Abstract
Adenosine diphosphate glucose pyrophosphorylase (AGPase) catalyzes a rate-limiting step in starch biosynthesis. The reaction produces ADP-glucose and pyrophosphate from glucose-1-P and ATP. Investigations from a number of laboratories have shown that alterations in allosteric properties as well as heat stability of this enzyme have dramatic positive effects on starch synthesis in the potato (Solanum tuberosum) tuber and seeds of important cereals. Here, we report the characterization of purified recombinant mosaic AGPases derived from protein motifs normally expressed in the maize (Zea mays) endosperm and the potato tuber. These exhibit properties that should be advantageous when expressed in plants. We also present an in-depth characterization of the kinetic and allosteric properties of these purified recombinant AGPases. These data point to previously unrecognized roles for known allosteric effectors.
Starch comprises one of the major Glc polymers in many living organisms. It is essential for plant life since it provides a mechanism to store the vast amounts of fixed carbon arising from photosynthesis. As the major component of many seeds, starch provides the important source of energy that is transferred from parent to progeny. Therefore, the rate-limiting enzyme in starch biosynthesis, ADP Glc pyrophosphorylase (AGPase), has been under much investigation. Increasing starch production by altering AGPase regulatory properties has been demonstrated in potato (Solanum tuberosum) tubers (Stark et al.,1992), maize (Zea mays; Giroux et al., 1996), wheat (Triticum aestivum; Smidansky et al., 2002), and rice (Oryza sativa) seeds (Smidansky et al., 2003).
AGPase catalyzes the formation of ADP-Glc and inorganic pyrophosphate (PPi) from ATP and Glc-1-P. In plants, AGPase is a tetramer, composed of two large and two small subunits. All plant AGPase small subunits are highly conserved, whereas the large subunits are more divergent (for review, see Hannah et al., 2001). In addition, the small and large subunits are similar in sequence and likely arose from primordial gene duplication. Although prokaryotic and eukaryotic AGPases have different quaternary structures and regulatory properties (Ballicora et al., 2003), their overall kinetic mechanisms appear to be similar (Paule and Preiss, 1971; Kleczkowski et al., 1993b).
Plant AGPases are tissue specific and are regulated by at least three different mechanisms. First, small molecules modulate AGPase activity; the most effective ones so far described are the allosteric activator 3-phosphoglyceric acid (3-PGA) and the inhibitor inorganic phosphate (Pi; Ghosh and Preiss, 1966; Dickinson and Preiss, 1969; Sowokinos, 1981; Sowokinos and Preiss, 1982; Kleczkowski et al., 1993a; Chen and Janes, 1997; Sikka et al., 2001). The degree of activation and inhibition is organism, tissue, and subcellular-localization specific. In addition to small-molecule effectors, reductive activation occurs at least for some plant AGPases in the absence of 3-PGA. This involves a Cys residue in the amino terminus of some, but not all, small subunits (Fu et al., 1998; Tiessen et al., 2002 ). A third type of regulation is thermal inactivation. Several plant AGPases are extremely resistant to heat inactivation, whereas the cereal endosperm AGPases are extremely heat labile (for review, see Greene and Hannah, 1998a, 1998b).
The significance of these forms of regulation is evident from analysis of the transgenic plants with enhanced starch synthesis. Stark et al. (1992) increased starch synthesis in the potato tuber by expressing an Escherichia coli AGPase with allosteric properties vastly different from that of the conventional potato tuber AGPase. Enhanced total starch synthesis in maize resulted from expression of an AGPase with less sensitivity to Pi (Giroux et al., 1996). An increased seed yield occurred in wheat and in rice following expression of a more heat-stable, less Pi-sensitive AGPase in the endosperm (Smidansky et al., 2002, 2003).
Despite differences in regulatory properties, active mosaic enzymes of plant AGPases have been successfully constructed using a combination of potato subunits and maize subunits in an E. coli expression system (Cross et al., 2004, 2005). In this article, we present the purification and kinetic properties of the purified recombinant maize endosperm AGPase and three maize/potato small-subunit mosaic proteins. This is the first time the wild-type enzyme expressed in E. coli has been purified to homogeneity. Each mosaic AGPase was subsequently purified and shown to be more active than the wild-type protein. One mosaic displays greater heat stability, while the other one has greater resistance to Pi inhibition. Both properties are extremely important in increasing starch development in maize endosperm.
RESULTS
Construction of Maize AGPase and Mosaic Subunits
Gene constructs analyzed here, as well as a sequence comparison between the maize and potato small subunit, are shown in Figure 1. The first mosaic, MPss, contained a maize amino acid sequence from 1 to 199 and a potato amino acid sequence from 200 to 475 (numbering convention is based on the maize amino acid sequence). Due to the highly conserved nature of the small subunits, this mosaic differs from the maize endosperm by only 34 amino acids. The second mosaic, P2M2Pss, contained amino acid residues 72 to 199 from maize and the remaining amino acids from potato. This differs from the maize small subunit at 47 amino acids. It also lacks the 25 amino-terminal amino acids of the maize small subunit. All small subunits were coexpressed with the maize large subunit. Recombinant maize AGPase expressed in E. coli contains the wild-type maize endosperm small and large subunits. Preliminary studies suggested that these mosaic AGPases had kinetic and stability properties superior to the wild-type maize AGPase. Accordingly, a detailed study, described below, was initiated.
Figure 1.
Top, Diagram of AGPase small-subunit mosaics analyzed in these studies. Maize (white) is the top subunit, while potato (black) is listed at the bottom. The mosaics are listed in the middle. Numbers between arrows (13, 9, and 34) signify the number of differing amino acids in that region. The first mosaic, MP, contains maize amino acids from position 1 to 199, while the second mosaic, P2M2P, contains maize amino acids from position 73 to 199. Maize contains 25 N-terminal amino acids not present on the potato small subunit. The numbering convention is based on the maize enzyme. The potato enzyme does not include the localization signals on the N terminus. Mosaics were expressed in E. coli with the maize endosperm large subunit, Mls. Bottom, Alignment of the maize and potato small-subunit amino acids. Sequences were aligned using the 5.4.1 Multalin version (Corpet, 1988).
Purification and Properties of Purified Recombinant Maize AGPase and AGPase Mosaics
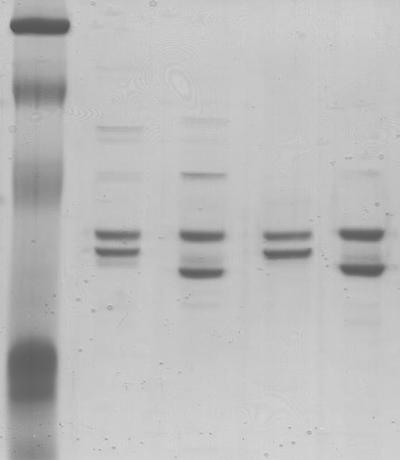
To characterize the recombinant maize and mosaic AGPases in detail, a purification procedure was developed. E. coli-expressed maize and mosaic AGPases were purified to homogeneity. Table I summarizes results obtained in a typical purification scheme. An SDS-PAGE analysis of the purified proteins from E. coli-expressed maize and mosaic AGPases is shown in Figure 2. Two major proteins are seen in each lane, corresponding to the size of the small (52 kD) and large (57 kD) subunits. Mosaic small subunits containing the N terminus from potato (Pss/Mls and P2M2Pss/Mls) are 25 amino acids smaller than the maize small subunit and hence migrate further on SDS gels (Fig. 2, lanes 3 and 5). Each enzyme preparation retained the majority of activity when stored at −80°C for several months. A vital step in the protocol was found to be the employment of protamine sulfate. Although this yielded only a 2-fold increase in activity, it was essential for enzymatic stability. This was especially critical in the latter stages of purification as the enzyme approached purity. The wild-type enzyme was purified over 50-fold, with a final specific activity of 41.4 units/mg (Table I). Mosaic AGPases were purified using similar procedures, and yields were typically greater than 25%. Increased yields may be due to enhanced stability, as described below. With the addition of an initial DEAE ion-exchange chromatography step, the purification procedure also yields purified AGPase from maize endosperm.
Table I.
Purification of maize endosperm AGPase expressed in E. colia
| Purification Step | Volume | Protein | Total | Specific Activity | Fold Purification | Total Units | % Recovery |
|---|---|---|---|---|---|---|---|
| mL | mg/mL | mg | μmol min−1 mg−1 | ||||
| Crude supernatant | 20 | 12.71 | 254.2 | 0.75 | 191 | 100 | |
| Protamine sulfate | 25 | 4.53 | 113.3 | 1.6 | 2 | 184 | 97 |
| 45% Ammonium sulfate | 1.2 | 17.57 | 21.1 | 6.5 | 8.7 | 137 | 72 |
| High Q | 18 | 0.45 | 8.2 | 11.5 | 15.3 | 94 | 44 |
| Concentrate | 2.5 | 1.61 | 4.0 | 10.9 | 14.5 | 44 | 23 |
| Hydroxyapetite | 16 | 0.07 | 1.0 | 31.9 | 42.5 | 33 | 18 |
| Concentrate | 0.15 | 3.49 | 0.5 | 41.1 | 54.9 | 21 | 11 |
Data correspond to a typical purification starting with approximately 250 mg of crude extract from E. coli. Assays were performed with assay A (reverse direction) using standard reaction conditions.
Figure 2.
Coomassie Blue-stained SDS-PAGE gel of purified maize and mosaic AGPase subunits. Lane 1, Molecular mass markers (207 kD, 129 kD, 85 kD, 39 kD); lane 2, maize AGPase; lane 3, P; lane 4, MP; lane 5, P2M2P. Approximately 2 μg of purified sample was loaded in each lane. All AGPases are expressed with the wild-type maize large subunit (SH2).
The identity of the small (52 kD) and large (57 kD) subunits was confirmed by western blots using antibodies specific for each subunit (data not shown). Only one protein was detected when the blot was probed with small- or large-subunit antibody. In addition, protein sequencing confirmed the presence of the proper N-terminal amino acids (data not shown).
Kinetic Analysis of Recombinant Maize AGPase
Since the recombinant maize AGPase has never been purified, an investigation into its kinetic properties was performed. Purified preparations were initially desalted into a HEPES buffer to remove Pi prior to kinetic analysis. Following protein quantification, bovine serum albumin (BSA) was added to a final concentration of 0.5 mg/mL to aid in enzyme stability. Preparations desalted and containing BSA were completely stable on ice for at least 8 h. BSA was especially needed to stabilize the recombinant maize AGPase during reactions performed in the absence of 3-PGA.
Initial velocity studies were carried out in the direction of ADP-Glc synthesis in the presence of 5 mm 3-PGA. Double reciprocal plots of 1/V versus 1/[ATP] at variable concentrations of Glc-1-P produced lines that intersected to the left of the ordinate axis. Likewise, double reciprocal plots of 1/V versus 1/[Glc-1-P] at variable concentrations of ATP gave rise to lines intersecting left of the ordinate axis (data not shown). All replots of the data were linear. Replots of 1/Vmax (intercept) versus 1/[Glc-1-P] yielded a Vmax of 11.6 μmol min−1 mg−1 and a Km for Glc-1-P of 0.041 mm, while replots 1/Vmax (intercept) versus 1/[ATP] yielded a Vmax of 11.8 μmol min−1 mg−1 and a Km for ATP of 0.15 mm. Similarly replots of slope 1/ATP versus 1/[Glc-1-P] and slope 1/Glc-1-P versus 1/[ATP] were linear and intersected above the horizontal axis. These kinetic parameters are similar to those reported in the literature for maize endosperm (Dickinson and Preiss, 1969; Hannah and Nelson, 1976; Fuchs and Smith, 1979; Plaxton and Preiss, 1987), Chlamydomonas reinhardtii (Iglesias et al., 1994), spinach (Spinacia oleracea) leaves (Ghosh and Preiss, 1966), potato tuber (Iglesias et al., 1993), and barley (Hordeum vulgare) endosperm (Kleczkowski et al., 1993a). Taken together, these initial velocity patterns indicate a sequential mechanism for substrate binding. A sequential mechanism has been reported for AGPases from barley leaf (Kleczkowski et al., 1993b) and the bacterium Rhodospirillum rubrum (Paule and Preiss, 1971). In addition, the closely related enzyme, Glc-1-P thymidylyltransferase, was recently shown to have a sequential mechanism through kinetic and crystallographic support (Zuccotti et al., 2001).
Kinetic Analysis of Mosaic AGPases
The Km and Vmax values were then determined for the mosaic AGPases and compared to those of the recombinant maize AGPase. The Km for ATP and Glc-1-P do not differ significantly for the mosaic enzymes in the presence of 3-PGA (Table II). The most notable difference is the 2-fold increase in Km for ATP when the potato small subunit is combined with the maize large subunit (Pss/Mls). Interestingly, the kcat value is increased for all mosaics. The kcat value for the MPss/Mls enzyme is approximately 1.4-fold greater, while the kcat values for P2M2Pss/Mls and Pss/Mls are increased by more than 2-fold. The specificity constant (kcat/Km) is also affected in the mosaic enzymes. While Mss/Mls and Pss/Mls have approximately the same specificity constant, MPss/Mls and P2M2Pss/Mls are twice as efficient as the recombinant maize endosperm enzyme. It is interesting that larger alterations in the kinetic constants are not seen in the mosaic subunits in the presence of 3-PGA, since 34 and 72 amino acid changes distinguish MPss/Mls and P2M2Pss/Mls, respectively, from recombinant maize endosperm AGPase. Most likely, this reflects the extreme conservation of catalytic residues within the potato and maize enzymes.
Table II.
Kinetic constants of wild-type AGPase and the small-subunit mosaics in the presence of 3-PGAa
| Mutant
|
ATP
|
Glc-1-P
|
||||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| mm | s−1 | m−1 s−1 | mm | s−1 | m−1 s−1 | |
| Maize AGPase | 0.12 ± 0.003 | 98 ± 1 | 8.46 × 105 ± 2.28 × 104 | 0.06 ± 0.001 | 85 ± 1 | 1.46 × 106 ± 2.98 × 104 |
| MPss/Mls | 0.10 ± 0.006 | 137 ± 2 | 1.41 × 106 ± 8.71 × 104 | 0.05 ± 0.002 | 116 ± 1 | 2.30 × 106 ± 9.41 × 104 |
| P2M2Pss/Mls | 0.13 ± 0.011 | 207 ± 5 | 1.61 × 106 ± 1.42 × 105 | 0.05 ± 0.003 | 165 ± 3 | 3.16 × 106 ± 1.98 × 104 |
| Pss/Mls | 0.26 ± 0.026 | 205 ± 7 | 7.95 × 105 ± 8.40 × 104 | 0.07 ± 0.002 | 169 ± 2 | 2.34 × 106 ± 7.24 × 104 |
Assays were performed in the forward direction (assay B) using standard reaction conditions in the presence of 10 mm 3-PGA.
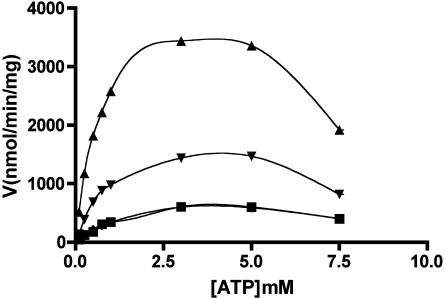
Plots of reaction rate (V) versus ATP or Glc-1-P concentration diverged from hyperbolic kinetics in the absence of 3-PGA. In addition, reaction rates were reduced by ATP concentration above 4 mm for each of the enzymes studied (Fig. 3). Similar results have been reported previously (Dickinson and Preiss, 1969). Although all mosaic and wild-type AGPase plots for reaction rate versus Glc-1-P diverged from hyperbolic kinetics, only the MPss/Mls seemed to show substrate inhibition with increasing concentrations of Glc-1-P (data not shown). Therefore, an observed maximum velocity (Vmax(obs)) and one-half saturation constant (S0.5) were calculated directly from V versus ATP or V versus Glc-1-P plots (Table III). The S0.5 for ATP was 3- to 7-fold higher and the S0.5 for Glc-1-P was 8- to 20-fold higher in the absence of 3-PGA. Interestingly, the MPss/Mls mosaic has a 3.25-fold lower S0.5 for Glc-1-P and an observed maximum velocity 5.6-fold greater than wild type in the absence of 3-PGA.
Figure 3.
Velocity versus ATP concentration in the absence of 3-PGA. Initial velocity was determined for each enzyme at varying concentrations of ATP. Shown are maize (▪), MP (▴), P2M2P (▾), and P(♦). A smooth curve was used to draw an arbitrary line through the points.
Table III.
Observed kinetic constants of wild-type AGPase and the small-subunit mosaics in the absence of 3-PGA
| Mutant
|
ATP
|
Glc-1-P
|
||||
|---|---|---|---|---|---|---|
| S0.5 | Vmax(obs) | kcat | S0.5 | Vmax(obs) | kcat | |
| mm | μmol min−1 mg−1 | s−1 | mm | μmol min−1 mg−1 | s−1 | |
| Maize AGPase | 0.84 | 1.4 | 5.1 | 1.3 | 1.6 | 5.9 |
| MPss/Mls | 0.44 | 7.9 | 30.0 | 0.4 | 8.8 | 32.3 |
| P2M2Pss/Mls | 0.60 | 3.1 | 11.4 | 0.9 | 3.5 | 12.8 |
| Pss/Mls | 0.82 | 1.3 | 4.8 | 0.9 | 1.4 | 5.1 |
3-PGA-induced-activation was then calculated from Vmax values obtained for the steady-state kinetics analysis for recombinant maize endosperm AGPase and the mosaics. The maximum velocity for each AGPase is listed in Table IV. Under the specified conditions, the recombinant maize endosperm enzyme was activated approximately 19-fold by 10 mm 3-PGA.
Table IV.
3-PGA activation of purified maize AGPase and the small-subunit mosaics
| Enzyme | Vmax (+3-PGA) | Vmax(obs) (−3-PGA) | Activation Folda |
|---|---|---|---|
| μmol min−1 mg−1 | μmol min−1 mg−1 | ||
| Maize AGPase | 26.7 | 1.4 | 19 |
| MPss/Mls | 37.3 | 7.9 | 4.7 |
| P2M2Pss/Mls | 56.6 | 3.1 | 18 |
| Pss/Mls | 55.6 | 1.3 | 43 |
The mosaic AGPases exhibited several noteworthy features. First, the observed maximum velocity of MPss/Mls in the absence of 3-PGA was 5.6-fold greater than that of the recombinant maize AGPase. Vmax values in the presence of the activator were also 1.4-fold greater than that of the recombinant maize AGPase. The activation fold (presence/absence of 3-PGA) was only 4.7 compared to the recombinant maize AGPase activation fold of 19. These data suggest that the presence of 3-PGA is not as critical for activity of this mosaic and, under limiting 3-PGA concentrations, the enzyme is more efficient than the recombinant maize AGPase. A 2-fold increase in the Vmax value in both the presence and absence of 3-PGA was seen with the P2M2Pss/Mls mosaic. Thus, the overall activation fold of this mosaic was similar to that of recombinant maize AGPase. In the absence of 3-PGA, the Pss/Mls mosaic exhibited a Vmax(obs) that was very similar to recombinant maize, but in the presence of 3-PGA it was approximately 2-fold greater. Hence, this enzyme was activated over 40-fold by 3-PGA.
Interestingly, the extent of 3-PGA activation increased steadily with enzyme purity (data not shown). In addition, desalted, purified AGPase preparations lacking BSA gave highly variable results, especially in the absence of 3-PGA. Velocities in the absence of 3-PGA were sometimes erroneously low. This led to higher apparent fold activations. It appears that 3-PGA acts not only as an activator but also as a stabilizer of activity in the absence of BSA.
We also note that concentrations of ATP above 4 mm in the absence of 3-PGA gave rise to apparent substrate inhibition (Fig. 3). This phenomenon was also reported previously (Dickinson and Preiss, 1969). Thus, true maximum velocities cannot be achieved in the absence of 3-PGA for the recombinant maize enzyme. These data may help explain reported discrepancies concerning 3-PGA activation of the maize endosperm enzyme (Dickinson and Preiss, 1969; Plaxton and Preiss, 1987).
Regulatory Properties of Recombinant Maize AGPase
To investigate the in vivo regulation of purified recombinant maize AGPase, different metabolites were analyzed in vitro. Specific activity was measured in the direction of ADP-Glc synthesis since it is the most physiologically relevant reaction and is more affected by allosteric activators. As seen in Table V, 3-PGA, Fru-6-P, and Glc-6-P stimulated the purified recombinant maize AGPase over 10-fold. This is similar to the increase found by Plaxton and Preiss (1987) for the partially purified maize endosperm enzyme, but the activation fold was much greater than that reported by Dickinson and Preiss (3- and 1.8-fold increases with Fru-6-P and Glc-6-P, respectively; Dickinson and Preiss, 1969). The data in Table V show that the combination of lactate with Pi or sugars and Pi, although similar in structure to 3-PGA, did not stimulate the activity. All reactions were performed in the presence of BSA to stabilize the enzyme.
Table V.
Activation of purified maize AGPase by various metabolitesa
| Compound | Stimulation Fold |
|---|---|
| Fru-6-P | 15 |
| Glc-6-P | 12 |
| 3-PGA | 16 |
| PO4− | 0.34 |
| Pi + Fru | 0.34 |
| Pi + Glc | 0.31 |
| Pi + sodium lactate | 0.34 |
| Pi + sodium acetate | 0.34 |
| Fru | 1.3 |
| Glc | 1.3 |
| Sodium lactate | 1.2 |
| Sodium acetate | 1.1 |
| No additive | – |
Assays were performed in the forward reaction (assay B) under standard reaction conditions. Each metabolite was added at a final concentration of 25 mm, except 3-PGA, which was added at 10 mm. Stimulation fold was calculated by dividing the activity in the presence of the metabolites by the activity in its absence.
The activation constant (Ka) for 3-PGA was determined for each of the mosaics. These constants are reported in Table VI. The Ka values for recombinant maize and Pss/Mls are 0.17 and 0.25 mm, respectively; the values for MPss/Mls and P2M2Pss/Mls are significantly less (0.03 and 0.06 mm, respectively). These data suggest that the mosaic enzymes are much more sensitive to 3-PGA and may be more efficient at lower 3-PGA concentrations.
Table VI.
Ka for 3-PGA and Ki for Pi of purified maize AGPase and the small-subunit mosaicsa
| Enzyme | Ka 3-PGA | Ki Pi |
|---|---|---|
| mm | mm | |
| Maize AGPase | 0.17 ± 0.015 | 2.96 ± 0.12 |
| MPss/Mls | 0.03 ± 0.008 | 12.28 ± 0.73 |
| P2M2Pss/Mls | 0.06 ± 0.007 | 2.99 ± 0.19 |
| Pss/Mls | 0.25 ± 0.032 | 0.68 ± 0.12 |
Assays were performed with assay B (forward direction) using standard reaction conditions.
The apparent inhibition constants (Ki app) for phosphate were then determined by varying the Pi concentration at a 2.5 mm 3-PGA. Phosphate inhibition is an important parameter in controlling starch synthesis in the maize endosperm, and understanding these kinetic parameters in vitro may aid enhancement of starch content in the endosperm. Accordingly, the Ki app (Pi level causing 50% reduction in activity) was measured with purified AGPase preparations in the presence of 2.5 mm 3-PGA (Table VI). In the absence of 3-PGA, the purified enzymes are not significantly inhibited by Pi, even at concentrations as high as 5.0 mm. In fact, small amounts of Pi actually stimulate the activity in the absence of 3-PGA. This is in contrast to barley leaf and spinach leaf chloroplast AGPases whereby Pi strongly inhibits the enzyme in the absence of 3-PGA (Ghosh and Preiss, 1966; Kleczkowski et al., 1993a). Furthermore, the addition of 3-PGA alone does not activate the wheat endosperm enzyme, but only derepresses inhibition caused by Pi (Gomez-Casati and Iglesias, 2002). By contrast, the addition of 2.5 mm 3-PGA causes Pi to inhibit recombinant maize AGPase activity with one-half maximal inhibition occurring at 2.96 mm. However, higher concentrations of 3-PGA greatly reduce the inhibition by Pi. It is interesting to note that the enzyme activity in the presence of 3-PGA and at high concentrations of Pi approaches the activity of the enzyme in the absence of 3-PGA. Thus, the inhibition by Pi brings the activity down to a baseline level, negating the activation caused by 3-PGA. This is evident in each of the mosaic enzymes as well as the wild-type AGPase. Interaction between 3-PGA and Pi has been noted earlier (Ghosh and Preiss, 1966; Plaxton and Preiss, 1987). A significant finding is that the mosaic MPss/Mls is significantly less sensitive to Pi at this particular 3-PGA concentration. Perhaps the insensitivity of MPss/Mls can be explained by its much greater affinity for 3-PGA, which more efficiently minimizes the effect of Pi.
Temperature Sensitivity
As reviewed earlier, temperature stress is a major environmental factor that greatly reduces grain yield in many cereal crops such as maize, wheat, and barley. Yield decreases due to a heat stress range from 7% to 35% in the cereals of worldwide importance. Since the maize endosperm AGPase is heat labile, and potato tuber AGPase is heat stable, it was hypothesized that these small-subunit mosaics may have increased heat stability. Preliminary experiments with partially purified preparations revealed substantial differences in heat stability. Therefore, purified preparations were used to determine enzymatic half-life at 42°C. The half-life for the recombinant maize endosperm AGPase was shown to be 4.5 min while that of MPss/Mls was approximately 18 min. Interestingly, the P2M2Pss/Mls and Pss/Mls mosaics had half-lives greater than 3 h at this temperature. Therefore, the half-life was then repeated at an increased temperature of 55°C (Table VII). Recombinant maize endosperm AGPase is very sensitive to this high temperature, with a half-life of less than 0.5 min. The MPss/Mls mosaic was slightly more stable, with a half-life of 1.4 min. The P2M2Pss/Mls and Pss/Mls mosaics had half-lives far exceeding that of recombinant maize: 27.3 and 19.3 min, respectively. Sequences common to the latter two mosaics and different from the former two AGPases lie in the N terminus of the small subunit. Hence, nonconserved amino acid(s) in the small-subunit N terminus plays a critical role in determining the heat stability of AGPase. Clearly, P2M2Pss/Mls has superior heat stability and may be useful in commercialization of heat-stable AGPase.
Table VII.
Heat stability of purified maize AGPase and the small-subunit mosaicsa
| Enzyme | t1/2 42°C | t1/2 55°C |
|---|---|---|
| min | min | |
| Maize AGPase | 4.5 | 0.35 |
| MPss/Mls | 18 | 1.4 |
| P2M2Pss/Mls | >180 | 27.4 |
| Pss/Mls | >180 | 19.3 |
Assays were performed using assay B (forward direction). Each enzyme was desalted into 50 mm HEPES, pH 7.4, 5 mm MgCl2, 0.5 mm EDTA at a protein concentration of 0.01 mg/mL, followed by the addition of BSA (0.5 mg/mL). Data were plotted as log % activity versus time, and the inactivation constant t1/2 was calculated from the slope of the straight line.
Blue Native Gel Electrophoresis
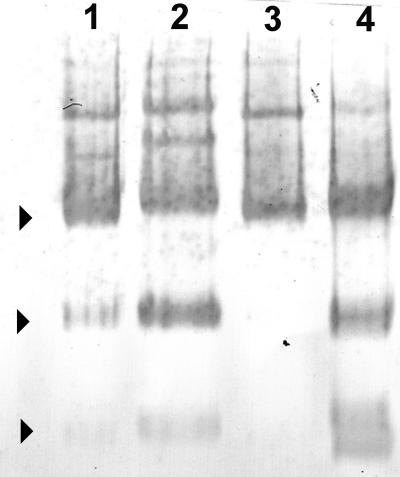
To determine whether differences in heat stability were associated with the aggregation state of AGPase, blue native gel electrophoresis (BN-PAGE) was performed on purified recombinant maize and mosaic AGPases to identify the aggregation states of each AGPase complex. This technique, originally designed to determine oligomeric states of large protein complexes, was modified here to identify aggregation states of nondenatured maize and mosaic AGPases. Since AGPase is water soluble, the protein solubilizer, aminocaproic acid, was removed from the standard gel running buffer. Its presence also caused a greater preponderance of monomers and dimers arising from the recombinant maize AGPase.
Three oligomeric states of AGPase were detected with the small-subunit polyclonal antibody (Fig. 4). Sizes corresponded to the monomeric, dimeric, and tetrameric forms of AGPase. The tetrameric state was most abundant. Several higher molecular mass species were also detected. Recombinant maize AGPase displayed all three oligomeric states, whereas the oligomeric structure of MPss/Mls is exclusively tetrameric and larger. This may imply that this mosaic forms a more stable tetramer and may be the reason for greater purification yields. Like recombinant maize, the recombinant potato and P2M2Pss/Mls AGPases exhibit several oligomeric states. Large-subunit polyclonal antibodies detected all aggregation states described above (data not shown). Interestingly, active maize endosperm AGPase of greater than 400 kD has been reported previously (Hannah and Nelson, 1975).
Figure 4.
BN-PAGE exhibiting multimeric states of AGPase. The gel was electroblotted and developed with polyclonal antibody raised against BT2. Arrows indicate the monomer, dimer, and tetrameric states of AGPase. All small subunits were expressed with the maize large subunit. Lane 1, Pss; lane 2, P2M2Pss; lane 3, MPss; lane 4, Mss.
DISCUSSION
Here, we report a purification protocol that routinely yields purified recombinant maize endosperm AGPase. Heretofore, problems of enzyme stability, particularly in dilute solution and accompanying low yields, have prevented purification to homogeneity. Two steps proved essential in successfully purifying AGPase. First, incorporation of a protamine sulfate-binding step early in the purification enhanced stability in later stages of purification. Second, it was found that BSA was needed to overcome enzyme instability following salt removal for kinetic analysis.
A comparison of kinetic and allosteric parameters of partially and totally purified AGPases revealed some interesting similarities and differences. While the differences in Pi inhibition noted here for purified recombinant maize and mosaic AGPases had also been observed for partially purified enzyme purifications (Cross et al., 2004), clear differences in 3-PGA activation are evident. Partially purified maize AGPase expressed in E. coli exhibited a 5-fold activation by 3-PGA (Cross et al., 2004). By contrast, we observed a 19-fold activation using the purified recombinant maize AGPase. A variety of different levels of 3-PGA activation have been reported for the maize endosperm enzyme (Dickinson and Preiss, 1969; Hannah and Nelson, 1976; Plaxton and Preiss, 1987). From our characterization of both partially and purified AGPases, we envisage at least three possibilities to account for these differences. First, purified AGPase is unstable, particularly in the absence of 3-PGA. Hence, velocity measurements in the absence of 3-PGA are underestimated due to loss of activity during the assay. While the presence of BSA reduces the instability, BSA-induced restoration of stability may not be complete. Second, we noted apparent substrate inhibition when AGPase is assayed in the absence of 3-PGA. Hence, the true turnover number is likely underestimated in the absence, but not in the presence, of 3-PGA. Third, we cannot exclude the possibility that 3-PGA is removed from the recombinant maize AGPase during purification or the possibility that the activity state of the enzyme induced by 3-PGA is lost during purification. In addition, removal of other proteins or enzymes may alter these allosteric parameters. Future experiments should shed light on these possibilities.
A comparison of the various AGPases not only delimits sequences important in each of the parameters but also highlights the complexity of the subunit interactions. For example, enhanced turnover number in the presence of the activator 3-PGA involves the substitution of potato sequences for the first 72 N-terminal maize small-subunit amino acids as seen in Pss/Mls and P2M2Pss/Mls. By contrast, enhanced turnover numbers in the absence of the activator involve substitution of potato sequences for maize in the distal amino acids of the small subunit from position 199 to 475 as seen in MPss/Mls. In addition, this enhanced small subunit must contain at least one maize-derived polymorphic amino acid from position 72 to 199. Surprisingly, the region with the greatest polymorphism density (position 1–72) as well as the Cys involved in redox regulation of potato (Fu et al., 1998; Tiessen et al., 2002) has no effect on kcat in the absence of 3-PGA. In addition, MPss/Mls is much less inhibited by Pi compared to the two parental small subunits. Because the mosaic differs from both parental small subunits, at least two important polymorphic sites must be involved. One site lies proximal to 199, while another site must be distal to 199. Because Pi inhibition of P2M2Pss/Mls is equal to that of potato, the polymorphic proximal site involved in Pi inhibition lies proximal to position 72.
A variant with heat stability enhanced almost 100-fold with respect to recombinant maize AGP was also identified. As reviewed above, this involves amino acids in the N terminus of the small subunit. Like a heat-stable variant described earlier in the large subunit (Greene and Hannah, 1998b), enhanced heat stability in these small-subunit variants also involves alterations in the aggregation states of the enzyme. Whether the enhanced heat stability reported here involves a new disulfide bridge (Fu et al., 1998; Tiessen et al., 2002) is a distinct possibility. Perhaps enhanced electrostatic interactions, as shown recently at the dimer-dimer interface in the tetrameric malate dehydrogenase (Bjork et al., 2004), are more prevalent in the interface of this enzyme, making it less susceptible to heat inactivation.
The allosteric effectors 3-PGA and Pi apparently have roles other than activation and inhibition. While 3-PGA is classically considered an activator of AGPase and Pi is considered an inhibitor, studies reported here point to other roles for these molecules as well. First, we note that Pi does not inhibit purified AGPase unless the enzyme is activated by 3-PGA. Accordingly, it may be more appropriate to consider Pi a modulator of 3-PGA activation, rather than an inhibitor of AGPase. A detailed investigation of the role of Pi is currently under investigation for maize AGPase. We presently envisage at least two mechanisms by which Pi insensitivity could mutationally arise. First, the Kd for Pi could be increased by mutation in the binding site, thus weakening the direct binding of Pi to the enzyme. Second, if Pi and 3-PGA share a common binding site, as the data above suggest, then enhanced 3-PGA binding might condition decreased Pi binding. Interestingly, results from analysis of the MPss/Mls mosaic are in agreement with this model. While the Ki for 3-PGA is 5-fold lower than recombinant maize, the Ki for Pi is 5-fold higher than maize. Salamone et al. (2002) isolated several mutants of a small-subunit, homotetrameric recombinant potato AGPase that had increased affinity for 3-PGA and reduced affinity for Pi. The activator-deactivator (PGA/Pi) balance is likely a way to control starch synthesis in vivo. An AGPase with altered activation-deactivation properties may be very useful in starch biosynthesis in planta.
Finally, we note that the AGPase variants described here likely are agriculturally useful. As reviewed earlier, plant AGPases are highly regulated enzymes, and variants affecting Pi inhibition and thermal stability have proven to be important in starch content and in yield. Here, we report AGPases with regulatory properties that are superior to those found in the maize endosperm. Enhanced parameters include turnover number in the presence and in the absence of 3-PGA, extent of 3-PGA activation, Pi inhibition, and heat stability. Future efforts will determine whether these have relevance in agriculture.
MATERIALS AND METHODS
Plasmids and Bacterial Strains
Construction of the small-subunit mosaics was performed by Cross et al. (2004, 2005). Construct MP contains the first 199 amino acids from the maize (Zea mays) small subunit and the remaining amino acids from the potato (Solanum tuberosum) small subunit. The mosaic P2M2P contains potato sequences from 1 to 72 and 100 to 475. The remaining sequence is from the maize small subunit. Mosaics were verified by sequence analysis.
Growth and Assay Conditions for Recombinant Maize and Mosaic AGPases
Escherichia coli AC70R1-504 (Iglesias et al., 1993) was transformed with the plasmids of interest, and the cells were stored as glycerol stocks. Expression vectors and conditions for growth and expression were performed according to Burger et al. (2003), with the following modifications. Induction of cells was performed for 3 h at room temperature. Following cell harvest by centrifugation, cell pellets were stored at −80°C.
Assay A (Reverse Direction)
A nonradioactive endpoint assay was used to determine the amount of Glc-1-P produced by coupling its formation to NADH production using phosphoglucomutase and Glc-6-P dehydrogenase (Sowokinos, 1976). The temperature of all the assays was 37°C unless otherwise specified. Standard reaction mixtures contained 100 mm MOPS-HCl, pH 7.4, 0.4 mg/mL BSA, 5 mm MgCl2, 1 mm ADP-Glc, 20 mm 3-PGA, 1 mm sodium pyrophosphate, and enzyme in a 100-μL reaction volume. Assay tubes were prewarmed to 37°C prior to the reactions, which were initiated by enzyme addition. Reactions were incubated at 37°C for 5 min and terminated by boiling in a water bath for 1 min. After termination, the amount of NADH was determined by the addition of a development mixture yielding a final concentration of 100 mm MOPS-HCl, pH 7.4, 0.1 mg/mL BSA, 7 mm MgCl2, 0.6 mm NAD, 1 unit Glc-6-P dehydrogenase, and 1 unit phosphoglucomutase in a 500-μL volume. Reactions were clarified by a 5-min centrifugation and the absorbance was determined at 340 nm. The amount of Glc-1-P produced was determined from a standard curve using freshly prepared Glc-1-P in complete reaction mixtures and omitting enzyme. Specific activity is defined as a unit/mg protein, whereby 1 unit is defined as 1 μmol/min. Protein assays were performed using the Bio-Rad protein reagent (Bio-Rad Laboratories, Hercules, CA), with BSA as a standard. Purification was always monitored using the reverse assay.
Assay B (Forward Reaction)
A nonradioactive endpoint assay was used to determine the amount of PPi produced by coupling it to a decrease in NADH concentration using PPi reagent (P-7275; Sigma, St. Louis). Standard reaction mixtures contained 50 mm HEPES, pH 7.4, 15 mm MgCl2, 4.0 mm ATP, and 4.0 mm Glc-1-P in a total volume of 200 μL. When 3-PGA was added to the reaction, its concentration is specified in tables or figure legends. Reaction tubes were prewarmed to 37°C and assays were initiated by enzyme addition. Reactions were performed at 37°C and were terminated by boiling for 1 min. The reactions were developed by adding 300 μL of PPi reagent (1 bottle dissolved in 22.5 mL of water) to each tube and determining the A340. Blank samples contained complete reaction mixtures without enzyme. The amount of PPi produced was determined from a standard curve using freshly prepared PPi in complete reaction mixtures and omitting enzyme. The change in absorbance between the blank and the reaction was used to calculate the amount of PPi. Reactions were linear with time and enzyme concentration.
Purification of AGPase
All steps were performed at 4°C and centrifugations were at 30,000g unless otherwise stated. Cell pellets from 5 L of E. coli were extracted in 20 mL of buffer A (50 mm KH2PO4, pH 7.0, 5 mm MgCl2, 0.5 mm EDTA), lysozyme (50 μg/mL), and protease inhibitors (1 μg/mL pepstatin, 0.1 mm PMSF, 10 μg/mL chymostatin, and 1 mm benzamidine). Cells were thawed on ice and sonicated until cell lysis was complete. The extract was clarified by centrifugation for 20 min. Three-tenths volume of a 1% protamine sulfate solution was then added and the mixture was stirred on ice for 20 min. Following a 20-min centrifugation, the supernatant was brought to 30% saturation with solid ammonium sulfate and allowed to stir on ice for 20 min. The solution was centrifuged for 20 min and the supernatant saved. The extract was then brought to 45% saturation with solid ammonium sulfate, stirred on ice for 20 min, centrifuged for 20 min, and the pellet saved. The pellet was resuspended in buffer A and stored at −80°C.
Approximately 13 mg of protein were desalted over a PD-10 column (Amersham Biosciences, Buckinghamshire, UK) equilibrated in buffer A. The desalted protein extract was then loaded onto a 5-mL strong anion-exchange column (macro-prep high Q support; Bio-Rad) equilibrated with buffer A at a flow rate of 1 mL/min. The column was washed in buffer A, followed by a gradient of 0% to 60% buffer B (buffer A + 0.5 m KCl) over 50 min. The gradient was followed by a 10-min extension at 60% buffer B. The most active fractions were pooled and concentrated in a Centricon-plus 20 concentrator, molecular weight cutoff 100,000 (Amicon, Toronto). The preparation was diluted 1:5 with cold water to decrease the Pi concentration and directly loaded onto two tandemly connected 5-mL econo-pac hydroxyapetite cartridges (Bio-Rad). This column was equilibrated in buffer C (10 mm KH2PO4, pH 7.0) at a flow rate of 1 mL/min. Unbound protein was eluted, followed by a wash step at 25% buffer D (400 mm KH2PO4, pH 7). A linear gradient from 25% to 80% buffer D was then applied to the column, followed by a 20-min extension at 80% buffer D. Fractions containing the highest specific activity during the linear gradient were combined and concentrated for further use. Concentrated, purified proteins were stored at −80°C for many months without appreciable loss of activity.
Prior to kinetic analysis, proteins were desalted using protein-desalting spin columns according to the manufacturer's instructions (Pierce, Rockford, IL). Proteins were routinely exchanged into 50 mm HEPES, 5 mm MgCl2, 0.5 mm EDTA.
SDS-PAGE and Electroblotting for N-Terminal Sequencing
Purified proteins were run on a 10% SDS-PAGE gel and transferred to a PVDF membrane using standard protocols. Transfer buffer for protein sequencing was 10 mm MES, pH 6, 20% MeOH. Gels were transferred for 1 h at 90 V. Membranes were washed in distilled water, stained for 30 s (0.02% Coomassie Blue in 40% methanol, 5% acetic acid), and destained (40% methanol, 5% acetic acid) for 1 min. Membranes were rinsed in distilled water and sequenced on an ABI (Sunnyvale, CA) protein sequencer (University of Florida Interdisciplinary Center for Biotechnology Research [ICBR] Protein Chemistry Core Facility).
Initial Velocity Reactions
Assay B was used for all initial velocity experiments. Reaction mixtures contained 50 mm HEPES, pH 7.0, 15 mm MgCl2, 5 mm 3-PGA, and varying concentrations of ATP (0.03–0.75 mm) at several fixed concentrations of Glc-1-P (0.03–0.75 mm) in a 300-μL volume. All reactions were initiated by the addition of 0.065 μg enzyme. Reactions proceeded at 37°C for 10 min and were terminated by boiling in a water bath for 1 min. Reactions were developed as described above.
Determination of the Kinetic Constants
All assays were performed using assay B (forward direction) using standard reaction conditions. Affinity constants (Km) for AGPase substrates were determined by incubating purified AGPase in reaction mixtures in which one substrate was held constant and saturating, while the other one was varied. All assays contained 50 mm HEPES, pH 7.4, 15 mm MgCl2, 10 mm 3-PGA. When ATP or Glc-1-P was varied, the concentration ranges used were 0.025 to 3.0 mm. When ATP or Glc-1-P was held constant, the concentration used was 4.0 mm. Assays were started with 0.04 μg of purified enzyme, incubated 10 min at 37°C, and inactivated by boiling for 2 min. The reactions were performed as above, except 0.4 μg of AGPase was used to determine S0.5 in the absence of 3-PGA. Kinetic constants were obtained by nonlinear regression using equations derived from the full kinetic expression using the Prism software program (Graph Pad, San Diego, CA).
Effect of Different Metabolites on the Activity of Purified AGPase
All assays were performed in the forward reaction (assay B) under standard reaction conditions. Reactions were started with 0.65 or 0.065 μg protein (+3-PGA, Fru-6-P, and Glc-6-P) of enzyme, incubated for 15 min at 37°C, and terminated by boiling for 1 min. Samples were run in duplicate. Each effector compound was added at a final concentration of 25 mm with the exception of 3-PGA, which was added at 10 mm.
Determination of the Ka and Ki
All assays were performed using assay B (forward direction) employing standard reaction conditions. 3-PGA was added at varying amounts, from 0 to 5 mm, to determine the Ka. The Ki for Pi was determined in the presence of 2.5 mm 3-PGA. Kinetic constants were obtained by nonlinear regression using equations derived from the full kinetic expression using the Prism software program (Graph Pad).
Heat Stability of Purified Recombinant Maize AGPase and the Small-Subunit Mosaics
All assays were performed using assay B (forward direction). Each enzyme was desalted into 50 mm HEPES, pH 7.4, 5 mm MgCl2, 0.5 mm EDTA at a concentration of 0.0043 mg protein/mL. BSA was then added to 0.5 mg/mL. Preparations were placed in a water bath at 55°C for varying times and then cooled on ice. All preparations were assayed for 10 min in the forward direction in the presence of 10 mm 3-PGA. Reactions were started with 0.017 μg of enzyme. Data were plotted as log % activity versus time, and the inactivation constant t1/2 was calculated as follows: slope = −k/(2.3); t1/2 is calculated from the equation k = 0.693/t1/2.
BN-PAGE Gel Electrophoresis
All buffers were prepared according to the procedure outlined by Schagger et al. (1994), with the following modifications. Linear gradient separating gels (5%–18% acrylamide) were prepared and could be stored for up to 1 week at 4°C. Stacking gels were prepared daily. The 3× gel buffer was prepared in the presence or absence of aminocaproic acid and used as noted in the figure. Two different cathode buffers were utilized; cathode buffer A contained 0.002% Serva Blue G, while cathode buffer B lacked Serva Blue G. The following modifications were made when gels were to be transferred. Gels were run at 100 V for 20 min at 4°C using cathode buffer A, and the gel tank was then emptied and refilled with cathode buffer B. The voltage was set at 200 V and run until the dye front had migrated to the bottom of the gel. The gel was then soaked in standard western transfer buffer (20% methanol, including 1% SDS) for 30 min, followed by a standard western transfer using immunoblot PVDF membranes (0.2 μm pore size; Bio-Rad). The electrophoresis calibration kit (Amersham-Pharmacia Biotech, Uppsala) containing the proteins thyroglobulin (669 kD), ferritin (440 kD), catalase (232 kD), lactate dehydrogenase (140 kD), and BSA (67 kD) was used for molecular mass standards. All BN-PAGE gels were developed with polyclonal antibodies raised against the maize AGPase small subunit (BT2).
This work was supported by the National Science Foundation (grant nos. IBN–9316887, IBN–960416, IBN–9982626, IBN–0444031, and MCB–9420422 to L.C.H.), the U.S. Department of Agriculture Competitive Grants Program (grant nos. 94–37300–453, 95–00836, 95–37301–2080, 97–01964, 97–36306–4461, 98–01006, and 2000–01488 to L.C.H.), and the Florida Agricultural Experiment Station (Journal Series No. R–10624).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.060699.
References
- Ballicora MA, Iglesias AA, Preiss J (2003) ADP-glucose pyrophosphorylase, a regulatory enzyme for bacterial glycogen synthesis. Microbiol Mol Biol Rev 67: 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork A, Dalhus B, Mantzilas D, Sirevag R, Eijsink VG (2004) Large improvement in the thermal stability of a tetrameric malate dehydrogenase by single point mutations at the dimer-dimer interface. J Mol Biol 341: 1215–1226 [DOI] [PubMed] [Google Scholar]
- Burger BT, Cross JM, Shaw JR, Caren JR, Greene TW, Okita TW, Hannah LC (2003) Relative turnover numbers of maize endosperm and potato tuber ADP-glucose pyrophosphorylases in the absence and presence of 3-phosphoglyceric acid. Planta 217: 449–456 [DOI] [PubMed] [Google Scholar]
- Chen BY, Janes HW (1997) Multiple forms of ADP-glucose pyrophosphorylase from tomato fruit. Plant Physiol 113: 235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchal clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross JM, Clancy M, Shaw JR, Boehlein SK, Greene TW, Schmidt RR, Okita TW, Hannah LC (2005) A polymorphic motif in the small subunit of ADP-glucose pyrophosphorylase modulates interactions between the small and large subunits. Plant J 41: 501–511 [DOI] [PubMed] [Google Scholar]
- Cross JM, Clancy M, Shaw JR, Greene TW, Schmidt RR, Okita TW, Hannah LC (2004) Both subunits of ADP-glucose pyrophosphorylase are regulatory. Plant Physiol 135: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DB, Preiss J (1969) ADP glucose pyrophosphorylase from maize endosperm. Arch Biochem Biophys 130: 119–128 [DOI] [PubMed] [Google Scholar]
- Fu Y, Ballicora MA, Leykam JF, Preiss J (1998) Mechanism of reductive activation of potato tuber ADP-glucose pyrophosphorylase. J Biol Chem 273: 25045–25052 [DOI] [PubMed] [Google Scholar]
- Fuchs RL, Smith JD (1979) The purification and characterization of ADP-glucose pyrophosphorylase A from developing maize seeds. Biochim Biophys Acta 566: 40–48 [DOI] [PubMed] [Google Scholar]
- Ghosh HP, Preiss J (1966) Adenosine diphosphate glucose pyrophosphorylase. A regulatory enzyme in the biosynthesis of starch in spinach leaf chloroplasts. J Biol Chem 241: 4491–4504 [PubMed] [Google Scholar]
- Giroux MJ, Shaw J, Barry G, Cobb BG, Greene T, Okita T, Hannah LC (1996) A single mutation that increases maize seed weight. Proc Natl Acad Sci USA 93: 5824–5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Casati DF, Iglesias AA (2002) ADP-glucose pyrophosphorylase from wheat endosperm. Purification and characterization of an enzyme with novel regulatory properties. Planta 214: 428–434 [DOI] [PubMed] [Google Scholar]
- Greene TW, Hannah LC (1998. a) Maize endosperm ADP-glucose pyrophosphorylase SHRUNKEN2 and BRITTLE2 subunit interactions. Plant Cell 10: 1295–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene TW, Hannah LC (1998. b) Enhanced stability of maize endosperm ADP-glucose pyrophosphorylase is gained through mutants that alter subunit interactions. Proc Natl Acad Sci USA 95: 13342–13347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah LC, Nelson OE (1975) Characterization of adenosine diphosphate glucose pyrophosphorylase from developing maize seeds. Plant Physiol 55: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah LC, Nelson OE Jr (1976) Characterization of ADP-glucose pyrophosphorylase from shrunken-2 and brittle-2 mutants of maize. Biochem Genet 14: 547–560 [DOI] [PubMed] [Google Scholar]
- Hannah LC, Shaw JR, Giroux MJ, Reyss A, Prioul JL, Bae JM, Lee JY (2001) Maize genes encoding the small subunit of ADP-glucose pyrophosphorylase. Plant Physiol 127: 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias AA, Barry GF, Meyer C, Bloksberg L, Nakata PA, Greene T, Laughlin MJ, Okita TW, Kishore GM, Preiss J (1993) Expression of the potato tuber ADP-glucose pyrophosphorylase in Escherichia coli. J Biol Chem 268: 1081–1086 [PubMed] [Google Scholar]
- Iglesias AA, Charng YY, Ball S, Preiss J (1994) Characterization of the kinetic, regulatory, and structural properties of ADP-glucose pyrophosphorylase from Chlamydomonas reinhardtii. Plant Physiol 104: 1287–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski LA, Villand P, Luthi E, Olsen OA, Preiss J (1993. a) Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate regulation. Plant Physiol 101: 179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski LA, Villand P, Preiss J, Olsen OA (1993. b) Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves. J Biol Chem 268: 6228–6233 [PubMed] [Google Scholar]
- Paule MR, Preiss J (1971) Biosynthesis of bacterial glycogen. X. The kinetic mechanism of adenosine diphosphoglucose pyrophosphorylase from Rhodospirillum rubrum. J Biol Chem 246: 4602–4609 [Google Scholar]
- Plaxton WC, Preiss J (1987) Purification and properties of nonproteolytic degraded ADP glucose pyrophosphorylase from maize endosperm. Plant Physiol 83: 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamone P, Kavakli I, Slattery C, Okita T (2002) Directed molecular evolution of ADP-glucose pyrophosphorylase. Proc Natl Acad Sci USA 99: 1070–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, Cramer WA, von Jagow G (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217: 220–230 [DOI] [PubMed] [Google Scholar]
- Sikka VK, Choi S, Kavakli IH, Sakulsingharoj C, Gupta S, Ito H, Okita TW (2001) Subcellular compartmentation and allosteric regulation of the rice endosperm ADP glucose pyrophosphorylase. Plant Sci 161: 461–468 [Google Scholar]
- Smidansky ED, Clancy M, Meyer FD, Lanning SP, Blake NK, Talbert LE, Giroux MJ (2002) Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc Natl Acad Sci USA 99: 1724–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidansky ED, Martin JM, Hannah LC, Fischer AM, Giroux MJ (2003) Seed yield and plant biomass increases in rice are conferred by deregulation of endosperm ADP-glucose pyrophosphorylase. Planta 216: 656–664 [DOI] [PubMed] [Google Scholar]
- Sowokinos JR (1976) Pyrophosphorylases in Solanum tuberosum. I. Changes in ADP glucose and UDP glucose pyrophosphorylase activities associated with starch biosynthesis during tuberization, maturation, and storage of potatoes. Plant Physiol 57: 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowokinos J (1981) Pyrophosphorylases in Solanum tuberosum. II. Catalytic properties and regulation of ADP-glucose and UDP-glucose pyrophosphorylase activities in potatoes. Plant Physiol 68: 924–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowokinos J, Preiss J (1982) Pyrophosphorylases in Solanum tuberosum. III. Purification, physical, and catalytic properties of ADP glucose pyrophosphorylase in potatoes. Plant Physiol 69: 1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark D, Timmerman K, Barry G, Preiss J, Kishore G (1992) Regulation of the amount of starch in plant tissues by ADP glucose pyrophosphorylase. Science 258: 287–291 [DOI] [PubMed] [Google Scholar]
- Tiessen A, Hendriks JH, Stitt M, Branscheid A, Gibon Y, Farre EM, Geigenberger P (2002) Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14: 2191–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccotti S, Zanardi D, Rosano C, Sturla L, Tonetti M, Bolognesi M (2001) Kinetic and crystallographic analyses support a sequential-ordered bi bi catalytic mechanism for Escherichia coli glucose-1-phosphate thymidylyltransferase. J Mol Biol 313: 831–843 [DOI] [PubMed] [Google Scholar]