gene shows higher sequence homology to animal calcineurin B than to any plant CBL. The predicted gene, OSJNBa0031I04.5304.t00002, from the TIGR whole rice genome automated annotation database was also not included in our list of CBL genes. This gene is phylogenetically distinct from other plant CBLs and also has higher homology to animal calcineurin B than plant CBLs. Thus, the spacing between the EF hands, which is absolutely conserved in Arabidopsis and rice CBLs (Kolukisaoglu et al., 2004), is very different in this predicted gene. Two other predicted CBL genes in the TIGR rice genome database, P0519B12.4294.t00009 and P0527E02.4884.t00007, are also not included in our list of rice CBLs. These genes lack several exons and the four predicted EF hand motifs found in the other Arabidopsis and rice CBL genes (Kolukisaoglu et al., 2004). Figure 2 shows an amino acid sequence alignment for the 10 rice genes that we believe encode authentic CBLs and for AtCBL1 from Arabidopsis. All OsCBLs contain several predicted EF hand motifs. Using Pfam software, three EF hands are predicted, but, using the 12-amino acid motif characterized by Lewit-Bentley and Rety (2000), four EF hand motifs are predicted and these are indicated in Figure 2 by dark lines over the amino acid sequences. The four EF hands identified in OsCBLs are also found in AtCBLs when the 12-amino acid motif (Lewit-Bentley and Rety, 2000) is used (Kolukisaoglu et al., 2004). Like their Arabidopsis homologs (Kolukisaoglu et al., 2004), OsCBLs show highly conserved amino acid sequence in regions that flank the EF hands. Structural motifs are not apparent in any of these regions, but the extent of amino acid conservation suggests functional importance. For example, OsCBL1 and AtCBL1 have 23 out of 24 identical amino acids immediately upstream of the first EF hand motif and 32 out of 34 identical amino acids at the carboxyl terminus (Fig. 2). The most divergent regions of the OsCBL sequences are in their amino-terminal regions. Whereas the amino termini of OsCBL1, 4, 5, 7, and 8 are almost identical to that of AtCBL1, OsCBL2, 3, 6, 9, and 10, all have amino-terminal extensions of varying lengths. The functional significance of these domains is not known.
Figure 2.
Rice CBLs contain extensive regions of high homology. A protein sequence alignment for OsCBLs is shown where black shading signifies identical amino acids and gray shading indicates similar amino acids. EF hand motifs are indicated by lines over the amino acid sequence. AtCLB1 from Arabidopsis and HvCBL2 from barley are included for comparison.
We used Motif Scan to search for sequences that might specify the intracellular location of the rice CBLs. None of the CBLs has a predicted signal peptide, but a putative myristoylation sequence (GMNLS) is found in OsCBL2, 3, and 6 at a position corresponding to amino acids 154 to 158 in OsCBL2 (Fig. 2). OsCBL1, 4, 5, 7, and 8 also have the consensus myristoylation sequence MGXXXS/T at their amino termini. The calcineurin A interaction domain found in Arabidopsis CBLs is absent from the rice genes.
Hormone and Tissue-Specific Expression of OsCBLs
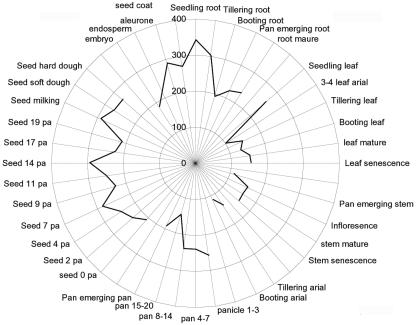
Only OsCBL2 contains the probe sequences found on the rice GeneChip microarray. It is therefore highly likely that the GA-regulated CBL identified in our microarray experiments (Fig. 1) is OsCBL2. We used the GeneChip microarray to quantitate the expression of OsCBL2 in the tissues of rice cv Nipponbare at all stages of development. These data are presented in Figure 3, where GeneChip intensity values for each tissue or organ are plotted with higher values farther from the center of the figure. OsCBL2 is expressed at high levels in roots of seedlings and tillering plants, during early stages of panicle and seed formation, and in the aleurone of mature grain. Expression of OsCBL2 was lowest in mature leaves and stems and in the emerging inflorescence shoot (Fig. 3).
Figure 3.
OsCBL2 is expressed in many rice organs and at all stages of rice plant development. Data are pooled from individual microarray experiments where each radius in the figure represents a separate experiment. RNA samples were pooled prior to hybridization to the chip, and the data are presented as normalized intensity values.
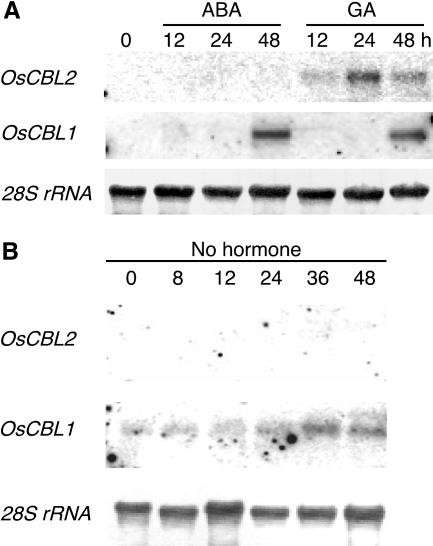
We confirmed and extended our microarray experiments by carrying out northern blotting with RNA isolated from rice half-grains incubated in the presence and absence of GA or ABA for up to 48 h and from rice seedling tissues. Four full-length cDNAs (OsCBL1–4) and a partial sequence (OsCBL7) were cloned by reverse transcription-PCR or PCR from genomic DNA. Partial sequences (OsCBL1, 3, and 7) or full-length cDNA sequences (OsCBL2 and 4) were used as gene-specific probes on northern blots since none cross-hybridized with each other (data not shown). Figure 4, A and B, shows that the OsCBL2 transcript is strongly up-regulated by GA for up to 48 h of incubation. In the absence of added hormone (Fig. 4B) or in the presence of ABA (Fig. 4A), there is almost no accumulation of OsCBL2 transcript. By contrast, OsCBL1 expression in rice aleurone is not specifically influenced by either ABA or GA treatment. In half-grains incubated in ABA, GA, or the absence of hormone, OsCBL1 expression increases with time (Fig. 4). We could not detect expression of OsCBL3, 4, or 7 in freshly prepared aleurone tissue, or in aleurone treated with GA or ABA for up to 48 h (data not shown).
Figure 4.
OsCBL2 but not OsCBL1 shows GA-specific up-regulation in embryoless rice half-grains. Total RNA was isolated from grains treated with ABA or GA (A) or no hormone (B) for the indicated times. Note that changes in mRNA abundance reflect changes occurring in the aleurone layer.
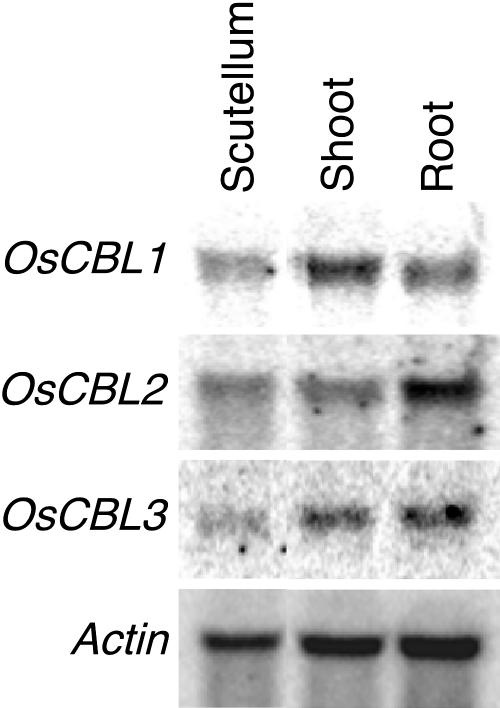
To investigate the expression of OsCBLs in germinating Nipponbare rice seedling tissues, RNA was isolated from scutellum, shoots, and roots of 7-d-old seedlings and northern blots were hybridized with gene-specific probes for OsCBL1 to 3 (Fig. 5). OsCBL2 is expressed in all rice seedling tissues and this confirmed the analysis made with the GeneChip array (Fig. 3). RNA blotting also confirmed that OsCBL2 mRNA was abundant in roots relative to shoots and scutella, whereas the OsCBL1 transcript was more abundant in shoots than in roots and the OsCBL3 transcript was abundant in both root and shoot tissue (Fig. 5). OsCBL4 and 7 were not expressed strongly enough in tissues of 7-d-old seedlings to be detected.
Figure 5.
OsCBLs are expressed in rice seedling tissues. Total RNA was isolated from scutella, shoots, and roots of 1-week-old rice seedlings. RNA blots were probed with gene-specific probes for OsCBL1 to 3. Hybridization to actin was used as a loading control.
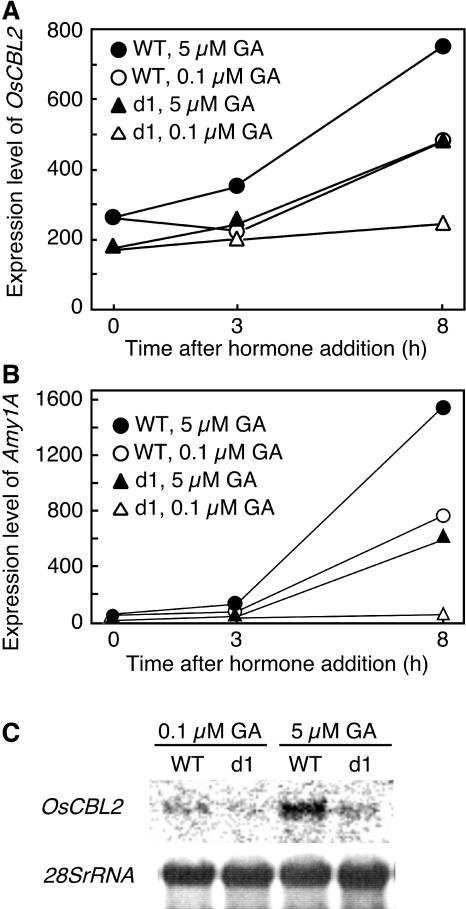
GA-Induced Expression of OsCBL2 Is Reduced in the Aleurone Layer of dwarf1 Mutant Rice
We also used RNA profiling and northern blotting to see whether GA-induced expression of OsCBL2 in aleurone cells was dependent on a signaling pathway that utilizes heterotrimeric G-proteins. For these experiments, RNA was isolated from half-grains of wild-type and dwarf1 (d1) mutant rice. The d1 rice mutant lacks the α-subunit of heterotrimeric G-proteins and shows a defective GA response, except at high GA concentrations (Ueguchi-Tanaka et al., 2000). In the experiment shown in Figure 6A, there was a 3-fold increase in OsCBL2 expression in wild-type rice aleurone after 8-h incubation at a high (5 μm) GA concentration. When wild-type half-grains were incubated with a low (100 nm) GA concentration, OSCBL2 expression was still almost twice as high as that at time zero (Fig. 6A). Expression of OsCBL2 in d1 half-grains, however, was much reduced at 5 μm GA compared to wild type, and transcript abundance was virtually unchanged following 8-h incubation with 100 nm GA (Fig. 6A). Similar changes in expression were observed for α-amylase in d1 and wild-type rice half-grains (Fig. 6B). Thus, there was virtually no change in the expression of the RAmy1A gene at low GA concentrations in d1 rice, whereas in wild-type rice grain low GA brought about a large change in RAmy1A expression (Fig. 6B). RNA blotting was used to confirm the microarray data on CBL expression as shown in Figure 6C. Expression of OsCBL2 was observed in wild-type aleurone and the d1 mutant at 5 μm GA, but OsCBL2 transcript could not be detected in the d1 mutant at 100 nm GA.
Figure 6.
Expression of OsCBL2 in wild-type rice grain is higher than expression in d1 mutant grain. Total RNA was extracted from embryoless wild-type rice grain or d1 mutant grain treated with 0.1 or 5 μm GA for 0, 3, or 8 h. RNA abundance of OsCBL2 was determined using microarray (A and B) or northern (C) analysis. The abundance of rice RAmy1A was also determined using the microarray (B).
OsCBLs Interact with AtCIPKs
In animals and fungi, calcineurin B is known to interact with the protein phosphatase calcineurin A (Klee et al., 1998). In Arabidopsis, however, AtCBLs interact with the AtCIPK family of protein kinases (Shi et al., 1999). Because the amino acid sequence of rice CBLs is similar to that of Arabidopsis CBLs, we hypothesized that rice CBLs also interact with CIPKs. This hypothesis is supported by an examination of rice genome databases. We have been unable to identify genes predicted to be calcineurin A, but we and others (Albrecht et al., 2001; Kolukisaoglu et al., 2004) have identified homologs of Arabidopsis CIPK genes. The rice protein kinases OsPK4 and 7, in particular, have high homology with the AtCIPKs, and they contain the NAF domain that in AtCIPKs is required for interaction with AtCBLs (Albrecht et al., 2001; Kolukisaoglu et al., 2004).
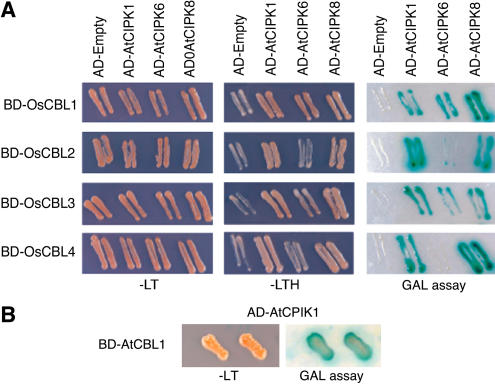
We used the yeast two-hybrid system to demonstrate that rice CBLs interact with AtCIPKs. OsCBL1 to 4 were fused to the binding domain of GAL4, whereas AtCIPK1, 6, and 8 were fused to the activation domain of GAL4. Figure 7A shows the growth of yeast on selection medium and the corresponding assay for β-galactosidase when these different OsCBLs and AtCIPKs were used as bait and prey. As expected, the positive control showed interaction between AtCBL1 and AtCIPK1 (Fig. 7B; Kim et al., 2000). OsCBL2, which has 74% amino acid similarity with AtCBL1 (Table II), also had a strong interaction with AtCIPK1. Like AtCBL1 (Kolukisaoglu et al., 2004), OsCBL2 interacted strongly with AtCIPK8 and weakly with AtCIPK6. OsCBL4 also interacted strongly with AtCIPK1 and 8, but unlike OsCBL2, it did not interact with AtCIPK6. OsCBL1 and 3 both interacted with all three of the Arabidopsis CIPKs examined. These data provide evidence that OsCBL1 to 4 proteins are functional homologs of Arabidopsis CBL proteins. They also suggest that the other rice CBL sequences identified in Figure 2, especially those based on sequenced ESTs, are likely to encode authentic CBL proteins.
Figure 7.
Yeast two-hybrid analysis demonstrates an interaction between OsCBLs and AtCIPKs. OsCBLs and AtCIPKs were translationally fused to the GAL4 DNA-binding domain (BD) and activation domain (AD) as indicated. Nutritional reporter systems minus Leu plus Trp (−LT) and minus Leu, Trp, and His (−LHT) and filter-lift GAL assays were employed to examine the interaction between OsCBLs and AtCIPKs (A). A positive control showing the interaction of AtCBL1 with AtCIPK1 is shown in B.
Subcellular Localization of OsCBLs
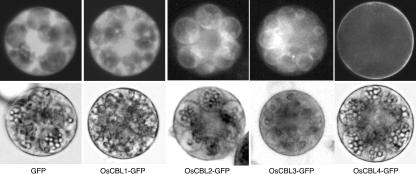
We examined the subcellular localization of OsCBL1 to 4 by transiently expressing translational gene fusion constructs of OsCBL1 to 4 with green fluorescent protein (GFP) in barley aleurone protoplasts. Barley aleurone protoplasts were used in these experiments because we have been unable to isolate protoplasts in high yield from rice aleurone. We and others have shown that barley and rice aleurone are structurally and functionally similar (Bechtel and Pomeranz, 1977; O'Neill et al., 1990). Figure 8 shows fluorescence and bright-field images of typical aleurone protoplasts 42 to 48 h after transfection. Transient expression of GFP alone resulted in bright fluorescence from the cytosol and nucleus of the aleurone cell and localization to intracellular membranes was not observed. When GFP was fused to the carboxyl terminus of OsCBL2 or 3 and expressed in aleurone protoplasts, the GFP fusion protein was localized largely to the TN of the protein storage vacuoles. By contrast, virtually all cells had a strong GFP signal from the plasma membrane (PM) when OsCBL4-GFP was transiently expressed. Protoplasts transformed with OsCBL1-GFP displayed yet another pattern of fluorescence, with no specific localization to either the TN or PM. The cytoplasm of many OsCBL1 transformants, however, appeared to be less organized than that of untransformed controls. This latter observation suggests that overexpression of the OsCBL1-GFP construct may have been toxic to aleurone cells.
Figure 8.
OsCBL2 to 4 are localized to membranes. OsCBL1 to 4 were translationally fused to GFP and transiently expressed in barley aleurone protoplasts. The figure shows representative epifluorescence images (top) and bright-field images (bottom) of single, transformed cells. The unmagnified width of each image is approximately 40 μm.
Antisense Expression of Barley and Rice CBLs Prevents GA-Induced Vacuolation, But Not GA-Induced Expression from an α-Amylase Promoter
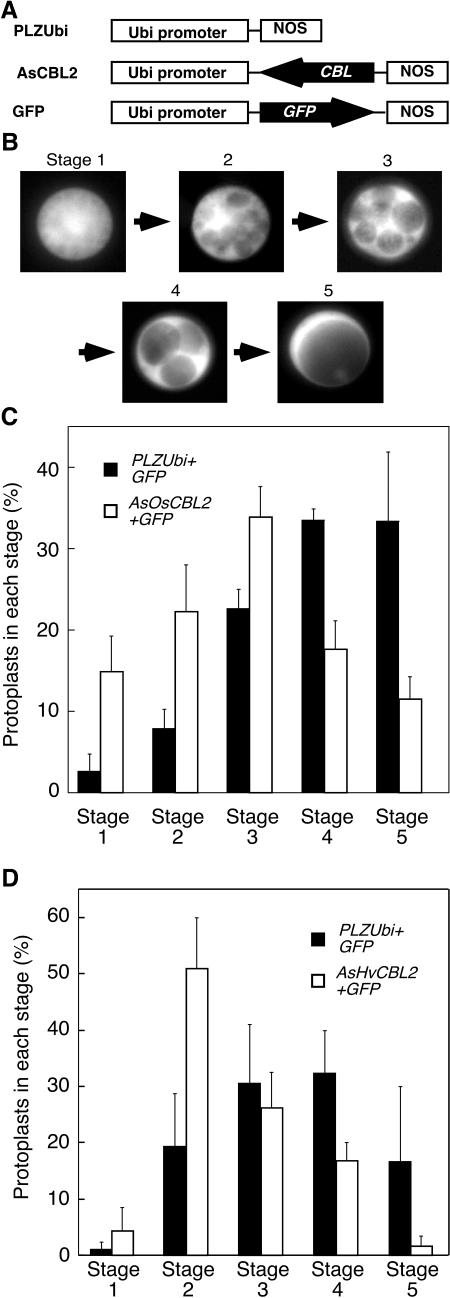
The vacuolation of aleurone protoplasts is hormonally regulated and strongly promoted by GA and inhibited by ABA (Bethke et al., 1998). Because OsCBL2 gene expression was hormonally regulated, and because OsCBL2-GFP was localized to the TN, we examined further the effects of rice and barley CBLs on vacuolation using antisense CBL constructs. For these experiments, we used antisense clones corresponding to the coding region of OsCBL2 and the 3′-untranslated region (UTR) of HvCBL2. HvCBl2 corresponds to a barley EST clone in the TIGR gene index database (TC 111766) that has 90.7% nucleotide sequence identity with OsCBL2, and 94.2% amino acid sequence identity. GA-treated barley aleurone protoplasts were cotransfected with GFP and either antisense CBL constructs (AsOsCBL2 or HvCBL2) or empty vector (pLZUbi) as diagrammed in Figure 9A. Cells were observed 48 h after transfection and fluorescent cells were scored from 1 to 5 for extent of vacuolation as indicated in Figure 9B. What the data in Figure 9, C and D, show is that expression of OsCBLs markedly decreased the percentage of cells that were highly vacuolate. Cells transfected with GFP alone underwent progressive vacuolation with many cells having only one (stage 5) or a few (stage 4) large vacuoles 42 h after GA treatment. Cells transfected with AsOsCBL2 or AsHvCBL2, on the other hand, had far fewer cells at stages 4 and 5 and many more cells at stages 1 to 3. Whereas 75% of protoplasts transformed with GFP were in stages 4 and 5, less than 35% of AsOsCBL2 transformants had reached this stage of vacuolation.
Figure 9.
Antisense OsCBL2 or HvCBL2 delays the GA-induced vacuolation of barley aleurone protoplasts. Barley protoplasts were cotransfected with GFP and AsOsCBL2, GFP, and AsHvCBL2, or with GFP and empty cassette (pLZUbi) using the constructs diagrammed in A. The extent of vacuolation for individual protoplasts was scored using the five categories indicated in B. Vacuoles are seen as dark regions surrounded by bright regions of cytoplasm. The number of protoplasts in each category 48 h after transfection and 42 h after treatment with GA are shown in C for AsOsCBL2 and in D for AsHvCBL2.
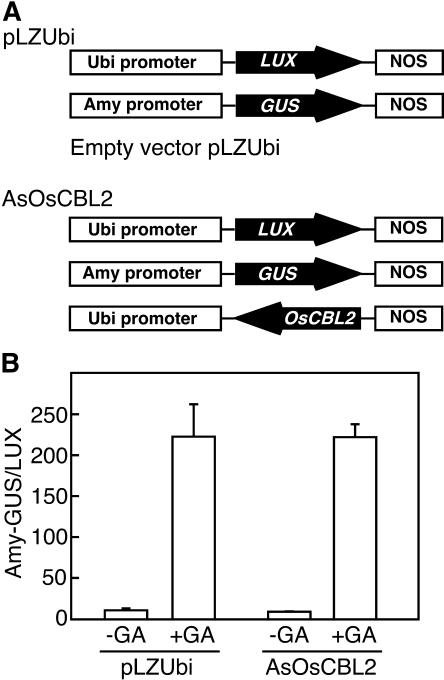
To test whether expression of AsOsCBL2 affected other GA-induced processes, we examined its effect on the expression of a GA-regulated β-glucuronidase (GUS) reporter gene relative to a constitutively expressed luciferase (LUX) gene. Rice half-grains were cotransfected by particle bombardment with GA-responsive Amy-GUS and constitutively expressed ubi-LUX and either AsOsCBL2 or empty vector, as diagrammed in Figure 10A. In these experiments, the same construct that inhibited GA-induced vacuolation of protoplasts had no effect on GA-induced Amy-GUS expression (Fig. 10B). The amount of GUS accumulating in aleurone layers was not significantly different when they were transformed with AsOSCBL2 or with empty vector.
Figure 10.
Antisense OsCBL2 does not delay GA-induced transcription of GUS from an α-amylase promoter in rice half-grain. A diagram of the constructs introduced by particle bombardment is shown in A. Transcription of GUS from a GA-regulated α-amylase promoter was measured relative to expression of LUX (GUS:LUX ratio) driven by a constitutive ubiquitin promoter (B). Half-grains were incubated for 24 h without hormone (−GA) or with GA and the ratio of GUS-to-LUX expression determined in the presence and absence of the antisense construct.
DISCUSSION
The data presented here contain important information about CBLs in plants. In particular, we show that three CBL genes are expressed in rice seedlings (Fig. 6) and two in rice aleurone (Figs. 1, 3, 4, and 6). The amount of OsCBL2 transcript was increased specifically by GA treatment in rice aleurone (Figs. 1, 4, and 6). Using microarray analyses and RNA blots, we show that the up-regulation of OsCBL2 expression occurs within 3 h of GA treatment and persists for at least 48 h (Figs. 1, 4, and 6). Data from experiments with the d1 mutant of rice strongly suggest that OsCBL2 transcription is part of a GA-signaling pathway that involves the α-subunit of heterotrimeric G-proteins (Fig. 6). Rice CBLs have extensive similarity to Arabidopsis CBLs (Fig. 2; Table II), and some rice CBLs can interact with CIPKs from Arabidopsis (Fig. 7). A search of rice databases shows that homologs of AtCIPKs are present in rice.
Specificity for rice CBL function is likely to arise from differences in intracellular localization and different timing of expression. We show here that OsCBL2 and 3 are targeted to the TN, and OsCBL4 to the PM (Fig. 8). Even though both OsCBL2 and 3 are targeted to the TN, their roles may be distinguished by the timing of their expression. For example, OsCBL2 is expressed in aleurone during germination, but OsCBL3 was not detectable in this tissue under the conditions that we have tested. OsCBL2 may be involved in vacuole function since transformation of aleurone protoplasts with an antisense construct of OsCBL2 or HvCBL2 slowed the rate of GA-induced vacuolation (Fig. 9), but not GA-induced transcription of an α-amylase reporter construct (Fig. 10).
Our data and those of Kolukisaoglu et al. (2004) show that the CBLs of rice belong to a multigene family. Based on our data and EST sequences, at least eight CBLs are expressed in rice. Members of a multigene family often have partially overlapping functions, but may be distinct in their cellular and subcellular distribution, regulatory properties, or affinity for ligands or interacting partners. The data presented here show clearly that the OsCBLs fit this paradigm. For example, OsCBL1 to 4 interact to different degrees with Arabidopsis AtCIPK1, 6, and 8. OsCBL2 interacted with all three AtCIPKs, but weakly with AtCIPK6. OsCBL4 interacted only with AtCIPK1 and 8. These data suggest that some rice CBLs may have overlapping functions in that they can partner with multiple CIPKs, as has been shown for Arabidopsis CBLs and CIPKs (Kolukisaoglu et al., 2004). This contention is further supported by an examination of Arabidopsis plants with mutations in individual CBLs (Kim et al., 2003). These single-mutant lines are phenotypically normal under favorable growth conditions and display only a mild growth phenotype when stressed. The rice genome contains a large number of Ser/Thr protein kinases similar to Arabidopsis CIPKs (Albrecht et al., 2001; Kolukisaoglu et al., 2004) that are designated OsCIPK1 to 30. Like their Arabidopsis counterparts, OsCIPKs have a highly conserved NAF domain that has been shown to be required for the binding of AtCBLs to AtCIPKs (Albrecht et al., 2001; Kolukisaoglu et al., 2004). It is likely, therefore, that rice has homologs of the Arabidopsis CIPKs that were used in these experiments.
If OsCBLs play roles in sensing changes in [Ca2+]cyt, then variability in the Ca2+ affinity of the individual OsCBLs might be expected and the Ca2+ affinity for all CBLs should approximate [Ca2+]cyt. OsCBL1 to 10 have four predicted calcium-binding EF hand motifs. The amino acid sequences for the motifs in one gene show high similarity to the EF hand motifs in the other genes, yet there are differences, and these could result in differing calcium affinities. To date, however, it has not been established that Arabidopsis or rice CBLs have affinities for Ca2+ compatible with a role in intracellular signaling, or that different CBLs show differences in their affinities for Ca2+.
We examined the regulation of five OsCBL genes (OsCBL1–4 and 7) and each shows a unique pattern of expression. Although both OsCBL1 and 2 were expressed in rice half-grains, OsCBL2 was specifically up-regulated by GA (Fig. 4). The OsCBL1 transcript, on the other hand, accumulated with time under all conditions of incubation of aleurone tissue (Fig. 4). GeneChip and RNA blotting experiments showed that OsCBL2 was most strongly expressed in aleurone and root and, using an expression intensity value of 50 as a cutoff, it is clear that OsCBL2 is expressed in most tissues of the rice plant. RNA blotting showed that OsCBL1 and 3 were also expressed in vegetative tissues of 1-week-old or 1-month-old rice seedlings (Fig. 5). OsCBL1 and 3 were up-regulated by NaCl treatment of seedlings (data not shown), and in this regard these rice CBLs are similar to the Arabidopsis CBLs AtCBL4/SOS3. We could not detect expression of OsCBL4 or 7 in half-grains or seedlings by northern analysis.
We used translational fusions with GFP to localize four OsCBLs in aleurone cells and large differences in spatial distribution were observed (Fig. 8). Because nonspecific localization of overexpressed proteins may occur, these data must be interpreted with caution. Our data strongly suggest, however, that OsCBL2 and 3 are targeted to the TN and OsCBL4 to the PM. Specific localization was not apparent for OsCBL1. The data do not allow us to conclude that these are the only sites occupied by the native OsCBLs. The localization of OsCBL4 at the PM is consistent with the existence of a possible myristoylation site in its amino acid sequence. We could not detect sequences in OsCBL2 or 3 that would direct it to the TN, but TN-targeting sequences are often difficult to identify (Neuhaus, 1996). None of the 10 putative OsCBLs contained a recognizable signal peptide.
Microarray analyses were used as a tool to identify novel GA-signaling components in cereal aleurone layers. OsCBL2 was identified in those gene discovery experiments (Fig. 1). Extensive research has shown that GA-signaling pathways in cereal aleurone cells can be separated into calcium-dependent and calcium-independent pathways. GA-stimulated transcription of α-amylase genes, for example, is on the calcium-independent pathway, and vacuolation is on the calcium-dependent pathway (Deikman and Jones, 1985; Gilroy, 1996). Because OsCBL1 to 10 have four motifs predicted to be the Ca2+-binding regions of EF hands, some rice CBLs may be signal transduction elements that participate in calcium-dependent signaling. The data presented here for OsCBL2, in particular, are consistent with this speculation.
OsCBL2 expression in aleurone is specifically up-regulated by GA (Figs. 1 and 4). Transcript abundance was unchanged when rice half-grains were incubated with ABA or no hormone, or when seedlings were exposed to various stresses. Perhaps more interesting is our observation that correct expression of OsCBL2 in aleurone protoplasts seems to be required for proper vacuolation (Fig. 9). When barley aleurone protoplasts were transiently transformed with antisense constructs for OsCBL2 or HvCBL2 (Fig. 9, C and D), vacuolation was retarded. This was a specific effect in that AsOsCBL2 did not inhibit transcription from an α-amylase promoter (Fig. 10). One interpretation of these data is that OsCBL2 interacts with one or more proteins in aleurone cells, and that an insufficient amount of OsCBL2 leads to a defect in vacuole function. For example, OsCBL2 may activate a CIPK and the OsCBL2/CIPK complex may promote vacuole fusion and enlargement. Antisense OsCBL2 would reduce the amount of OsCBL2 and prevent the formation of the active OsCBL/CIPK complex. This speculation is consistent with our previous data showing that a Ser/Thr protein kinase present on the TN in barley aleurone protoplasts is involved in the gating of a Ca2+-regulated ion channel (Bethke and Jones, 1997).
MATERIALS AND METHODS
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requester.
Plant Material
Oryza sativa L. cv Nipponbare was used in all rice experiments. Rice grains were dehusked and half-grains prepared by cutting off the end containing the embryo. Half-grains were surface sterilized in 1% sodium hypochlorite and 0.01% Tween X-20 for 10 min with shaking, washed with water 10 times, incubated with 0.01 n HCl for 10 min, and washed with water 10 times. Approximately 250 half-grains were placed in 10-cm petri dishes containing 10 mL of 10 mm CaCl2 alone (no hormone) or with 5 μm ABA or 5 μm GA and incubated at 30°C with shaking at 100 rpm for the times indicated.
Seedlings for total RNA isolation were grown from rice grains that were dehusked and surface sterilized, as described above. Three hundred grains were placed in 10-cm petri dishes containing two layers of 3MM Whatman paper moistened with 10 mL of 10 mm CaCl2 and placed in a growth chamber at 25°C under long-day (16-h light and 8-h dark) conditions. Barley (Hordeum vulgare cv Himalaya) aleurone layers were used to prepare aleurone protoplasts as described in detail in Bethke and Jones (2001).
Rice Oligonucleotide Chip Experiments
RNA was prepared from rice half-grains imbibed in 10 mm CaCl2 with or without 5 μm GA or 5 μm ABA for 0, 0.5. 1, 3, 6, and 8 h as described in Hwang et al. (1999). Total RNA was further cleaned up using the Qiagen Rneasy mini spin column from the Rneasy plant mini kit (Qiagen, Valencia, CA) based on the Qiagen Rneasy mini protocol for RNA cleanup. RNA was used to prepare cDNA from which labeled cRNA was synthesized as described in Zhu et al. (2001). Samples were hybridized to a custom-designed rice genome GeneChip oligonucleotide microarray (Zhu et al., 2003). The microarray contains probe sets for approximately 21,000 rice genes. Validation of the GeneChip design, the algorithm for calculating fluorescence intensity of a probe set, and performance of this GeneChip microarray have been described previously (Zhu et al., 2003). The data were scaled globally to a target intensity of 100, and fluorescence intensity values of probe sets, which represent transcript level for each putative gene on the array, were examined to identify hormonally regulated genes that might be involved in signaling using protocols similar to that described in Hudson et al. (2003).
Plasmid Construction
Total RNA was isolated from 1-week-old rice (cv Nipponbare) or barley (cv Himalaya) seedlings as described in Hwang et al. (1999) and used for cDNA first-strand synthesis. The first strand of cDNA was synthesized from oligo(dT) primer using SuperScript II RNaseH− reverse transcriptase (Invitrogen, Carlsbad, CA) based on their recommended protocol. Full-length cDNAs for OsCBL1 (642 bp), 2 (678 bp), 3 (678 bp), and 4 (633 bp) were amplified from the cDNA first strand using the primer sets OsCBL1FW1/RV1, OsCBL2FW1/RV1, OsCBL3FW1/RV1, and OsCBL4FW1/RV1, respectively, and cloned into a pCR2.1 vector, resulting in each pCR 2.1-OsCBL construct. The 3′-UTR (379 bp) of HcCBL2 was amplified from barley cDNA first strand using the primer set HvCBL2FW/RV and cloned into a pCR2.1 vector, resulting in a pCR2.1-HvCBL2 construct. The sequences of the antisense constructs for OsCBL and HvCBL2 were confirmed by DNA sequencing. The PCR cycling conditions were 2 min predenaturing at 94°C, 35 cycles of denaturing at 94°C for 45 s, annealing at 58°C for 45 s, and extension at 72°C for 1 min, and were carried out using a thermal controller (PTC-100; MJ Research, Watertown, MA). A 50°C annealing temperature was used for OsCBL4.
Full-length coding regions of OsCBL1 to 4 were cut out of each pCR2.1-OsCBL construct using BglII and EcoRI, and subcloned into pGBT9.BS (Elledge et al., 1991) through BamHI and EcoRI sites to make pGBT-OsCBL1–4, which are GAL4 DNA-binding domain translational fusion constructs for yeast (Saccharomyces cerevisiae) two-hybrid analysis.
A pH-sensitive GFP (Phluorin) optimized for plant expression (Moseyko and Feldman, 2001) was transferred to pLZUbi, an expression cassette containing the promoter and first intron from maize (Zea mays) ubiquitin and a nopaline synthase terminator, using BamHI and SacI, and this resulted in pLZUbi-GFP. The coding regions of OsCBL1 to 4 with the translation stop codon deleted were produced by PCR amplification using the primer sets OsCBL1FW1/RV2, OsCBL2FW1/RV2, OsCBL3FW1/RV2, and OsCBL4FW1/RV2, respectively. The PCR cycling conditions for each OsCBL were as described above. PCR products without a translation stop codon were extracted with phenol:chloroform:isoamylalcohol and precipitated with ethanol. The redissolved pellets were digested with BglII and inserted in front of the GFP translation start codon using the BamHI site of pLZUbi-GFP to produce the GFP translational fusion constructs. The coding region of OsCBL2 was cut from pCR2.1-OsCBL2 with BamHI and SacI and inserted into pLZUbi to make AsOsCBL2, an OsCBL2 antisense construct.
The HvCBL2 antisense construct, AsHvCBL2, was produced by cloning the 3′-UTR from pCR2.1-HvCBL2 into pLZUbi through SacI and BamHI.
Primers
The primers described above are as follows (restriction enzyme sites are underlined): Oligo(dT), 5′-GGAATTCTAGATTTTTTTTTTTTTTTTT-3′; OsCBL1FW1, 5′-AGATCTCATGGGGTGCTTCCAGTCGACGGCGAGG-3′; OsCBL1RV1, 5′-TCATGTGACGAGATCATCA-3′; OsCBL1RV2, 5′-TTCAGATCTCTGTGACGAGATCATC-3′; OsCBL1FW2, 5′-ATGCAGGAAGAGTTCCAAC-3′; OsCBL2FW1, 5′-GCTGCAGATCTCATGGTGCAGTGTCTCGAC-3′; OsCBL2RV1, 5′-TCAGGTGTCATCGACCTGGGAATGGAAGAC-3′; OsCBL2 RV2, 5′-AGATCTCGGTGTCATCGACCTGGGAATGGAAGAC-3′; OsCBL3FW1, 5′-AGATCTCATGTTGCAGTGTCTGGAGG-3′; OsCBL3RV1, 5′-TCAAGTATCGTCGACTTGAGAATGGAAGAC-3′; OsCBL3RV2, 5′-TTCAGATCTCAGTATCGTCGACTTGAG-3′; OsCBL3FW2, 5′-ATCACAACTACATTTCCAAGC-3′; OsCBL3RV3, 5′-CAACTAGGGAATTCCTGAGCC-3′; OsCBL4FW1, 5′-TCGCCAGATCTCATGGGATGCGCGTCGTCG-3′; OsCBL4RV1, 5′-ATTCGAATTCTCAGTCATGGGCTTCTGAAT-3′; OsCBL4RV2, 5′-TCTGTAAGATCTGTCATGGGCTTCTGAATGC-3′; OsCBL7FW, 5′-GTTCAAGCAAGCAGACTTAAACAG-3′; OsCBL7RV, 5′-TATCCTTCCAAGTGCTGACAGCTGG-3′; HvCBL2FW, 5′-GGTGCTCCGATACCTGAAATTCTTGCAG-3′; HvCBL2RV, 5′-GGGATCCTAGATGACAAACAATATCAAG-3′; RAC1FW, 5′-CCTGCTATGTACGTCGCCAT-3′; and RAC1RV, 5′-AGGCTGGAAGAGGACCTCAG-3′.
Yeast Two-Hybrid Analysis
Yeast strain Y190 was cotransformed with each pBD-CBL (AtCBL1, OsCBL1–4) and with each pAD-AtCIPK (AtCIPK1, 6, and 8) by the lithium acetate method (Ito et al., 1983). Cotransformants were first selected on synthetic medium lacking Leu and Trp. Two colonies from the first selection medium were picked and transferred to medium lacking Leu, Trp, and His (SC-Leu-Trp-His). Twenty-five millimolars of 3-amino-1, 24-aminotriazole was supplemented to the medium to suppress background growth of yeast cells. Interactions between OsCBLs and AtCIPKs were confirmed by the HIS3 nutritional reporter and expression of β-galactosidase by His+ cells in a filter lift assay as described (Breeden and Nasmyth, 1985).
Northern Analysis
Rice half-grains or rice seedlings were harvested at the times indicated and ground in liquid nitrogen with a mortar and pestle. Total RNA was isolated as described (Hwang et al., 1999). For northern analysis, partial sequences covering most of the coding region for OsCBL1 or 3-′UTR for OsCBL3 were amplified from pCR2.1-OsCBL1 or the first-strand cDNA using the primer set OsCBL1FW2/RV1 and OsCBL3FW2/RV3, respectively, and used as probes. Full-length cDNA sequences were used for detecting the expression of OsCBL2 and 4. Rice genomic DNA was prepared using the potassium acetate method (Dellaporta et al., 1983). A 349-bp fragment of OsCBL7 covering the third and fourth exons, including a short intron (79 bp), were amplified from genomic DNA using the primer set OsCBL7FW/RV. A rice Actin1 gene probe was amplified from the cDNA first strand using the primer set RAC1FW/RV. PCR cycling conditions were as described above. Probes were labeled by random priming (Prime-It II random primer labeling kit; Stratagene, La Jolla, CA) and purified by column chromatography (Micro Bio-spin P-30 chromatography column; Bio-Rad, Hercules, CA). DNA dot blots containing 1 and 0.1 fmol of OsCBL1, 2, and 4 were included in hybridization tubes during hybridization of northern blots to test for cross-hybridization.
Hybridization and washing conditions were as described in Hwang et al. (1999). Nylon membrane was exposed to a phosphorimager screen and analyzed by laser densitometry (Molecular Dynamics, Sunnyvale, CA).
Quantification of expression was carried out with ImageQuant software (Molecular Dynamics). RNA-blot experiments were repeated at least twice and a typical result is presented.
Transformation of Barley Aleurone Protoplasts and Rice Half-Grains
Barley aleurone protoplasts were prepared as described (Bethke and Jones, 2001). Each flask of protoplasts was made from 40 to 50 quarter-grains and the Arg and cellulase solution volumes were 3 mL. Freshly prepared protoplasts were released into 3 to 5 mL of Gamborg's medium (Gamborg's B-5 salts with minimal organics [Sigma, St. Louis] augmented with 5 mm KNO3, 58 mm Suc, 10 mm l-Arg-HCl, 0.68 m mannitol, 111 mm Glc, and 10.9 mm MES monohydrate, pH 5.4). The protoplast suspension from one to four flasks was filtered through one layer of 33-μm nylon mesh in order to remove large debris, and then transferred to a sterile 12-mL, round-bottom culture tube and centrifuged at 60g for 3 min at 4°C in a HN-SII centrifuge (International Equipment). The supernatant was discarded and the protoplast pellet resuspended by gentle rocking in 10 mL of MW5 (200 mm mannitol, 154 mm NaCl, 125 mm CaCL2, 5 mm KCl, 5 mm Glc, pH 5–6). The tube was kept on ice for 30 to 60 min, then centrifuged as before. The pellet was resuspended in MaMg (0.5 m mannitol, 15 mm MgCl2, 5 mm MES, pH 5.6) at 0.5 mL per flask of protoplasts. Protoplast density was at least 106 protoplasts per mL. For transformations, 0.3 mL of protoplast suspension was transferred to a fresh culture tube and 30 μg of purified plasmid DNA was added. Plasmid DNA was isolated using Qiagen midiprep columns prior to precipitating in isopropanol, washing in 75% ethanol, drying, and resuspending in sterile water to at least 1 μg/μL. After 5-min incubation, protoplasts were swirled gently to resuspend, 0.3 mL of polyethylene glycol (PEG)-4-diphosphocytidyl methylerythriol synthase [40% PEG 4,000 or 6,000, 0.4 m mannitol, 0.1 m Ca(NO3)2, pH 7 to 9] were added and the protoplast suspension was mixed gently. Cells were incubated at room temperature for 30 min. The PEG-containing solution was added with MW5 over approximately 10 min by repeatedly (eight times) adding 1 mL of MW5 and mixing. Protoplasts were centrifuged as before, resuspended in 1.5 mL Gamborg's medium containing an additional 0.08 m Glc, transferred to 25-mL sterile flasks, and treated with hormones as indicated. Transient expression assays were repeated three times and a typical result is presented.
Transient expression of candidate constructs by particle bombardment of rice half-grains was as described in detail (Gubler et al., 1997).
Fluorescence Microscopy
GFP fluorescence from living barley aleurone protoplasts was visualized using a Nikon Axiophot microscope equipped with a GFP filter set (set no. 41017; Chroma Technology, Rockingham, VT). Digital images were captured with a Q Imaging micropublisher 5.0 camera (Q Imaging, Burnaby, British Columbia, Canada) and Q Capture software (Q Imaging) and images were saved as TIF files. Bright-field images were captured immediately without adjusting the focus and before fluorescence images were captured. Adjustments to image brightness and contrast were made using Adobe Photoshop (San Jose, CA).
Acknowledgments
We thank Jeorg Kudla and Uener Kolukisaoglu for their helpful discussions about the rice CBL nomenclature and also for sharing their unpublished data. The help of Sheng Luan is gratefully acknowledged. We are also grateful to Frank Gubler, CSIRO, Canberra, Australia, for plasmid pLZUbi. Fluorescence microscopy was done in the College of Natural Resources Biological Imaging Facility at the University of California, Berkeley, campus.
This work was supported in part by grants from the National Science Foundation, the Division of Natural Resources of the University of California, and the Torrey Mesa Research Institute.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.062703.
References
- Albrecht V, Ritz O, Linder S, Harter K, Kudla J (2001) The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J 20: 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtel DB, Pomeranz Y (1977) Ultrastructure of the mature rice (Oryza sativa) caryopsis. The caryopsis coat and the aleurone cells. Am J Bot 64: 966–973 [Google Scholar]
- Bethke PC, Gilroy S, Jones RL (1995) Calcium and plant hormone action. In PJ Davies, ed, Plant Hormones. Physiology, Biochemistry and Molecular Biology, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 298–317
- Bethke PC, Jones RL (1997) Reversible protein phosphorylation regulates the activity of the slow-vacuolar ion channel. Plant J 11: 1227–1235 [Google Scholar]
- Bethke PC, Jones RL (2001) Cell death of barley aleurone protoplasts is mediated by reactive oxygen species. Plant J 25: 19–29 [DOI] [PubMed] [Google Scholar]
- Bethke PC, Schuurink RC, Jones RL (1997) Hormonal signaling in cereal aleurone. J Exp Bot 48: 1337–1356 [Google Scholar]
- Bethke PC, Swanson SJ, Hillmer S, Jones RL (1998) From storage compartment to lytic organelle: the metamorphosis of the aleurone protein storage vacuole. Ann Bot (Lond) 82: 399–412 [Google Scholar]
- Breeden L, Nasmyth K (1985) Regulation of the yeast HO gene. Cold Spring Harb Symp Quant Biol 50: 643–650 [DOI] [PubMed] [Google Scholar]
- Chen X, Chang M, Wang B, Wu R (1997) Cloning of a Ca2+-ATPase gene and the role of cytosolic Ca2+ in the gibberellin-dependent signaling pathway in aleurone cells. Plant J 11: 363–371 [DOI] [PubMed] [Google Scholar]
- Deikman J, Jones R (1985) Control of α-amylase mRNA accumulation by gibberellic acid and calcium in barley aleurone layers. Plant Physiol 78: 192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S, Wood J, Hicks J (1983) A plant minipreparation version 11. Plant Mol Biol Rep 1: 19–21 [Google Scholar]
- Elledge S, Muligan J, Ramer S, Spottswood M, Davis R (1991) Lambda YES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutants. Proc Natl Acad Sci USA 88: 1731–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S (1996) Signal transduction in barley aleurone protoplasts is calcium dependent and independent. Plant Cell 8: 2193–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Watts RJ, Kalla R, Matthews P, Keys M, Jacobsen JV (1997) Cloning of a rice cDNA encoding a transcription factor homologous to barley GAMyb. Plant Cell Physiol 38: 362–365 [DOI] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97: 3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Gubrium E, Harper JF (2001) The CDPK superfamily of protein kinases. New Phytol 151: 175–183 [DOI] [PubMed] [Google Scholar]
- Hudson ME, Lisch DR, Quail PH (2003) The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J 34: 453–471 [DOI] [PubMed] [Google Scholar]
- Hwang YS, Thomas BR, Rodriguez RL (1999) Differential expression of rice alpha-amylase genes during seedling development under anoxia. Plant Mol Biol 40: 911–920 [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S (2003) CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15: 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KN, Cheong YH, Gupta R, Luan S (2000) Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol 124: 1844–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee CB, Ren H, Wang XT (1998) Regulation of the calmodulin-stimulated protein phosphatase calcineurin. J Biol Chem 273: 13367–13370 [DOI] [PubMed] [Google Scholar]
- Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134: 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Xu Q, Harter K, Gruissem W, Luan S (1999) Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc Natl Acad Sci USA 96: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewit-Bentley A, Rety S (2000) EF-hand calcium-binding proteins. Curr Opin Struct Biol 10: 637–643 [DOI] [PubMed] [Google Scholar]
- Liu J, Zhu J-K (1998) A calcium sensor homolog required for plant salt tolerance. Science 280: 1943–1945 [DOI] [PubMed] [Google Scholar]
- Lovegrove A, Hooley R (2000) Gibberellin and abscisic acid signaling in aleurone. Trends Plant Sci 5: 102–110 [DOI] [PubMed] [Google Scholar]
- Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell (Suppl) 14: S389–S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseyko N, Feldman LJ (2001) Expression of pH-sensitive green fluorescent protein in Arabidopsis thaliana. Plant Cell Environ 24: 557–563 [DOI] [PubMed] [Google Scholar]
- Neuhaus JM (1996) Protein targeting to the plant vacuole. Plant Physiol Biochem 34: 217–221 [Google Scholar]
- O'Neill S, Kumagai M, Majumdar A, Huang N, Sutliff T, Rodriguez R (1990) The alpha-amylase genes in Oryza sativa: characterization of cDNA clones and mRNA expression during germination. Mol Gen Genet 221: 235–244 [DOI] [PubMed] [Google Scholar]
- Ritchie S, Gilroy S (1998) Calcium-dependent protein phosphorylation may mediated the gibberellic acid response in barley aleurone. Plant Physiol 116: 765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie S, Swanson SJ, Gilroy S (2002) From common signaling components to cell specific responses: insights from the cereal aleurone. Physiol Plant 115: 342–351 [DOI] [PubMed] [Google Scholar]
- Sanders D, Brownlee C, Harper JF (1999) Communicating with calcium. Plant Cell 11: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell 14: S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurink RC, Chan PV, Jones RL (1996) Modulation of calmodulin mRNA and protein levels in barley aleurone. Plant Physiol 111: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JR, Kim KN, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J (1999) Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11: 2393–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden WA, Fromm H (2001) Calmodulin as a versatile calcium signal transducer in plants. New Phytol 151: 35–66 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, Matsuoka M (2000) Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci USA 97: 11638–11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lu YT (2003) Calmodulin-binding protein kinases in plants. Trends Plant Sci 8: 123–127 [DOI] [PubMed] [Google Scholar]
- Zhu T, Budworth P, Chen W, Provart N, Chang H-S, Guimil S, Su W, Estes B, Zou GXW (2003) Transcriptional control of nutrient partitioning during rice grain filling. Plant Biotechnol J 1: 59–70 [DOI] [PubMed] [Google Scholar]
- Zhu T, Budworth P, Han B, Brown D, Chang H-S, Zou Z, Wang X (2001) Towards elucidating global gene expression in developing Arabidopsis: parallel analysis of 8300 genes. Plant Physiol Biochem 39: 221–242 [Google Scholar]
- Zielinski RE (1998) Calmodulin and calmodulin-binding proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 697–726 [DOI] [PubMed] [Google Scholar]