Abstract
Acetylcholinesterase (AChE) has been increasingly recognized in plants by indirect evidence of its activity. Here, we report purification and cloning of AChE from maize (Zea mays), thus providing to our knowledge the first direct evidence of the AChE molecule in plants. AChE was identified as a mixture of disulfide- and noncovalently linked 88-kD homodimers consisting of 42- to 44-kD polypeptides. The AChE hydrolyzed acetylthiocholine and propyonylthiocholine, but not S-butyrylthiocholine, and the AChE-specific inhibitor neostigmine bromide competitively inhibited its activity, implying that maize AChE functions in a similar manner as the animal enzyme. However, kinetic analyses indicated that maize AChE showed a lower affinity to substrates and inhibitors than animal AChE. The full-length cDNA of maize AChE gene is 1,471 nucleotides, which encode a protein having 394 residues, including a signal peptide. The deduced amino acid sequence exhibited no apparent similarity with that of the animal enzyme, although the catalytic triad was the same as in the animal AChE. In silico screening indicated that maize AChE homologs are widely distributed in plants but not in animals. These findings lead us to propose that the AChE family, as found here, comprises a novel family of the enzymes that is specifically distributed in the plant kingdom.
A well-known biological function of acetylcholine (ACh) is as a neurotransmitter in animals and insects. ACh serves to propagate an electrical stimulus across the synaptic junction. At the presynaptic neuron end, an electrical impulse triggers release of ACh, which accumulates in vesicles into the synaptic cleft via exocytosis. ACh then binds to an ACh receptor (AChR) on the postsynaptic neuron surface, and the ACh-AChR binding induces subsequent impulses to the postsynaptic neuron. Finally, ACh, which is released again by the receptor into the synaptic cleft, is rapidly degraded by acetylcholinesterase (AChE; E.C.3.1.1.7; Kelly et al., 1979; Dunant et al., 1980; MacIntosh, 1981). Meanwhile, ACh has been recently identified in a number of non-neuronal tissues in animals, fungi, bacteria, and plants (Wessler et al., 2001; Horiuchi et al., 2003). Although plants lack a nervous system, both ACh and ACh-hydrolyzing activity have been widely recognized in the plant kingdom (Evans, 1972; Fluck and Jaffe, 1974a, 1974b; Hartmann and Kilbinger, 1974; Verbeek and Vendrig, 1977; Lees et al., 1978; Momonoki and Momonoki, 1991). The ACh-hydrolyzing activities in plant tissues can be measured by the appearance of radioactive acetate following [1-14C]ACh hydrolysis (Momonoki, 1992), by the appearance of thiol group following acetylthiocholine (ASCh) hydrolysis (Momonoki, 1997), and by histochemical detection of ASCh hydrolysis (Momonoki, 1997; Momonoki et al., 1998). Moreover, the ACh-hydrolyzing activity in plant tissue is inhibited by neostigmine bromide, which is known as a specific inhibitor of the animal AChE (Momonoki, 1992). Thus, it is widely accepted that an ACh-hydrolyzing enzyme, namely AChE, is widely distributed in the plant kingdom.
When a plant is moved from the vertical orientation to a horizontal position, the stem shows curvature as it returns to a vertical position by response acting against gravistimulus. According to investigations of asymmetrically growing maize (Zea mays) shoots, endogenous indole-3-acetic acid (IAA; Bandurski, 1980; Bandurski et al., 1986), d-Glc (Momonoki, 1988), and IAA-inositol (Momonoki, 1988) were all localized mainly in the lower half of the horizontally oriented shoots. The IAA accelerates the growth of the lower side of the shoot, and, as a result, the shoot curves to a vertical position. These findings implied the probable existence of a potential-gating system, which would allow the asymmetric hormone and substance distributions following the stimulus. On the other hand, maize seedlings that were treated first with neostigmine bromide did not respond to the gravistimulus (Momonoki, 1997). Furthermore, after the stimulus, the AChE activity was distributed asymmetrically at the interface between the stele and the cortex of the gravistimulated maize seedlings (Momonoki, 1992). An asymmetric solute distribution of the AChE activities in maize seedlings (Momonoki et al., 1998) and in rice (Oryza sativa) seedlings (Momonoki et al., 2000) was also inhibited by neostigmine bromide. The role of AChE in animal nervous systems has been clearly demonstrated by the effects of its inhibitors. Neostigmine bromide is an AChE inhibitor having no inhibitory effect on IAA metabolism in plants (Ballal et al., 1993). Therefore, these findings strongly suggest that AChE plays a role in the gravity response of plants, and a hypothetical ACh-mediated system was proposed as a candidate for the potential-gating regulator. This system might achieve cell-to-cell transport of the hormone and other substances between the plant cells and might propagate a membrane depolarization. Significant increases of AChE activity in plant tissues appeared not only after gravity stimuli but also in response to heat stress (Momonoki and Momonoki, 1993). Heat tolerant plants, such as yard-long bean (Vigna sinensis var. sesquipedalis), cucumber (Cucumis sativus), and radish (Raphanus sativus), which are native tropical zone plants, showed high AChE activity in their node, stem, pulvini, and root after heat stress (Momonoki and Momonoki, 1993).
At the animal neuromuscular junction, an electrical impulse causes an influx of Ca2+ from outside the membrane via voltage-gated channels, and then the Ca2+ acts as a trigger for ACh release. Based on fluorescence analysis, Ca2+ was localized in the cortex cells surrounding the vascular system, in the epidermis, and in adhering peripheral cortical cells. A simultaneous increase of AChE activity and Ca2+ in the endodermal cells between the stele and the cortex in the maize seedlings was observed following heat stress, indicating that the plant ACh-mediated system might respond to Ca2+ signals in a manner similar to the animal system (Momonoki et al., 1996). Recently, it was reported that the bamboo (Phyllostachys bambusoides) shoot (especially the upper portion), whose growth rate is very rapid, contains significant levels of ACh and ACh synthesis capacity (Horiuchi et al., 2003). Most reports concerning plant ACh and its related molecules have suggested that an ACh-mediated system might play a role in the response to environmental stimuli.
The existence and biological functions of AChE as a constituent of an ACh-mediated system in plant tissues has been hypothesized based on indirect evidence such as measurement of ACh-hydrolyzing activity and inhibitory tests using anti-AChE reagents. However, direct evidence of the molecule(s) responsible for ACh hydrolysis was missing, and the corresponding gene had not yet been identified. In this study, the AChE protein was purified from etiolated maize seedlings, and the cDNA encoding the maize AChE was cloned and sequenced. Moreover, the enzymatic properties of the purified maize AChE were characterized and compared with those of electric eel AChE as a representative of animal AChE. The purified maize AChE exhibited enzymatic properties similar to those of the electric eel AChE, except in the relative affinities to substrates and inhibitors. The maize AChE possesses a consensus sequence from the lipase Gly-Asp-Ser-Leu (GDSL) family, which is distinct from the carboxylesterase family consensus sequence that characterizes the animal AChE. Homologous maize AChE genes are widely distributed in the plant kingdom, based on in silico screening results, but similar genes were not found in any animal, fungal, or bacterial databases. Therefore, the AChE family, as found in this study, comprises a novel AChE family that is specifically distributed in the plant kingdom.
RESULTS
Purification of AChE from Maize Seedlings
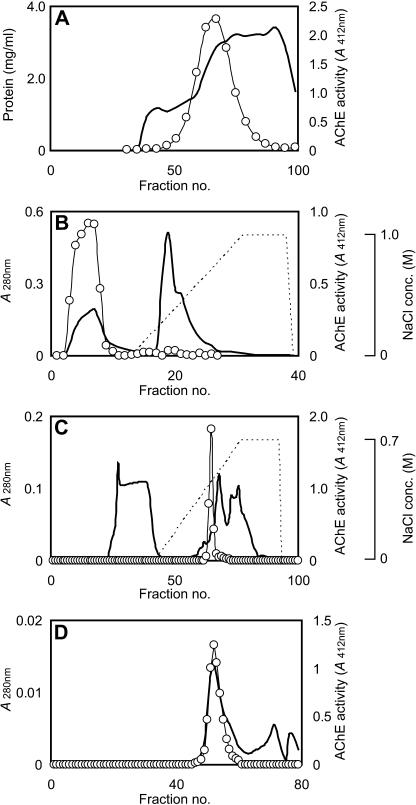
Ten batches of the crude extract of the maize seedlings were combined and subjected to the following chromatographic purification procedure. The extract was applied to a Sephadex G-200 gel filtration column (2.5 × 75 cm; Amersham Biosciences, Uppsala; Fig. 1A). The active fractions were collected and applied to a Poros HQ/20 column (4.6 × 100 mm; PE-Applied Biosystems, Foster City, CA) equilibrated with 20 mm phosphate buffer, pH 7.0. The AChE activity was found in flow-through fraction, as shown in Figure 1B. The active fractions were collected and applied to a Poros HS/20 column (4.6 × 100 mm; PE-Applied Biosystems) equilibrated with 20 mm phosphate buffer, pH 5.0, and the adsorbed proteins were eluted with a linear NaCl gradient ranging from 0 to 0.7 m, as shown in Figure 1C. The active fractions were eluted as the major peak. The fractions were further purified with a Hiload Superdex 200 pg 16/60 column (1.6 × 60 cm; Amersham Biosciences) equilibrated with 20 mm phosphate buffer, pH 7.0, as shown in Figure 1D. The purification procedure parameters are summarized in Table I. We could not assay to determine the precise AChE activities for the crude enzyme and the ammonium sulfate fractions because these samples were colored, probably due to a pigment from the plant tissues, and this pigment would interfere with the colorimetric assay. After this series of purification procedures, the yield from 2 kg of maize seedlings was 0.8 μg of protein having a total activity of 0.07 units.
Figure 1.
Purification of the maize AChE by column chromatography. Line, Absorbance at 280 nm; line with circles, AChE activity. A, A typical elution profile of maize AChE on a Sephadex G-200 column equilibrated with phosphate-buffered saline. B, A typical elution profile of maize AChE on a Poros HQ/20 column equilibrated with 20 mm phosphate buffer, pH 7.0. C, A typical elution profile of maize AChE on a Poros HS/20 column equilibrated with 20 mm phosphate buffer, pH 5.0. D, A typical elution profile of maize AChE on a Hiload Superdex 200 pg 16/60 column equilibrated with 20 mm phosphate buffer, pH 7.0.
Table I.
Summary of the purification of maize AChE
| Purification Steps | Activity | Total Protein | Specific Activity | Purification-fold | Yield |
|---|---|---|---|---|---|
| units | mg | units/mg | % | ||
| Sephadex G-200 | 5.45 | 15.59000 | 0.4 | 1.0 | 100 |
| Poros HQ | 3.33 | 5.37000 | 0.6 | 1.8 | 61.09 |
| Dialysis | 3.12 | 3.71000 | 0.8 | 2.4 | 57.27 |
| Poros HS | 0.80 | 0.06530 | 12.3 | 35.0 | 14.67 |
| Superdex 200 | 0.07 | 0.00079 | 82.5 | 236.0 | 1.20 |
Molecular Composition of Maize AChE
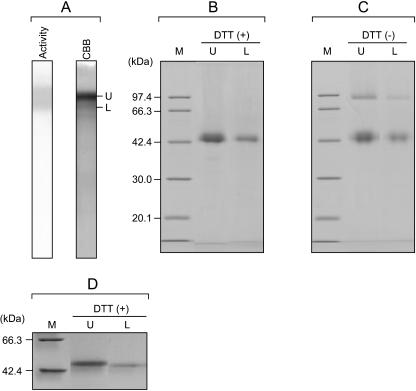
The protein was then analyzed by PAGE under nondenaturing conditions (native PAGE), and the purified AChE preparation showed two predominant bands having different Coomassie Brilliant Blue staining intensities (designated as the U and L forms, Fig. 2A) and several minor bands. Both the U and L bands showed AChE activity, as indicated by staining using ASCh as a substrate (Fig. 2A), and production of the colored product was inhibited by adding an AChE-specific inhibitor, neostigmine bromide, indicating that each band certainly contained AChE. The native PAGE gel slices corresponding to AChE protein bands, which appeared as U and L forms, were boiled with SDS-PAGE sample buffer in the presence of dithiothreitol (DTT) for 5 min and then subjected to SDS-PAGE using 12.5% polyacrylamide gels. As shown in Figure 2B, each extract migrated as a diffuse band corresponding to a molecular mass of between 42 and 44 kD on SDS-PAGE. On the other hand, each extract displayed two bands on SDS-PAGE in the absence of DTT, one sharp band with an apparent molecular mass of 88 kD and another diffuse band with a molecular mass of approximately 42 to 44 kD, as shown in Figure 2C. These results seemed to indicate that both the U and L forms of AChE exist as a mixture of 88-kD dimers consisting of disulfide-linked 42- to 44-kD polypeptides and a 42- to 44-kD monomer. Diffuse bands are often observed in the electrophoretic banding patterns of glycoproteins on SDS-PAGE, and sharpness of the bands may vary with the polyacrylamide concentration of the gels. Indeed, both of the 42- to 44-kD polypeptides corresponding to the U and L forms seen on reduced SDS-PAGE were stained with the periodic acid Schiff reagent, indicating that both polypeptides are certainly glycosylated (data not shown). To compare the electrophoretic mobilities of the bands in more detail, the U and L forms of AChE extracted from the native PAGE gels were applied to reducing SDS-PAGE using 15% polyacrylamide gels. As shown in Figure 2D, each extract exhibited a single band on the gels, as with the 12.5% gels, but these bands were sharper. The L form of AChE migrated farther than did the U form on the 15% gel, indicating that the L form of AChE consists of a smaller subunit, compared with the U form of AChE. The N-terminal amino acid sequences for the 42- to 44-kD U- and L-form proteins were identical, NH2-AGAGGDCHFPAVFNFGDSNS. This result implied that the U and L forms of AChE are the same gene products with different posttranslational modifications, which may consist of the deletion of several residues from the C termini and/or different degrees of the glycosylation.
Figure 2.
Native and SDS-PAGE analysis of the purified maize AChE. A, Native PAGE of the partially purified maize AChE. Separated proteins were stained with Coomassie Brilliant Blue (CBB) and AChE activity stain (Activity). B, SDS-PAGE (12.5%) of the extractions (U and L, which correspond to bands in A) from the native PAGE gels in the presence of DTT. The lane indicated by M is molecular mass standards. C, SDS-PAGE (12.5%) of the extractions from the native PAGE gels in the absence of DTT. D, SDS-PAGE (15%) of the extractions from the native PAGE gels in the presence of DTT.
Thus far, we have presumed that the purified AChE existed as mixture of the monomeric and dimeric forms of the 42- to 44-kD glycosylated polypeptides. However, the maize AChE eluted as a single peak during all chromatographic purification procedures, and both the U and L forms of the AChE protein ran as a single band on the native PAGE. These behaviors showed an obvious discrepancy with the original postulation. The purified enzyme was subjected to analytical gel filtration on a Superdex 200 HR10/30 column equilibrated with phosphate buffer, pH 7.0, containing 0.15 m NaCl and 10% glycerol. As a result, the AChE activity eluted as a single peak with an elution volume of 14.2 mL, and its apparent molecular mass was estimated as 67 kD. The AChE preparation did not display any other active peaks eluting in ranges corresponding to either twice or half the size of the 67-kD protein on gel filtration, indicating that the maize AChE exists as a single molecular species without any differently sized forms. Therefore, it was concluded that the U and L forms of AChE were composed of a mixture of different dimeric forms: a disulfide-linked homodimer and a noncovalently linked homodimer, each of which consists of 42- to 44-kD polypeptides. Because the U-form protein was identified as the predominant form by Coomassie Brilliant Blue staining on the native PAGE, we speculated that the U-form protein is a mature enzyme, and the L form is a Golgi-mediated processing intermediate form or a partially digested form of the mature enzyme, resulting from proteolytic digestion in the plant tissues or during the purification procedures. The molecular mass of the enzyme was estimated by analytical gel filtration, and this estimate did not agree with the 88-kD band observed on SDS-PAGE in the absence of DTT. The apparent molecular mass (67 kD) of the enzyme on analytical gel filtration was probably a low estimate resulting from a specific interaction between the enzyme and the gel filtration resin and/or a compact conformation of the dimer forms.
Characterization of Purified Maize AChE
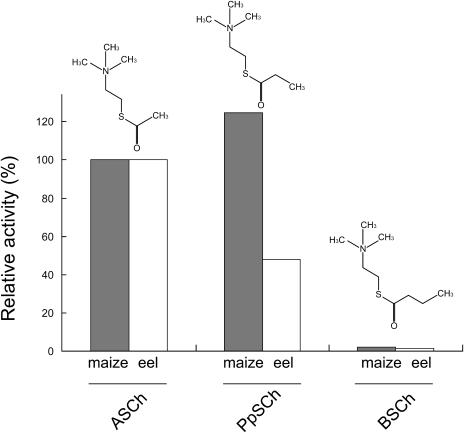
The cholinester-hydrolyzing enzymes are usually classified into AChE and butyrylcholinesterase (BChE), according to their substrate and inhibitor specificity. To examine the substrate specificity of the maize AChE among thiocholinesters containing acyl chains of different lengths, the enzymatic activity was tested using ASCh, propionylthiocholine (PpSCh), and S-butyrylthiocholine (BSCh) as substrates with the 5-5′-dithiobis-2-nitrobenzoic acid (DTNB) method. To compare the enzymatic characteristics of the maize enzyme with those of the animal enzyme, the commercially purchased electric eel AChE (Sigma-Aldrich, St. Louis) was used. The activity of maize and electric eel AChE against ASCh was taken to be 100%. As shown in Figure 3, the maize AChE exhibited 1.2-fold increased enzymatic activity against PpSCh compared with that against ASCh, while the electric eel AChE showed 0.5-fold activity against PpSCh than against ASCh. Both of the AChEs exhibited extremely low enzymatic activity against BSCh. Thus, the substrate selectivity of the maize AChE was estimated as PpSCh > ASCh ≫ BSCh, and that of the electric eel AChE was ASCh > PpSCh ≫ BSCh. Furthermore, the kinetics of the maize and eel AChEs were examined by using ASCh and PpSCh as substrates (Table II). The maize AChE showed a slightly higher Km value of 4.7 mm for ASCh, compared with 3.1 mm for PpSCh. Moreover, these values were higher than the Km values of 0.16 mm for ASCh and 0.84 mm for PpSCh, as determined for the electric eel AChE. Furthermore, the maize AChE displayed relatively low Vmax values of 1.7 μmol min−1 mg−1 for ASCh and PpSCh, compared with the Vmax values of 312.5 μmol min−1 mg−1 for ASCh and 263.2 μmol min−1 mg−1 for PpSCh on the electric eel AChE.
Figure 3.
Comparison of substrate selectivity of the maize and eel AChE. ASCh, PpSCh, and BSCh were used as substrates, and the activities were measured by the DTNB method as described in “Materials and Methods.” The activity of maize and electric eel AChE against ASCh was taken to be 100%. The structural diagrams of the substrates are illustrated at the top of each column.
Table II.
Kinetic data of the maize and electric eel AChE against substrate using ASCh and PpSCh
| ASCh
|
PpSCh
|
|||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| mm | μmol min−1 mg−1 | mm | μmol min−1 mg−1 | |
| Maize | 4.7 | 1.7 | 3.1 | 1.7 |
| Eel | 0.2 | 312.5 | 0.8 | 263.2 |
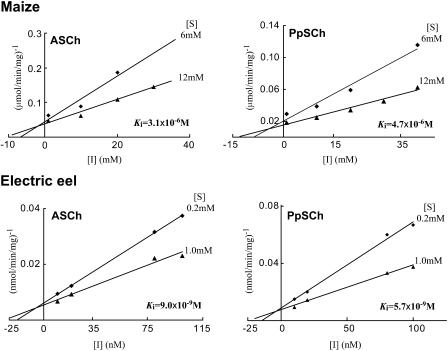
Neostigmine bromide is known as a specific inhibitor of the animal AChE, as it binds to the catalytic site of the enzyme (Standaert, 1990), and it also inhibits plant AChE (Momonoki, 1992; Ballal et al., 1993). To examine the inhibitory effects of neostigmine bromide on the hydrolyzing activities of maize AChE, the kinetics of the maize and electric eel enzymes against neostigmine bromide were examined by Dixon plots (Dixon, 1953), as shown in Figure 4. As a result, the neostigmine bromide inhibited the hydrolysis of the ASCh and PpSCh in a typically competitive manner, with apparent inhibition constant values (Ki) of 3.1 × 10−6 m and 4.7 × 10−6 m, respectively, for maize AChE. The maize AChE exhibited relatively high Ki values, compared with Ki values for the electric eel AChE, which were 9.0 × 10−9 m for the ASCh hydrolysis and 5.7 × 10−9 m for PpSCh hydrolysis.
Figure 4.
Inhibitory efficiency of neostigmine bromide on ASCh and PpSCh by the maize and eel AChE. The Ki values were determined by Dixon plot, as described in “Materials and Methods.”
Nucleotide Sequence Coding for the Maize AChE cDNA
The 42- to 44-kD polypeptide of the U-form AChE, which was separated by SDS-PAGE in the presence of DTT, was digested with Staphylococcus aureus V8 protease and generated proteolytic fragments were applied for N-terminal amino acid sequence analyses. Of the several derivative fragments, two probable internal sequences were yielded, NH2-YFSQALYTFDIGQNDITSSY and NH2-VEAIIPDLME. A tBLASTn search against the maize database using these amino acid sequences identified a partial-length maize mRNA nucleotide sequence (accession no. AY107095) as a candidate. A partial open reading frame, which covers all short amino acid sequences determined for the purified AChE protein, was identified in the mRNA sequence. However, the sequence does not contain termination codon, although it possesses a presumed initiating ATG codon. Hence, we determined the complete nucleotide sequence corresponding to its full-length cDNA in this study. The cDNA library was prepared by 5′- and 3′-RACE using total RNA extracted from 5-d-old seedlings, prepared according to the same procedure used for the AChE protein purification. The primers used were Mmn-2 for the 5′-RACE (5′-ATCCAGAAGTACCTTCCTCC-3′) and Sgn-1 for the 3′-RACE (5′-ACAGAAGCCAACTCGTCTAC-3′), and these were designed to obtain fragments that overlapped with each other in a 235-bp region. The purified 5′-RACE (770 bp) and 3′-RACE (940 bp) PCR products were subcloned, and then each nucleotide sequence was determined using the primers designed from the partial-length mRNA sequence (Table III). The determined nucleotide sequence consists of 1,471 bp. The sequence contains a putative entire open reading frame, which consists of 1,182 bp and encodes 394 amino acid residues. The N-terminal amino acid sequence for the purified AChE corresponds to the sequence starting from Ala-30 of the amino acid sequence deduced from the nucleotide sequence of the cDNA. The N-terminal amino acid sequences determined for the V8 protease-derived fragments were identical to those deduced from the DNA sequence for Tyr-171 to Tyr-190 and for Val-199 to Glu-208, indicating that this cDNA certainly encodes the maize AChE protein. The sorting server TargetP (Emanuelsson et al., 2000) predicts that the N terminus of the deduced sequence of the maize AChE precursor protein contains a signal sequence consisting of 29 residues, which routes the protein to secretory pathway with a probability of 93%. This prediction is supported by the observation that 29 amino acid residues are removed from the mature AChE. The calculated molecular mass of AChE without these 29 N-terminal amino acids is 39,722.3 D, with an pI of 7.18. The apparent molecular masses of the AChE subunits were estimated to be approximately 42 to 44 kD by SDS-PAGE in the presence of DTT, which differed from the calculated value. In the deduced sequence, six consensus sequences indicating N-glycosylation sites, N-X-(S/T), were identified, and this prediction was supported by the periodic acid Schiff staining results. Furthermore, 10 Ser, five Thr, and five Tyr residues were predicted to be probable phosphorylation sites by the NetPhos 2.0 server (http://www.cbs.dtu.dk/services/NetPhos/). Thus, the discrepancy observed between estimated and calculated molecular masses might be due to posttranslational modifications such as glycosylation.
Table III.
Nucleotide sequences of primers used for this study
| Forward | Sequence | Target | Reverse | Sequence | Target |
|---|---|---|---|---|---|
| Sgn-1 | 5′-ACAGAAGCCAACTCGTCTAC-3′ | 531–550 | Mmn-2 | 5′-ATCCAGAAGTACCTTCCTCC-3′ | 746–765 |
| Sgn-3 | 5′-CAAAGGCGGTATCTACAAGG-3′ | 556–575 | Mmn-4 | 5′-TAGATGTGAGCCTCTCCATC-3′ | 701–722 |
| Sgn-5 | 5′-TTCTCCCAGGCACTTTACACCTTCG-3′ | 599–623 | Mmn-6 | 5′-CGGTCAAAACGACATCACCTCAAGC-3′ | 1,306–1,330 |
| Sgn-7 | 5′-GCAGGGAGAGGGAAATGG-3′ | 72–89 | Mmn-8 | 5′-GTCAGCAGAGGGTCGTCG-3′ | 1,006–1,023 |
| Sgn-9 | 5′-ACCCTCTGCTGACATGCTG-3′ | 1,011–1,029 | Mmn-10 | 5′-GCACTGAGGTGAGTCAGTC-3′ | 360–378 |
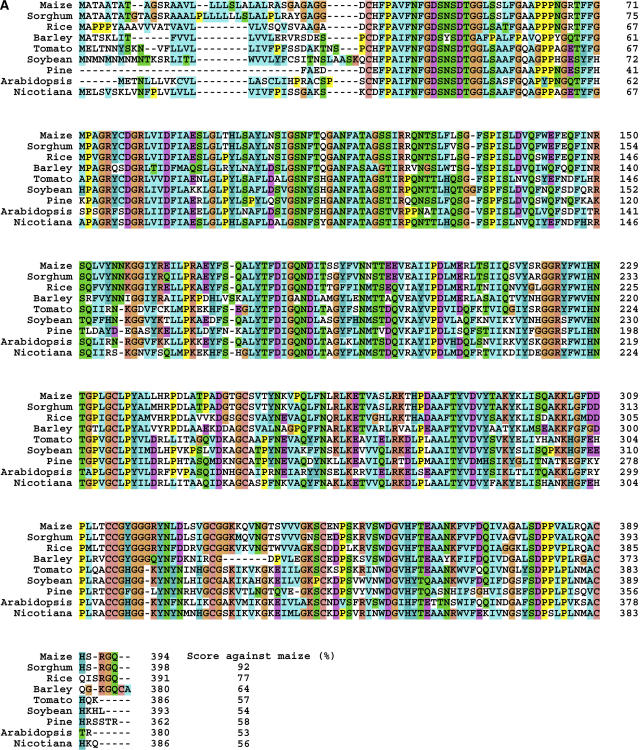
The protein domain architecture derived from the deduced amino acid sequence of the maize AChE was examined by computer-assisted protein family analysis using the protein family database Pfam (http://www.sanger.ac.uk/Software/Pfam/). The result showed that the maize AChE contains a conserved putative lipase GDSL family domain at residues 41 to 373. To search for genes homologous to maize AChE in other eukaryotes, the deduced amino acid sequence of the maize AChE was submitted to the tBLASTn search against The Institute for Genomic Research (TIGR) Unique Gene Indices (http://tigrblast.tigr.org/tgi/), which contain tentative consensus and singleton EST sequences from 29 animals, 24 plants, 15 protists, and eight fungi. Significantly related sequences were found only in plant indices containing the sorghum (Sorghum bicolor), rice, barley (Hordeum vulgare), tomato (Lycopersicon esculentum), tobacco (Nicotiana tabacum), soybean (Glycine max), pine (Pinus ponderosa), and Arabidopsis (Arabidopsis thaliana) plants. Comparison of the amino acid sequences deduced from these sequences using a ClustalX multiple sequence alignment analysis indicated that the amino acid sequences of probable AChE gene products in these plants are highly conserved, and they exhibit 53% to 92% identity against the amino acid sequence of maize AChE (Fig. 5). tBLASTn analysis against the bacterial genomic sequence database was also performed on the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/BLAST/), but no homologous genes were found in this database.
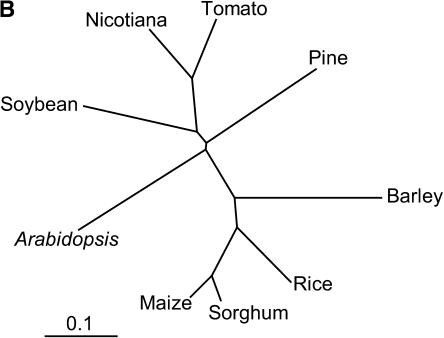
Figure 5.
ClustalX analysis of the amino acid sequences of the products of maize AChE gene homologs found in plant gene indices. The AChE homologs were identified by a tBLASTn analysis against TIGR Unique Gene Indices. A, Alignment of the amino acid sequences deduced from the AChE homologous genes. The amino acid sequence identities between the maize AChE and each gene product are indicated as percentages in the bottom column. B, Phylogenetic tree based on the ClustalW analysis. The tentative consensus sequence numbers used in this analysis are as follows: sorghum, SbGI TC95805; rice, OsGI TC237385; barley, HvGI TC120185; tomato, LeGI TC116906; soybean, GmGI TC195186; pine, PGI TC39002; Arabidopsis, AtGI TC209133; Nicotiana, NbGI TC2824.
To analyze the exon and intron organization of the maize AChE in the genomic DNA, the 28 nucleotide sequences of the genomic DNA fragments partially encoding the maize AChE precursor were gathered by tBLASTn analysis using the deduced amino acid sequence from the maize genomic DNA library database, and then the sequence fragments were assembled into single contiguous sequences by the CAP3 Sequence Assembly Program (Huang and Madan, 1999). By comparing the sequences of cDNA and genomic DNA, the AChE gene was found to consist of five exons and four introns on the maize genomic DNA, according to the GT-AG rule.
DISCUSSION
The AChE protein was purified from the crude extract of 5-d-old etiolated maize seedlings. Consequently, 0.8 μg of AChE protein having a specific activity of 82.5 units/mg was purified from 2 kg of seedlings. The purified AChE separated into two forms having different electrophoretic mobilities and different Coomassie Brilliant Blue and AChE-activity staining intensities on native PAGE. There were dominant U and minor L forms. The U and L forms of AChE ran as a broadly diffuse band with an apparent molecular mass of 42 to 44 kD on reducing SDS-PAGE, while on the nonreducing SDS-PAGE, it ran as two bands with apparent molecular masses of 42 to 44 and 88 kD, respectively. Because the purified AChE eluted as a single active peak with an apparent molecular mass of 67 kD upon analytical gel filtration, the native AChE protein apparently has only one molecular size, rather than several differently sized forms. Therefore, the U and L forms of AChE might exist as two types of dimeric forms: disulfide-linked and noncovalently linked dimers, each consisting of 42- to 44-kD subunits. Simultaneous existence of the disulfide-linked and noncovalently linked homodimeric enzyme has also been observed in cyanide-resistant alternative oxidase from the mitochondria of the voodoo lily (Amorphophallus kiusianus), soybean, and mung bean (Vigna radiate; Umbach and Siedow, 1993), and their oxidation reduction states are responsible for their activities. In animals, the active AChE was found in monomeric, disulfide-linked dimeric, and tetrameric forms, depending upon the animal species. For example, Sipunculida (Talesa et al., 1993) and Insecta (Gnagey et al., 1987; Toutant 1989) produce only the disulfide-linked dimeric form. The oxidation reduction states of the disulfide bond between the subunits in the maize AChE dimeric form could dominate an enzymatic activity, similar to the cyanide-resistant alternative oxidase. However, we could not determine which form was responsible for the AChE activity in this study because of the small yield of enzyme that was purified.
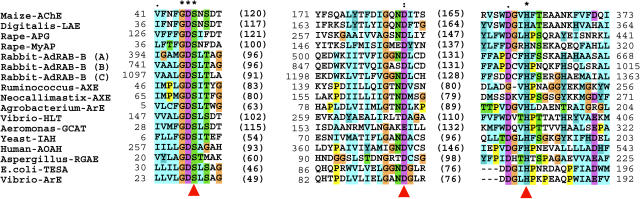
In general, cholinester-hydrolyzing enzymes are classified into the AChE and BChE classes, depending on their substrate and inhibitor specificity. AChE preferably hydrolyzes ACh or propionylcholine more readily than butyrylcholine. In contrast, BChE hydrolyzes butyrylcholine more quickly. The purified maize AChE displayed relatively high affinity to PpSCh and was followed in affinity by ASCh among the thiocholinesters employed, while BSCh was apparently a poor substrate. This indicates that the maize enzyme was characterized as an AChE, not a BChE. In addition, neostigmine bromide is a specific AChE inhibitory reagent that binds to the catalytic site of the animal AChE protein via a covalent carbamyl bond (Standaert, 1990), but its inhibition mode is nearly competitive probably because the enzyme and inhibitor initially forms reversible complex, and then the binding mode slowly changes to covalent (Kishibayashi et al., 1994). Furthermore, the neostigmine bromide was previously found to inhibit the ACh-hydrolyzing activity in the plant tissues, as well as animal AChE (Momonoki, 1992). Neostigmine bromide inhibited the hydrolytic activity of the purified maize AChE in a typically competitive manner. This means that neostigmine bromide competes with the substrates for binding to the active site of the maize AChE, in the same manner as it inhibited the animal AChE. These findings strongly indicate that the maize AChE has an active site similar to that of the animal AChE protein. The amino acid sequences of the maize and electric eel AChE precursors, however, shared extremely low homology with scores of 11.7% on ClustalW analysis. Computer-assisted protein family analysis using the Pfam database server implied that the maize AChE contains a consensus sequence of the lipase GDSL family at residues 41 to 373, and this region possesses a characteristic consensus Gly-Asp-Ser-Leu-Ser sequence. Most proteins with this consensus sequence are involved in removal of the acyl chain from esters or lipids by hydrolysis. The catalytic Ser/Asp (Glu)/His triads of the enzymes belonging to lipase GDSL family were characterized in some of these proteins, and the sequences surrounding the consensus residues are well conserved (Upton and Buckley, 1995). Therefore, the putative catalytic triad for the maize AChE, Ser-47/Asp-185/His-360, can be deduced by superimposing the sequences on those of the lipase GDSL family enzymes (Fig. 6). Although the animal AChE also has a catalytic Ser/Asp (Glu)/His triad (Shafferman et al., 1992), it possesses a distinct carboxylesterase consensus domain, but not the lipase GDSL consensus, based on Pfam protein family analysis. Thus, the maize AChE could not belong to the well-known AChE family, which is distributed throughout the animal kingdom. Based on in silico screening against the plant gene database, the genes homologous to maize AChE are widely distributed in higher plants (Fig. 5). On the other hand, a tBLASTn search of the maize AChE sequence against animal, fungi, and bacterial genomic sequence databases revealed no homologs. Therefore, we appear to have discovered a novel AChE family that is specifically distributed in the plant kingdom and that might be evolved from a different family of ancestor protein of the well-known AChE class. According to a report by Fluck and Jaffe (1975), the cholinesterase purified from eggplant (Solanum melongena), but not that from maize, was inhibited by superoptimal substrate concentration. Because the inhibition by the excess of the substrate was regarded as characteristic and common feature of the animal AChE, they had concluded that the maize cholinesterase was not real AChE, despite the enzyme specifically hydrolyzed ASCh among thiocholinesters employed. In this study, we also found that the maize AChE purified did not exhibit the inhibition by superoptimal substrate (data not shown). Among the animal AChE, the inhibitory effects of the excess of the substrate molecules on the activity might be variable depending on the source organisms and tissues, probably due to the structural difference around the active site (Tõugu, 2001). Indeed, an AChE, whose activity is not affected under the superoptimal substrate condition, was recently found from an oriental fruit fly Bactrocera dorsalis (Hsiao et al., 2004). Therefore, the discrepancy of the inhibition at high substrate concentration may be reflected by such structural difference also among the plant enzymes. As an alternative possibility, two genes in the published Arabidopsis database were suggested to have similarity with animal AChE (Fluck et al., 2000), although their gene products have not yet been recognized from the plant tissues. Thereby, multiple classes of enzymes associating with ACh-hydrolyzing activity could be distributed in the plant kingdom, which might result in the difference of the substrate inhibition manner as observed in the enzymes purified from eggplant and maize. On the other hand, the maize AChE showed a low affinity for the substrate, compared with the electric eel AChE. In addition, the maize AChE required a high concentration of neostigmine bromide for complete inhibition and displayed a low affinity to this reagent, compared with the electric eel AChE. ACh has been recognized even in evolutionally primitive organisms, such as bacteria (Horiuchi et al., 2003); thus, its function as neurotransmitter might be specialized in the animal kingdom. The animal AChE is known to be one of the fastest enzymes. In animal nerve systems, AChE degrades the neurotransmitter, ACh, to terminate signaling by preventing adsorption of ACh to the receptor. The quick reactivity of AChE could be essential to rapid control of synaptic activity, so animal AChE may have acquired a high affinity to the substrate and hydrolytic efficiency through molecular evolution (Shen et al., 2002). The difference in the affinity to the substrate between the plant and animal AChE, thereby, might be dominantly related to the different biological roles of the enzyme in each kingdom, rather than the loss of the affinity due to partial inactivation of the maize AChE during purification.
Figure 6.
Alignment of the amino acid sequences of maize AChE and selected members of the GDSL lipase family. The alignment was generated by a ClustalX analysis. Dashes indicate gaps introduced to improve the alignment. Numbers in brackets indicate intervening amino acid numbers, and the numbers without brackets indicate the positions in the sequences. The putative catalytic triad Ser/Asp (Glu)/His residues in the GDSL family enzymes, which were presumed previously (reference in Upton and Buckley, 1995), are indicated by arrowheads. The abbreviations and accession numbers of the cDNA nucleotide sequences used for this analysis are follows: Maize-AChE, Zea mays AChE (determined in this study); Digitalis-LAE, Digitalis lanata lanatoside 15′-O-acetylesterase (AJ011567); Rape-APG, Brassica napus anter-specific Pro-rich protein APG (P40603); Rape-MyAP, Brassica napus myrosinase-associated protein (U39289); Rabbit-AdRAB-B, Oryctolagus cuniculus phospholipase (Z12841); Ruminococcus-AXE, Ruminococcus flavefaciens acetyl xylan esterase (AJ238716); Neocallimastix-AXE, Neocallimastix patriciarum acetylxylan esterase bnaIII (U66253); Agrobacterium-ArE, Agrobacterium radiobacter arylesterase (AF044683); Vibrio-HLT, Vibrio parahaemolyticus thermolabile hemolysin (M36437); Aeromonas-GCAT, Aeromonas hydrophila glycerophospholipid-cholesterol acyltransferase (X07279); Yeast-IAH, Saccharomyces cerevisiae isoamyl acetate hydrolytic enzyme (X92662); Human-AOAH, Homo sapiens acyloxyacyl hydrolase (BC025698); Aspergillus-RGAE, Aspergillus aculeatus rhamnogalacturon acetylesterase (X89714); E. coli-TESA, Escherichia coli acyl-coA thioesterase I (L06182); Vibrio-ArE, Vibrio mimicus arylesterase (X71116).
The computer-assisted cellular sorting prediction program TargetP presumed that the maize AChE is targeted to the secretory pathway via the endoplasmic reticulum. Furthermore, the SOSUI program (http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0.html), which discriminates between membrane and soluble proteins, showed that the maize AChE does not contain any likely transmembrane helical regions, which are features of proteins that associate with lipid bilayers of the cell membrane. Histochemical analysis of maize seedlings showed that the AChE activities were localized at peripheral regions of the coleoptile, coleoptile node, and mesocotyl cells (Momonoki, 1997). According to Fluck and Jaffe (1974b), most of the AChE activities in the maize root were associated with the cell wall materials, based on AChE assays of the cell organelle fractions. Moreover, they demonstrated that a salt solution could effectively solubilize the AChE activities from cell wall materials. In this study, we used a high salt buffer containing 4% ammonium sulfate for the AChE extraction from the maize seedling homogenates, while a low salt buffer without 4% ammonium sulfate yielded relatively low AChE activity from the homogenate (data not shown). Therefore, the maize AChE, which is glycosylated through the secretory pathway, could be localized in the cell walls via a probable interaction between the sugar chain on the molecule and polysaccharides in the cell wall materials. In a previous report (Momonoki, 1997), it was speculated that AChE is localized at the plasmodesmatal cell-to-cell interface and that it plays a role in regulation of the plasmodesmatal channel as a constituent of the ACh-mediated system. However, it was speculated that the maize AChE might be localized at the cell wall, based on findings in this study. Thus, we can suggest improvements to the model of the ACh-AChR-AChE systems proposed in the original report (Momonoki, 1997). In particular, the system might be localized to the extracellular region around the plasmodesmatal channel and might conduct cell-to-cell trafficking by a channel gating regulation. In plant tissues, the two adjoining cells are interconnected via plasmodesmata, which allow the trafficking of low-molecular-mass materials across the cell wall between two cells. According to a recent model (Lucas, 1995), transport of these substances could be regulated by the opening or/and closing of conductive channels to prevent infection by pathogens and to selectively control trafficking through the plasmodesmata. Moreover, it has been speculated that morphoregulatory proteins around the plasmodesmata could be involved in channel regulation (Cassab, 1998). The ACh-mediated system might regulate the opening and/or closing of channels by interaction with morphoregulatory proteins at the cell wall matrix surrounding the plasmodesmata.
Recently, the ACh molecule has been definitively identified not only in animal tissues, but also in fungi and plant tissues. Furthermore, recent reports suggest that the plant ACh is involved in the regulation of water homeostasis and photosynthesis (Wessler et al., 2001). However, nothing had been known about ACh-synthesizing enzyme(s), AChRs, and ACh-hydrolyzing enzyme(s) in the plant kingdom; thus, knowledge of plant ACh has remained primitive. According to Ballal et al. (1993), indol-3-ACh, the choline conjugate of the IAA, stimulates the elongation of the pea (Pisum sativum) stem segments. More recently, Fluck et al. (2000) suggested a model as the pea cholinesterase might release the free IAA from the conjugates, but they failed to obtain the direct molecule evidence. Therefore, the discovery of an AChE gene here is the first direct evidence, to our knowledge, of the AChE molecule in the plant kingdom, and this will surely provide us with a better understanding of the ACh-AChR-AChE systems in this kingdom. Further characterization of its precise structure, e.g. via crystallography, will be required to clarify the relationship between the molecular conformation and the catalytic mechanism, which still remains unclear. In addition, the expression patterns and cellular localization of the AChE gene products and identification of additional ACh-related proteins would be required to clarify the precise role of the ACh-AChR-AChE systems in the plant kingdom.
MATERIALS AND METHODS
Plant Materials
Kernels of Zea mays L. cv Silver Honey Bantam sweet corn were rinsed with 10% sodium hypochlorite and then soaked overnight in running water. After stabilizing with sodium hypochlorite (1.2% of the available chloride) and rinsing with distilled water, the kernels were placed in a plastic container (diameter, 12.5 cm; height, 19.5 cm) and germinated in rolled moist paper towels for 5 d in a darkroom at 28°C and 80% humidity. The seedlings' roots were then harvested and stored at −30°C until use for AChE purification.
Purification of AChE from Maize Seedlings
Two hundred grams of 5-d-old etiolated maize seedlings were homogenized in a 4-fold volume (v/w) of extraction buffer consisting of 10 mm potassium phosphate buffer, pH 7.0, 10 mm EDTA, and 4% ammonium sulfate. Subsequent purification steps were carried out at 4°C or on ice. The homogenate was left for 1 h in the dark, and then the supernatant was collected by centrifugation at 6,000g for 15 min and stored at 4°C. The precipitate was resuspended in a 3-fold volume (v/w) of the extraction buffer, along with an equal weight of quartz pieces (size 0.3 to 0.5 mm, 30 to 50 mesh; Kishida Chemical Co., Osaka), and was stirred for 1 h. This procedure was repeated twice. The extracts were combined and brought to 80% saturation with ammonium sulfate and left overnight. The resulting precipitate was collected by centrifugation at 15,000g for 15 min, resuspended in 3 mL of phosphate-buffered saline, pH 7.4, and then dialyzed against the same buffer. The precipitate that formed during dialysis was removed by centrifugation, and the supernatant was stored at −30°C until use for the chromatographic purification procedure.
AChE Assay
One hundred microliters of the AChE preparation were added to 150 μL of distilled water or neostigmine bromide (8 mm) in 100 mm sodium phosphate buffer, pH 7.0, as a control. After preincubation for 10 min at 30°C, 250 μL of 12.5 mm ASCh chloride in 100 mm sodium phosphate buffer, pH 7.0, was added to the vials, and incubation was continued for 120 min at 30°C. After incubation, 300 μL of solution was transferred to a new vial, and 1,425 μL of 100 mm sodium phosphate buffer, pH 7.0, and 75 μL of DTNB in 100 mm sodium phosphate, pH 7.0, were added to the vial. The A412 was read after 1 min. All experiments were adjusted using the control described above. One enzyme unit was defined as 1 μmol of thiol group appearance/min.
Substrate specificities of the maize and electric eel AChEs were examined by using PpSCh chloride and BSCh chloride as substrates instead of ASCh chloride, following the same procedure described above. To determine Ki values for neostigmine bromide inhibition of ASCh and PpSCh hydrolysis by the maize and electric eel AChE, the parameters for Dixon plots (1/v and [I]) were measured by the ASCh and PpSCh hydrolysis assay according to the same procedure described above. Experiments were carried out in the presence of several sets of concentrations of neostigmine bromide and the substrates, ASCh and PpSCh. The Ki values were calculated according to the method of Dixon (1953). All measurements were repeated three times, and the results represent the arithmetic means.
PAGE Analysis
SDS-PAGE was performed as described by Laemmli (1970) using a 12.5% or 15% gel. Phosphorylase b (97.4 kD), bovine serum albumin (66.3 kD), aldolase (42.4 kD), carbonic anhydrase (30.0 kD), and soybean trypsin inhibitor (20.1 kD) were used for protein standards. Native PAGE, using a slab minigel (10.8%), was performed according to the procedure described by Davis (1964). The activity staining solution was adjusted to 6 mm ASCh and 3 mm DTNB in 100 mm sodium phosphate buffer, pH 7.0. The native-PAGE gel resolved AChE preparation was soaked into the activity staining solution at room temperature, until the colored band was visualized.
Analytical Gel Filtration for Estimation of Molecular Mass
The sample was applied to Superdex 200 HR10/30 column equilibrated with phosphate buffer, pH 7.0, containing 0.15 m NaCl and 10% glycerol. Molecular size markers were β-amylase (200 kD), alcohol hydrase (150 kD), bovine serum albumin (66 kD), carbonic anhydrase (29 kD), and cytochrome C (12.4 kD). Molecular size was estimated from the mean of three experiments from a plot of the log of molecular mass versus elution volume.
Partial Amino Acid Sequence Analysis of the AChE Protein
The N-terminal amino acid sequences of the AChE protein and its derivative polypeptides were determined by the direct protein sequencing method described by Hirano and Watanabe (1990). The derivative polypeptides of the AChE protein were prepared by proteolytic digestion with Staphylococcus aureus V8 protease (Wako Pure Chemical, Osaka) during electrophoresis according to the method described by Cleveland et al. (1977). The AChE protein separated by SDS-PAGE was electroblotted onto a polyvinylidene difluoride membrane using a semidry blotting apparatus (Nippon Eido, Tokyo). After the membrane was stained, visible bands were cut out. The N-terminal amino acid sequence of the parent AChE protein and its derivative polypeptides were determined using a pulsed-liquid phase sequencer (model 477A; PE-Applied Biosystems) equipped with online HPLC (model 120A; PE-Applied Biosystems).
Determination of the Nucleotide Sequence of the cDNA Encoding AChE
Frozen 5-d-old seedlings were ground into powder with an adequate volume of liquid nitrogen. Total RNA was isolated from the seedling powder (3 g) using the RNeasy plant mini kit (Qiagen, KJ Venlo, The Netherlands) according to the manufacturer's instructions. The cDNAs were synthesized from the total RNA preparation by 3′- and 5′-RACE using the SMART RACE cDNA amplification kit (MD Biosciences, CLONTECH, Palo Alto, CA). Amplification of the RACE fragments corresponding to the AChE gene was performed by PCR using gene-specific primers, Mmn-2 for the 5′-RACE and Sgn-1 for the 3′-RACE. PCR was performed in a 50-μL reaction mixture containing 2.5 μL of cDNA solution, 0.5 μm of each primer, 50 mm KCl, 10 mm Tris-HCl, pH 8.3, 1.5 mm MgCl2, 0.2 mm each deoxynucleotide triphosphate, 5% dimethylsulfoxide, and 1.25 units of Gene Taq Polymerase (Nippon Gene, Osaka). The PCR reaction was carried out for 35 cycles consisting of 1 min at 94°C, annealing for 1 min at 57°C, and 2 min at 72°C using a Gene-Amp automated thermal cycler (model 9700; PE-Applied Biosystems). The PCR products were separated by agarose gel electrophoresis and then extracted from the gels using an EasyTrap DNA extraction kit (Takara, Ohtsu, Japan). The purified fragments were subcloned into a pT7Blue T-vector (Novagen, Madison, WI) according to the manufacturer's instructions. The purified DNA was sequenced on both strands using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (PE-Applied Biosystems). The primers indicated in Table III and the attached primers in the SMART RACE cDNA amplification kit were used for cycle sequencing. If necessary, additional primers for the cycle sequencing were designed based on the determined sequence. MacVector nucleotide and protein analysis software (Oxford Molecular) was utilized for the sequence analysis.
Sequence data from this article have been deposited with the DDBJ/EMBL/GenBank data libraries under accession number AB093208.
This work was supported by a grant from the Basic Research Project, Bio-oriented Technology Research Advancement Institution.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.062927.
References
- Ballal S, Ellias R, Fluck R, Jameton R, Leber P, Lirio R, Salama D (1993) The synthesis and bioassay of indole-3-acetylcholine. Plant Physiol Biochem 31: 249–255 [Google Scholar]
- Bandurski RS (1980) Homeostatic control of concentrations of indole-3-acetic acid. In F, Skoog, ed, Plant Growth Substances. Springer-Verlag, Heidelberg, pp 37–49
- Bandurski RS, Schulze A, Domagalski W (1986) Possible effects of organelle charge and density on cell metabolism. Adv Space Res 6: 47–54 [DOI] [PubMed] [Google Scholar]
- Cassab GI (1998) Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol 49: 281–309 [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Fischer SG, Kirschner MW, Laemmli UK (1977) Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem 252: 1102–1106 [PubMed] [Google Scholar]
- Davis BJ (1964) Disc electrophoresis. II. Method and application to human serum proteins. Ann NY Acad Sci 121: 404–427 [DOI] [PubMed] [Google Scholar]
- Dixon M (1953) The determination of enzyme inhibitor constants. Biochem J 55: 170–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunant Y, Babel-Guerin E, Droz D (1980) Calcium metabolism and acetylcholine release at the nerve-electroplaque junction. J Physiol 76: 471–478 [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of protein based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Evans ML (1972) Promotion of cell elongation in Avena coleopltiles by acetylcholine. Plant Physiol 50: 414–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluck RA, Jaffe MJ (1974. a) Cholinesterase from plant tissues. III. Distribution and subcellular localization in Phaseolus aureus Roxb. Plant Physiol 53: 752–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluck RA, Jaffe MJ (1974. b) The distribution of cholinesterases in plant species. Phytochemistry 13: 2475–2480 [Google Scholar]
- Fluck RA, Jaffe MJ (1975) Cholinesterase from plant tissues. VI. Preliminary characterization of enzymes from Solanum melongena L. and Zea mays L. Biochim Biophys Acta 410: 130–134 [DOI] [PubMed] [Google Scholar]
- Fluck RA, Leber PA, Lieser JD, Szczerbicki SK, Varnes JG, Vitale MA, Wolfe EE (2000) Choline conjugates of auxins. I. Direct evidence for the hydrolysis of choline-auxin conjugates by pea cholinesterase. Plant Physiol Biochem 38: 301–308 [Google Scholar]
- Gnagey AL, Forte M, Rosenberry TL (1987) Isolation and characterization of acetylcholinesterase from Drosophila. J Biol Chem 262: 13290–13298 [PubMed] [Google Scholar]
- Hartmann E, Kilbinger H (1974) Occurrence of light-dependent acetylcholine concentration in higher plants. Experientia 30: 1387–1388 [DOI] [PubMed] [Google Scholar]
- Hirano H, Watanabe T (1990) Microsequencing of proteins electrotransferred onto immobilizing matrices from polyacrylamide gel electrophoresis: application to an insoluble protein. Electrophoresis 11: 573–580 [DOI] [PubMed] [Google Scholar]
- Horiuchi Y, Kimura R, Kato N, Fujii T, Seki M, Endo T, Kato T, Kawashima K (2003) Evolutional study on acetylcholine expression. Life Sci 72: 1745–1756 [DOI] [PubMed] [Google Scholar]
- Hsiao YM, Lai JY, Liao HY, Feng HT (2004) Purification and characterization of acetylcholinesterase from oriental fruit fly [Bactrocera dorsalis (Hendel)] (Diptera: Tephritidae). J Agric Food Chem 52: 5340–5346 [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RB, Deutsch JW, Carlson SS, Wagner JA (1979) Biochemistry of neurotransmitter release. Annu Rev Neurosci 2: 399–466 [DOI] [PubMed] [Google Scholar]
- Kishibayashi A, Ishii A, Karasawa A (1994) Inhibitory effects of KW-5092, a novel gastroprokinetic agent, on the activity of acetylcholinesterase in guinea pig ileum. Jpn J Pharmacol 66: 397–403 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lees GL, Lahue R, Thompson JE (1978) Changes in acetylcholine titer of sensing cotyledons. J Exp Bot 29: 1117–1124 [Google Scholar]
- Lucas WJ (1995) Plasmodesmata: intercellular channels for macromolecular transport in plants. Curr Opin Cell Biol 7: 673–690 [DOI] [PubMed] [Google Scholar]
- MacIntosh FC (1981) Acetylcholine. In GJ Siegel, RW Albers, BW Agranoff, R Kazman, eds, Basic Neurochemistry. Little Brown and Co., Boston, pp 183–204
- Momonoki YS (1988) Asymmetric distribution of glucose and indole-3-myo-inositol in geostimulated Zea mays seedlings. Plant Physiol 87: 751–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momonoki YS (1992) Occurrence of acetylcholine-hydrolyzing activity at the stele-cortex interface. Plant Physiol 99: 130–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momonoki YS (1997) Asymmetric distribution of acetylcholinesterase in gravistimulated maize seedlings. Plant Physiol 114: 47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momonoki YS, Hineno C, Noguchi K (1998) Acetylcholine as a signaling system to environmental stimuli in plants. III. Asymmetric solute distribution controlled by ACh in gravistimulated maize seedlings. Plant Prod Sci 1: 83–88 [DOI] [PubMed] [Google Scholar]
- Momonoki YS, Kawai N, Takamure I, Kowalczyk S (2000) Gravitropic response of acetylcholinesterase and IAA-inositol synthase in lazy rice. Plant Prod Sci 3: 17–23 [Google Scholar]
- Momonoki YS, Momonoki T (1991) Changes in acetylcholine levels following leaf wilting and leaf recovery by heat stress in plant cultivars. Jpn J Crop Sci 60: 283–290 [Google Scholar]
- Momonoki YS, Momonoki T (1993) Changes in acetylcholine-hydrolyzing activity in heat-stressed plant cultivars. Jpn J Crop Sci 62: 438–446 [Google Scholar]
- Momonoki YS, Momonoki T, Whallon JH (1996) Acetylcholine as a signaling system to environmental stimuli in plants. I. Contribution of Ca2+ in heat-stressed Zea mays seedlings. Jpn J Crop Sci 65: 260–268 [Google Scholar]
- Shafferman A, Kronman C, Flashner Y, Leitner M, Grosfeld H, Ordentlich A, Gozes Y, Cohen S, Ariel N, Barak D, et al (1992) Mutagenesis of human acetylcholinesterase. J Biol Chem 267: 17640–17648 [PubMed] [Google Scholar]
- Shen T, Tai K, Henchman RH, Mccamon JA (2002) Molecular dynamics of acetylcholinesterase. Acc Chem Res 35: 332–340 [DOI] [PubMed] [Google Scholar]
- Standaert F (1990) Neuromuscular physiology. In R Miller, ed, Anesthesia. Churchill-Livingstone, New York, pp 650–684
- Talesa V, Principato GB, Giovamini E, DiGiomanni MV, Rosi G (1993) Dimeric forms of cholinesterase in Sipunculus nudus. Eur J Biochem 215: 267–275 [DOI] [PubMed] [Google Scholar]
- Tõugu V (2001) Acetylcholinesterase: mechanism of catalysis and inhibition. Curr Med Chem 1: 155–170 [Google Scholar]
- Toutant JP (1989) Insect acetylcholinesterase: catalytic properties, tissue distribution and molecular forms. Prog Neurobiol 32: 423–446 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN (1993) Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol 103: 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton C, Buckley JT (1995) A new family of lipolytic enzymes? Trends Biochem Sci 20: 178–179 [DOI] [PubMed] [Google Scholar]
- Verbeek M, Vendrig JC (1977) Are acetylcholine-like cotyledon-factors involved in the growth of the cucumber hypocotyl? Z Pflanzenphysiol 83: 335–340 [Google Scholar]
- Wessler I, Kilbinger H, Bittinger F, Kirkpatrick CJ (2001) The biological role of non-neuronal acetylcholine in plants and humans. Jpn J Pharmacol 85: 2–10 [DOI] [PubMed] [Google Scholar]