Abstract
Histidine kinases (Hiks) in Synechocystis sp. PCC 6803 are involved in the transduction of signals associated with various kinds of environmental stress. To examine the potential role in thermotolerance of Hiks, we used genome microarray analysis to screen a Hik knockout library for mutations that affected the expression of genes for heat shock proteins. Mutation of the hik34 gene enhanced the levels of transcripts of a number of heat shock genes, including htpG and groESL1. Overexpression of the hik34 gene repressed the expression of these heat shock genes. In addition, the cells with a mutant gene for Hik34 (ΔHik34 cells) survived incubation at 48°C for 3 h, while wild-type cells and cells with mutations in other Hiks were killed. However, mutation of the hik34 gene had only an insignificant effect on the global expression of genes upon incubation of the mutant cells at 44°C for 20 min. Quantitative two-dimensional gel electrophoresis revealed that levels of GroES and HspA were elevated in ΔHik34 cells after incubation of cells at 42°C for 60 min. We overexpressed recombinant Hik34 protein in Escherichia coli and purified it. We found that the protein was autophosphorylated in vitro at physiological temperatures, but not at elevated temperatures, such as 44°C. These results suggest that Hik34 might negatively regulate the expression of certain heat shock genes that might be related to thermotolerance in Synechocystis.
The cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis) is an aquatic photoautotrophic prokaryote that has been used extensively for studies of genome-wide responses to stress (Suzuki et al., 2001; Kanesaki et al., 2002; Inaba et al., 2003). These studies have led to the identification of specific His kinases (Hiks) and response regulators (Rres) that are involved in the perception and transduction of signals due to low temperature, salt stress, osmotic stress, and metal ion deficiency (Suzuki et al., 2001; Yamaguchi et al., 2002; Marin et al., 2003; Paithoonrangsarid et al., 2004).
Heat shock genes and heat shock proteins (HSPs) are conserved in plants, animals, and microorganisms. In plant cells, HSPs have been found in the cytosol, chloroplasts, mitochondria, endoplasmic reticulum, and peroxisomes (Wang et al., 2004). The HSP Hsp101/ClpB is essential for thermotolerance in plants (Hong and Vierling, 2000), while Hsp90 participates in normal morphogenesis in both Arabidopsis (Arabidopsis thaliana; Queitsch et al., 2002) and Drosophila melanogaster (Rutherford and Lindquist, 1998). Thus, HSPs are involved in development and play important roles in physiological responses to a variety of environmental stresses, such as drought, salinity, cold, and heat (Lindquist, 1986; Lindquist and Craig, 1988).
In cyanobacteria, the expression of heat shock genes is regulated at the transcriptional level under heat shock and other stress conditions (Webb et al., 1990; Kanesaki et al., 2002; Nakamoto et al., 2003), and HSPs play important roles in the tolerance of cyanobacterial cells to heat, cold, and oxidative stress (Nakamoto et al., 2001; Hossain and Nakamoto, 2002, 2003). The genome of Synechocystis includes the sll1670 gene (Nakamoto et al., 2003), which encodes an ortholog of HrcA, a negative regulator of the expression of the grpE-dnaK-dnaJ and groESL operons for chaperonins in Bacillus subtilis (Hecker et al., 1986). However, in Synechocystis, HrcA only regulates the groESL1 operon and the groEL2 gene, suggesting that other heat shock genes might be regulated by different mechanisms (Nakamoto et al., 2003).
It seems plausible that, in Synechocystis, two-component systems, consisting of a Hik and an Rre, might regulate the expression of heat shock genes just as they regulate responses to high salt and osmotic stress (Marin et al., 2003; Paithoonrangsarid et al., 2004). The chromosome of Synechocystis includes 44 putative genes for Hiks that are candidates for sensors of environmental signals (Kaneko et al., 1996; Mizuno et al., 1996). It has already been demonstrated that the Hiks in Synechocystis are involved in the perception and transduction of signals due to cold stress (Suzuki et al., 2000, 2001), phosphate deprivation (Hirani et al., 2001; Suzuki et al., 2004), high osmolarity (Mikami et al., 2002; Paithoonrangsarid et al., 2004), heavy metal ions (López-Maury et al., 2002; Ogawa et al., 2002; Yamaguchi et al., 2002), and high concentrations of NaCl (Marin et al., 2003). In this article, we provide evidence that Hik34, one of the 44 putative Hiks in Synechocystis, is involved in the control of the expression of heat shock genes and the regulation of thermotolerance in Synechocystis.
RESULTS
Screening of an Hik Knockout Library Revealed That Mutation of hik34 Enhanced the Expression of Heat Shock Genes
Each of the putative hik genes in Synechocystis was inactivated previously by insertion of a spectinomycin-resistance gene cassette to create a gene knockout library of ΔHik cells (Suzuki et al., 2000; CyanoMutants, http://www.kazusa.or.jp/cyano/Synechocystis/mutants). We examined the genome-wide expression of genes in wild-type cells and in all the lines of Δhik mutant cells using DNA microarrays that covered 3,075 of the 3,267 genes in the Synechocystis chromosome in an attempt to identify Hiks that might be involved in the regulation of expression of heat shock genes (Fig. 1).
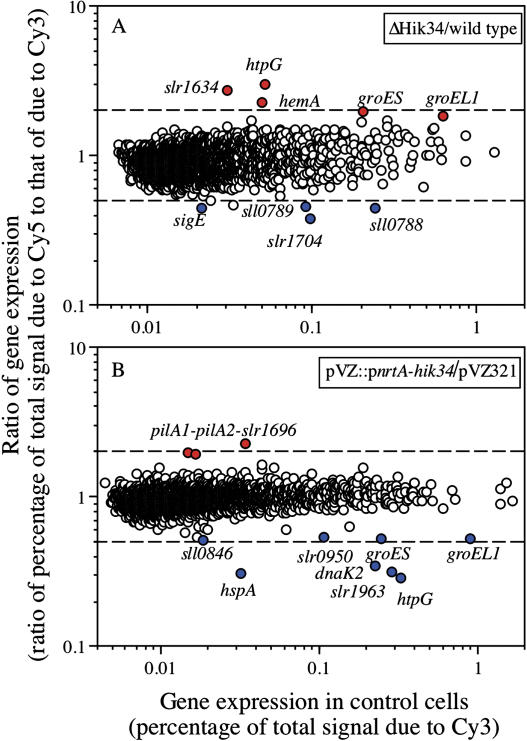
Figure 1.
DNA microarray analysis of changes in gene expression under standard growth conditions upon mutation (A) and overexpression (B) of the hik34 gene in Synechocystis. RNA extracted from ΔHik34 cells and from hik34-overexpressing cells that had been grown at 34°C for 16 h was labeled with Cy5 and RNA extracted from wild-type and pVZ321-transformed cells (control) that had been grown at 34°C for 16 h were labeled with Cy3. Experiments were repeated at least twice with independently prepared samples and essentially the same respective results were obtained in every case.
We first compared the genome-wide expression of genes in all 44 mutant lines with that in wild-type cells under our standard growth conditions at 34°C. Most of the ΔHik mutants yielded profiles of gene expression similar to that of wild-type cells, suggesting that these Hiks were inactive under normal growth conditions. We only found clear changes in gene expression in ΔHik27 and ΔHik34 cells. In ΔHik27 cells, as we published previously, expression of the mntCAB operon that encodes an ATP-binding cassette-type Mn2+ transporter was enhanced (Yamaguchi et al., 2002). By contrast, in ΔHik34 cells, the level of the transcript of the heat shock gene htpG was approximately 3-fold higher than that in wild-type cells (Fig. 1A; Table I). Levels of transcripts of other heat shock genes, such as groES and groEL1, were also elevated (Fig. 1A; Table I). These results suggested that Hik34 might act as a negative regulator of the expression of these genes under standard growth conditions.
Table I.
DNA microarray analysis of changes in gene expression upon mutation of the hik34 gene in Synechocystis
Genes whose expression was induced (induction factor >3.0) by heat shock at 44°C for 20 min are underlined. The values are averages of results of three independent experiments. Genes marked by the same Greek letter are located in tandem on the Synechocystis genome.
| Gene ID | Gene | Product | Δhik34/Wild Type |
|---|---|---|---|
| Genes whose expression was enhanced by mutation of Hik34 | |||
| sll0430 | htpG | HSP 90 | 2.94 ± 0.19 |
| slr1634 | Protein of unknown function | 2.72 ± 1.05 | |
| slr1808 | hemA | tRNA-Gln reductase | 2.25 ± 0.72 |
| slr2075α | groES | 10-kD chaperonin | 1.94 ± 0.75 |
| sll1579 | cpcC | Phycocyanin-associated linker | 1.81 ± 0.67 |
| slr2076α | groEL1 | 60-kD chaperonin 1 | 1.70 ± 0.50 |
| sll1453 | nrtD | Component of nitrate transporter | 1.69 ± 0.41 |
| sll1324 | atpF | Subunit-β of ATP synthase | 1.66 ± 0.40 |
| ssl3044 | Ferredoxin-like protein | 1.55 ± 0.32 | |
| slr0927 | psbD2 | D2 protein of photosystem II | 1.54 ± 0.58 |
| sll1867 | psbA3 | D1 protein of photosystem II | 1.50 ± 0.48 |
| sll1514 | hspA | Small HSP | 1.50 ± 0.32 |
| Genes whose expression was repressed by mutation of Hik34 | |||
| slr1704 | Protein of unknown function | 0.39 ± 0.19 | |
| sll0788β | Protein of unknown function | 0.45 ± 0.08 | |
| sll0789β | rre34 | Rre34 | 0.46 ± 0.05 |
| sll0856 | sigE | RNA polymerase σ-E factor | 0.47 ± 0.16 |
| sll0790β | hik31 | His kinase 31 | 0.48 ± 0.01 |
| sll1336 | Protein of unknown function | 0.53 ± 0.06 | |
| sll1070 | tktA | Transketolase | 0.59 ± 0.06 |
| slr1198 | Antioxidant protein | 0.61 ± 0.13 | |
| ssr1375 | Protein of unknown function | 0.62 ± 0.12 | |
| ssl2245 | Protein of unknown function | 0.62 ± 0.04 | |
| slr1261 | Protein of unknown function | 0.64 ± 0.09 | |
The mutation in hik34 also enhanced the expression of some genes that are related to photosynthesis (Table I). These genes included the hemA gene, which encodes the tRNA-Gln reductase that catalyzes the initial step in the biosynthesis of heme and chlorophyll. In addition, levels of transcripts of the cpcC, atpF, psbD2, and psbA genes, which encode the phycocyanin linker protein, a subunit of ATPase, and the D2 and D1 proteins of PSII, respectively, were elevated at room temperature in ΔHik34 cells. The implications of the enhanced expression of photosynthesis-related genes remain to be clarified. Mutation of hik34 also repressed the expression of several genes, such as rre34, sigE, hik31, and tktA, whose products have been identified, as well as the expression of genes for proteins of unknown function, such as slr1704, sll0788, sll1336, ssr1375, ssl2245, and slr1261. The sll0788, rre34, and hik31 genes are adjacent to one another and might be transcribed from a single promoter.
Mutation of Hik34 Enhanced the Thermotolerance of Synechocystis
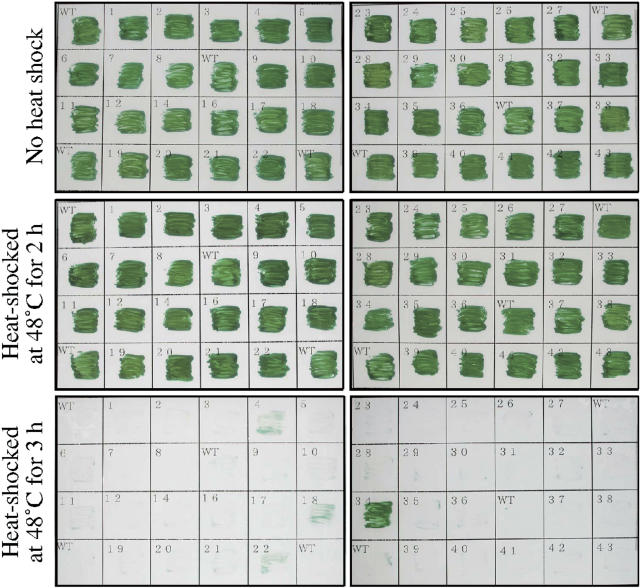
Since mutation of the hik34 gene enhanced the expression of some heat shock genes, such as htpG and groESL1, under normal growth conditions, we screened the library of ΔHik cells for survival at a high temperature, namely, 48°C. All lines of cells were tolerant to heat stress at 48°C for 2 h. However, when cells were exposed to 48°C for 3 h, only Δhik34 cells survived (Fig. 2). These results suggested that the enhanced expression of heat shock genes, caused by mutation of Hik34 under normal growth conditions, might have contributed to the acquisition of thermotolerance by ΔHik34 cells.
Figure 2.
Screening of the knockout library of His kinases for survival of Synechocystis cells after heat shock. Cells of individual mutant lines were inoculated separately on plates of agar-solidified BG-11 medium (Stanier et al., 1971) and incubated at 30°C for 3 d. After heat shock at 48°C for either 2 or 3 h, as indicated, plates were incubated at 30°C for a further 7 d. Among the 41 lines with mutation in individual His kinases, only ΔHik34 cells survived.
Overexpression of hik34 Repressed the Expression of Heat Shock Genes
Since Hik34 appeared to be a negative regulator of heat shock-responsive genes, we postulated that overexpression of this Hik should result in strong repression of the expression of these genes. Therefore, we engineered the overexpression of Hik34 in Synechocystis to examine the effect of an elevated level of this protein on genome-wide gene expression by DNA microarray analysis.
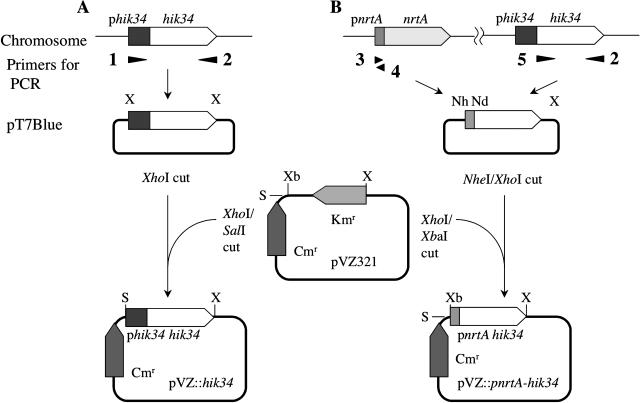
We first attempted to overexpress the hik34 gene under the control of the native promoter of the gene from Synechocystis using the replicable multicopy plasmid pVZ321 (Zinchenko et al., 1999). The DNA fragment, including the promoter region and the coding region of hik34, were amplified by PCR and cloned into pVZ321 (Fig. 3A). We attempted to introduce the resultant plasmid pVZ::hik34 into Synechocystis cells by triparental conjugation (Zinchenko et al., 1999). However, we failed to obtain transformants that harbored pVZ::hik34 (data not shown). Overexpression of the hik34 gene might have resulted in excessive repression of heat shock genes that are required for cell viability. Therefore, we weakened the promoter activity of the gene by replacing the promoter of the hik34 gene in pVZ::hik34 by the promoter of the nrtA operon, which encodes the nitrate/nitrite transporter (Aichi et al., 2001). When Synechocystis is cultured in BG-11 medium that contains 17.5 mm sodium nitrate as the sole source of nitrogen, the activity of the promoter of the nrtA operon remains relatively low (Aichi et al., 2001). We introduced the plasmid pVZ::pnrtA-hik34, which included the promoter of the nrtA operon and the coding region of the hik34 gene (Fig. 3B), into Synechocystis and confirmed that the plasmid propagated stably in the cells. Then we examined the effects of the introduction of pVZ::pnrtA-hik34 using the whole-genome DNA microarray.
Figure 3.
Schematic representation of the construction of pVZ::hik34 (A) and pVZ::pnrtA-hik34 (B). White and dotted broad arrows indicate the coding sequences of the hik34 and nrtA genes, respectively, and black and hatched boxes indicate the promoter of the hik34 and the nrtA gene, respectively. Numbered arrowheads indicate primers used to amplify DNA fragments that corresponded to the promoters and coding regions of the genes, as follows: 1, Hik34up; 2, Hik34dn; 3, NrtANheI; 4, NrtANdeI; and 5, Hik34NheINdeI (for details, see “Materials and Methods”). Kmr and Cmr indicate the kanamycin- and chloramphenicol-resistance gene cassettes in pVZ321, respectively. X, Nh, Nd, Xb, and S indicate restriction sites recognized by XhoI, NheI, NdeI, XbaI, and SalI, respectively, which were utilized to cleave and ligate the plasmids.
We cultured control cells, which harbored the empty vector pVZ321, and transformed cells, which harbored pVZ::pnrtA-hik34, under standard growth conditions and then compared the genome-wide expression of genes in transformed cells with that in control cells (Fig. 1B; Table II). Overexpression of the hik34 gene clearly depressed the expression of several heat shock genes, such as the htpG, hspA, groESL1, dnaK2, and groEL2 genes (Table II). These results support our hypothesis that Hik34 negatively regulates the expression of heat shock genes. The overexpression of the hik34 gene enhanced the expression of the pilA1-pilA2-sll1965 operon, which encodes components of type IV pili and a protein of unknown function. In addition, the expression of several genes for proteins of unknown function, which included sll1774, sll1862, sll0359, and sll1201, was enhanced in the cells that overexpressed Hik34. The expression of these genes was unaffected by heat shock and the mechanism responsible for the enhanced expression of these genes remains to be clarified.
Table II.
DNA microarray analysis of changes in genome-wide expression upon overexpression of the hik34 gene in Synechocystis
Genes whose expression was induced (induction factor >3.0) by heat shock at 44°C for 20 min are underlined (see also Table I). The values are averages of results of two independent experiments. Genes marked by the same Greek letter are located in tandem on the Synechocystis genome.
| Gene ID | Gene | Product | Ratio (pVZ::pnrtA-hik34/pVZ321) |
|---|---|---|---|
| Genes whose expression was enhanced by overexpression of the hik34 gene | |||
| sll1695α | pilA2 | Component of type IV pili, pilin 2 | 2.26 ± 0.08 |
| sll1696α | Protein of unknown function | 1.97 ± 0.04 | |
| sll1694α | pilA1 | Component of type IV pili, pilin 1 | 1.94 ± 0.17 |
| sll1774 | Protein of unknown function | 1.64 ± 0.01 | |
| sll1862 | Protein of unknown function | 1.56 ± 0.15 | |
| sll0788 | Protein of unknown function | 1.55 ± 0.03 | |
| sll0359 | Protein of unknown function | 1.54 ± 0.10 | |
| sll1201 | Protein of unknown function | 1.52 ± 0.07 | |
| Genes whose expression was repressed by overexpression of the hik34 gene | |||
| sll0430 | htpG | HSP 90 | 0.29 ± 0.01 |
| sll1514 | hspA | Small HSP | 0.31 ± 0.01 |
| slr1963 | Water-soluble carotenoid protein | 0.32 ± 0.01 | |
| sll0170 | dnaK2 | HSP 70 | 0.34 ± 0.01 |
| sll0846 | Protein of unknown function | 0.51 ± 0.01 | |
| slr2076β | groEL1 | 60-kD chaperonin 1 | 0.52 ± 0.03 |
| slr2075β | groES | 10-kD chaperonin | 0.53 ± 0.01 |
| slr0959 | Protein of unknown function | 0.53 ± 0.03 | |
| slr1516 | sodB | Superoxide dismutase | 0.54 ± 0.01 |
| slr0958 | cysS | Cysteinyl-tRNA synthetase | 0.58 ± 0.01 |
| slr1674 | Protein of unknown function | 0.61 ± 0.03 | |
| sll0416 | groEL2 | 60-kD chaperonin 2 | 0.63 ± 0.02 |
Inactivation of the hik34 gene and introduction of the hik34 gene via the pVZ vector had opposing effects on the expression of heat shock genes at the standard growth temperature, as shown in Figure 1 and Tables I and II. These findings were consistent with the hypothesis that Hik34 plays an important role in the regulation of the expression of heat shock genes. However, it was still unclear whether ΔHik34 cells could also exhibit a temperature-sensitive heat shock response with the accumulation of even more HSPs or whether expression of the HSPs under standard conditions was sufficient for the observed enhanced tolerance to elevated temperatures of the ΔHik34 cells. Therefore, we investigated the heat shock response in ΔHik34 cells.
The Hik34 Mutation Had an Insignificant Effect on Heat-Induced Gene Expression
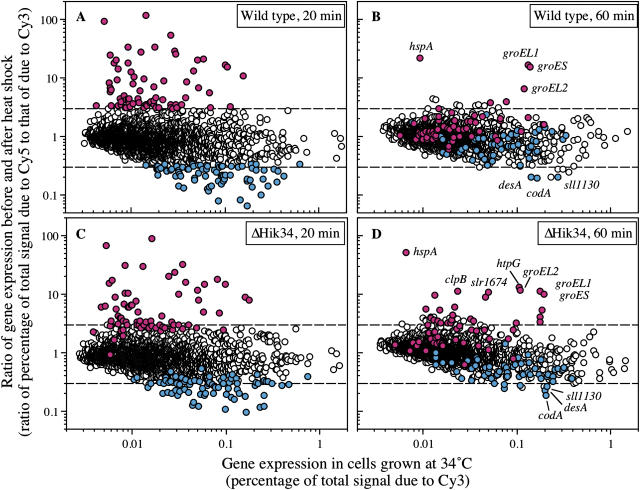
We transferred wild-type Synechocystis cells from 34°C to 44°C to induce a heat shock response. We withdrew samples at 20 and 60 min and isolated total RNA from them for analysis of genome-wide transcription with the DNA microarray. The level of expression of 59 genes rose more than 3-fold and expression of 58 genes was suppressed to less than one-third of each initial respective level (Fig. 4A). Levels of expression of 123 genes more than doubled and the expression of 232 genes was suppressed to less than one-half of the initial respective levels (data not shown). Levels of expression of heat shock genes, such as clpB1, hspA, groESL, htpG, dnaJ, and dnaK2 genes, which encode chaperonins, rose more than 6-fold during the first 20 min at 44°C (Table III). The expression of 10 genes that encode proteins of unknown function was also enhanced by a factor of more than 6. In particular, the expression of the hik34 gene was enhanced 19-fold by heat shock treatment (Table III). The expression of the sigB gene for an RNA polymerase group 2 σ factor was also elevated, and we plan to examine this phenomenon and its implications in future experiments. The induction factors (elevated level divided by basal level) of almost all the heat shock-inducible genes fell to below 3 after incubation at 44°C for 60 min (Fig. 4B; Table III). These findings suggest that the expression of certain genes that respond to heat shock, such as clpB1 and dnaJ, was enhanced only transiently and decreased within 60 min of the exposure to heat shock. Some heat shock-responsive genes, such as hspA, groEL1, groES, and groEL2, were still expressed at high levels after incubation of cells at 44°C for 60 min (Fig. 4B; Table III). The full data set is available (see supplemental data) and has also been deposited on the Web site at http://www.genome.jp/kegg/expression.
Figure 4.
DNA microarray analysis of changes in genome-wide expression of genes in response to heat shock at 44°C in wild-type cells for 20 min (A) and 60 min (B) and in ΔHik34 cells for 20 min (C) and 60 min (D). RNA extracted from cells that had been grown for 16 h at 34°C and then incubated at 44°C for 20 min or 60 min was labeled with Cy5 (as the sample) and RNA extracted from the respective cells that had not been exposed to heat shock (as the control) was labeled with Cy3. Genes whose levels of expression rose more than 3-fold and genes whose expression was suppressed to less than one-third of the control level in wild-type cells after incubation at 44°C for 20 min are indicated in red and blue, respectively. Upper and lower dashed lines indicate a 3-fold increase and a decrease to one-third, respectively. Note that the scale of the ordinate in this figure is different from that in Figure 1.
Table III.
DNA microarray analysis of changes in the genome-wide expression of genes in response to heat shock at 44°C for 20 and 60 min in wild-type and ΔHik34 mutant cells
Genes whose expression was enhanced more than 6-fold in wild-type cells at 20 min are listed. The listed values (ratios of levels of transcripts) were calculated from two to four independent experiments. Genes marked by the same Greek letter are located in tandem on the Synechocystis genome. An apostrophe indicates localization on opposite strands of DNA.
| Gene ID
|
Gene
|
Product
|
Ratio (44°C/34°C, Value ± sd)
|
|||
|---|---|---|---|---|---|---|
| Wild Type
|
ΔHik34
|
|||||
| 20 min | 60 min | 20 min | 60 min | |||
| slr1641α | clpB1 | ClpB protein | 92.5 ± 9.1 | 1.9 ± 0.1 | 74.9 ± 6.4 | 11.1 ± 0.2 |
| sll1514β | hspA | Small HSP | 68.1 ± 2.9 | 21.6 ± 0.1 | 50.7 ± 3.7 | 50.7 ± 1.9 |
| slr2075γ | groES | 10-kD chaperonin | 44.7 ± 2.3 | 15.5 ± 1.0 | 26.9 ± 1.2 | 10.0 ± 0.6 |
| sll0416 | groEL2 | 60-kD chaperonin 2 | 42.1 ± 7.7 | 6.5 ± 0.2 | 14.7 ± 0.1 | 11.5 ± 0.4 |
| slr0093δ | dnaJ | HSP 40 | 36.9 ± 3.6 | 2.2 ± 0.0 | 39.6 ± 0.9 | 3.3 ± 0.2 |
| sll0430 | htpG | HSP 90 | 34.6 ± 5.6 | 3.7 ± 0.2 | 6.3 ± 0.2 | 13.1 ± 0.1 |
| slr1674 | Protein of unknown function | 28.3 ± 0.8 | 1.7 ± 0.1 | 17.3 ± 1.3 | 10.6 ± 0.9 | |
| slr0095δ | O-Methyltransferase | 24.4 ± 4.6 | 0.9 ± 0.1 | 16.4 ± 0.7 | 1.6 ± 0.0 | |
| sll0306 | sigB | RNA polymerase group 2 σ factor | 22.9 ± 0.7 | 1.8 ± 0.2 | 10.9 ± 0.3 | 8.8 ± 0.8 |
| slr1963 | Water-soluble carotenoid protein | 22.8 ± 0.0 | 1.2 ± 0.0 | 2.5 ± 0.1 | 3.4 ± 0.2 | |
| sll0170 | dnaK2 | HSP 70 | 22.3 ± 1.5 | 2.1 ± 0.1 | 13.7 ± 2.7 | 5.3 ± 0.2 |
| sll0528 | Protein of unknown function | 19.5 ± 0.9 | 1.4 ± 0.1 | 13.7 ± 0.8 | 5.3 ± 0.7 | |
| slr1285 | hik34 | His kinase 34 | 18.9 ± 2.6 | 0.6 ± 0.1 | – | – |
| ssl2971α' | Protein of unknown function | 15.1 ± 1.7 | 1.0 ± 0.1 | 15.1 ± 1.9 | 3.2 ± 0.6 | |
| sll0939ɛ | Protein of unknown function | 11.9 ± 0.2 | 0.7 ± 0.1 | 9.1 ± 0.1 | 1.7 ± 0.4 | |
| sll0846 | Protein of unknown function | 11.7 ± 3.0 | 1.8 ± 0.2 | 8.8 ± 0.3 | 5.9 ± 0.6 | |
| sll1621 | AhpC/TSA family protein | 11.0 ± 0.6 | 3.8 ± 0.2 | 12.1 ± 0.4 | 3.2 ± 0.2 | |
| ssl3044ζ | Ferredoxin-like protein | 10.3 ± 0.0 | 1.3 ± 0.1 | 6.5 ± 0.7 | 2.3 ± 0.0 | |
| slr0967ɛ' | Protein of unknown function | 9.8 ± 0.6 | 1.2 ± 0.1 | 8.4 ± 0.1 | 2.2 ± 0.2 | |
| slr2076γ | groEL1 | 60-kD chaperonin 1 | 8.7 ± 1.5 | 16.4 ± 2.4 | 3.7 ± 0.0 | 11.0 ± 1.8 |
| slr1915 | Protein of unknown function | 8.5 ± 0.8 | 1.2 ± 0.0 | 6.0 ± 0.3 | 3.4 ± 0.0 | |
| slr0589 | Protein of unknown function | 7.8 ± 0.5 | 1.8 ± 0.1 | 9.6 ± 0.7 | 1.4 ± 0.1 | |
| slr1686ζ' | Protein of unknown function | 7.6 ± 0.5 | 0.9 ± 0.0 | 3.5 ± 0.1 | 1.4 ± 0.1 | |
| ssl1633 | hliC | CAB/ELIP/HLIP superfamily | 6.9 ± 0.5 | 1.5 ± 0.1 | 3.6 ± 0.2 | 5.4 ± 1.1 |
| slr1597β' | Protein of unknown function | 6.7 ± 1.0 | 1.0 ± 0.0 | 4.1 ± 0.4 | 1.5 ± 0.0 | |
| sll1433 | Protein of unknown function | 6.1 ± 0.3 | 2.2 ± 0.3 | 6.5 ± 0.0 | 2.4 ± 0.1 | |
| slr1204 | htrA | Ser protease | 6.0 ± 0.7 | 2.5 ± 0.1 | 3.1 ± 0.6 | 3.0 ± 0.3 |
We next examined the effects of mutation of the hik34 gene on expression of heat shock genes under heat shock conditions. The extent of the induction of most heat-inducible genes was somewhat reduced by mutation of hik34. For some genes, such as clpB1, the extent of the reduction was small, but for some genes, such as htpG, it was very marked (Fig. 4C; Table III).
These results suggest that mechanisms unrelated to Hik34 might also contribute to the regulation, in response to heat shock, of the above-mentioned genes. To our surprise, the levels of transcripts in Δhik34 cells of most heat shock genes after incubation at 44°C for 60 min remained higher than those in wild-type cells, suggesting that Hik34 might also be involved in maintaining a high steady-state level of these transcripts either via synthesis de novo or via stabilization of the mRNA. This effect was particularly evident in the case of the hspA gene. Since Hik34 appeared to repress the expression of heat shock genes, the induction by heat shock of the expression of the hik34 gene in wild-type cells might have had a negative effect on the stability or synthesis de novo of such transcripts.
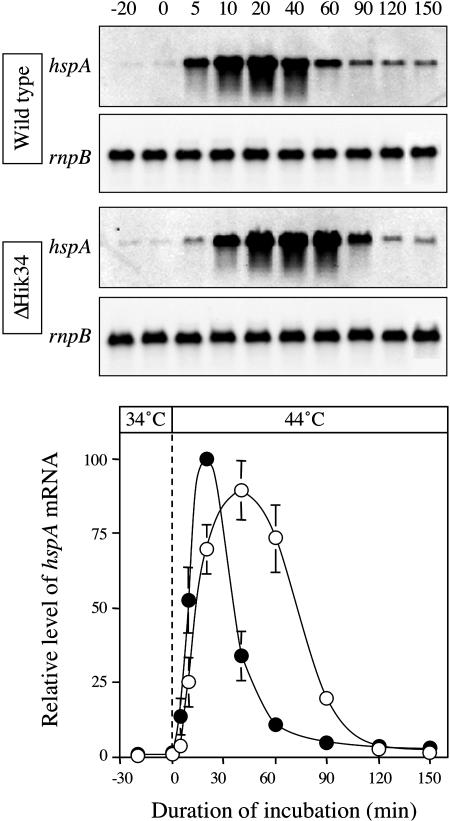
The Elevated Expression in Response to Heat Shock of the hspA Gene in ΔHik34 Cells Was of a Longer Duration Than in Wild-Type Cells
We examined changes in the relative level of the hspA transcript over a 150-min period (Fig. 5). Northern-blot analysis demonstrated that the level of the hspA transcript in wild-type cells increased 5 min after the transfer of cells from 34°C to 44°C and reached a maximum after 20 min. It then declined rapidly to a steady-state level that was approximately 5% of the maximum. By contrast, in ΔHik34 cells, the level of the hspA transcript was very low at 5 min. The transcript started to appear at 10 min and reached a maximum level 40 min after the temperature had been raised. Thereafter, the level of the hspA transcript decreased gradually and reached a steady state at 120 min. As a result, the level of hspA mRNA in ΔHik34 cells was higher than that in wild-type cells from 30 to 120 min after the start of heat shock. These results suggest that ΔHik34 cells might produce a larger amount of the small HSP than do wild-type cells.
Figure 5.
Kinetics of the heat-induced expression of the hspA gene in wild-type (black circles) and ΔHik34 (white circles) cells. Cultures that had been prepared by incubation at 34°C were incubated at 44°C for 150 min. At the indicated times, aliquots were withdrawn and cells were harvested for extraction of total RNA. Aliquots of 10 μg of total RNA were fractionated on a 1.2% agarose gel that contained 0.44 m formaldehyde. The experiments were repeated three times with independently prepared samples. The graph shows means ± ses of the means of results from three independent experiments.
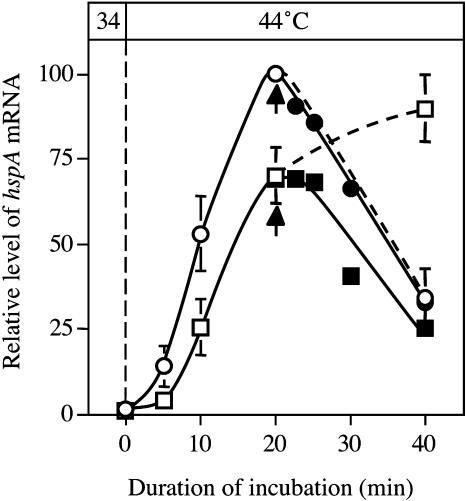
Changes in the level of any transcript depend on the rate of transcription and the rate of degradation of the transcript or its stability. To examine whether the difference in the kinetics of the changes in level of the hspA transcript were due to the regulation of transcription or to the stability of the transcript, we monitored changes in the level of the transcript during incubation in the presence of rifampicin (an inhibitor of transcription), which was added to the culture medium 20 min after the initiation of heat shock (Fig. 6). In wild-type cells, the inhibition of transcription by rifampicin did not affect the rapid decline in the level of the transcript of hspA, suggesting that the rapid decline depended on the degradation of the transcript with no contribution from transcription. By contrast, in ΔHik34 cells, the addition of rifampicin had a drastic effect on the changes in the levels of the hspA transcript, such that the decline in the level of the transcript in the presence of rifampicin occurred at the same rate as in wild-type cells. These observations suggested that the stability of the hspA transcript was unaffected by the mutation of Hik34 and that, in the absence of rifampicin, transcription of the hspA gene continued for more than 20 min during incubation at 44°C in ΔHik34 cells but not in wild-type cells. These findings, illustrated in Figures 5 and 6, are consistent with the hypothesis that Hik34 enhances the expression of heat shock genes, such as hspA, just after the initial heat shock but negatively regulates the transcription of heat shock genes after peak levels of transcripts have been reached.
Figure 6.
Stability of the hspA transcript after heat induction. Cultures that had been prepared at 34°C were incubated at 44°C for 20 min. Rifampicin was added at 50 μg mL−1 to the cultures 20 min after the temperature shift (as indicated by arrows). Squares and circles indicate wild-type and ΔHik34 cells, respectively. Black symbols and white symbols indicate with and without addition of rifampicin, respectively.
Analysis of HSPs in Wild-Type and ΔHik34 Cells
There is often little correlation between levels of an mRNA and the corresponding protein in a cell. To investigate whether the results of our analysis of transcripts reflected changes in the levels of corresponding proteins, we examined levels of protein quantitatively.
In a previous study of proteins in Synechocystis, we separated more than 300 protein spots and identified more than 100 soluble proteins from Synechocystis cells by two-dimensional (2D) electrophoresis and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Simon et al., 2002). These proteins included HSPs such as GroEL, DnaK2, HspA, and ClpB. Therefore, we examined the effects of mutation of Hik34 on the levels of soluble proteins using 2D differential gel electrophoresis (2D-DIGE) technology (Tonge et al., 2001; Table IV). This technology allows rapid detection of differences in levels of expression of individual proteins in two samples by labeling them with different fluorescent dyes and separating them on a single gel (Tonge et al., 2001). After electrophoresis, we used DeCyder image analysis software (GE Healthcare Amersham, Buckinghamshire, UK) to compare samples. We routinely analyzed five independent biological samples in each experiment and used a statistical confidence limit of 95% to quantify the differences in levels of respective proteins in cells under different conditions.
Table IV.
Results of analysis by 2D-DIGE of changes in levels of proteins upon mutation of the hik34 gene and heat shock in Synechocystis
The first column of numbers indicates ratios of levels of individual proteins in wild-type and ΔHik34 cells grown at 30°C. The second and third columns show changes in relative levels of proteins after heat shock due to incubation at 42°C for 60 min. This table lists six proteins that we identified in previous studies (Simon et al., 2002) and that we also identified as products of heat-inducible genes by DNA microarray analysis (Table III). The values are averages of results of five independent experiments, which gave a statistical confidence limit of 95%.
| Gene Product
|
Protein
|
ΔHik34/Wild Type
|
Heat Shock Induction at 42°C, 60 min
|
|
|---|---|---|---|---|
| 30°C | Wild Type (42°C/30°C) | ΔHik34 (42°C/30°C) | ||
| GroEL | 60-kD chaperonin | 2.5 | 5.3 | 3.8 |
| DnaK2 | HSP 70 | 2.8 | 2.5 | 1.8 |
| HspA | Small HSP | 1.6 | 19.9 | 21.0 |
| ClpB | Subunit of Clp protease | 1.8 | 2.6 | 3.6 |
| HtpG | HSP 90 | 3.8 | 2.4 | 1.9 |
| GroES | 10-kD chaperonin | 2.0 | 3.2 | 2.6 |
When we compare the electrophoretic patterns of proteins from wild-type and ΔHik34 cells that had been grown at 30°C, we found a marked increase in the levels of GroEL, DnaK2, HspA, ClpB, HtpG, and GroES in ΔHik34 cells (Table IV). It was evident, therefore, that not only did the levels of mRNAs for these proteins increase in ΔHik34 cells under non-heat shock conditions, but the levels of the proteins also increased. This observation is important since changes detected at the mRNA level do not necessarily reflect differences in levels of proteins, and there are several notable examples where an absence of any correlation has been observed (Scherl et al., 2005).
When we incubated wild-type cells at 42°C for 60 min, the intensities of the spots that corresponded to GroEL, DnaK2, HspA, ClpB, HtpG, and GroES all increased relative to the total amount of protein (Table IV). When we performed the same experiment with ΔHik34 cells, the level of change of these proteins was comparable to that of wild-type cells (Table IV). These results were consistent with our finding that the levels of mRNAs for these HSPs were similar between wild-type and ΔHik34 cells.
Overexpression of Hik34 in Escherichia coli as a His-Tagged Protein
Among the mutations in each of the 44 Hiks encoded by the chromosome of Synechocystis, only mutation of Hik34 affected the expression of some heat shock genes (Fig. 1A) and allowed cells to survive a sudden increase in growth temperature from 30°C to 48°C (Fig. 3). These observations suggest that this Hik might regulate the expression of certain heat shock genes. To examine the activity of Hik34 in vitro, we expressed the recombinant Hik34 protein in E. coli and purified it.
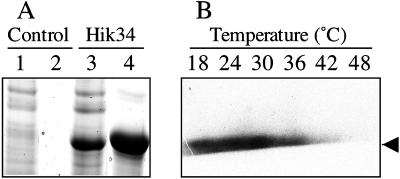
We introduced a hik34 gene that contained an amino-terminal His-tag into a pET expressional system (Novagen, Madison, WI), which consists of the pET28a vector and a T7 RNA polymerase lysogenic strain of E. coli BL21(DE3). After treatment of cells with isopropyl-1-thio-β-d-galactopyranoside, we looked for expression of the recombinant protein. Figure 7A shows a Coomassie Blue-stained gel after electrophoretic fractionation of the soluble protein from E. coli cells that had been transformed with the empty pET28a vector (lane 1) and from cells that harbored pET28a::hik34 (lane 3). An additional intense band of a protein of approximately 52 kD is clearly visible in the case of the soluble protein from the cells that harbored pET28a::hik34. This protein corresponds in terms of mobility to the expected His-tagged Hik34. We purified the recombinant His-tagged protein on a Ni affinity column (lane 4) and isolated it in an electrophoretically pure form (data not shown).
Figure 7.
Purification and autophosphorylation in vitro of recombinant Hik34. A, Coomassie Brilliant Blue-stained gel. Total soluble protein (100 μg; lanes 1 and 3) and proteins eluted from the Ni-NTA column (lanes 2 and 4) originated from E. coli cells that harbored pET28a (control) and pET28a-Hik34 (Hik34); proteins were fractionated on an SDS-polyacrylamide gel. B, Autoradiography of phosphorylated proteins. Purified recombinant Hik34 was incubated with [γ-32P]ATP at the indicated temperatures for 10 min and then each reaction mixture was fractionated by SDS-PAGE. The gel was dried and exposed to x-ray film. The arrow indicates the mobility of autophosphorylated recombinant Hik34 (approximately 52 kD).
The Hik proteins identified to date all function via autophosphorylation of a His residue within each respective Hik. We postulated that, under non-heat shock conditions, Hik34 might be phosphorylated and transfer the phosphate group to a Rre that would repress the expression of heat shock genes. Under heat shock conditions, such phosphorylation might not occur, preventing subsequent repression of gene expression. It has been demonstrated that Hiks contain a conserved phosphorylatable His residue in a motif known as the HisKA motif (Pfam database; Bateman et al., 2004). A second conserved domain, HATPase_C, has also been found in Hiks and is an ATP-binding domain. The bound ATP donates the phosphate group for the autophosphorylation (Bateman et al., 2004). We analyzed the amino acid sequence of Hik34 (slr1285; Kaneko et al., 1996; Mizuno et al., 1996) and identified a conserved HisKA motif. However, we found no HATPase_C domain. Nonetheless, Hik34 might still be capable of autophosphorylation. Accordingly, we examined the phosphorylation in vitro of the recombinant Hik34 and the dependence on temperature of this reaction.
Recombinant Hik34 Was Autophosphorylated In Vitro But Not at Heat Shock Temperatures
We examined the dependence on temperature of the autophosphorylation of Hik34 in the presence of ATP in vitro using purified recombinant Hik34. With [γ-32P]ATP as the substrate, the recombinant Hik34 was phosphorylated in vitro (Fig. 7B), confirming that Hik34 can utilize ATP as a phosphate donor for autophosphorylation, as can other Hiks, even in the absence of a conserved ATP-binding domain. Our results suggest that the signature motifs of Hik domains, which are conserved in all the Hiks of cyanobacteria with the exception of Hik34, might not be essential for autophosphorylation. The profile of the temperature-dependent phosphorylation of Hik34 in vitro revealed that autophosphorylation occurred at 18°C, 24°C, and 30°C; it occurred to a much lesser extent at 36°C; and it did not occur at all at 42°C and 48°C (Fig. 7B). These results suggest that Hik34 might respond directly to a change in temperature by a change in the extent of autophosphorylation and that, while Hik34 is in an active form at the normal growth temperature, it is in an inactive form at elevated temperatures.
DISCUSSION
A Possible Scheme for Regulation of Gene Expression by Hik34
At the physiological temperature of 34°C, mutation of the hik34 gene in Synechocystis cells enhanced the expression of some heat shock genes, such as htpG, groESL, and hspA (Table I), whereas overexpression of the hik34 gene repressed the expression of these heat shock genes (Table II). The extent of autophosphorylation of recombinant Hik34 was high at normal physiological temperatures (Fig. 7). These results suggest that Hik34 might produce a signal that regulates the expression of heat shock genes under normal growth conditions.
Mutation of the hik34 gene enhanced the expression of heat shock genes, such as the htpG, groESL, and hspA genes, at the transcriptional level (Table I; Fig. 1). Moreover, the levels of HSPs, such as GroEL, GroES, and HspA, were also elevated at the normal growth temperature (Table IV). These changes, caused by the mutation of Hik34, might explain the enhanced thermotolerance of ΔHik34 cells (Fig. 3).
In cells grown at an elevated temperature, DNA microarray analysis and northern-blot analysis, with the hspA gene as probe, indicated that mutation of the hik34 gene suppressed the heat shock response only partially over the course of the first 20 min at this temperature (Table III; Fig. 5), suggesting that Hik34 might not be the only regulator of expression of heat shock genes under heat shock conditions. The kinetics of induction of the expression of the hspA gene under heat shock conditions indicated that Hik34 is not involved in induction of the hspA gene at the early stages of induction, while the major role of Hik34 might be that of a strong repressor of the expression of this gene at the later stages of the transient induction (Fig. 5). The effects of an inhibitor of transcription, rifampicin, indicated that Hik34 regulates the expression of certain heat shock genes at the transcriptional level (Fig. 6). DNA microarray analysis also suggested that the response of most heat shock genes was prolonged in the Hik34 mutant, as was the duration of induction of expression of hspA (Fig. 4; Table III). The effect of Hik34 on the regulation of genes induced by heat shock might not be specific to the hspA gene and might be a rather general phenomenon. At present, we do not have any clear explanation for the effect of the Hik34 mutation on the kinetics of the heat shock-induced expression of genes.
Temperature-Dependent Autophosphorylation of Hik34
The autophosphorylation of Hik34 was temperature dependent, clearly supporting our hypothesis that Hik34 might be important at elevated temperatures. We do not have any clear idea how Hik34 might function under heat stress conditions. However, identification of the mechanism responsible for the regulation of autophosphorylation by temperature and identification of downstream components, such as Rres that might accept a phosphate group from this particular Hik, should help us to understand the functioning of this Hik, which is strongly conserved within cyanobacteria. The sensing by living organisms of changes in environmental temperature is very poorly understood. The identification of Hiks whose activity is regulated by temperature should help to shed some light on this phenomenon.
Orthologs of Hik34 in Other Cyanobacteria
Genes that are homologous to hik34 have been found in several cyanobacteria whose genomes have been sequenced, such as Anabaena sp. PCC 7120 (Kaneko et al., 2001), Thermosynechococcus elongatus BP-1 (Nakamura et al., 2002), Synechococcus sp. WH8102 (Palenik et al., 2003), Prochlorococcus marinus MIT9313 (Rocap et al., 2003), P. marinus MED4 (Rocap et al., 2003), and P. marinus SS120 (Dufresne et al., 2003), as described by Mary and Vaulot (2003). Genes homologous to hik34 have also been found in the genomes of Gloeobacter violaceus PCC 7421 (Nakamura et al., 2003), Nostoc punctiforme (http://genome.jgi-psf.org/draft_microbes/nospu/nospu.home.html), and Trichodesmium erythraeum (http://genome.jgi-psf.org/draft_microbes/trier/trier.home.html). Each of these cyanobacteria has a single copy of the ortholog of hik34 in its chromosome.
Hik34 and its orthologs in cyanobacteria include a conserved signature motif of His kinases, H1, which includes a phosphorylatable His residue. Nevertheless, they lack other signature motifs of Hiks, such as the N, G1 (DXGXG), F, and G2 (GXGXG) motifs, which probably play important roles in the binding of ATP as the phosphate donor for the autophosphorylation of Hiks (Parkinson and Kofoid, 1992; Mizuno et al., 1996). Hik34 is an unusual Hik and appears to be unique among the Hiks encoded by the chromosome and plasmids of Synechocystis. Moreover, to date, it has been found only in cyanobacteria. Modification of the ATP-binding motif in the Hik domain of Hik34 and its orthologs might be related to the way in which this kinase functions.
MATERIALS AND METHODS
Strains and Culture Conditions
Synechocystis sp. PCC 6803, a Glc-tolerant strain, was kindly provided by Dr. J. G. K. Williams (Du Pont de Nemours, Wilmington, DE) and served as the wild type. Wild-type cells and cells with a mutation in Hik34 were grown at 34°C in BG-11 medium (Stanier et al., 1971) with aeration with CO2-enriched air (1%, v/v) under continuous illumination at 70 μmol photons m−2 s−1 (Wada and Murata, 1989). For examination of thermotolerance, cells of wild type and cells of Hik mutants were spread on plates of agar-solidified BG-11 medium and incubated at 30°C under light at 70 μmol photons m−2 s−1 for 3 d. Then, after heat shock at 48°C for either 2 or 3 h at the same light intensity, they were incubated at 30°C for a further 7 d.
Construction of Hik34 Mutant Cells and Cells That Harbored the Vectors pVZ::hik34 and pVZ::pnrtA-hik34
Construction of a mutant Hik library has been described elsewhere (Suzuki et al., 2000; http://www.kazusa.or.jp/cyanobase/Synechocystis/mutants).
A DNA fragment containing the hik34 gene was obtained by PCR with Hik34F, GGGTGGAGTAATCAGCAACCTTCC, and Hik34R, CAAGGGGGGTGGAGGCAGTTTATC, as primers. This DNA fragment was digested with EcoRV and SpeI at restriction sites near the ends of the amplified sequence and then cloned into pBluescriptII SK(+) (Stratagene, La Jolla, CA) that had been cleaved with EcoRV and XbaI. The resultant plasmid, containing the hik34 gene, was digested with HindIII within the fragment that included the hik34 gene at a site that corresponded to nucleotide position 1,882,399 bp in the Synechocystis chromosome. A spectinomycin-resistance gene cassette was inserted at this site after the cassette had been cleaved with HindIII. The plasmid with the spectinomycin-resistance gene cassette within the cloned hik34 gene fragment was used to transform Synechocystis cells (Williams, 1988). Spectinomycin-resistant colonies were isolated and cells with a fully segregated mutation were obtained after growth for several generations in BG-11 media that contained the antibiotic, as previously reported (Suzuki, et al., 2000, 2004; Yamaguchi et al., 2002).
To introduce a native hik34 gene or a hik34 gene driven by the promoter of the nrtA operon (Aichi et al., 2001) from Synechocystis into Synechocystis cells via the multicopy plasmid pVZ321 (Zinchenko et al., 1999), we amplified DNA fragments that corresponded to the native hik34 gene, the coding sequence of the hik34 gene, and the promoter region of the nrtA gene with the following pairs of primers: Hik34up, CTCGAGTAGTTGACACCACC, and Hik34dn, CTCGAGCTAGACCATGGTGAA; Hik34NheINdeI, GCTAGCAGCATATGAATGAAGTTTGCCTAAAGTTGA, and Hik34dn; and NrtANheI, GCTAGCAACGGCGATCGCCAAAGCGG, and NrtANdeI, CATATGGTAGGCAATAACTAAGGCCA, respectively. The three amplified fragments were cloned separately into pT7Blue (Merck, Darmstadt, Germany) and their nucleotide sequences were determined. The promoter sequence of the nrtA operon was removed by cleavage with NheI and NdeI from pT7Blue that contained the promoter fragment and inserted into the plasmid that contained the coding region of the hik34 gene that had been digested with the same restriction enzymes. The native hik34 gene was isolated by digestion with XhoI and cloned into pVZ321 that had been digested with XhoI and SalI. Then the hik34 coding sequence, linked to the promoter of the nrtA gene, was isolated by cleavage with NheI and XhoI and cloned into pVZ321 that had been digested with XbaI and XhoI. The resultant plasmids, namely, pVZ::hik34 and pVZ::pnrtA-hik34, and the original plasmid, pVZ321, were introduced into Synechocystis cells via triparental conjugation, as described previously (Zinchenko et al., 1999; Yamaguchi et al., 2002) and transformants were selected with resistance to chloramphenicol as the selectable marker.
DNA Microarray Analysis
A 50-mL aliquot of culture was mixed with an equal volume of an ice-cold mixture of 5% phenol in ethanol (w/v) to stop cellular metabolism. Cells were harvested from the mixture by centrifugation at 4,000g, for 5 min at 4°C, and then frozen immediately and stored at −80°C. Total RNA was extracted with hot phenol as described previously (Suzuki et al., 2000). A Synechocystis DNA microarray (CyanoCHIP) was purchased from TaKaRa Bio Co. Ltd. (Shiga, Japan). This microarray covered 3,079 of the 3,264 open reading frames of the Synechocystis genome (Kaneko et al., 1996), excluding several open reading frames for transposases. Synthesis of cDNA labeled with Cy3 and Cy5 was achieved with an MMLV RNA fluorescent labeling kit (TaKaRa Bio) and hybridization and washing were performed as described previously (Kanesaki et al., 2002). Hybridization of the dye-labeled cDNAs was evaluated with an array scanner (GMS418; Affymetrix, Santa Clara, CA). For quantification with the ImaGene version 5.5 program (BioDiscovery, Marina del Rey, CA), the local background of each spot was subtracted and each signal was normalized by calculating the ratio of the spot-specific intensity to the total intensity of signals from all genes, with the exception of genes for rRNAs. Thus, a change in the amount of the transcript of each gene could be calculated, relative to the total amount of mRNA. Each gene was included twice on the microarray, allowing for adequate evaluation of signals and exclusion of errors. A gene was regarded as heat inducible or heat repressible when the induction factor for this gene was greater than 2.0 or smaller than 0.5, respectively (Suzuki et al., 2001).
2D Gel Electrophoresis
Soluble proteins from wild-type cells and ΔHik34 mutant cells were extracted and prepared for 2D electrophoresis as described previously (Simon et al., 2002). 2D electrophoresis was carried out using immobilized pH gradient strip and the IPGphor and Ettan DALT system (GE Healthcare Amersham, Buckinghamshire, UK). 2D-DIGE (GE Healthcare Amersham) was performed according to the manufacturer's standard protocols. Cy3 and Cy5 dyes were used with pairs of samples to compare the amounts of protein in each spot on the 2D gels and the signals from Cy2-labeled pooled standard were used for normalization of intensities of fluorescence. Comparison between samples and statistical validation of changes in expression of proteins were performed using the differential in-gel analysis and biological variation analysis modules of DeCyder image analysis software (GE Healthcare Amersham).
Phosphorylation of Hik34 In Vitro
The entire sequence of the hik34 gene was amplified by PCR with oligonucleotides Hik34NdeI, CATATGAATGAAGTTTGCCTAAAGTTGAGTG, and Hik34PstI, GACGTCCTAGACCATGGTGAACTGCCTATCG, as primers. These primers include an NdeI and a PstI site, respectively. The amplified fragment was cloned first into pT7Blue (Novagen, Madison, WI) and its nucleotide sequence was determined. The resultant plasmid was digested with PstI and an expression plasmid pET28a (Novagen) was cleaved with XhoI. The linearized plasmids were blunted with a DNA blunting kit (TaKaRa Bio). Then both plasmids were further digested with NdeI. The DNA fragment containing the coding sequence of hik34 and the vector, pET28a, were recovered and ligated to construct pETHik34. Then pET28a and pETHik34 were introduced separately into Escherichia coli strain BL21(DE3) pLysS (Novagen). The transformed E. coli cells were treated with 0.5 mm isopentenyl thiogalactoside for 3 h at 25°C. Proteins were extracted from cells with a French press (SLM Instruments, Urbana, IL) and the His-tagged protein was purified with Ni-NTA resin (Novagen).
Assays of protein phosphorylation were performed in 30 μL of a solution that contained 100 mm Tris-HCl, pH 8.0, 100 mm KCl, 5% (v/v) glycerol, 10 mm MgCl2, 5 mm CaCl2, 5 μg of protein, and 100 μm [γ-32P]ATP (approximately 6,000 Ci/mmol; Amersham Pharmacia) at 18°C, 24°C, 30°C, 36°C, 42°C, and 48°C for 15 min. Each reaction was terminated by gel filtration on a Centri-Sep spin column (Applied Biosystems, Foster City, CA) that had been equilibrated with a solution of 100 mm NaCl plus 5 mm EDTA. Each eluate was mixed with an equal volume of 2× SDS gel loading buffer (100 mm Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 0.1% bromphenol blue, 200 mm 2-mercaptoethanol) and incubated at 65°C for 10 min. The proteins were separated by SDS-PAGE (8% polyacrylamide) and phosphorylated proteins were detected by autoradiography.
Supplementary Material
Acknowledgments
The authors thank Ms. Akiko Okada and Ms. Yukari Koike for their skilled technical assistance. Access to the DNA sequences of cyanobacterial genomes in the databases of the Department of Energy-Joint Genome Institute and the Kazusa DNA Research Institute is gratefully acknowledged.
This work was supported by Grants-in-Aid for Scientific Research for Priority Areas (C) Genome Biology (grant no. 16013249 to I.S.) and for Priority Areas (grant no. 14086207 to N.M.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by a Grant-in-Aid for Scientific Research (S; grant no. 13854002 to I.S. and N.M.) from the Japan Society for the Promotion of Science. This work was also supported by the Salt Science Research Foundation and by the National Institute for Basic Biology Cooperative Research Program on Stress-Tolerant Plants. A.R.S., J.J.H., and W.J.S. are supported by the Biotechnology and Biological Sciences Research Council (BBSRC). J.J.H. wishes to thank Amersham Biosciences, United Kingdom, for supporting a BBSRC Ph.D. Cooperative Awards in Science and Engineering studentship.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.059097.
References
- Aichi M, Takatani N, Omata T (2001) Role of NtcB in activation of nitrate-assimilation genes in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 183: 5840–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL, et al (2004) The Pfam protein families database. Nucleic Acids Res 32: D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A, Salanoubat M, Partensky F, Artiguenave F, Axmann IM, Barbe V, Dupra TS, Galperin MY, Koonin EV, Le Gall F, et al (2003) Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc Natl Acad Sci USA 100: 10020–10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M, Schumann W, Völker U (1986) Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol 19: 417–428 [DOI] [PubMed] [Google Scholar]
- Hirani TA, Suzuki I, Murata N, Hayashi H, Eaton-Rye JJ (2001) Characterization of a two-component signal transduction system involved in the induction of alkaline phosphatase under phosphate-limiting conditions in Synechocystis sp. PCC 6803. Plant Mol Biol 45: 133–144 [DOI] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high-temperature stress. Proc Natl Acad Sci USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MM, Nakamoto H (2002) HtpG plays a role in cold acclimation in cyanobacteria. Curr Microbiol 44: 291–296 [DOI] [PubMed] [Google Scholar]
- Hossain MM, Nakamoto H (2003) Role for the cyanobacterial HtpG in protection from oxidative stress. Curr Microbiol 46: 70–76 [DOI] [PubMed] [Google Scholar]
- Inaba M, Suzuki I, Szalontai B, Kanesaki Y, Los DA, Hayashi H, Murata N (2003) Gene-engineered rigidification of membrane lipids enhances the cold inducibility of gene expression in Synechocystis. J Biol Chem 278: 12191–12198 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T, et al (2001) Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res 8: 205–213, 227–253 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3: 109–136 [DOI] [PubMed] [Google Scholar]
- Kanesaki Y, Suzuki I, Allakhverdiev SI, Mikami K, Murata N (2002) Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem Biophys Res Commun 290: 339–348 [DOI] [PubMed] [Google Scholar]
- Lindquist S (1986) The heat-shock response. Annu Rev Biochem 55: 1151–1191 [DOI] [PubMed] [Google Scholar]
- Lindquist S, Craig EA (1988) The heat-shock proteins. Annu Rev Genet 22: 631–677 [DOI] [PubMed] [Google Scholar]
- López-Maury L, García-Domínguez M, Florencio FJ, Reyes JC (2002) A two-component signal transduction system involved in nickel sensing in the cyanobacterium Synechocystis sp. PCC 6803. Mol Microbiol 43: 247–256 [DOI] [PubMed] [Google Scholar]
- Marin K, Suzuki I, Yamaguchi K, Ribbeck K, Yamamoto H, Kanesaki Y, Hagemann M, Murata N (2003) Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc Natl Acad Sci USA 100: 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mary I, Vaulot D (2003) Two-component systems in Prochlorococcus MED4: genomic analysis and differential expression under stress. FEMS Microbiol Lett 226: 135–144 [DOI] [PubMed] [Google Scholar]
- Mikami K, Kanesaki Y, Suzuki I, Murata N (2002) The histidine kinase Hik33 perceives osmotic stress and cold stress in Synechocystis sp. PCC 6803. Mol Microbiol 46: 905–915 [DOI] [PubMed] [Google Scholar]
- Mizuno T, Kaneko T, Tabata S (1996) Compilation of all genes encoding bacterial two-component signal transducers in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803. DNA Res 3: 407–414 [DOI] [PubMed] [Google Scholar]
- Nakamoto H, Suzuki M, Kojima K (2003) Targeted inactivation of the hrcA repressor gene in cyanobacteria. FEBS Lett 549: 57–62 [DOI] [PubMed] [Google Scholar]
- Nakamoto H, Tanaka N, Ishikawa N (2001) A novel heat-shock protein plays an important role in thermal stress management in cyanobacteria. J Biol Chem 276: 25088–25095 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kaneko T, Sato S, Ikeuchi M, Katoh H, Sasamoto S, Watanabe A, Iriguchi M, Kawashima K, Kimura T, et al (2002) Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res 9: 123–130 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kaneko T, Sato S, Mimuro M, Miyashita H, Tsuchiya T, Sasamoto S, Watanabe A, Kawashima K, Kishida Y, et al (2003) Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res 10: 137–145 [DOI] [PubMed] [Google Scholar]
- Ogawa T, Bao DH, Katoh H, Shibata M, Pakrasi HB, Bhattacharyya-Pakrasi M (2002) A two-component signal transduction pathway regulates manganese homeostasis in Synechocystis 6803, a photosynthetic organism. J Biol Chem 277: 28981–28986 [DOI] [PubMed] [Google Scholar]
- Paithoonrangsarid K, Shoumskaya MA, Kanesaki Y, Satoh S, Tabata S, Los DA, Zinchenko VV, Hayashi H, Tanticharoen M, Suzuki I, et al (2004) Five histidine kinases perceive osmotic stress and regulate distinct sets of genes in Synechocystis. J Biol Chem 297: 53078–53086 [DOI] [PubMed] [Google Scholar]
- Palenik B, Brahamsha B, Larimer FW, Land M, Hauser L, Chain P, Lamerdin J, Regala W, Allen EE, McCarren J, et al (2003) The genome of a motile marine Synechococcus. Nature 424: 1037–1042 [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Kofoid EC (1992) Communication modules in bacterial signaling proteins. Annu Rev Genet 26: 71–112 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624 [DOI] [PubMed] [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, et al (2003) Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424: 1042–1047 [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S (1998) Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342 [DOI] [PubMed] [Google Scholar]
- Scherl A, François P, Bento M, Deshusses JM, Charbonnier Y, Converset V, Huyghe A, Walter N, Hoogland C, Appel RD, et al (2005) Correlation of proteomic and transcriptomic profiles of Staphylococcus aureus during the post-exponential phase of growth. J Microbiol Methods 60: 247–257 [DOI] [PubMed] [Google Scholar]
- Simon WJ, Hall JJ, Suzuki I, Murata N, Slabas AR (2002) Proteomic study of the soluble proteins from the unicellular cyanobacterium Synechocystis sp. PCC 6803 using automated matrix-assisted laser desorption/ionization-time of flight peptide mass fingerprinting. Proteomics 2: 1735–1742 [DOI] [PubMed] [Google Scholar]
- Stanier RY, Kunisawa R, Mandel M, Cohen-Bazire G (1971) Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol Rev 35: 171–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Ferjani A, Suzuki I, Murata N (2004) The SphS-SphR two-component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in Synechocystis. J Biol Chem 279: 13234–13240 [DOI] [PubMed] [Google Scholar]
- Suzuki I, Kanesaki Y, Mikami K, Kanehisa M, Murata N (2001) Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol Microbiol 40: 235–244 [DOI] [PubMed] [Google Scholar]
- Suzuki I, Los DA, Kanesaki Y, Mikami K, Murata N (2000) The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J 19: 1327–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M (2001) Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 1: 377–396 [DOI] [PubMed] [Google Scholar]
- Wada H, Murata N (1989) Synechocystis PCC6803 mutants defective in desaturation of fatty acids. Plant Cell Physiol 30: 971–978 [Google Scholar]
- Wang WX, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9: 244–252 [DOI] [PubMed] [Google Scholar]
- Webb R, Reddy KJ, Sherman LA (1990) Regulation and sequence of the Synechococcus sp. strain PCC 7942 groESL operon, encoding a cyanobacterial chaperonin. J Bacteriol 172: 5079–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JGK (1988) Construction of specific mutants in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. In L Packer, AN Glazer, eds, Methods in Enzymology, Vol. 167. Academic Press, San Diego, pp 766–778
- Yamaguchi K, Suzuki I, Yamamoto H, Lyukevich A, Bodrova I, Los DA, Piven I, Zinchenko V, Kanehisa M, Murata N (2002) A two-component Mn2+-sensing system negatively regulates expression of the mntCAB operon in Synechocystis. Plant Cell 14: 2901–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinchenko VV, Piven IV, Melnik VA, Shestakov SV (1999) Vectors for the complementation analysis of cyanobacterial mutants. Genetika 35: 291–296 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.