Abstract
A basic β-galactosidase with high specificity toward β-(1→3)- and β-(1→6)-galactosyl residues was cloned from radish (Raphanus sativus) plants by reverse transcription-PCR. The gene, designated RsBGAL1, contained an open reading frame consisting of 2,532 bp (851 amino acids). It is expressed in hypocotyls and young leaves. RsBGAL1 was highly similar to β-galactosidases having exo-β-(1→4)-galactanase activity found in higher plants and belongs to family 35 of the glycosyl hydrolases. Recombinant RsBGAL1 was expressed in Pichia pastoris and purified to homogeneity. The recombinant enzyme specifically hydrolyzed β-(1→3)- and β-(1→6)-galactooligosaccharides, the same substrates as the native enzyme isolated from radish seeds (Sekimata et al., 1989). It split off about 90% of the carbohydrate moieties of an arabinogalactan protein extracted from radish roots in concerted action with microbial α-l-arabinofuranosidase and β-glucuronidase. These results suggest that RsBGAL1 is a new kind of β-galactosidase with different substrate specificity than other β-galactosidases that exhibit exo-β-(1→4)-galactanase activity. The C-terminal region (9.6 kD) of RsBGAL1 is significantly similar to the Gal lectin-like domain, but this region is not retained in the native enzyme. Assuming posttranslational processing of RsBGAL1 with elimination of the Gal lectin-like domain results in a protein consisting of two subunits with molecular masses of 46 and 34 kD (calculated from the RsBGAL1 gene sequence). This is in good agreement with the SDS-PAGE and matrix-assisted laser desorption/ionization-time-of flight mass spectrometry measurements for subunits of the native enzyme (45 and 34 kD) and may thus partially explain the formation process of the native enzyme.
The enzyme group of β-galactosidases (EC 3.2.1.23) is widely distributed in higher plants. Plant β-galactosidases can be divided into at least two classes according to substrate specificity: one class that comprises exo-β-(1→4)-galactanases that specifically act on pectic β-(1→4)-galactan (sugars in this study are D series unless designated otherwise), and a second class that prefers p-nitrophenyl-β-galactoside (PNP-β-Gal) and lacks hydrolytic activity toward β-(1→4)-galactan (in this study, we call the former enzymes β-galactosidase/exo-β-(1→4)-galactanases). In the tomato (Lycopersicon esculentum), β-galactosidase/exo-β-(1→4)-galactanase activity significantly increases due to specific expression of the enzyme proteins during fruit ripening. This indicates their role in the degradation of β-(1→4)-galactan side chains of pectins as part of the ripening process. On the other hand, the activity level of the second class of β-galactosidase does not markedly change during ripening (Carey et al., 1995; Smith et al., 1998; Smith and Gross, 2000). Since the in vivo substrates for the second class of β-galactosidase is not yet identified, their functions in plant growth and development remain elusive.
Arabinogalactan proteins (AGPs), a family of proteoglycans found in higher plants, consist of a Hyp-rich core protein and carbohydrate moieties attached to the Hyp, Ser, and/or Thr residues. The carbohydrate moieties of AGPs have a common structure of β-(1→3)-galactosyl backbones to which side chains of β-(1→6)-linked galactosyl residues are attached through O-6. The β-(1→6)-linked galactosyl chains are further substituted with l-arabinofuranose (l-Ara) and lesser amounts of other auxiliary sugars, such as GlcUA, 4-O-methyl-GlcUA (4-Me-GlcUA), l-rhamnose, and l-Fuc. AGPs are implicated in many physiological processes, such as cell-to-cell signaling, cell adhesion, cell elongation, cell death, and stress responses (Fincher et al., 1983; Nothnagel, 1997; Majewska-Sawka and Nothnagel, 2000; Shi et al., 2003). In tobacco (Nicotiana tabacum), transmitting tissue-specific (TTS) protein that belongs to the AGP family is incorporated into pollen tube walls and undergoes degradation of the carbohydrate moieties, leading to stimulation of elongation growth (Cheung et al., 1995; Wu et al., 1995). Although the enzymes involved in the degradation of TTS protein have not yet been identified, the incorporated carbohydrate moieties of TTS protein are thought to serve as nutrients for the pollen tubes. It is also known that the sugar composition and structure of the carbohydrate moieties of AGPs are organ specific and regulated depending on the stage of development, implying that the carbohydrate moieties undergo rapid metabolism (Tsumuraya et al., 1988). Indeed, the rate of turnover of the carbohydrate moieties of AGPs in proso millet cells is extremely high (Gibeaut and Carpita, 1991). This means that a substantial portion of liberated sugars must be reutilized for the synthesis of new polymers.
It seems likely that hydrolysis of the carbohydrate moieties of AGPs is the result of concerted action of several glycosidases, such as β-galactosidase, α-l-arabinofuranosidase (α-l-arafase), and β-glucuronidase (β-GlcUAase). We have previously found β-galactosidases in radish (Raphanus sativus) seeds and spinach (Spinacia oleracea) leaves, which were capable of hydrolyzing β-(1→3)- and β-(1→6)-galactosyl sequences of AGPs but not pectic β-(1→4)-galactan (Sekimata et al., 1989; Hirano et al., 1994). These enzymes belong to the second class of β-galactosidase. The monomeric sugars released by β-galactosidase and other glycosidases may be utilized as nutrients during the development of plant tissues, as stated above, and/or salvaged via incorporation into the cytoplasm followed by conversion to nucleotide sugars by the successive actions of respective monosaccharide kinases and pyrophosphorylases (Reiter and Vanzin, 2001). This last metabolic process has been partially verified in Pisum sativum by our recent finding of a novel pyrophosphorylase that catalyzes conversion of various monosaccharide-1 phosphates (including Gal, Glc, GlcUA, l-arabinopyranose, and Xyl-1 phosphates) in the presence of UTP to the respective UDP sugars (Kotake et al., 2004).
In this article, we report the isolation of a cDNA clone encoding a β-galactosidase from radish and the properties of the recombinant protein expressed in Pichia pastoris. Based on the substrate specificity of the recombinant protein, we suggest that the β-galactosidase has specificity toward β-(1→3)- and β-(1→6)-galactosyl residues and plays a key role in the degradation of the carbohydrate moieties of AGPs.
RESULTS
Nature of the Native β-Galactosidase Protein
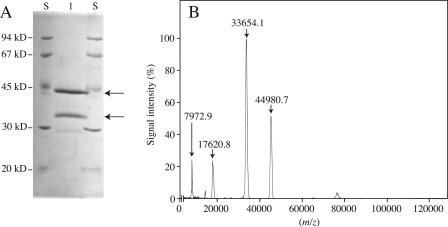
A native β-galactosidase was purified from radish seeds by the methods employed in our previous study (Sekimata et al., 1989) and analyzed on SDS-PAGE (Fig. 1A). The purified enzyme was resolved into two subunits with relative molecular masses of 45 and 34 kD on SDS-PAGE. Determination of the molecular mass of the fragments by matrix-assisted laser desorption/ionization-time-of flight mass spectrometry (MALDI-TOF/MS) gave 44,981 and 33,654 D, respectively (Fig. 1B). The small signals corresponding to molecular masses of 7,973 and 17,621 D seem to be caused by contaminations and could not be detected on SDS-PAGE. The faint band at molecular mass 29 kD on SDS-PAGE seems to be a contaminant that appears only on SDS-PAGE; it was not detectable by MALDI-TOF/MS. The dimeric nature and the molecular masses of the two subunits of the radish enzyme are similar to those observed for a persimmon β-galactosidase (44 and 34 kD; Kang et al., 1994) and an apple (Malus domestica) β-galactosidase (44 and 32 kD; Ross et al., 1994). The N-terminal amino acid sequences of the radish β-galactosidase were ASVTYDHRAL VIDGKRKILI SGSIHY for the 45-kD subunit and AELGSQWSYP KEPVGADAFD VKP for the 34-kD subunit. The peptide sequences of both subunits are highly similar to those of the corresponding subunits of β-galactosidase/exo-β-(1→4)-galactanase and the second class of β-galactosidases found in higher plants.
Figure 1.
Molecular masses of the subunits of a native β-galactosidase from radish seeds. A, Enzyme protein (0.5 μg) purified from imbibed radish seeds by several chromatographic procedures was separated on SDS-PAGE. Lane S, Molecular mass markers; lane 1, purified enzyme. Protein in the gel was stained with Coomassie Brilliant Blue R-250. The protein bands subjected to peptide sequence analysis are indicated by arrows. B, Molecular masses of the subunits were determined by MALDI-TOF/MS. The numbers at the top of the peaks indicate the observed molecular mass.
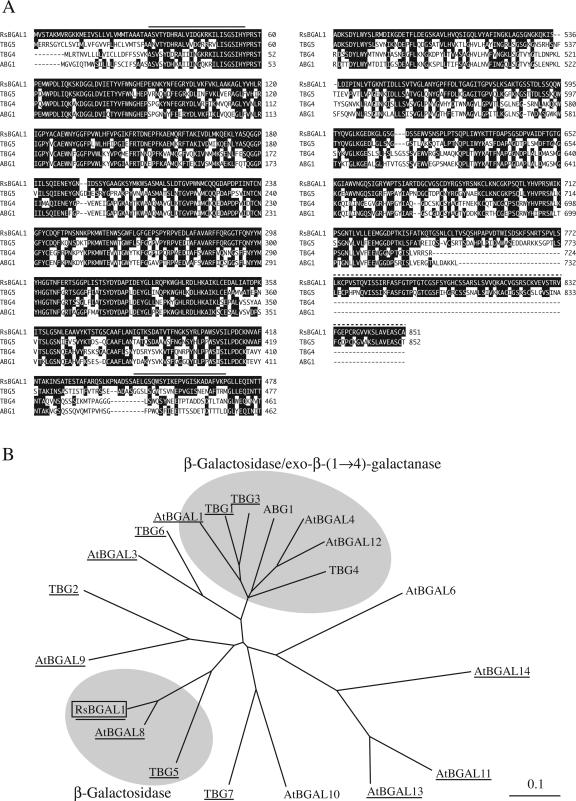
Based on the sequence determined for the purified β-galactosidase, reverse transcription-PCR followed by 3′ and 5′ RACE procedures were performed. The cloned cDNA, designated RsBGAL1 (Raphanus sativus beta-galactosidase 1), appeared to encode a polypeptide of 851 amino acids (molecular mass 92,534 D; Fig. 2A). The calculated pI value for RsBGAL1 (from Ala-31 to Ala-851) was 9.72. The N-terminal sequence of the 45-kD subunit of the native enzyme coincided with the region from Ala-31 to Tyr-56 of RsBGAL1, but that for the 34-kD subunit differed considerably from the corresponding sequence from Ala-445 to Pro-469. The reason for this discrepancy is not clear, but it may be a consequence of low purity of the native enzyme and/or a difference between the cultivars used for the purification of the native enzyme and for the cDNA cloning. The cDNA contained a putative signal sequence (30 amino acid residues) preceding the N-terminal sequence of the native enzyme, and this signal sequence agreed well with the prediction by the SignalP program (Bendtsen et al., 2004). Two putative N-glycosylation sites were found in the deduced protein sequence. The molecular mass (45 kD) determined for the larger fragment by MALDI-TOF/MS was almost identical to the calculated molecular mass (46.2 kD) of the region from Ala-31 to Ser-444; the mass of the smaller (34 kD) fragment, however, did not agree with that expected for the region from Ala-445 to Ala-851 (43.2 kD). This discrepancy between the relative molecular mass observed and the calculated molecular mass from the cDNA sequence is likely a result of posttranslational processing occurring around the Ser-759 residue and at Ser-444. Assuming that the posttranslational processing occurred between Ser-444 and Ala-445 and between Ser-759 and Asp-760, the polypeptide (from Ala-31 to Ala-851) would be divided into three fragments with molecular masses of 46.2 (from Ala-31 to Ser-444), 33.6 (from Ala-445 to Ser-759), and 9.6 kD (from Asp-760 to Ala-851). Since we could not find the small C-terminal fragment (9.6 kD) in the native enzyme, we surmise that the missing C-terminal fragment has likely been removed in the posttranslational processing, but we cannot rule out the possibility that it has been lost during the purification procedures.
Figure 2.
Amino acid sequence of RsBGAL1. A, Amino acid sequence of RsBGAL1 was aligned with plant β-galactosidase and β-galactosidase/exo-β-(1→4)-galactanase sequences by the pairwise method using the ClustalW program. The amino acid residues are numbered from the first Met. Gaps (-) were introduced to achieve maximum similarity. Residues identical to RsBGAL1 are highlighted in black. The solid lines indicate amino acid sequences corresponding to those determined for the native enzyme purified from radish seeds and the dotted line indicates a domain with similarity to Gal lectin. B, Phylogenetic relationships of RsBGAL1, β-galactosidases, and β-galactosidases/exo-β-(1→4)-galactanases were analyzed using ClustalW. Clones containing a Gal lectin-like domain are underlined. Accession numbers for the clones are as follows: ABG1, AAA62324; TBG1, CAA58734; TBG2, AAF70821; TGB3, CAA10173; TBG4, AAC25984; TBG5, AAF70824; TBG6, AAF70825; TBG7, AAF70823; AtBGAL1, At3g13750; AtBGAL3, At4g36360; AtBGAL4, At5g56870; AtBGAL6, At5G63800; AtBGAL8, At2g28470; AtBGAL9, At2g32810; AtBGAL10, At5g63810; AtBGAL11, At4g35010; AtBGAL12, At4g26140; AtBGAL13, At2g16730; AtBGAL14, At4g38590; RsBGAL1, AB180725.
Amino Acid Sequence of RsBGAL1
Based on amino acid sequence and structural similarities, glycoside hydrolases are classified into more than 90 families (Henrissat, 1991; Henrissat and Bairoch, 1993). RsBGAL1 is quite similar to other plant β-galactosidases and β-galactosidase/exo-β-(1→4)-galactanases, such as TBG5 (AF154423; 67% identical) and TBG4 (AF023847; 54% identical) from tomato (Smith et al., 1998), indicating that RsBGAL1 is a member of family 35 of the glycoside hydrolases (Fig. 2A). Phylogenetic analysis of plant β-galactosidases revealed that RsBGAL1 forms, together with Arabidopsis (Arabidopsis thaliana) BGAL8 (AtBGAL8, At2g28470) and tomato TBG5, a small subgroup apart from plant β-galactosidase/exo-β-(1→4)-galactanases, such as tomato TBG4 and apple ABG1 (Ross et al., 1994; Fig. 2B). These results suggest that RsBGAL1, AtBGAL8, and TBG5 have properties and functions distinct from those of β-galactosidase/exo-β-(1→4)-galactanases in the cell wall metabolism. As in a previous study on β-galactosidase genes from strawberry (Trainotti et al., 2001), RsBGAL1 was found to possess a Gal-binding lectin-like domain at the C terminus (Leu-773-Ala-851), which shows significant similarities with a Gal-specific lectin from Anthocidaris crassispina (32% identical; A37961) and an l-rhamnose-binding lectin from Silurus asotus (31% identical; Q9PVW8). Interestingly, the lectin-like domain is a structural characteristic of the RsBGAL1 subfamily, but not common in members of the β-galactosidases/exo-β-(1→4)-galactanase family, such as TGB4 (Fig. 2, A and B).
Organization and Expression Pattern of the RsBGAL1 Gene
The expression pattern of RsBGAL1 in young radish seedlings was analyzed. The transcript of RsBGAL1 was detected in hypocotyls and leaves, but not in roots. Relatively strong expression of RsBGAL1 was observed in the uppermost part of the hypocotyls (Fig. 3).
Figure 3.
Northern-blot analysis of RsBGAL1. Total RNA was extracted from young leaves, hypocotyls, and roots and then subjected to northern hybridization using the labeled RsBGAL1 fragment excised from RsBGAL1 cDNA as the probe. The position of the segment excised from the hypocotyls for RNA preparation is indicated in parentheses (distance from the cotyledons). The methylene blue-stained 18S rRNA used as a loading control is shown at the bottom.
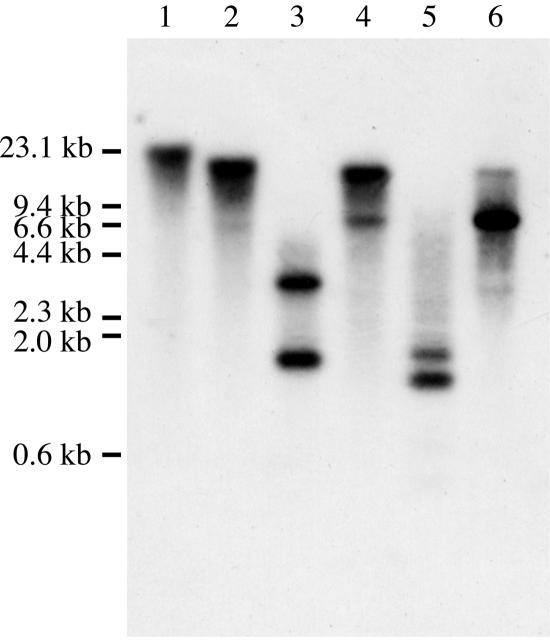
The number of RsBGAL1-related genes in the radish genome was determined by Southern-blot analysis. A labeled cDNA probe hybridized to several restriction fragments of the genomic DNA digested with DraI, EcoRI, or HindIII. A few faint bands were also detected in the genomic DNA digested with BamHI or XbaI (Fig. 4). These results suggest that several related genes exist in the radish genome.
Figure 4.
Southern-blot analyses of RsBGAL1 in the radish genome. Genomic DNA (10 μg) of radish was digested with ApaI (lane 1), BamHI (lane 2), DraI (lane 3), EcoRI (lane 4), HindIII (lane 5), and XbaI (lane 6) and then subjected to Southern hybridization using the labeled RsBGAL1 fragment excised from RsBGAL1 cDNA as the probe. Locations of λDNA markers digested with HindIII are shown on the left side.
Heterologous Expression of RsBGALI in P. pastoris
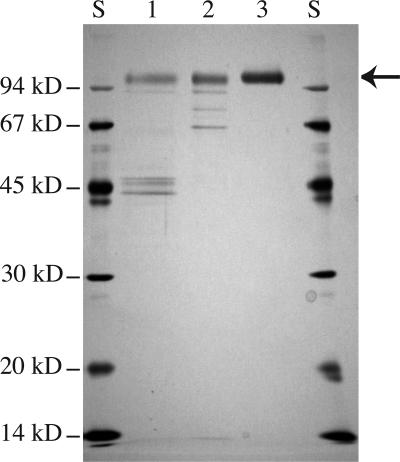
The RsBGAL1 open reading frame, except for the signal peptides, was fused to a yeast secretion signal sequence (α-factor) and introduced into the methylotrophic yeast P. pastoris. Recombinant RsBGAL1 (rRsBGAL1) was induced under the control of the alcohol oxidase promotor and purified from the culture medium by conventional chromatography (Table I). The specific activity (64.9 units mg protein−1) of rRsBGAL1 was more than 10 times (5.6 units mg protein−1) that of the native enzyme (Sekimata et al., 1989). The purified rRsBGAL1 appeared as a single band, with a relative molecular mass of approximately 100 kD on SDS-PAGE, indicating that the recombinant enzyme had not undergone posttranslational proteolysis in the yeast (Fig. 5). The molecular mass for rRsBGAL1 was determined as 106,449 D by MALDI-TOF/MS analysis (data not shown). The higher molecular mass of the recombinant enzyme compared to the value (89.4 kD) expected from the cDNA sequence is likely the result of a difference in the N-glycosylation of the protein between plants and yeast. It is known that yeast attaches large high-Man-type glycans to secreted proteins (Gemmill and Trimble, 1999).
Table I.
Purification of rRsBGAL1 expressed in P. pastoris
| Total Protein | Total Activitya | Specific Activity | Recoveryb | Purification Factorb | |
|---|---|---|---|---|---|
| mg | units | units mg protein−1 | % | -fold | |
| Culture medium | 8.17 | 39.8 | 4.87 | 100.0 | 1.0 |
| CM-cellulose | 0.98 | 49.1 | 50.1 | 123.4 | 10.3 |
| Hydroxyapatite | 0.35 | 22.7 | 64.9 | 57.0 | 13.3 |
Activity determined with PNP-β-Gal as the substrate.
Recoveries are expressed as a percentage of initial activity, and purification factors are calculated on the basis of specific activities.
Figure 5.
SDS-PAGE of rRsBGAL1 at different purification steps. The rRsBGAL1 proteins (0.5 μg) obtained after different purification steps of the recombinant protein were analyzed by SDS-PAGE. Lane S, Molecular mass markers; lane 1, supernatant from P. pastoris culture medium; lane 2, rRsBGAL1 purified on CM-cellulose column; lane 3, rRsBGAL1 purified on hydroxyapatite column. Protein in the gel was stained with Coomassie Brilliant Blue R-250. The arrow indicates the purified rRsBGAL1.
Properties of rRsBGAL1
The properties of rRsBGAL1 were examined using PNP-β-Gal as the substrate. The recombinant enzyme showed maximum activity between pH 3.5 and pH 4.0 and became almost inactive on PNP-β-Gal below pH 2.0 and above pH 6.0. The optimum temperature for enzyme action was 50°C, and the enzyme lost about 90% of its activity at 65°C. The native β-galactosidase from radish seeds shows maximal activity at 40°C and completely loses activity when exposed to 55°C for 10 min (Sekimata et al., 1989). The greater stability of rRsBGAL1 under high temperature may be attributed to the N-glycosylation performed by P. pastoris. The effects of heavy metal ions were examined at a final concentration of 1 mm. The metal ions Fe3+, Mn2+, Cu2+, Mg2+, Ba2+, Ca2+, Co2+, Zn2+, and Cd2+ did not significantly affect enzyme activity, whereas Hg2+ almost completely inactivated the enzyme. Iodoacetic acid and SDS also inhibited more than 80% of the enzyme activity when applied at a concentration of 1 mm.
Substrate Specificity of rRsBGAL1 toward Oligosaccharides
The activity of rRsBGAL1 toward oligosaccharides was examined using various β-(1→3)-, β-(1→4)-, and β-(1→6)-galactooligosaccharides. Whereas rRsBGAL1 extensively hydrolyzed β-(1→3)- and β-(1→6)-galactooligosaccharides, it failed to act on β-(1→4)-galactooligosaccharides. The action of this enzyme seems thus specific to β-(1→3)- and β-(1→6)-linked galactosyl residues (Table II). The hydrolysis rates of these β-(1→3)- and β-(1→6)-galactooligomers tended to increase with increasing degree of polymerization. The weak action on methyl-β-galactoside and lactose [β-Gal-(1→4)-Glc], and the failure of hydrolysis of β-Gal-(1→3)-GalNAc and β-Gal-(1→3)-GlcNAc, suggest that adjacent residues linked to the galactosyl residues also affect the enzymatic action of rRsBGAL1. On heterooligosaccharides substituted with α-l-arabinofuranosyl, β-glucuronosyl, or 4-O-methyl-β-glucuronosyl, residues at the nonreducing terminals of galactooligomers, rRsBGAL1 did not act (Table II).
Table II.
Substrate specificity of rRsBGAL1 toward oligosaccharides
| Substratea | Relative Activityb |
|---|---|
| β-(1→3)-Galactooligosaccharides | |
| β-(1→3)-Galactobiose | 32 |
| β-(1→3)-Galactotriose | 44 |
| β-(1→4)-Galactooligosaccharides | |
| β-(1→4)-Galactobiose | 0 |
| β-(1→4)-Galactotriose | 0 |
| β-(1→4)-Galactotetraose | 0 |
| β-(1→6)-Galactooligosaccharides | |
| β-(1→6)-Galactobiose | 27 |
| β-(1→6)-Galactotriose | 49 |
| β-(1→6)-Galactotetraose | 68 |
| β-(1→6)-Galactopentaose | 50 |
| Heterooligosaccharides | |
| β-Gal-(1→6)-β-Gal-(1→3)-Gal | 40 |
| β-Gal-(1→3)[β-Gal-(1→6)]-Gal | 42 |
| β-Gal-(1→3)-Arap | 21 |
| β-Gal-(1→3)-GalNAc | 0.5 |
| β-Gal-(1→3)-GlcNAc | 0.5 |
| β-Gal-(1→4)-Glc (lactose) | 11 |
| β-Gal-(1→4)-Man | 0.4 |
| α-l-Ara-(1→3)-β-Gal-(1→6)-Gal | 0 |
| β-GlcUA-(1→6)-Gal | 0 |
| β-GlcUA-(1→6)-β-Gal-(1→6)-Gal | 0 |
| 4-Me-β-GlcUA-(1→6)-Gal | 0 |
| 4-Me-β-GlcUA-(1→6)-β-Gal-(1→6)-Gal | 0 |
| 4-Me-β-GlcUA-(1→6)- β-Gal-(1→6)- β-Gal-(1→3)-Gal | 0 |
| Others | |
| PNP-β-Gal | 100 |
| Methyl-β-galactoside | 4 |
| Laminaribiose [β-Glc-(1→3)-Glc] | 0 |
| Cellobiose [β-Glc-(1→4)-Glc] | 0 |
| Gentiobiose [β-Glc-(1→6)-Glc] | 0 |
The enzyme was incubated with substrates at a concentration of 5 mm, and 2 mm was employed for PNP-β-Gal.
Activity is expressed as percent of that (64.9 units mg protein−1) of PNP-β-Gal.
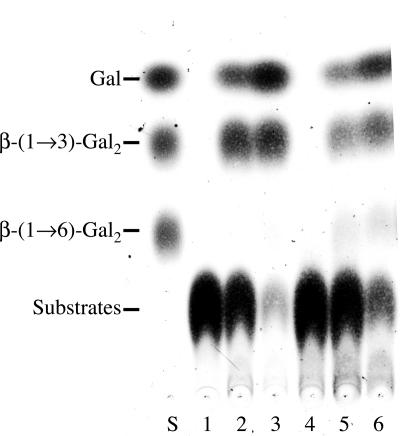
Since the carbohydrate moieties of AGPs commonly have branched β-(1→3)(1→6)-galactan structures as their backbones, the action of rRsBGAL1 on mixed-linkage galactotrioses, β-Gal-(1→6)-β-Gal-(1→3)-Gal and β-Gal-(1→3)[β-Gal-(1→6)]-Gal, was examined in order to explore the mechanism for the complete digestion of the galactan backbones by plant β-galactosidases. These galactotrioses were good substrates for rRsBGAL1, almost to the same extent as β-(1→3)- and β-(1→6)-galactotrioses (Table II). In the case of β-Gal-(1→3)[β-Gal-(1→6)]-Gal, the hydrolysate contained more β-(1→3)-galactobiose than β-(1→6)-galactobiose as intermediate products (Fig. 6), which indicates that the enzyme preferred the β-(1→6)-linked galactosyl residues to the β-(1→3)-linked galactosyl residues of β-Gal-(1→3)[β-Gal-(1→6)]-Gal. Since the intermediate products from mixed-linkage galactotrioses, β-(1→3)- and β-(1→6)-galactobioses are, in turn, good substrates for rRsBGAL1, it seems that not only the linear β-(1→3)- and β-(1→6)-galactosyl stretches but also the branch points of β-(1→3)(1→6)-galactans of AGPs can be completely degraded by this enzyme.
Figure 6.
Hydrolysis of mixed-linkage galactooligosaccharides. Hydrolysis products of β-Gal-(1→6)-β-Gal-(1→3)-Gal and β-Gal-(1→3)[β-Gal-(1→6)]-Gal by action of rRsBGAL1 were analyzed on TLC. Lane S, Standard Gal, β-(1→3)-galactobiose, and β-(1→6)-galactobiose; lane 1, β-Gal-(1→6)-β-Gal-(1→3)-Gal before hydrolysis; lane 2, hydrolysis products of β-Gal-(1→6)-β-Gal-(1→3)-Gal after 1 h; lane 3, those after 12 h; lane 4, β-Gal-(1→3)[β-Gal-(1→6)]-Gal before hydrolysis; lane 5, hydrolysis products of β-Gal-(1→3)[β-Gal-(1→6)]-Gal after 1 h; lane 6, those after 12 h. Localization of the standard sugars is indicated on the left side.
The effect of substrate concentration on the activity of rRsBGAL1 was examined using PNP-β-Gal, β-(1→3)-galactobiose, and β-(1→6)-galactobiose. The resulting Km, kcat, and catalytic efficiency (kcat/Km) values are listed in Table III. Although the Km value (0.30 mm) of rRsBGAL1 for PNP-β-Gal was comparable to that (0.46 mm) of the native enzyme (Sekimata et al., 1989), the catalytic efficiency (4.10 × 105) of rRsBGAL1 was much higher than that (0.16 × 105, calculated from the Km value, 0.46 mm, and the Vmax value, 5.36 μmol min−1 mg protein−1) of the native enzyme (Sekimata et al., 1989). The lower Km value (i.e. higher affinity) of rRsBGAL1 toward β-(1→6)-galactobiose than toward β-(1→3)-galactobiose is consistent with the preference of the enzyme for the β-(1→6)-galactosyl residues over the β-(1→3)-galactosyl residues of β-Gal-(1→3)[β-Gal-(1→6)]-Gal (Fig. 6).
Table III.
Kinetic values of rRsBGAL1
| Substratea | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | s−1 | m−1s−1 | |
| PNP-β-Gal | 0.30 | 123.0 | 4.10 × 105 |
| β-(1→3)-Galactobiose | 1.77 | 28.6 | 0.16 × 105 |
| β-(1→6)-Galactobiose | 0.77 | 32.5 | 0.42 × 105 |
To examine the effects of galactooligosaccharides on the activity, reactions were carried out with varying concentrations of PNP-β-Gal (0.1–5 mm), β-(1→3)-galactobiose (0.5–10 mm), and β-(1→6)-galactobiose (0.5–10 mm). The Km and kcat values were calculated from a Hanes-Woolf plot using the obtained activities.
Substrate Specificity of rRsBGAL1 toward Polysaccharides
Compared to the activity against PNP-β-Gal, which is taken as 100 in Table IV, β-(1→3)-galactan degradation proceeded slowly. Under exhaustive digestion, rRsBGAL1 released about 30% of Gal from β-(1→3)-galactan, whereas β-(1→4)-galactan and Prototheca zopfii β-(1→3)(1→6)-galactan were essentially resistant to the enzyme. Native root and leaf AGPs were resistant to enzymatic hydrolysis even after prolonged incubation. However, partial removal of l-arabinosyl residues from the carbohydrate moieties made the modified AGPs more accessible to rRsBGAL1, resulting in the release of 16% and 25% of Gal based on total sugars in the modified AGPs, respectively. The lower activity of rRsBGAL1 toward α-l-arafase-treated root AGP is likely attributable to obstruction by l-arabinosyl residues remaining even after treatment. The microbial α-l-arafase used releases almost all l-arabinosyl residues from the leaf AGP, whereas 10% to 30% of total l-arabinosyl residues remain in root AGP even after exhaustive digestion (see Table V; Tsumuraya et al., 1984, 1988). Overall, the action pattern of rRsBGAL1 on AGPs and galactans was quite similar to that of the native β-galactosidase (Sekimata et al., 1989). Other polysaccharides, such as β-(1→3)(1→4)-glucan from barley, β-(1→3)(1→6)-glucan from Laminaria digitata, β-(1→6)-glucan from Umbilicaria papullosa, CM-curdlan [β-(1→3)-glucan], CM-cellulose [β-(1→4)-glucan], galactomannans from guar and locust bean, β-(1→4)-xylan from birchwood (Betula spp.), debranched arabinan, and chitosan from crab shells were not hydrolyzed by rRsBGAL1 at all.
Table IV.
Substrate specificity of rRsBGAL1 toward polysaccharides
| Substratea | Relative Activityb | Limit of Hydrolysisc |
|---|---|---|
| β-(1→3)-Galactan | 8 | 29 |
| β-(1→3)(1→6)-Galactan from P. zopfii | 2 | 3 |
| β-(1→4)-Galactan from lupin | 0.8 | 1 |
| Native AGP from radish roots | 3 | 3 |
| α-l-Arafase-treated AGP from radish roots | 4 | 16 |
| Native AGP from radish leaves | 4 | 1 |
| α-l-Arafase-treated AGP from radish leaves | 8 | 25 |
| PNP-β-Gal | 100 | –d |
The enzyme was incubated with polymers at a concentration of 5 mg mL−1 and 2 mm was employed for PNP-β-Gal.
Activity is expressed as percent of that (64.9 units mg protein−1) of PNP-β-Gal.
The reaction was carried out under the standard conditions, except for the concentration of the substrate (1 mg mL−1) and the amount (20 mU) of the enzyme for 16 h, followed by further incubation with the addition of an equal amount of the enzyme for another 10 h. The limit of hydrolysis was determined after the liberation of reducing sugars reached a plateau, and expressed as Gal equivalent against the total sugars in each substrate.
Not determined.
Table V.
Characterization of α-l-arafase-treated AGP and high-Mr products obtained by digestion with rRsBGAL1 with or without the presence of β-GlcUAase and α-l-arafase
|
α-l-Arafase-Treated AGP
|
High-Mr Product
|
||
|---|---|---|---|
| With rRsBGAL1 Alone | With rRsBGAL1, β-GlcUAase, and α-l-Arafase | ||
| Yield (percent, based on sugar content) | 100 | 85 | 15 |
| Sugar composition (mol%) | |||
| l-Ara | 6 | 8 | 25 |
| Gal | 83 | 81 | 67 |
| 4-Me-GlcUA | 11 | 11 | 8 |
| Mode of glycosidic linkages (mol%)a | |||
| Araf1→ | 3b | 3 | 9 |
| →2Araf1→ | 3 | 2 | 9 |
| →5Araf1→ | –c | +d | 2 |
| Galp1→ | 14 | 18 | 18 |
| →3Galp1→ | 7 | 7 | 20 |
| →6Galp1→ | 54 | 54 | 26 |
| →3,6Galp1→ | 19 | 16 | 16 |
Methylation analysis was done without carboxyl reduction of uronosyl residues. The amounts of nonreducing terminal 4-Me-GlcUA groups are thus not accounted for.
Calculated from data in Tsumuraya et al. (1988).
Not detectable.
Less than 1%.
Degradation of AGP by rRsBGAL1 with Other Glycosidases
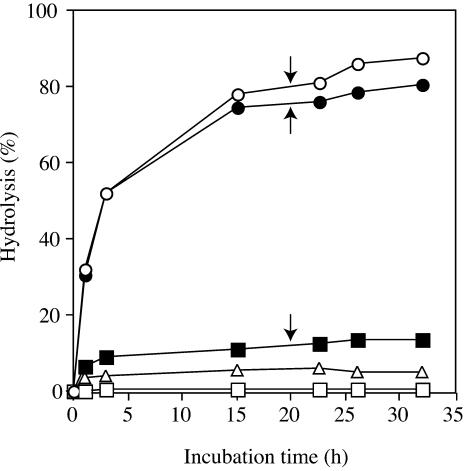
To gain an insight into the mechanism of turnover of AGPs in vivo, the synergistic action of rRsBGAL1 with other glycosidases on an AGP was investigated. As shown in Figure 7, the action of rRsBGAL1 alone liberated only a limited amount (approximately 14% of the total sugars) of Gal as the sole hydrolysis product from α-l-arafase-treated radish root AGP under exhaustive digestion (Table IV). This relatively limited hydrolysis of the carbohydrate moieties of the modified AGP can be attributed to the presence of uronic acids at the nonreducing ends of β-(1→6)-linked galactosyl side chains of most of the AGPs (Fincher et al., 1983; Nothnagel, 1997), which renders galactosyl residues inaccessible to the enzyme. Indeed, more than 70% of the total side chains of radish root AGP are known to carry 4-Me-GlcUA groups at their nonreducing ends (Tsumuraya et al., 1990). As anticipated, the simultaneous action of rRsBGAL1 and a microbial β-GlcUAase on the modified AGP liberated much more (approximately 80%) reducing sugar by exposing nonreducing galactosyl residues, making the side chains susceptible to rRsBGAL1. Further addition of a microbial α-l-arafase to the above reaction mixture liberated nearly 90% of total sugar from the modified AGP. Finally, a large portion (91%) of Gal, together with 4-Me-GlcUA (8%) and l-Ara (1%), was liberated as free monosaccharides. The control experiment using β-GlcUAase alone liberated only very little 4-Me-GlcUA, and α-l-arafase alone did not act on the modified AGP. Since α-l-arafase-treated radish root AGP still retains a small portion (approximately 5%) of l-arabinosyl residues in the carbohydrate moiety, possibly at the inner parts of the side chains (Tsumuraya et al., 1988), our results imply that the l-arabinosyl residues become accessible to α-l-arafase during stepwise elimination of galactosyl and 4-O-methyl-glucuronosyl residues by the action of both rRsBGAL1 and β-GlcUAase, leading to liberation of most of the sugar residues. These conclusions are consistent with the substrate specificity of rRsBGAL1 toward oligosaccharides (Table II) in that the substitution with l-Ara or uronic acids at nonreducing ends of β-(1→6)-galactooligosaccharides renders those galactooligomers impervious to the enzyme.
Figure 7.
Hydrolysis of α-l-arafase-treated radish root AGP by rRsBGAL1. α-l-Arafase-treated radish root AGP was digested with rRsBGAL1 (black square), β-GlcUAase (white triangle), or α-l-arafase (white square), each acting alone or by simultaneous action of rRsBGAL1 and β-GlcUAase (black circle), or simultaneously by rRsBGAL1, β-GlcUAase, and α-l-arafase (white circle). The amounts of sugars released were determined reductometrically and the extent (percentage) of hydrolysis was calculated based on the total sugar content of the modified AGP. Arrows indicate the addition of additional rRsBGAL1 into three reaction mixtures as described in “Materials and Methods.”
Digestion of α-l-arafase-treated AGP with rRsBGAL1 in the presence of β-GlcUAase and α-l-arafase in a large-scale reaction yielded a small portion (15%) of a high-Mr component possibly representing a core part of the AGP consisting of a polypeptide with short oligosaccharide remnants that have an increased proportion (25% of total sugars) of l-arabinosyl residues resistant to α-l-arafase attack (Table V). However, the chain lengths and numbers of the remnants along the single polypeptide backbone are unknown. Structural analysis of the high-Mr component indicated an increase in the proportion of nonreducing terminal and O-2-linked l-arabinosyl residues, as well as O-3-linked galactosyl residues and fewer O-6-linked galactosyl residues, when compared with the data obtained for the initial AGP. These observations suggest that the β-(1→3)- and β-(1→6)-linked galactosyl sequences in the α-l-arafase-treated AGP are removed in a concerted stepwise enzymatic degradation process with rRsBGAL1 in the presence of β-GlcUAase and α-l-arafase, leaving only the core protein of the root AGP. On the other hand, a clearly different high-Mr component obtained after digestion of the modified AGP with rRsBGAL1 alone showed sugar composition and structure similar to those of the initial AGP. Only a small amount (15% of total sugar) of galactosyl residues was eliminated by the action of rRsBGAL1 in this case.
DISCUSSION
To date, hundreds of cDNA sequences for β-galactosidase/exo-β-(1→4)-galactanases and β-galactosidases from higher plants have been cloned. Although these enzymes are widely distributed in higher plants, until now their functions have been discussed primarily with respect to the degradation of pectic β-(1→4)-galactan. The reason for this is that the structure of pectic β-(1→4)-galactan is regulated spatially during the development of plant tissues, and pectin thus plays an important role in the architecture of the cell wall and intercellular attachment (McCartney et al., 2000; Sørensen et al., 2000). On the other hand, we have previously shown that β-galactosidase specimens isolated from radish seeds and spinach leaves (spinach β-Gal I) hydrolyze specifically β-(1→3)- and β-(1→6)-galactooligosaccharides besides PNP-β-Gal, and are thereby able to degrade the β-(1→3)(1→6)-galactan backbones of AGPs, but not pectic β-(1→4)-galactan (Sekimata et al., 1989; Hirano et al., 1994). These enzymes can be classified into the second class of β-galactosidases, the β-galactosidase/exo-β-(1→3)(1→6)-galactanases. However, we cannot rule out the possibility that this second group of β-galactosidases participates in the degradation of both pectic β-(1→4)-galactan and the carbohydrate moieties of AGPs, because a β-galactosidase specimen (β-Gal II) purified from spinach leaves shows a broad substrate specificity, acting not only on β-(1→3)- and β-(1→6)-galactooligosaccharides but also weakly on β-(1→4)-galactooligosaccharides (Hirano et al., 1994). Further studies are required to clarify the functional differences of the second class of β-galactosidases from β-galactosidase/exo-β-(1→4)-galactanases. For this study, we have cloned a gene (RsBGAL1) encoding radish β-galactosidase and expressed the recombinant enzyme (rRsBGAL1) in P. pastoris. We then investigated the nature of the RsBGAL1 protein and compared the enzymatic properties of rRsBGAL1 with those of the native enzyme.
Substrate specificity was quite similar to that of the native enzyme (Sekimata et al., 1989); that is, rRsBGAL1 preferred β-(1→3)- and β-(1→6)-galactooligosaccharides, and hydrolyzed β-(1→3)-galactan and β-(1→3)(1→6)-galactan backbones of AGPs. Although the amino acid sequence of the smaller subunit deduced from RsBGAL1 cDNA was not completely identical to that determined for the corresponding 34-kD subunit of the native enzyme, RsBGAL1 likely encodes the native radish enzyme or, at least, a β-galactosidase with properties quite similar to the native enzyme. Purification of rRsBGAL1 yielded a single polypeptide with a molecular mass of 106 kD, including a Gal lectin-like domain at the C terminus, whereas the native RsBGAL1 consisted of two subunits with molecular masses of 45 and 34 kD and lacked the Gal lectin-like domain. These observations clearly indicate that the RsBGAL1 polypeptide undergoes posttranslational processing in radish plants. A similar case of posttranslational processing in the C-terminal region of the native enzyme has been observed for α-l-arabinofuranosidase and β-xylosidase from barley (Lee et al., 2003). Elucidating the precise mechanism of posttranslational processing and its significance to kinetic properties and functions of plant glycosidases is an important problem, but we are not yet able to give a satisfactory explanation concerning rRsBGAL1. In our experiments, even though the substrate specificities of recombinant and native enzymes were quite similar, several kinetic parameters differed considerably. The kcat value (123.0 s−1) of rRsBGAL1 for PNP-β-Gal was much higher than that (7.48 s−1) of the native enzyme (Sekimata et al., 1989). This difference affected other kinetic data obtained in this study. For example, the relatively lower hydrolysis rates (corresponding to 8% of that for PNP-β-Gal) of β-(1→3)-galactan and α-l-arafase-treated radish leaf AGP by rRsBGAL1 (Table IV) when compared with those (17 and 49%, respectively) by the native enzyme seem to reflect the high preference of rRsBGAL1 for PNP-β-Gal. Nevertheless, the limits of hydrolysis are almost the same for both enzymes. In addition, the recombinant enzyme showed a lower Km value (1.77 mm) for β-(1→3)-galactobiose compared with that (7.79 mm) of the native enzyme (Sekimata et al., 1989). It seems likely that these catalytic differences result from the posttranslational processing of the enzyme protein in the radish plant. However, the processing does not affect the essential recognition of β-(1→3)- and β-(1→6)-linked galactosyl sequences.
Previous studies have explored the physiological roles of AGPs by application of β-glycosyl-Yariv reagent, chemical name 1,3,5-tri(p-glycosyloxyphenylazo)-2,4,6-trihydroxybenzene, that specifically binds to the carbohydrate moieties of AGPs and perturbs their functions (Komalavilas et al., 1991; Majewska-Sawka and Nothnagel, 2000). In cultured cells of Arabidopsis, β-glycosyl-Yariv reagent induces programmed cell death, possibly by disrupting the plasma membrane-cell wall connections (Gao and Showalter, 1999). Recently, Motose et al. (2004) have reported that xylogen, a nonclassical AGP, induces differentiation of zinnia (Zinnia elegans) mesophyll cells. The inductive function of xylogen, however, was suppressed when the zinnia cells were treated with β-glycosyl-Yariv reagent. It was also lost when the carbohydrate moieties were removed from the xylogen by chemical treatment. We think it likely that the carbohydrate moieties of AGPs are required for intercellular communication and that various glycosidases, such as β-galactosidase, are involved in the structural modification of the carbohydrate moieties of AGPs in response to the developmental stage. In this study, rRsBGAL1 was not very active toward native AGP from radish roots, but removed nearly 90% of the carbohydrate moieties of the AGP when aided by other microbial glycosidases (Fig. 7). It is highly probable that the carbohydrate moieties of AGPs are degraded by the concerted action of β-galactosidase, α-l-arafase, and β-GlcUAase, together with other auxiliary glycosidases, depending on the sugar compositions of the various AGPs in vivo. This has been postulated previously in a study on both radish β-galactosidase and α-l-arafase (Hata et al., 1992). Here, we have shown that β-galactosidase plays a key role in the degradation of the carbohydrate moieties of AGPs, leading to their structural modification. There have been only a small number of studies of plant β-GlcUAases, such as that found in suspension cells of skullcap (Scutellaria baicalensis), which participates in the metabolism of β-GlcUA groups in flavones (Sasaki et al., 2000), and no plant β-GlcUAases capable of hydrolyzing plant cell wall polysaccharides have been found so far. However, a preliminary experiment we performed showed that radish plants contain low, but detectable, levels of enzyme activity hydrolyzing both PNP-β-GlcUA and β-GlcUA-(1→6)-Gal. It is possible that the radish β-GlcUAase(s) are not unimportant as degrading enzymes for the recycling of AGPs in vivo.
We found that rRsBGAL1 hydrolyzed α-l-arafase-treated radish leaf AGP in preference to α-l-arafase-treated radish root AGP (Table IV). Except for the occurrence of l-Fuc residues as a minor (5%–6% of total sugar) constituent in leaf AGP, the structures of the carbohydrate moieties of both AGPs are quite similar. Both native AGPs possess a common β-(1→3)(1→6)-galactan backbone, to which a considerable amount of α-l-arabinofuranosyl and other minor sugar residues are attached, and have comparable relative molecular masses (88 kD for the root AGP, 75 kD for the leaf AGP; Nakamura et al., 1984; Tsumuraya et al., 1984, 1988). We are not able, at the moment, to give a detailed explanation of the particular structural features that influence the susceptibility to β-galactosidase.
The physiological functions of most β-galactosidase and β-galactosidase-like genes cloned so far from higher plants have not yet been established. Arabidopsis BGAL8 and tomato TBG5 are close homologs of RsBGAL1 and form a putative subfamily of β-galactosidases distinct from that of the β-galactosidase/exo-β-(1→4)-galactanases, which includes TBG4 (Fig. 2B). The substrate specificity of these two homologs has not yet been examined, but they seem to behave similarly to RsBGAL1; that is, they recognize specifically β-(1→3)- and β-(1→6)-linked galactosyl sequences. We are planning to further investigate the physiological functions of plant β-galactosidases using T-DNA knockout lines of Arabidopsis.
MATERIALS AND METHODS
Plant Material
Seeds of radish (Raphanus sativus L. var hortensis cv aokubi-miyashige-nagajiri) were purchased from Tokita Seed and Plant (Saitama, Japan). For DNA and RNA preparations, the radish seeds were sown on moist plastic mesh and grown at 25°C for 6 d.
Oligo- and Polysaccharides
The β-(1→3)- and β-(1→6)-linked galactobioses and -trioses used were prepared from larch wood (Larix decidua) arabinogalactan (Aspinall et al., 1958b), β-(1→6)-galactotetraose was prepared from gum ghatti (Aspinall et al., 1958a), and β-(1→4)-galactooligosaccharides with degree of polymerization 2 to 4 were prepared from soybean arabinan galactan (Sekimata et al., 1989). The α-l-Ara-(1→3)-Gal-β-(1→6)-Gal was prepared from enzymatic hydrolysate of the Smith degradation product of acacia (Acacia senegal) gum by incubation with exo-β-(1→3)-galactanase (Tsumuraya et al., 1990), β-GlcUA-(1→6)-Gal, β-GlcUA-(1→6)-β-Gal-(1→6)-Gal, 4-Me-β-GlcUA-(1→6)-Gal, 4-Me-β-GlcUA-(1→6)-β-Gal-(1→6)-Gal, and 4-Me-β-GlcUA-(1→6)-β-Gal-(1→6)-β-Gal-(1→3)-Gal were prepared from acacia gum and the sap of the lac tree (Rhus vernicifera; Kuroyama et al., 2001). The β-(1→6)-galactopentaose was a gift from Dr. Miura of the Chiba Institute of Science (Chiba, Japan) and Prof. Inazu of Tokai University (Kanagawa, Japan). The β-galactan [essentially a β-(1→3)(1→6)-galactan with galactofuranosyl residues attached] from P. zopfii (Okemoto et al., 2003), β-(1→3)-galactan (Sekimata et al., 1989), was prepared in our laboratories. Radish leaf AGP (AGP R-II) and root AGP (AGP IV) were extracted from mature leaves and roots of radish, respectively, with 14.5 mm sodium phosphate buffer, pH 7.2, containing 130 mm NaCl. The AGPs were purified on a DEAE-cellulose (DEAE 23 SH-Cellulose; SERVACEL, Heidelberg) column (HCO3− form), then on a Sepharose 6B column (Amersham Biosciences, Buckinghamshire, UK), as described previously (Tsumuraya et al., 1984, 1988). The α-l-arafase-treated AGPs were prepared by digestion of the AGPs (10 mg) with α-l-arafase (1 unit) from Rhodotorula flava in 10 mm citrate phosphate buffer (pH 3.0) at 37°C for 20 h, followed by separation from free l-Ara on a Sephadex G-15 (Amersham Biosciences) column (Nakamura et al., 1984; Tsumuraya et al., 1984, 1988). The relative molecular mass of the root AGP was 88 kD and that of the leaf AGP was 75 kD. Laminaribiose and cellobiose were purchased from Seikagaku (Tokyo). Gentiobiose, lactose, β-Gal-(1→3)-arabinopyranose, β-Gal-(1→3)-GalNAc, β-Gal-(1→3)-GlcNAc, β-Gal-(1→4)-Man, PNP-β-Gal, PNP-β-GlcUA, chitosan from crab shells, guar gum, locust bean gum, laminarin from Laminaria digitata, and xylan from birchwood (Betula spp.) were from Sigma (St. Louis). Native and debranched arabinans from sugar beet, β-(1→4)-galactan from lupin, β-(1→3)(1→4)-glucan from barley (low viscosity), and CM-cellulose 4 m were purchased from Megazyme (Wicklow, Ireland); β-(1→6)-glucan (pustulan) from Umbilicaria papullosa was from Calbiochem (San Diego); CM-curdlan was from Wako (Osaka).
Preparation and Identification of Mixed-Linkage Galactotrioses
From a trisaccharide fraction in the partial acid hydrolysates of larch wood arabinogalactan (Aspinall et al., 1958b), mixed-linkage galactotrioses 1 and 2 were purified, which were later identified as β-Gal-(1→6)-β-Gal-(1→3)-Gal and β-Gal-(1→3)[β-Gal-(1→6)]-Gal, respectively (see below). The fraction was analyzed on an HPLC system with a Waters Alliance 2695 fitted with a refractive index indicator and a column (4.6 × 100 mm) of TSKgel Carbon-500 (Tosoh, Tokyo). The column was eluted with acetonitrile:water (5:95; v/v) at a flow rate of 0.5 mL min−1 and at 50°C, resulting in partial separation of α- and β-anomers of 1 and 2 eluted at 6.8 (1), 7.7 (an overlap peak of 1 and 2), and 8.9 min (2). For preparation of these two trioses, a portion of the fraction (74 mg) was separated repeatedly by using a large column (20 × 100 mm) with acetonitrile:water (4:96; v/v) at a flow rate of 15 mL min−1 and at room temperature, yielding purified 1 (18 mg) and 2 (10 mg). Both oligosaccharides gave the same signal at m/z 527.4 on MALDI-TOF/MS (see below), which exactly coincides with the expected mass of [Gal3 + Na]+.
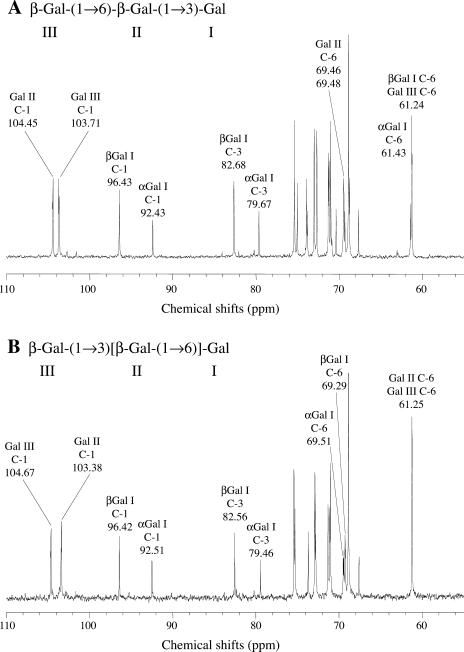
The structures of the trioses were determined by NMR. 1H-NMR and proton-decoupled carbon NMR spectra were recorded at 500 and 125 MHz, respectively, on a JEOL (Tokyo) ECA-500 spectrometer equipped with an inverse 5-mm gradient probe in D2O at 30°C. Acetone (2.22 ppm for 1H- or 35.0 ppm for 13C-NMR) was used as the internal reference. JEOL standard pulse sequences were used for DEPT135, 1D-total correlation spectroscopy , FG-heteronuclear single quantum correlation, FG-heteronuclear multiple-bond correlation, and rotating frame overhauser enhancement spectroscopy experiments. Typical 13C-NMR spectra of galactotriose 1 and 2 are shown in Figure 8. Glycosidic linkages of the galactotrioses were determined by downfield shift of signals in the 13C-NMR spectra caused by substitution with other galactosyl residues. Signals at 69.46, 69.48, 79.67, and 82.68 ppm in 1, and 69.29, 69.51, 79.46, and 82.56 in 2 were shifted downfield when compared with free Gal. The signals at 69 to 70 ppm were assigned to the methylene groups at substituted C-6 by DEPT135 spectra, while those of unsubstituted groups were 61 to 62 ppm. Signals at 79 to 83 ppm were assigned to C-3 of reducing-end galactosyl residues by 1D-total correlation spectroscopy and heteronuclear single quantum correlation spectra. Therefore, both galactotrioses appeared to consist of (1→6)-linked galactosyl residues and (1→3)-linked reducing-end galactosyl residues. The difference between chemical shifts of two C-6 signals of 2 was 0.22 ppm, attributed to the presence of α- and β-forms in the reducing-end galactosyl residues, whereas that (0.02 ppm) of 1 was much smaller. Galactotriose 1 and 2 were, thus, identified as β-Gal-(1→6)-β-Gal-(1→3)-Gal and β-Gal-(1→3)[β-Gal-(1→6)]-Gal, respectively. These structures were also confirmed by heteronuclear multiple-bond correlation and rotating frame overhauser enhancement spectroscopy spectra.
Figure 8.
13C-NMR spectra of galactotriose 1 (A) and 2 (B).
Preparation of Glycosidases
The α-l-arafase (specific activity 29 units mg protein−1; EC 3.2.1.55) and β-GlcUAase (specific activity 19.3 units mg protein−1; EC 3.2.1.31) were purified from a culture of R. flava (Uesaka et al., 1978) and Pectinex Ultra SP-L, a commercial pectolytic enzyme from Aspergillus niger (Kuroyama et al., 2001), respectively. One unit of α-l-arafase and β-GlcUAase released 1 μmol of l-Ara or PNP from arabinan and PNP-β-GlcUA, respectively, per minute under the respective standard assay conditions (Uesaka et al., 1978; Kuroyama et al., 2001).
Analytical Methods
The concentration of protein was determined by the method of Bradford with bovine serum albumin as the standard (Bradford, 1976). Reducing sugars were estimated by the method of Nelson (1944) and Somogyi (1952). Total sugars were determined by the phenol-sulfuric acid method (Dubois et al., 1956). Mono- and oligosaccharides in enzymatic hydrolysates were separated by thin-layer chromatography (TLC) on silica gel 60F254 (Merck, Darmstadt, Germany) using 7:1:2 (v/v/v) 1-propanol:ethanol:water as solvent and detected by charring after spraying TLC plates with 20% (v/v) H2SO4-methanol. The hydrolysis products were also analyzed on paper chromatography with 6:4:3 (v/v/v) 1-butanol:pyridine:water and 5:2:3 (v/v/v) 1-butanol:acetic acid:water as the solvents and detected with an alkaline AgNO3 reagent. Quantification of monosaccharides was carried out by high-performance anion-exchange chromatography (HPAEC) using a Dionex DX-500 liquid chromatograph fitted with a CarboPac PA-1 column and a pulsed amperometric detector as described previously (Ishikawa et al., 2000). Methylation of polysaccharides was performed by the method of Hakomori (1964). Gas-liquid chromatography of sugars as alditol acetates was performed with a Shimadzu gas chromatograph GC-6A fitted with a column (0.28 mm × 50 m) of Silar-10C, according to the method of Albersheim et al. (1967). MALDI-TOF/MS was performed with a KOMPACT MALDI IV tDE (Shimadzu, Kyoto). Proteins dissolved in 0.5 μL of water were crystallized by adding 0.5 μL of matrix solution containing 1% (w/v) sinapinic acid and 0.1% (v/v) trifluoroacetic acid, and 0.5 μL of 1% (w/v) NaCl, which was then allowed to dry. For the mass calibration, bovine serum albumin (molecular mass 66,431 D) was used. For oligosaccharide samples, 2,5-dihydroxybenzoic acid was used as the matrix in 10% (v/v) ethanol at a concentration of 10 mg mL−1. Samples were mixed with 0.5 μL of the matrix solution and 0.5 μL of 1% (w/v) NaCl solution. The masses of sugars were determined mainly as pseudomolecular ions (sodium adduct, [M + Na]+).
Peptide Sequencing
Native β-galactosidase was purified from radish seeds by conventional chromatographic techniques as described (Sekimata et al., 1989). The purified β-galactosidase was separated on SDS-PAGE (Laemmli, 1970), blotted onto a PVDF-Plus-membrane (Osmonics, Moers, Germany), and the protein bands with relative molecular masses of 44 and 35 kD were subjected to an N-terminal amino acid analysis with a protein sequencer (HP G1000A; Hewlett-Packard, Palo Alto).
Isolation of cDNA
The cDNA encoding radish β-galactosidase was cloned by reverse transcription-PCR. Total RNA was extracted from 6-d-old dark-grown seedlings. The seedlings were frozen in liquid nitrogen, homogenized with mortar and pestle, and extracted with a kit of Isogen (Nippon Gene, Tokyo) according to the manufacturer's instructions. Single-strand cDNA was synthesized from 1 μg of total RNA from the seedlings using a reverse transcriptase, ReverTra Ace-α- (Toyobo, Osaka) and oligo(dT)-adaptor primer (5′-GTTTTCCCAGTCACGAC(T)12–18-3′; TaKaRa, Tokyo). A set of degenerate primers, F-1 (5′-ACNTAYGAYCAYCGNGC-3′) and R-1 (5′-TTNACRAANGCRTCNGC-3′), was designed based on the determined amino acid sequences of the purified β-galactosidase. PCR was performed with the set of degenerate primers using the single-strand cDNA as a template under the following conditions: 0.5 min denaturing at 94°C, 0.5 min annealing at 50°C, and 1.5 min amplification at 72°C, 35 cycles. The amplified cDNA fragment encoding the 1.3-kb region of the β-galactosidase gene was subcloned into a pGEM T-Easy vector (Promega, Madison, WI) and the nucleotide sequence was determined with an ABI PRISM 310 genetic analyzer (Applied Biosystems, Foster City, CA). The 3′ region of the cDNA was amplified with an internal specific primer, F-2 (5′-AATTCTGCAACCGAGTCCAC-3′) and an M13M4 adaptor primer (5′-GTTTTCCCAGTCACGAC-3′; TaKaRa), using the single-strand cDNA as a template under the following conditions: 0.5 min denaturing at 94°C, 0.5 min annealing at 55°C, and 2.0 min amplification at 72°C, 35 cycles. The 5′ region of the cDNA was cloned with a 5′-RACE kit (Invitrogen, Carlsbad, CA) using the internal specific primers R-2 (5′-CTATAACGTCCAAGCCACCG-3′) and R-3 (5′-CATCTCAGGAGTACTGCGAG-3′). The coding region for the radish β-galactosidase was amplified with proofreading polymerase (KOD-plus-; Toyobo) and the nucleotide sequence was determined.
Northern- and Southern-Blot Analyses
Genomic DNA was extracted from cotyledons of radish by the method described by Murray and Thompson (1980). The genomic DNA (20 μg) was digested with restriction enzymes, separated on a gel containing 0.7% agarose, and blotted onto nylon membrane (Hybond N+; Amersham Biosciences). The blotted membrane was baked at 80°C for 2 h and then hybridized with a digoxygenin (DIG)-labeled cDNA probe prepared with a DIG high-prime DNA-labeling and detection kit (Roche Diagnostics, Basel). The cDNA probe was the 714-bp fragment excised from RsBGAL1 cDNA with restriction enzyme HindIII. Probe labeling, hybridization, and signal detection were carried out according to the manufacturer's instructions. Total RNA was extracted from light-grown hypocotyls, leaves, and roots, as described above, with a kit of Isogen. Approximately 20 μg of total RNA were separated on 1.2% formaldehyde agarose gel and blotted onto nylon membrane. The blotted membrane was baked and then hybridized with the DIG-labeled cDNA probe. To verify the amount of loaded RNA, ribosomal RNA was stained with 1% (w/v) methylene blue.
Expression of β-Galactosidase in Pichia pastoris
Partial RsBGAL1 cDNA corresponding to the region from Ala-31 to Ala-851 (Fig. 2A) was amplified with specific primers and subcloned into a pGEM5zf+ vector (Promega). After confirmation of the nucleotide sequence, the cDNA fragment was inserted between the EcoRI and SpeI sites that are preceded by yeast α-factor of pPICZαC (Invitrogen). The methylotrophic yeast P. pastoris strain KM71 was transformed with the linearized plasmid construct with a multicopy Pichia expression kit (Invitrogen). The transformants resistant to zeocin were screened according to the manufacturer's instructions. The zeocin-resistant colony was cultured in 1,600 mL of YPG medium containing 1% (w/v) yeast extract, 2% (w/v) peptone, and 1% (w/v) glycerol at 28°C with shaking at 100 rpm for 2 d. The cells were harvested by centrifugation (15 min, 3,200g), washed with ice-cold distilled water, then suspended in 50 mL of YMP medium containing 1% (w/v) yeast extract, 2% (w/v) peptone, and 1% (v/v) methanol. The yeast cells were cultured for another 5 d at 28°C, during which time 0.5 mL of methanol were added each day to induce recombinant RsBGAL1.
Purification of Recombinant Enzyme
All operations were carried out at 0°C to 4°C. The culture medium of the Pichia cells treated with 1% (v/v) methanol for 5 d was centrifuged (15 min, 8,100g) and the supernatant was collected. After dialysis against 20 mm sodium acetate buffer, pH 5.0, for 2 d, the sample was adsorbed onto a 2.8 × 73-cm CM-cellulose (CM-32; Whatman, Clifton, NJ) column that had been equilibrated with the buffer. The column was eluted with a linear gradient of 0 to 500 mm KCl in the buffer (flow rate 1 mL min−1, total volume 150 mL, 6 mL per fraction). The active fractions were collected, dialyzed against 10 mm potassium phosphate buffer (pH 6.8), and applied onto a 2 × 19.5-cm hydroxyapatite (Bio-Gel HTP; Bio-Rad, Richmond, CA) column equilibrated with the same buffer. The column was first eluted with 400 mm KCl in the buffer (flow rate 0.3 mL min−1, total volume 10 mL, 1 mL per fraction), then with a linear gradient of 10 to 400 mm potassium phosphate buffer (pH 6.8; total volume 54 mL). The active fractions were combined and dialyzed against 20 mm sodium acetate buffer (pH 5.0) for 2 d before their properties were determined.
Enzyme Assay
The activity of β-galactosidase was determined in a reaction mixture (0.3 mL) consisting of the enzyme, 2 mm PNP-β-Gal, and 50 mm acetate buffer (pH 4.0). After incubation at 37°C, the reaction was terminated by addition of 200 mm Na2CO3 (800 μL) and monitored at 420 nm. One unit of enzyme activity liberates 1 μmol of PNP per minute.
For the determination of the substrate specificity of the purified recombinant enzyme (rRSBGAL1), enzyme activity was measured using reaction mixtures (100 μL) consisting of the enzyme, 0.5% (w/v) polysaccharide or 5 mm galactooligosaccharide, and 100 mm sodium acetate buffer (pH 4.0). After incubation at 37°C for the appropriate reaction time, the liberated sugars were determined reductometrically. Mono- and oligosaccharides in enzymatic hydrolysates were separated by TLC or paper chromatography.
Hydrolysis of AGPs by Recombinant Enzyme
In order to examine the synergistic action of rRsBGAL1 and other glycosidases on AGPs, aliquots (0.1 mg) of radish root AGP pretreated with α-l-arafase were digested by incubation with rRsBGAL1 (20 mU), β-GlcUAase (4 mU), and α-l-arafase (4 mU), or with the same amounts of rRsBGAL1 and β-GlcUAase in 100 mm acetate buffer, pH 4.0 (0.1 mL) at 37°C for 20 h under a drop of toluene. This was followed by the addition of an equal amount of rRsBGAL1 and incubation for an additional 12 h. Reducing sugars released were determined reductometrically at appropriate time intervals with Gal as the standard. For the controls, equal amounts of rRsBGAL1, β-GlcUAase, and α-l-arafase acted separately on the modified AGP under the same conditions. Only to the rRsBGAL1 mixture a further 20 mU of rRsBGAL1 were added after 20 h of incubation. The hydrolysis products were analyzed by paper chromatography and HPAEC.
A large-scale reaction was conducted in a reaction mixture (2 mL) containing 10 mg of the enzymatically modified AGP with increasing amounts of the three enzymes or rRsBGAL1 alone under conditions similar to those specified above. After the reactions were terminated by heating followed by desalting with Dowex 50W (H+) resins, the reaction products were chromatographed on a 2 × 90-cm Bio-Gel P-2 (Bio-Rad) column equilibrated and eluted with 1% (v/v) acetic acid. The products emerged at void volume (Vo) and inner volume (Vi) of the column: The yields of total sugars in high- and low-Mr components were 1.4 and 7.5 mg for the reaction with the three enzymes, and 6.2 and 1.1 mg for the reaction with rRsBGAL1 alone. Parts of the high-Mr components were subjected to acid hydrolysis by heating with 2 n H2SO4 at 100°C for 4 h and the hydrolysates were analyzed by paper chromatography and HPAEC. The structures of the high-Mr components were determined by methylation analysis.
Acknowledgments
We are grateful to Dr. T. Miura (Chiba Institute of Science, Chiba, Japan) and Prof. T. Inazu (Tokai University, Kanagawa, Japan) for providing an oligosaccharide substrate.
This work was supported in part by a Grant for Ground Research for Space Utilization (to T.K.) from the Japan Space Forum.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.062562.
References
- Albersheim P, Nevins DJ, English PD, Karr A (1967) A method for the analysis of sugars in plant cell-wall polysaccharides by gas liquid chromatography. Carbohydr Res 5: 340–345 [Google Scholar]
- Aspinall GO, Auret BJ, Hirst EL (1958. a) Gum ghatti (Indian gum). Part III. Neutral oligosaccharides found on partial acid hydrolysis of the gum. J Chem Soc 4408–4414
- Aspinall GO, Hirst EL, Ramstad E (1958. b) The constitution of larch ɛ-galactan. J Chem Soc 593–601
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Carey AT, Holt K, Picard S, Wilde R, Tucker GA, Bird CR, Schuch W, Seymour GB (1995) Tomato exo-(1→4)-β-D-galactanase. Isolation, changes during ripening in normal and mutant tomato fruit, and characterization of a related cDNA clone. Plant Physiol 108: 1099–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Wang H, Wu H-M (1995) A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82: 383–393 [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28: 350–356 [Google Scholar]
- Fincher GB, Stone BA, Clarke AE (1983) Arabinogalactan-proteins: structure, biosynthesis, and function. Annu Rev Plant Physiol 34: 47–70 [Google Scholar]
- Gao M, Showalter AM (1999) Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan protein involvement. Plant J 19: 321–331 [DOI] [PubMed] [Google Scholar]
- Gemmill TR, Trimble RB (1999) Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim Biophys Acta 1426: 227–237 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Carpita NC (1991) Tracing cell wall biogenesis in intact cells and plants. Selective turnover and alteration of soluble and cell wall polysaccharides in grasses. Plant Physiol 97: 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakomori S (1964) A rapid permethylation of glycolipid, and polysaccharide catalyzed by methylsulfinyl carbanion in dimethyl sulfoxide. J Biochem (Tokyo) 55: 205–208 [PubMed] [Google Scholar]
- Hata K, Tanaka M, Tsumuraya Y, Hashimoto Y (1992) α-L-Arabinofuranosidase from radish (Raphanus sativus L.) seeds. Plant Physiol 100: 388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280: 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Bairoch A (1993) New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 293: 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Tsumuraya Y, Hashimoto Y (1994) Characterization of spinach leaf α-L-arabinofuranosidases and β-galactosidases and their synergistic action on an endogenous arabinogalactan-protein. Physiol Plant 92: 286–296 [Google Scholar]
- Ishikawa M, Kuroyama H, Takeuchi Y, Tsumuraya Y (2000) Characterization of pectin methyltransferase from soybean hypocotyls. Planta 210: 782–791 [DOI] [PubMed] [Google Scholar]
- Kang I-K, Suh S-G, Gross KC, Byun J-K (1994) N-terminal amino acid sequence of persimmon fruit β-galactosidase. Plant Physiol 105: 975–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komalavilas P, Zhu J-K, Nothnagel EA (1991) Arabinogalactan-proteins from the suspension culture medium and plasma membrane of rose cells. J Biol Chem 266: 15956–15965 [PubMed] [Google Scholar]
- Kotake T, Yamaguchi D, Ohzono H, Hojo S, Kaneko S, Ishida HK, Tsumuraya Y (2004) UDP-sugar pyrophosphorylase with broad substrate specificity toward various monosaccharide 1-phosphates from pea sprouts. J Biol Chem 279: 45728–45736 [DOI] [PubMed] [Google Scholar]
- Kuroyama H, Tsutsui N, Hashimoto Y, Tsumuraya Y (2001) Purification and characterization of a β-glucuronidase from Aspergillus niger. Carbohydr Res 333: 27–39 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lee RC, Hrmova M, Burton RA, Lahnstein J, Fincher GB (2003) Bifunctional family 3 glycoside hydrolases from barley with α-L-arabinofuranosidase and β-D-xylosidase activity. Characterization, primary structures, and COOH-terminal processing. J Biol Chem 278: 5377–5387 [DOI] [PubMed] [Google Scholar]
- Majewska-Sawka A, Nothnagel EA (2000) The multiple roles of arabinogalactan proteins in plant development. Plant Physiol 122: 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney L, Ormerod AP, Gidley MJ, Knox JP (2000) Temporal and spatial regulation of pectic (1→4)-β-D-galactan in cell walls of developing pea cotyledons: implications for mechanical properties. Plant J 22: 105–113 [DOI] [PubMed] [Google Scholar]
- Motose H, Sugiyama M, Fukuda H (2004) A proteoglycan mediates inductive interaction during plant vascular development. Nature 429: 873–878 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Tsumuraya Y, Hashimoto Y, Yamamoto S (1984) Arabinogalactan-proteins reacting with eel anti-H agglutinin from leaves of cruciferous plants. Agric Biol Chem 48: 753–760 [Google Scholar]
- Nelson N (1944) A photometric adaptation of the Somogyi method for the determination of glucose. J Biol Chem 153: 375–380 [Google Scholar]
- Nothnagel EA (1997) Proteoglycans and released components in plant cells. Int Rev Cytol 174: 195–291 [DOI] [PubMed] [Google Scholar]
- Okemoto K, Uekita T, Tsumuraya Y, Hashimoto Y, Kasama T (2003) Purification and characterization of an endo-β-(1→6)-galactanase from Trichoderma viride. Carbohydr Res 338: 219–230 [DOI] [PubMed] [Google Scholar]
- Reiter W-D, Vanzin GF (2001) Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol Biol 47: 95–113 [PubMed] [Google Scholar]
- Ross GS, Wegrzyn T, MacRae EA, Redgwell RJ (1994) Apple β-galactosidase. Activity against cell wall polysaccharides and characterization of a related cDNA clone. Plant Physiol 106: 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Taura F, Shoyama Y, Morimoto S (2000) Molecular characterization of a novel β-glucuronidase from Scutellaria baicalensis Georgi. J Biol Chem 275: 27466–27472 [DOI] [PubMed] [Google Scholar]
- Sekimata M, Ogura K, Tsumuraya Y, Hashimoto Y, Yamamoto S (1989) A β-galactosidase from radish (Raphanus sativus L.) seeds. Plant Physiol 90: 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu J-K (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Gross KC (2000) A family of at least seven β-galactosidase genes is expressed during tomato fruit development. Plant Physiol 123: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Starrett DA, Gross KC (1998) A gene coding for tomato fruit β-galactosidase II is expressed during fruit ripening. Cloning, characterization, and expression pattern. Plant Physiol 117: 417–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi N (1952) Notes on sugar determination. J Biol Chem 195: 19–23 [PubMed] [Google Scholar]
- Sørensen SO, Pauly M, Bush M, Skjøt M, McCann MC, Borkhardt B, Ulvskov P (2000) Pectin engineering: modification of potato pectin by in vivo expression of an endo-1,4-β-D-galactanase. Proc Natl Acad Sci USA 97: 7639–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainotti L, Spinello R, Piovan A, Spolaore S, Casadoro G (2001) β-Galactosidases with a lectin-like domain are expressed in strawberry. J Exp Bot 52: 1635–1645 [PubMed] [Google Scholar]
- Tsumuraya Y, Hashimoto Y, Yamamoto S, Shibuya N (1984) Structure of L-arabino-D-galactan-containing glycoproteins from radish leaves. Carbohydr Res 134: 215–228 [Google Scholar]
- Tsumuraya Y, Mochizuki N, Hashimoto Y, Kovác P (1990) Purification of an exo-β-(1→3)-galactanase of Irpex lacteus (Polyporus tulipiferae) and its action on arabinogalactan-proteins. J Biol Chem 265: 7207–7215 [PubMed] [Google Scholar]
- Tsumuraya Y, Ogura K, Hashimoto Y, Mukoyama H, Yamamoto S (1988) Arabinogalactan-proteins from primary and mature roots of radish (Raphanus sativus L.). Plant Physiol 86: 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesaka E, Sato M, Raiju M, Kaji A (1978) α-L-Arabinofuranosidase from Rhodotorula flava. J Bacteriol 133: 1073–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-M, Wang H, Cheung AY (1995) A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell 82: 395–403 [DOI] [PubMed] [Google Scholar]