Abstract
Nucleosome assembly protein 1 (NAP1) is conserved from yeast to human and facilitates the in vitro assembly of nucleosomes as a histone chaperone. Inconsistent with their proposed function in the nucleus, however, many NAP1 proteins had been reported to localize in the cytoplasm. We investigated the subcellular localization of tobacco (Nicotiana tabacum) and rice (Oryza sativa) NAP1 family proteins first by identification of interacting partners and by direct examination of the localization of green fluorescent protein-tagged proteins. Through treatment of tobacco cells with leptomycin B and mutagenesis of nuclear export signal, we demonstrated that Nicta;NAP1;1 and Orysa;NAP1;1 shuttle between the cytoplasm and the nucleus. Together with the demonstration that tobacco NAP1 proteins bind histone H2A and H2B, our results support the current model and provide additional evidence that function of NAP1 as histone chaperones appears to be conserved in plants. In addition, we show that tobacco NAP1 proteins interact with tubulin and the mitotic cyclin Nicta;CYCB1;1, suggesting a role for NAP1 in microtubule dynamics. Interestingly, in spite of their high homology with the above NAP1 proteins, the other three tobacco proteins and Orysa;NAP1;2 did not show nucleocytoplasmic shuttling and were localized only in the cytoplasm. Moreover, Orysa;NAP1;3 that lacks a typical nuclear localization signal sequence was localized in both the cytoplasm and the nucleus. Finally, we show that only Orysa;NAP1;3 could be phosphorylated by casein kinase 2α in vitro. However, this phosphorylation was not responsible for nuclear import of Orysa;NAP1;3 as being demonstrated through mutagenesis studies. Together, our results provide an important step toward elucidating the molecular mechanism of function of the NAP1 family proteins in plants.
Nucleosome assembly protein 1 (NAP1) family proteins are considered as histone chaperones, which are conserved from yeast (Saccharomyces cerevisiae) to human and are proposed to facilitate assembly of newly synthesized histone H2A and H2B into a dimmer, transfer the dimmer to replicating DNA in the nucleus, and act synergistically with ATP-dependent chromatin remodeling factors to facilitate assembly and remodeling of chromatin (Ishimi et al., 1984; Yoon et al., 1995; Ito et al., 1997; McQuibban et al., 1998; Nakagawa et al., 2001; Levenstein and Kadonaga, 2002). Consistent with this view, genetic studies reveal important functions of NAP1 in several organisms. In yeast, deletion of NAP1 altered the expression of about 10% of all nuclear genes, suggesting the requirement of NAP1 for correct chromatin function (Ohkuni et al., 2003). Knocking out of NAP1 in Drosophila melanogaster dramatically reduced viability of the fly, which became more severe and penetrant in offspring, a phenotype characteristic of nucleosome remodeling factor deficient mutants (Lankenau et al., 2003). In mouse, knocking out of the neuron-specific NAP1-homolog-2 gene resulted in embryonic lethality at the mid-gestation stage, suggesting a role of NAP1 in neuronal cell proliferation (Rogner et al., 2000).
In addition, it has been shown that yeast NAP1 plays important roles during mitotic progression, possibly via interactions with several mitotic factors including Clb2 (a mitotic cyclin), Gin4 (a kinase), and NBP1 (a nuclear protein; Kellogg and Murray, 1995; Kellogg et al., 1995; Altman and Kellogg, 1997; Shimizu et al., 2000). More recently, Miyaji-Yamaguchi et al. showed that the yeast NAP1 protein is a nucleocytoplasmic shuttling protein and its shuttling is important for its function in mitotic progression (Miyaji-Yamaguchi et al., 2003). Some animal NAP1 proteins showed a cell cycle phase-dependent localization in either cytoplasm or nucleus (Ito et al., 1996; Rodriguez et al., 1997; Rogner et al., 2000), suggesting that they might also be nucleocytoplasmic shuttling proteins. Furthermore, animal NAP1 proteins form complexes with the casein kinase 2 (CK2) and the transcriptional coactivator p300/CREB (Li et al., 1999; Ito et al., 2000; Rodriguez et al., 2000; Shikama et al., 2000; Asahara et al., 2002), suggesting that they are also involved in functions possibly independent of their histone chaperone activity.
In higher plants, several NAP1 homologs have been identified in a given species including tobacco (Nicotiana tabacum), rice (Oryza sativa), maize (Zea mays), and Arabidopsis (Arabidopsis thaliana; Dong et al., 2003; http://www.chromdb.org/). We have previously shown that tobacco and rice NAP1 family proteins could bind in vitro animal histones and that the tobacco proteins Nicta;NAP1;3 and Nicta;NAP1;4 were localized in the cytoplasm, whereas the rice protein Orysa;NAP1;1 was localized in both the cytoplasm and the nucleus (Dong et al., 2003). A cell cycle dependent translocation of plant NAP1 proteins between the cytoplasm and the nucleus was, however, not observed by the use of partially synchronized tobacco suspension cells (Dong et al., 2003).
Here, we show that tobacco NAP1 proteins bind plant histones H2A and H2B. More interestingly, we show that tobacco NAP1 proteins can also interact with tubulin and the mitotic cyclin Nicta;CYCB1;1; suggesting that plant NAP1 proteins may play a role in microtubule dynamics. We further addressed the subcellular localization of the tobacco and rice NAP1 family proteins through identification and functional characterization of nuclear export signal (NES) and nuclear localization signal (NLS). Our results provide important information about the intracellular localization and signals involved in nucleocytoplasmic shuttling of the tobacco and rice NAP1 proteins, suggesting specific functions of the NAP1 family in higher plants.
RESULTS
Production and Specificity of Antibodies against Tobacco NAP1 Proteins
The tobacco NAP1 family proteins Nicta;NAP1;L1, Nicta;NAP1;2, and Nicta;NAP1;3 share approximately 81% to 85% amino acid sequence identity to each other, but only approximately 67% to 68% identity with Nicta;NAP1;4. We used Nicta;NAP1;3 and Nicta;NAP1;4 proteins to immunize mice for antibody production. While the antiserum against Nicta;NAP1;4 (αNicta;NAP1;4) recognized all four tobacco proteins, the antiserum against Nicta;NAP1;3 (αNicta;NAP1;3) only recognized Nicta;NAP1;1, Nicta;NAP1;2, and Nicta;NAP1;3 (Fig. 1A), suggesting that αNicta;NAP1;3 specifically recognizes the C-terminal region that is lacking from Nicta;NAP1;4 but is conserved in the other three tobacco proteins (Dong et al., 2003).
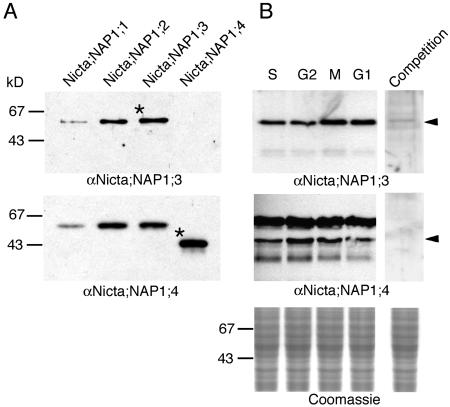
Figure 1.
Western-blot analyses of tobacco NAP1 proteins. A, A total of 0.1 μg of recombinant Nicta;NAP1;1, Nicta;NAP1;2, Nicta;NAP1;3, and Nicta;NAP1;4 proteins were analyzed using antibodies raised against Nicta;NAP1;3 (αNicta;NAP1;3) or Nicta;NAP1;4 (αNicta;NAP1;4). The asterisks indicate bands of the proteins used in antibody production. B, Twenty micrograms of total protein extracts of synchronized tobacco BY2 cells at S, G2, M, and G1 phases of the cell cycle were analyzed using indicated antibodies. For competition, recombinant Nicta;NAP1;3 or Nicta;NAP1;4 proteins were added to a final concentration of 20 μg/mL together with antibodies in the incubation solution. The arrowheads indicate positions corresponding to the size of antigens.
Western-blot analyses on total protein extracts of tobacco cells revealed a single major band recognized by αNicta;NAP1;3, which can be competed by increasing concentrations of recombinant Nicta;NAP1;3 protein (Fig. 1B, top). This band most likely comprises Nicta;NAP1;1, Nicta;NAP1;2, and Nicta;NAP1;3 proteins that are similar in molecular mass and therefore would migrate at a same position in the gel. Alternatively, the single band could also reflect cell cycle phase-specific expression of the different tobacco NAP1 proteins. Our experiments currently cannot distinguish these possibilities. The αNicta;NAP1;4 antibody recognized a band corresponding to the size of Nicta;NAP1;4, but also an additional band corresponding to the size of the other three NAP1 proteins (Fig. 1B, bottom). A weaker band of lower molecular mass was also revealed by both antibodies, particularly visible after extensive exposure (Fig. 1B, bottom), which could be a degradation product or unrelated cross-reactive protein. Although slight variations (less than 2-fold) were detected in some experiments, the level of the tobacco NAP1 proteins did not change significantly and consistently with a specific phase of the cell cycle. This observation is in agreement with our previous northern data showing that the different tobacco NAP1 genes are expressed relatively constantly during the cell cycle (Dong et al., 2003).
Tobacco NAP1 Proteins Bind Histone H2A and H2B as Well as Tubulins and the Mitotic Cyclin Nicta;CYCB1;1
Using pulldown assays we could demonstrate that Nicta;NAP1;4 specifically interacts with H2B-yellow fluorescent protein (YFP) as well as H2A-YFP but not YFP alone (Fig. 2A). Similar results were obtained for Nicta;NAP1;3 (data not shown). These results are consistent with the proposed role for NAP1 proteins as H2A/H2B chaperones and with our previous ELISA data performed with animal histones (Dong et al., 2003). The same fractions were also analyzed for the presence of other binding proteins. These experiments showed that tobacco NAP1 proteins can interact with each other (Fig. 2B, left). The detection of different tobacco NAP1 proteins in the pulldown assays clearly indicates their potential to form heteromeric complexes. Formation of homo- or heterodimeric complexes has already been reported for mammalian NAP1 homologs (Shikama et al., 2000). Our results provide additional evidence that heteromeric complex formation appears to be conserved among NAP1 proteins from different organisms.
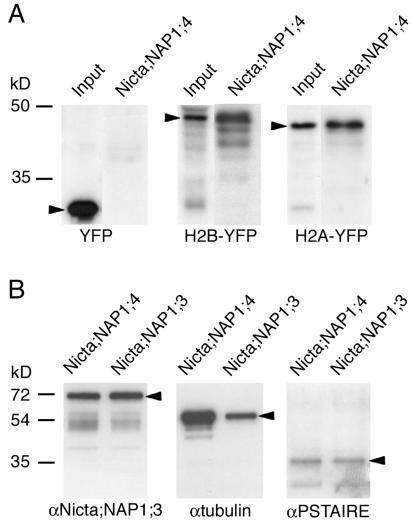
Figure 2.
Identification of proteins binding to tobacco NAP1 proteins by pulldown assays. A, The pulldown fractions by Nicta;NAP1;4-coated beads from total protein extracts of transgenic Arabidopsis plants expressing YFP, H2B-YFP, and H2A-YFP were analyzed by western blotting with a polyclonal anti-GFP antibody. The input fraction represents 5% of total proteins used in pulldown. Arrowheads mark the positions of YFP, H2B-YFP, and H2A-YFP proteins. B, The pulldown fractions by Nicta;NAP1;4- or Nicta;NAP1;3-coated beads from total protein extracts of tobacco BY2 cells were analyzed by western blotting with indicated antibodies. Arrowheads mark the positions of Nicta;NAP1;3, β-tubulin, and CDKA proteins.
Our previous observation that tobacco NAP1 proteins colocalize with the mitotic spindle and the phragmoplast (Dong et al., 2003) prompted us to analyze their potential interactions with tubulins and cell cycle regulatory proteins. As shown in Figure 2B (middle), tubulin was found to interact with both Nicta;NAP1;3 and Nicta;NAP1;4. The results also suggest that Nicta;NAP1;4 has a higher binding activity for tubulin than Nicta;NAP1;3. We did not detect any interaction between tobacco NAP1 proteins and the plant-specific cyclin-dependent kinase CDKB (Porceddu et al., 2001). In contrast, a weak signal could be detected for CDKA (Fig. 2B, right), a PSTAIRE-containing cyclin-dependent kinase that associates with different cyclins to control cell cycle progression (Stals and Inze, 2001).
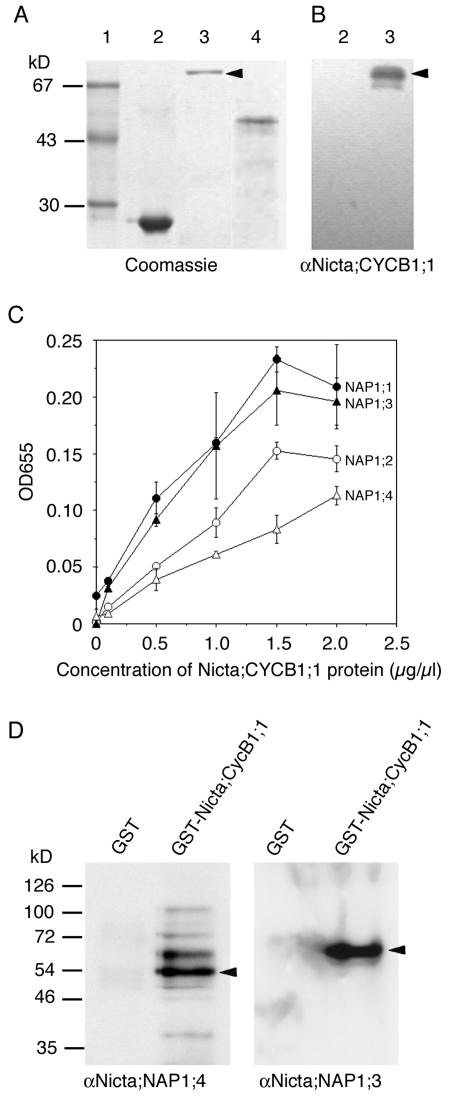
Yeast and Xenopus NAP1 interact with B-type cyclins (Kellogg and Murray, 1995; Kellogg et al., 1995). The weak signal for CDKA detected in our pulldown assay could have therefore resulted from the interaction between tobacco NAP1 proteins, CDKA, and B-type cyclins. To test this possibility directly, we expressed the tobacco B-type cyclin Nicta;CYCB1;1 fused to a 6 × His or glutathione S-transferase (GST) tag in Escherichia coli (Fig. 3A) and used the 6 × His-Nicta;CYCB1;1 protein for antibody production. The αNicta;CYCB1;1 antibody specifically recognized GST-Nicta;CYCB1;1 but not GST (Fig. 3B). ELISA analysis showed that Nicta;CYCB1;1 bound to all four tobacco NAP1 family proteins. The binding affinity of Nicta;CYCB1;1 was higher for Nicta;NAP1;1 and Nicta;NAP1;3 than for Nicta;NAP1;2 and Nicta;NAP1;4 (Fig. 3C). Furthermore, Figure 3D shows that NAP1 proteins were specifically precipitated from tobacco total protein extracts by the GST-Nicta;CYCB1;1 but not by GST beads. Together, our results demonstrate that tobacco NAP1 proteins bind to Nicta;CYCB1;1.
Figure 3.
Tobacco NAP1 proteins bind Nicta;CYCB1;1. A, Purified recombinant proteins were analyzed by SDS-PAGE gel. Lane 1, Mr marker; lane 2, GST; lane 3, GST-Nicta;CYCB1;1; lane 4, 6 × His-Nicta;CYCB1;1. B, Western-blot analysis of GST (lane 2) and GST-Nicta;CYCB1;1 (lane 3) with the polyclonal antibody raised against 6 × His-Nicta;CYCB1;1. Note that only GST-Nicta;CYCB1;1 (indicated by an arrowhead in A as well as in B) but not GST was specifically recognized by the antibody. C, Binding of recombinant GST-Nicta;CYCB1;1 to recombinant tobacco NAP1 proteins was determined by ELISA using the antibody against 6 × His-Nicta;CYCB1;1. The mean values with error bars were derived from three independent experiments. D, Pulldown assays. Total protein extract from tobacco BY2 cells was subdivided into two and incubated with GST or GST-Nicta;CYCB1;1 coated beads. The pulldown fractions were analyzed by western blotting with indicated antibodies. Arrowheads indicate bands corresponding to sizes of Nicta;NAP1;4 (left) and Nicta;NAP1;3 (right) proteins.
Inhibitor Treatment and Mutagenesis Reveal Nucleocytoplasmic Shuttling of Nicta;NAP1;1 But Not the Other Three Tobacco NAP1 Proteins
The green fluorescent protein (GFP)-Nicta;CYCB1;1 fusion protein could be detected in both the cytoplasm and nucleus (Criqui et al., 2001; A. Dong, unpublished data), but the GFP-Nicta;NAP1;3 and GFP-Nicta;NAP1;4 fusion proteins appeared to localize only to the cytoplasm (Dong et al., 2003). GFP-Nicta;NAP1;1 and GFP-Nicta;NAP1;2 were also localized in the cytoplasm (Fig. 4, A and D). To test if the tobacco NAP1 proteins could dynamically shuttle between the nucleus and the cytoplasm, we first examined their localization in cells after treatment with leptomycin B (LMB), a specific nuclear export inhibitor (Toyoshima et al., 1998). As shown in Figure 4B, LMB treatment resulted in accumulation of GFP-Nicta;NAP1;1 in the nucleus in more than 90% of cells. In contrast, GFP-Nicta;NAP1;2 (Fig. 4E) as well as GFP-Nicta;NAP1;3 and GFP-Nicta;NAP1;4 (data not shown) fusion proteins remained localized in the cytoplasm in the presence of LMB.
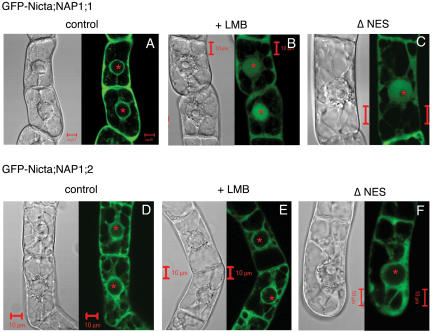
Figure 4.
Intracellular localization and nuclear export of the tobacco NAP1 family proteins. Transgenic tobacco BY2 cells expressing GFP-fused NAP1 proteins were visualized by fluorescence confocal microscopy. GFP fluorescence (green) images are shown together with bright-field differential interference contrast images. Cells treated with the nuclear export inhibitor (+LMB) or expressing mutant proteins containing deletion of the nuclear export signal sequences (ΔNES) were compared with untreated cells expressing wild-type proteins (control). Both GFP-Nicta;NAP1;1 (A) and GFP-Nicta;NAP1;2 (D) were localized only in the cytoplasm. Either LMB treatment (B) or NES deletion (C) resulted in nuclear accumulation of Nicta;NAP1;1, but not Nicta;NAP1;2 (E and F). GFP-Nicta;NAP1;3 and GFP-Nicta;NAP1;4 gave similar results as GFP-Nicta;NAP1;2. For space reasons, images on Nicta;NAP1;3 and Nicta;NAP1;4 are not shown. The asterisks mark positions of nucleoli inside the spherical nuclei. Bars = 10 μm.
Tobacco NAP1 proteins contain a 16-amino acid sequence in their N-terminal regions that is similar to the NES of the yeast NAP1 protein (Fig. 5). To examine whether these sequences function as active NES signals, we constructed mutant proteins lacking the corresponding regions. As shown in Figure 4C, deletion of the NES-like sequence in GFP-Nicta;NAP1;1 resulted in nuclear accumulation of the protein. However, deletion of the NES-like sequence in Nicta;NAP1;2 did not alter the cellular localization pattern of the GFP-Nicta;NAP1;2 protein (Fig. 4F). Similar results were obtained for GFP-Nicta;NAP1;3 and GFP-Nicta;NAP1;4. These results are consistent with the previous observations of the localization of all four NAP1 proteins after treatment with LMB. The data suggest that under our experimental conditions, only Nicta;NAP1;1 is a nucleocytoplasmic shuttling protein whose nuclear export is mediated by an NES-dependent transport in tobacco BY2 cells. The absence of nuclear accumulation of the other three tobacco NAP1 proteins suggests that they are either transported by a temporally or functionally different mechanism that did not operate in BY2 cells under our experimental conditions or that they are exclusively localized in the cytoplasm.
Figure 5.
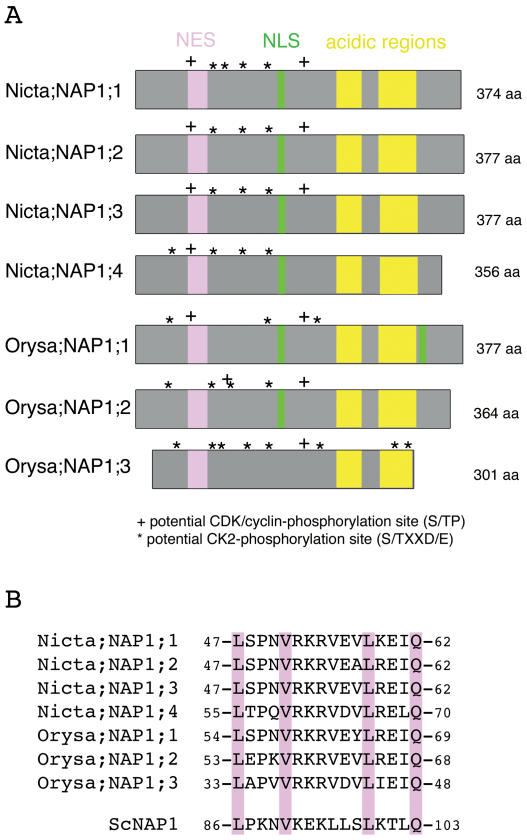
Schematic presentation and comparison of nuclear transport signal sequences of the tobacco and the rice NAP1 family proteins. A, Two acidic regions involved in histone binding are highly conserved in all NAP1 proteins and are shown by yellow boxes. An NLS sequence (shown as green box) was identified in the central region for all but except Orysa;NAP1;3 proteins. Only Orysa;NAP1;1 contains a second NLS, which is located in the C-terminal region of the protein. A region (shown as pink box) showing high sequence homology to the NES of the yeast NAP1 protein is conserved in all tobacco and rice NAP1 proteins. Sites matching to the consensus sequence of the CDK/cyclin- and CK2-phosphorylation sites are indicated. B, Sequence alignment of NES sequence of tobacco, rice, and yeast NAP1 proteins.
Rice NAP1 Family Proteins Also Exhibit Specific Nucleocytoplasmic Shuttling
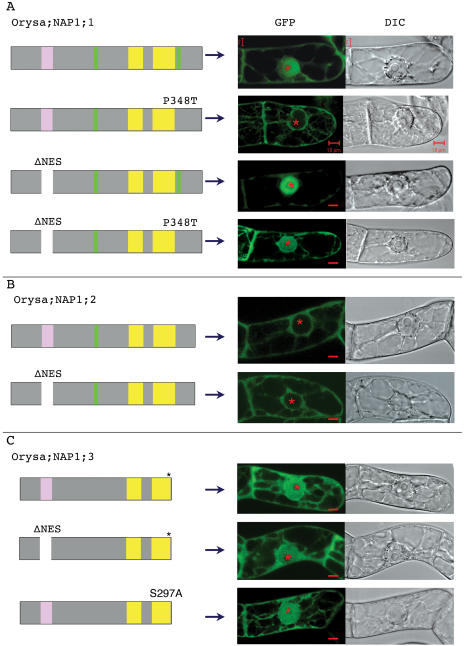
To investigate whether the different activity in nucleocytoplasmic shuttling observed with the tobacco NAP1 family is common in plants, we studied the rice NAP1 family proteins. The rice genome contains three genes encoding NAP1 proteins (Fig. 5). Like the tobacco proteins, all three rice proteins contain the conserved NES sequence. The rice protein Orysa;NAP1;1 contains, in addition to the conserved central NLS, a second NLS at its C-terminal region (Fig. 5), and the GFP-Orysa;NAP1;1 fusion protein was localized in both the cytoplasm and the nucleus (Dong et al., 2003; Fig. 6A). To test if this C-terminal NLS is responsible for the observed nuclear localization, we substituted the inside hydrophobic Pro by a Thr. The resulting mutant protein Orysa;NAP1;1 P348T showed an exclusive cytoplasmic localization (Fig. 6A), indicating that this C-terminal sequence is a bona fide NLS and is responsible for the nuclear accumulation of GFP-Orysa;NAP1;1. Deletion of the NES sequence of Orysa;NAP1;1 resulted in an enhanced nuclear localization as compared to the wild-type protein (Fig. 6A), indicating that the identified NES plays a role in the nuclear export of the protein. This conclusion is further supported by the observations that Orysa;NAP1;1 P348T was localized in both cytoplasm and nucleus in cells treated with LMB (data not shown) and that the mutant protein containing double mutations of the NLS and NES was localized in both cytoplasm and nucleus (Fig. 6A). These later results further indicate that in addition to the C-terminal NLS other signals exist and are involved in nuclear import of the Orysa;NAP1;1 protein. Together, our results clearly show that Orysa;NAP1;1 is a nucleocytoplasmic shuttling protein.
Figure 6.
Intracellular localization and functional characterization of nuclear transport signals of the rice NAP1 family proteins. Intracellular localization of wild-type and mutant proteins of Orysa;NAP1;1 (A), Orysa;NAP1;2 (B), and Orysa;NAP1;3 (C) was examined as described in Figure 4. Mutations include Pro-to-Thr substitution within the C-terminal NLS (P348T), NES deletion (ΔNES), and the combination of both mutations in Orysa;NAP1;1, NES deletion in Orysa;NAP1;2 and Orysa;NAP1;3, as well as Ser-to-Ala substitution within the CK2-phosphorylation site at the C-terminal end of Orysa;NAP1;3 (S297A). See legend of Figure 5 for details of schematic presentation of protein structures and that of Figure 4 for cell images.
Orysa;NAP1;2 has an NES and NLS organization similar to the tobacco NAP1 proteins (Fig. 5), and GFP-Orysa;NAP1;2 was localized only in cytoplasm (Fig. 6B). Deletion of the NES sequence in Orysa;NAP1;2 did not affect the intracellular localization of the protein; the mutant protein like the wild-type Orysa;NAP1;2 was localized exclusively in the cytoplasm (Fig. 6B). Moreover, treatment of cells with LMB did not result in nuclear localization of Orysa;NAP1;2 (data not shown). Thus, Orysa;NAP1;2 shows very similar localization activity as the tobacco proteins Nicta;NAP1;2, Nicta;NAP1;3, and Nicta;NAP1;4.
Typical NLS could not be identified from the Orysa;NAP1;3 sequence. Interestingly, GFP-Orysa;NAP1;3 was localized in both the cytoplasm and the nucleus (Fig. 6C). Deletion of the conserved NES sequence in Orysa;NAP1;3 did not increase nuclear accumulation of the protein (Fig. 6C).
Together, our results on the rice NAP1 family further confirm the previous data on tobacco proteins, which reveal the different importance of the NES sequence in the localization of different NAP1 proteins. It is particularly striking that Orysa;NAP1;3 does not carry any typical NLS sequence but was nevertheless localized in both the cytoplasm and nucleus.
The Orysa;NAP1;3 Protein Could Be Specifically Phosphorylated by CK2
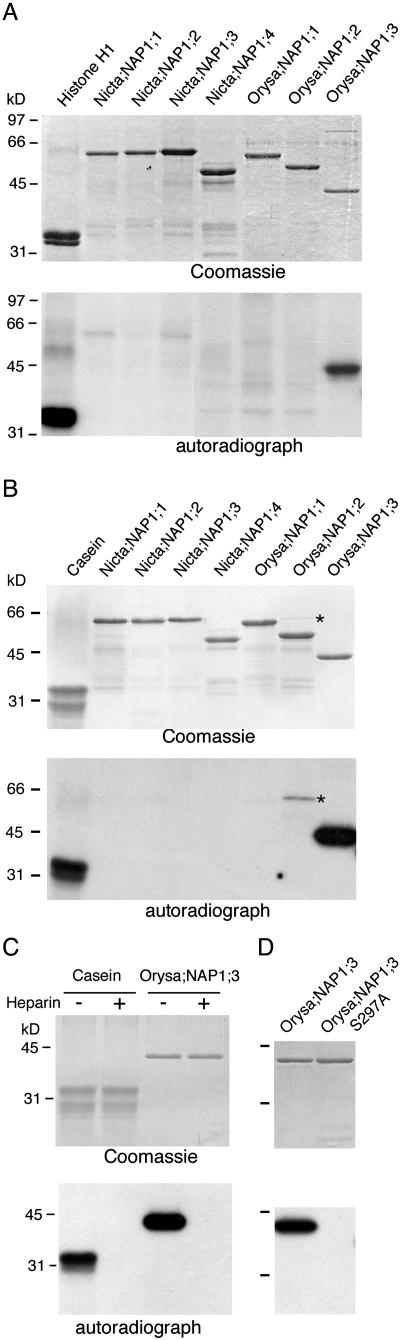
Many examples show that posttranslational modifications can regulate intracellular localization of proteins (Jans and Hubner, 1996; Kaffman et al., 1998; Dong et al., 2002). Our findings that NAP1 proteins bind B-type cyclin prompted us to test phosphorylation of NAP1 proteins. The pulldown fraction from plant protein extracts by p13SUC1-beads, which specifically bind CDK/cyclin kinase complexes (Bögre et al., 1997), could efficiently phosphorylate Orysa;NAP1;3 but not the other rice and tobacco NAP1 proteins (Fig. 7A). In this experiment, however, we could not exclude some possible contaminations by other types of kinases that might be copurified with CDK/cyclin complexes in the fraction. We next used purified recombinant casein kinase 2α (CK2α) protein produced in E. coli to test the phosphorylation of NAP1 proteins. As shown in Figure 7B, CK2α specifically phosphorylated Orysa;NAP1;3 but not the other rice and tobacco NAP1 proteins. The specificity of CK2α kinase was further confirmed by the use of its specific inhibitor, heparin, which inhibited phosphorylation of Orysa;NAP1;3 (Fig. 7C). Together, these results show that Orysa;NAP1;3 but not the other examined NAP1 proteins could be phosphorylated by CK2 kinase.
Figure 7.
In vitro phosphorylation of the tobacco and rice NAP1 family proteins. A, Phosphorylations of NAP1 proteins by p13SUC1-bound CDK/cyclin kinases. The p13SUC1-Sepharose pulldown fraction from total protein extract of tobacco BY2 cells was used to test phosphorylation of histone H1, tobacco (Nicta;NAP1;1 to Nicta;NAP1;4), and rice (Orysa;NAP1;1 to Orysa;NAP1;3) NAP1 proteins. B, Phosphorylation of NAP1 proteins by CK2α kinase. Purified recombinant GST-CK2α produced in E. coli was tested in phosphorylation of casein and NAP1 proteins. C, Heparin inhibits the phosphorylation of Orysa;NAP1;3 by CK2α. Phosphorylation of Orysa;NAP1;3 by CK2α was tested in the absence (−) or presence (+) of heparin. D, The mutant Orysa;NAP1;3 S297A protein is defective in phosphorylation by CK2α. The top sections show the Coomassie Brilliant Blue R250 stained proteins on the SDS-PAGE gels and the bottom sections show the corresponding autoradiography for phosphorylated (radioactive labeled) proteins. An E. coli protein that was copurified with Orysa;NAP1;2 and phosphorylated by CK2α is indicated by asterisks in B.
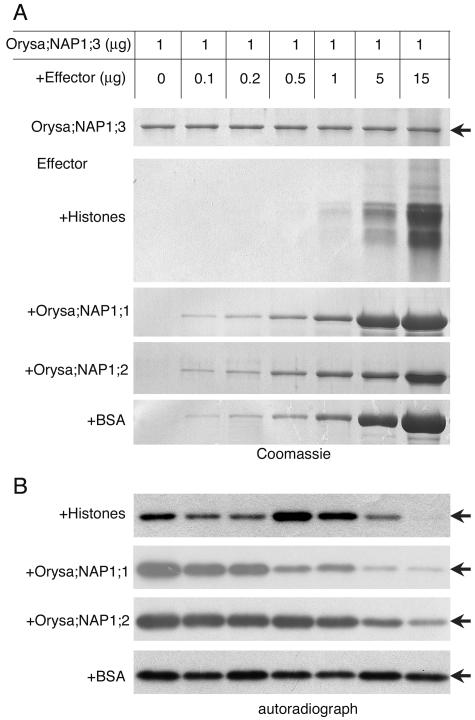
Our previous data show that NAP1 proteins can form heterodimers and bind histones. We next tested whether the phosphorylation of Orysa;NAP1;3 by CK2α could be influenced by histones and other NAP1 proteins. At approximately 1:1 molar ratio, histones stimulated phosphorylation of Orysa;NAP1;3, whereas a higher amount of histones inhibited the phosphorylation (Fig. 8). This result indicates that histone-associated Orysa;NAP1;3 is a good substrate of CK2α. Increasing amounts of either Orysa;NAP1;1 or Orysa;NAP1;2 protein progressively inhibited phosphorylation of Orysa;NAP1;3 (Fig. 8). Binding of CK2α by Orysa;NAP1;1 or Orysa;NAP1;2 possibly impaired its kinase activity toward Orysa;NAP1;3.
Figure 8.
Phosphorylation of Orysa;NAP1;3 by CK2α can be influenced by histones and other NAP1 proteins. Different quantities of core histones (+Histone), NAP1 proteins (+Orysa;NAP1;1, +Orysa;NAP1;2), and bovine serum albumin protein (+BSA) were tested as effectors in the in vitro phosphorylation of Orysa;NAP1;3 by CK2α. Coomassie Brilliant Blue R250-stained proteins on the SDS-PAGE gels (A) and autoradiography of Orysa;NAP1;3 phosphorylation (B) were shown. Phosphorylation of effector proteins was not observed. Arrows indicate positions of the Orysa;NAP1;3 protein.
The CK2-Phosphorylation Site Is Located at the C-Terminal End and Is Not Required for the Nuclear Localization of Orysa;NAP1;3
CK2 kinases catalyze the phosphorylation of the Ser or Thr residue within an S/TxxD/E consensus sequence (Meggio and Pinna, 2003). A number of sequences matching the CK2 consensus phosphorylation site could be identified in the tobacco and rice NAP1 proteins (Fig. 5). A comparison between different NAP1 proteins revealed that two of these sites are uniquely present at the C-terminal region of the Orysa;NAP1;3 protein. We tested the most C-terminal located site, 297SDEE300, as a possible target of CK2-phosphorylation. Substitution of the Ser by Ala within this site, Orysa;NAP1;3 S297A, abolished the phosphorylation of the protein by CK2α (Fig. 7D), indicating that this site is responsible for Orysa;NAP1;3 phosphorylation by CK2α. Then we tested the role of phosphorylation in protein localization by analyzing the intracellular localization of the Orysa;NAP1;3 S297A mutant protein. As shown in Figure 6C, the Orysa;NAP1;3 S297A mutant protein was, like wild-type protein, localized in both the cytoplasm and nucleus. Therefore, it is unlikely that phosphorylation of Orysa;NAP1;3 is responsible for nuclear import of this protein.
DISCUSSION
Our experimental data showing that the tobacco NAP1 family proteins interact with H2A/H2B and that the tobacco protein Nicta;NAP1;1 as well as the rice protein Orysa;NAP1;1 shuttles between the cytoplasm and the nucleus are consistent with the current model in which yeast and animal NAP1 proteins are proposed to bind newly synthesized H2A and H2B in the cytoplasm, transport the histones to the nucleus, and deposit H2A/H2B on preformed H3/H4-DNA complex during nucleosome formation (Krude and Keller, 2001; Loyola and Almouzni, 2004). Interestingly, the other three tobacco NAP1 proteins as well as the rice protein Orysa;NAP1;2 could not be detected in the nucleus under our experimental conditions, suggesting that they have specific functions in the cytoplasm. Our observation that tobacco NAP1 proteins interact with tubulin and with the B-type cyclin Nicta;CYCB1;1 suggests that plant NAP1 proteins may play a role in microtubule dynamics.
Microtubule dynamics is essential during assembly of the mitotic spindle, which is induced specifically by B-type cyclins during mitosis. Sea urchin cyclin B but not cyclin A induces destabilization of microtubules when cells enter into mitosis (Verde et al., 1992). Ectopic expression of a nondegradable Nicta;CYCB1;1 mutant in tobacco cells alters microtubule organization and dynamics and impairs formation of the phragmoplast, a plant specific microtubule array (Weingartner et al., 2004). Nicta;CYCB1;1 is localized in the cytoplasm in early G2 phase and translocated to the nucleus during G2/M transition (Criqui et al., 2001). The presence of NLS- and NES-like sequences in the N-terminal region of Nicta;CYCB1;1 is consistent with nucleocytoplasmic shuttling of the protein. It is known that low doses of microtubule-binding drugs that do not affect the tubulin monomer-polymer equilibrium can functionally cause mitotic arrest (Jordan et al., 1993). In NAP1-deficient yeast mutant, microtubules are more resistant to benomyl, which destabilizes microtubules (Kellogg and Murray, 1995). Furthermore, it was reported that in the absence of NAP1, Clb2 was unable to efficiently induce mitotic events and yeast cells underwent a prolonged delay at the short spindle stage in spite of a normal level of CDK/Clb2 kinase activity (Kellogg and Murray, 1995). The physical interaction between NAP1 and B-type cyclin, which is conserved in yeast, Xenopus, and tobacco (Kellogg et al., 1995; this study), together with our identification of tubulin as a NAP1-binding protein strongly suggests that tobacco NAP1 proteins function together with B-type cyclins in the regulation of microtubule dynamics. The localization of both plant NAP1 proteins (Dong et al., 2003) and Nicta;CYCB1;1 (Criqui et al., 2001) with mitotic spindle and phragmoplast further supports this view.
Animal NAP1 proteins are phosphorylated at multiple sites by CDK/cyclin B and CK2 kinases, and it is hypothesized that phosphorylation may alter intracellular localization and/or the stability of the NAP1 proteins (Kellogg et al., 1995; Li et al., 1999; Rodriguez et al., 2000). Using either p13SUC1-pulldown tobacco CDK/cyclin kinase fraction or purified recombinant rice CK2α kinase, we could detect phosphorylation only on Orysa;NAP1;3 but not on the other rice and tobacco NAP1 proteins. We further identified the CK2-phosphorylation site and tested its function by mutagenesis on Orysa;NAP1;3. Surprisingly, the mutant Orysa;NAP1;3 S297A protein, which is defective in phosphorylation by CK2, showed a similar pattern of intracellular localization as wild-type protein, suggesting that phosphorylation at this site had no effect on Orysa;NAP1;3 nuclear transport. Moreover, similar intensity of fluorescence was observed on transgenic cells for GFP-tagged wild type and the mutant Orysa;NAP1;3 S297A proteins, suggesting that phosphorylation also did not affect the protein stability. Precise function of this phosphorylation is currently unknown. In addition to this characterized CK2-phosphorylation site on Orysa;NAP1;3, we identified several other potential CDK/cyclin- and CK2-phosphorylation site sequences on Orysa;NAP1;3 as well as on the other plant NAP1 proteins (Fig. 5). Although these sites could not be phosphorylated by CK2α in vitro in our experimental conditions, it is not excluded that they could be phosphorylated in vivo, particularly because plants contain multiple types of CDK/cyclin and CK2 kinases (Vandepoele et al., 2002; Riera et al., 2004). Future research will be needed to resolve these different issues and it is likely that additional roles of phosphorylation in NAP1 function still await discovery.
Our studies identify a bona fide function for the C-terminal located NLS and the N-terminal located NES in the transport of Orysa;NAP1;1. Although the N-terminal located NES sequence is conserved in the other rice and tobacco NAP1 proteins (Fig. 5), only Nicta;NAP1;1 but not the other NAP1 proteins showed an NES-dependent transport in our experimental conditions. Furthermore, Orysa;NAP1;3, which does not contain any typical NLS sequences, could enter the nucleus. Together, our data suggest existence of additional but uncharacterized signals that are involved in nuclear transport of plant NAP1 proteins. Although NLS and NES sequences have yet to be characterized, human NAP1 family proteins also exhibit a complex pattern of intracellular localization. While the human NAP1 was localized exclusively in the cytoplasm (Marheineke and Krude, 1998), the human NAP1-homolog-2 protein was localized in the cytoplasm during G1 phase and onto the condensing chromatin within the nucleus during S phase (Rogner et al., 2000) and the human NAP1-homolog-4 was localized in the cytoplasm during G0/G1 phase and in the nucleus during S phase (Rodriguez et al., 1997). It is different in yeast, where only one NAP1 protein exists, which is statically localized in the cytoplasm but with ability of nucleocytoplasmic shuttling (Miyaji-Yamaguchi et al., 2003). It is possible that intracellular localization provides a means of functional specialization of the NAP1 family proteins in plants and animals.
MATERIALS AND METHODS
Production of Recombinant Proteins
The entire coding region of Nicta;CYCB1;1 cDNA (Criqui et al., 2001) was amplified by PCR using 5′-GCCATGGATAACAATAGTGTTGG-3′ and 5′-CAAGCTTATTAGTCTAAGCTAGAGAGATC-3′ primers. The PCR product was subsequently cloned between the NcoI and HindIII sites in pET-30a (Novagen), resulting in pET-30a-Nicta;CYCB1;1. To produce recombinant 6 × His-Nicta;CYCB1;1, freshly pET-30a-Nicta;CYCB1;1-transformed E. coli BL21(DE-3) cells were grown at 37°C to an A600 of 1.0, then induced by 0.1 mm isopropylthio-β-galactoside for 3 h. The bacteria were collected by centrifugation, subsequently resuspended in buffer A (20 mm Tris-Cl, 0.5 m NaCl, 0.2% Tween 20, 0.5 mm EDTA, 1 mm dithiothreitol [DTT], 1 mm phenylmethylsulfonyl fluoride [PMSF], 5% glycerol, pH 9.5), and lysed by sonication. After centrifugation at 8,000g for 20 min to remove inclusion bodies in which the majority of 6 × His-Nicta;CYCB1;1 proteins are found, the precipitate was washed twice with buffer B (buffer A in the presence of 2 m urea) and then solubilized in buffer C (buffer A in the presence of 8 m urea). After centrifugation at 20,000g for 30 min, the supernatant was loaded onto a Ni2+-Sepharose Fast Flow column (Pharmacia Biotech Europe, Freiburg, Germany), which had been treated with 0.1 m Ni2SO4 and preequilibrated with buffer D (buffer C in the absence of EDTA). After extensive wash with buffer E (buffer D in the presence of 60 mm imidazole), the retained proteins were eluted from the column with buffer F (20 mm Tris-Cl, 0.15 m NaCl, 0.3 m imidazole, 1 mm DTT, 8 m urea, pH 9.5). The eluted fraction was dialyzed against buffer G (20 mm Tris-Cl, 8 m urea, pH 9.5) and used directly to produce antibody.
To obtain soluble recombinant GST-fused Nicta;CYCB1;1 protein, the Nicta;CYCB1;1 cDNA was cloned into pGEX-KG from pET-30a-Nicta;CYCB1;1 resulting in pGEX-KG-Nicta;CYCB1;1. Freshly pGEX-KG-Nicta;CYCB1;1 transformed Escherichia coli DH5α cells were grown at 30°C to an A600 of 0.6, then induced by 0.1 mm isopropylthio-β-galactoside for 2 h. The bacteria were collected by centrifugation, resuspended in buffer 1 (1× phosphate-buffered saline [PBS], 0.1% Triton X-100, 1 mm EDTA, 1 mm DTT, 1 mm PMSF, and 5% glycerol), and lysed by sonication. After centrifugation at 20,000g for 30 min, the supernatant was loaded onto a glutathione-Sepharose-4B column (Amersham-Pharmacia Biotech, Uppsala), which had been pre equilibrated with buffer 2 (1× PBS, 1 mm DTT, and 1 mm PMSF). The column was washed first with 5 bed volumes of buffer 3 (1× PBS, 0.1% Triton X-100, 1 mm DTT, and 1 mm PMSF) then with 5 bed volumes of buffer 2. The retained GST-Nicta;CYCB1;1 was eluted from the column with buffer 4 (50 mm Tris-Cl, 10 mm glutathione, pH 7.5). After dialysis against 1× PBS, the purified GST-Nicta;CYCB1;1 protein was stored at −80°C.
Recombinant GST-fused CK2α protein was produced and purified according to the same procedure ascribed for GST-Nicta;CYCB1;1 protein. Production and purification of recombinant tobacco (Nicotiana tabacum) and rice (Oryza sativa) NAP1 proteins were as previously described (Dong et al., 2003).
Production of Antibodies and Western-Blot Analyses
The purified recombinant 6 × His-fused Nicta;NAP1;3, Nicta;NAP1;4, and Nicta;CYCB1;1 proteins were used to raise antibodies in mice. The antiserums were used at a dilution of 1:2,000 for western blot and of 1:5,000 for ELISA. Western-blot analyses were performed as described (Criqui et al., 2001).
Protein Binding Assays between Tobacco NAP1 and Nicta;CYCB1;1 Proteins
ELISA was used to check the binding of recombinant tobacco NAP1 proteins to Nicta;CYCB1;1, according to Rodriguez et al. (1997). Purified recombinant tobacco NAP1 proteins were used to coat the wells of microtiter plates (Nunc, New York). The purified recombinant GST-Nicta;CYCB1;1 was assayed at different concentrations to bind the NAP1 proteins-coated wells. The retained GST-Nicta;CYCB1;1 was determined by using the polyclonal antibody directed against 6 × His-Nicta;CYCB1;1 as the primary antibody, the horseradish peroxidase-conjugated anti-mouse antibody as the second antibody, and 3,3′,5,5′-tetramethylbenzidine as substrate. Reactions were quantitated by recording the A655 using a Bio-Rad model 450 Microplate Reader.
Pulldown Assays
Ten milligrams of purified recombinant proteins Nicta;NAP1;3 and Nicta;NAP1;4 were coupled to 2 mL of BrCN-activated Sepharose 4B resin (Amersham-Pharmacia Biotech). The resulting resin was used in pulldown assays according to a previously described protocol (Yu et al., 2004). Total protein extracts from both tobacco BY2 cells and transgenic Arabidopsis (Arabidopsis thaliana) plants expressing YFP, H2A-YFP, or H2B-YFP (Benvenuto et al., 2002; Yu et al., 2003) were used. The pulldown fractions were analyzed by western blot using specific antibodies: the anti-Nicta;NAP1;3 and −Nicta;NAP1;4 mice polyclonal antibodies at a dilution of 1:2,000, the anti-GFP rabbit polyclonal antibody (Molecular Probes, Leiden, The Netherlands) at a dilution of 1:5,000, the anti-β-tubulin mice monoclonal antibody (Amersham) at a dilution of 1:5,000, and the anti-PSTAIRE rabbit polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at a dilution of 1:2,000.
GFP-Fusion NAP1 Constructs
Construction of GFP-Nicta;NAP1;3, GFP-Nicta;NAP1;4, and GFP-Orysa;NAP1;1 under the control of the dexamethasone-inducible promoter in pTA7002 (Aoyama and Chua, 1997) has been previously described (Dong et al., 2003). GFP-Nicta;NAP1;1, GFP-Nicta;NAP1;2, GFP-Orysa;NAP1;2, and GFP-Orysa;NAP1;3 in pTA7002 were constructed through similar cloning procedures. Constructs encoding mutant proteins with NES deletion or amino acid substitutions within NLS or CK2-phosphorylation sites were obtained by the use of a mutation kit (TaKaRa Bio, Kyoto).
Tobacco Cell Culture, Transformation, and Localization of GFP-Fused NAP1 Proteins
Transgenic tobacco BY2 cells for different GFP-NAP1 fusion constructs were obtained by Agrobacterium-mediated transformation (Shen, 2001). They were maintained by weekly subculture as described for untransformed cells (Nagata et al., 1992).
Dexamethasone induction of transgene expression and microscopy analyses of transgenic tobacco cells were performed as previously described (Shen, 2001). LMB-treatment experiments were performed by adding 20 nm LMB to tobacco cells, which had been induced by 24-h incubation with dexamethasone. Effects of LMB on protein localization were examined every 5 to 10 min following 30 min to 2 h after its addition.
p13SUC1-Sepharose Affinity Binding and Kinase Assays
p13SUC1-Sepharose affinity binding was used to precipitate CDK/cyclin complex according to Bögre et al. (1997). The precipitated fraction was used to test phosphorylation of histone H1 and recombinant NAP1 proteins as described (Bögre et al., 1997; Criqui et al., 2001).
Phosphorylation of NAP1 Proteins by Recombinant CK2α
CK2α phosphorylation was performed essentially as described (Hériché et al., 1997). Briefly, 2 μg of recombinant NAP1 protein was incubated with 2 ng of purified recombinant GST-OsCK2α in 20 μL kinase buffer (50 mm Tris-Cl, 10 mm MgCl2, 0.1 m NaCl, 1 mm DTT, pH 7.5) in the presence of 5 μCi 32P-γ-ATP at 30°C for 20 min. Casein (Sigma-Aldrich Chimie, Saint Quentin Fallavier, France) was used as a control substrate. For inhibitor assays, heparin (Sigma) was added at a final concentration of 20 μg/mL to the reaction. Reaction products were separated on 12% to 15% SDS-PAGE gels. Proteins on the gel were visualized by Coomassie Brilliant Blue R250 staining and radioactive products by autoradiography.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Tobacco and rice NAP1 family genes are named according to the guide by the Commission on Plant Gene Nomenclature and are with the following sequence accession numbers: AJ438613 for Nicta;NAP1;1 (formerly named NtNAP1_L1), AJ438614 for Nicta;NAP1;2 (formerly named NtNAP1_L2), AJ438615 for Nicta;NAP1;3 (formerly named NtNAP1_L3), AJ438616 for Nicta;NAP1;4 (formerly named NtNAP1_L4), AJ438611 for Orysa;NAP1;1 (formerly named OsNAP1_L1), AJ438612 for Orysa;NAP1;2 (formerly named OsNAP1_L2), and AY830122 for Orysa;NAP1;3 (formerly named OsNAP1_L3).
Acknowledgments
We thank Professor Hai Huang (SIPPE, Shanghai, China) and Professor Leon Otten (IBMP, Strasbourg, France) for excellent comments on the manuscript.
This work was supported by the Centre National de la Recherche Scientifique (PICS no. 2391 to W.-H.S.), by the National Natural Science Foundation of China (grant no. NSF30100094), by the Scientific and Technological Council Foundation of Shanghai (grant no. 04JC14017), and by the Shanghai Rising Star Program (to A.D.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.060509.
References
- Altman R, Kellogg D (1997) Control of mitotic events by Nap1 and the Gin4 kinase. J Cell Biol 138: 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua N-H (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Asahara H, Tartare-Deckert S, Nakagawa T, Ikehara T, Hirose F, Hunter T, Ito T, Montminy M (2002) Dual roles of p300 in chromatin assembly and transcriptional activation in cooperation with nucleosome assembly protein 1 in vitro. Mol Cell Biol 22: 2974–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto G, Formiggini F, Laflamme P, Malakhov M, Bowler C (2002) The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr Biol 12: 1529–1534 [DOI] [PubMed] [Google Scholar]
- Bögre L, Zwerger K, Meskiene I, Binarova P, Csizmadia V, Planck C, Wagner E, Hirt H, Heberle-Bors E (1997) The cdc2Ms kinase is differently regulated in the cytoplasm and in the nucleus. Plant Physiol 113: 841–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criqui MC, Weingartner M, Capron A, Parmentier Y, Shen W-H, Heberle-Bors E, Bögre L, Genschik P (2001) Subcellular localization of GFP-tagged tobacco mitotic cyclins during the cell cycle and after spindle checkpoint activation. Plant J 28: 569–581 [DOI] [PubMed] [Google Scholar]
- Dong A, Xin H, Yu Y, Sun CR, Cao K, Shen W-H (2002) The subcellular localization of an unusual rice calmodulin isoform, OsCaM61, depends on its prenylation status. Plant Mol Biol 48: 203–210 [DOI] [PubMed] [Google Scholar]
- Dong A, Zhu Y, Yu Y, Cao K, Sun C, Shen W-H (2003) Regulation of biosynthesis and intracellular localization of rice and tobacco homologues of nucleosome assembly protein 1. Planta 216: 561–570 [DOI] [PubMed] [Google Scholar]
- Hériché J, Lebrin F, Rabbilloud T, Leroy D, Chambaz EM, Goldberg Y (1997) Regulation of protein phosphatase 2A by direct interaction with casein kinase 2α. Science 276: 952–955 [DOI] [PubMed] [Google Scholar]
- Ishimi Y, Hirosumi J, Sato W, Sugasawa K, Yokota S, Hanaoka F, Yamada M (1984) Purification and initial characterization of a protein which facilitates assembly of nucleosome-like structure from mammalian cells. Eur J Biochem 142: 431–439 [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Kobayashi R, Kadonaga JT (1996) Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol Cell Biol 16: 3112–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT (1997) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90: 145–155 [DOI] [PubMed] [Google Scholar]
- Ito T, Ikehara T, Nakagawa T, Kraus WL, Muramatsu M (2000) p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev 14: 1899–1907 [PMC free article] [PubMed] [Google Scholar]
- Jans DA, Hubner S (1996) Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol Rev 76: 651–685 [DOI] [PubMed] [Google Scholar]
- Jordan MA, Toso RJ, Thrower D, Wilson L (1993) Mechanism of mitotic block and inhibition of cell proliferation by taxol at low concentrations. Proc Natl Acad Sci USA 90: 9552–9556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A, Rank NM, O'Neill EM, Huang LS, O'Shea EK (1998) The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396: 482–486 [DOI] [PubMed] [Google Scholar]
- Kellogg DR, Kikuchi A, Fujii-Nakata T, Turck CW, Murray AW (1995) Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J Cell Biol 130: 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Murray AW (1995) NAP1 acts with Clb2 to perform mitotic functions and to suppress polar bud growth in budding yeast. J Cell Biol 130: 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T, Keller C (2001) Chromatin assembly during S phase: contributions from histone deposition, DNA replication and the cell division cycle. Cell Mol Life Sci 58: 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankenau S, Barnickel T, Marhold J, Lyko F, Mechler BM, Lankenau DH (2003) Knockout targeting of the Drosophila nap1 gene and examination of DNA repair tracts in the recombination products. Genetics 163: 611–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenstein ME, Kadonaga JT (2002) Biochemical analysis of chromatin containing recombinant Drosophila core histones. J Biol Chem 277: 8749–8754 [DOI] [PubMed] [Google Scholar]
- Li M, Strand D, Krehan A, Pyerin W, Heid H, Neumann B, Mechler BM (1999) Casein kinase 2 binds and phosphorylates the nucleosome assembly protein-1 (NAP1) in Drosophila melanogaster. J Mol Biol 293: 1067–1084 [DOI] [PubMed] [Google Scholar]
- Loyola A, Almouzni G (2004) Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta 1677: 3–11 [DOI] [PubMed] [Google Scholar]
- Marheineke K, Krude T (1998) Nucleosome assembly activity and intracellular localization of human CAF-1 changes during the cell division cycle. J Biol Chem 273: 15279–15286 [DOI] [PubMed] [Google Scholar]
- McQuibban GA, Commisso-Cappelli CN, Lewis PN (1998) Assembly, remodeling, and histone binding capabilities of yeast nucleosome assembly protein 1. J Biol Chem 273: 6582–6590 [DOI] [PubMed] [Google Scholar]
- Meggio F, Pinna LA (2003) One-thousand-and-one substrates of protein kinase CK2? FASEB J 17: 349–368 [DOI] [PubMed] [Google Scholar]
- Miyaji-Yamaguchi M, Kato K, Nakano R, Akashi T, Kikuchi A, Nagata K (2003) Involvement of nucleocytoplasmic shuttling of yeast Nap1 in mitotic progression. Mol Cell Biol 23: 6672–6684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S (1992) Tobacco BY-2 cell line as the “HeLa” cells in the biology of higher plants. Int Rev Cytol 132: 1–30 [Google Scholar]
- Nakagawa T, Bulger M, Muramatsu M, Ito T (2001) Multistep chromatin assembly on supercoiled plasmid DNA by nucleosome assembly protein-1 and ATP-utilizing chromatin assembly and remodeling factor. J Biol Chem 276: 27384–27391 [DOI] [PubMed] [Google Scholar]
- Ohkuni K, Shirahige K, Kikuchi A (2003) Genome-wide expression analysis of NAP1 in Saccharomyces cerevisiae. Biochem Biophys Res Commun 306: 5–9 [DOI] [PubMed] [Google Scholar]
- Porceddu A, Stals H, Reichheld JP, Segers G, De Veylder L, Barroco RP, Casteels P, Van Montagu M, Inze D, Mironov V (2001) A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J Biol Chem 276: 36354–36360 [DOI] [PubMed] [Google Scholar]
- Riera M, Figueras M, Lopez C, Goday A, Pages M (2004) Protein kinase CK2 modulates developmental functions of the abscisic acid responsive protein Rab17 from maize. Proc Natl Acad Sci USA 101: 9879–9884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P, Munroe D, Prawitt D, Chu LL, Bric E, Kim J, Reid LH, Davies C, Nakagama H, Loebbert R, et al (1997) Functional characterization of human nucleosome assembly protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics 44: 253–265 [DOI] [PubMed] [Google Scholar]
- Rodriguez P, Pelletier J, Price GB, Zannis-Hadjopoulos M (2000) NAP-2: histone chaperone function and phosphorylation state through the cell cycle. J Mol Biol 298: 225–238 [DOI] [PubMed] [Google Scholar]
- Rogner UC, Spyropoulos DD, Le Novere N, Changeux JP, Avner P (2000) Control of neurulation by the nucleosome assembly protein-1-like 2. Nat Genet 25: 431–435 [DOI] [PubMed] [Google Scholar]
- Shen W-H (2001) NtSET1, a member of a newly identified subgroup of plant SET-domain-containing proteins, is chromatin-associated and its ectopic overexpression inhibits tobacco plant growth. Plant J 28: 371–383 [DOI] [PubMed] [Google Scholar]
- Shikama N, Chan HM, Krstic-Demonacos M, Smith L, Lee C-W, Cairns W, La Thangue NB (2000) Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol Cell Biol 20: 8933–8943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Akashi T, Okuda A, Kikuchi A, Fukui K (2000) NBP1 (Nap1 binding protein 1), an essential gene for G2/M transition of Saccharomyces cerevisiae, encodes a protein of distinct sub-nuclear localization. Gene 246: 395–404 [DOI] [PubMed] [Google Scholar]
- Stals H, Inze D (2001) When plant cells decide to divide. Trends Plant Sci 6: 359–364 [DOI] [PubMed] [Google Scholar]
- Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E (1998) Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J 17: 2728–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouze P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Dogterom M, Stelzer E, Karsenti E, Leibler S (1992) Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J Cell Biol 118: 1097–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartner M, Criqui MC, Meszaros T, Binarova P, Schmit AC, Helfer A, Derevier A, Erhardt M, Bogre L, Genschik P (2004) Expression of a nondegradable cyclin B1 affects plant development and leads to endomitosis by inhibiting the formation of a phragmoplast. Plant Cell 16: 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HW, Kim MC, Lee SY, Hwang I, Bahk JD, Hong JC, Ishimi Y, Cho MJ (1995) Molecular cloning and functional characterization of a cDNA encoding nucleosome assembly protein 1 (NAP-1) from soybean. Mol Gen Genet 249: 465–473 [DOI] [PubMed] [Google Scholar]
- Yu Y, Dong A, Shen W-H (2004) Molecular characterization of the tobacco SET-domain protein NtSET1 unravels its role in histone methylation, chromatin binding and segregation. Plant J 40: 699–711 [DOI] [PubMed] [Google Scholar]
- Yu Y, Steinmetz A, Meyer D, Brown S, Shen W-H (2003) The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. Plant Cell 15: 2763–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]