Abstract
A recent study has demonstrated that phototropins act as blue light receptors in stomatal guard cells. However, the downstream components responsible for phototropin signaling are largely unknown. In this study, using a yeast two-hybrid system, we isolated a Vicia faba protein that has a high similarity to dynein light chain in the C terminus, which interacts with Vicia faba phototropin 1a (Vfphot1a). Protein-blot and two-hybrid analyses revealed that Vfphot1a interacting protein (VfPIP) bound to the C-terminal region of Vfphot1a but did not bind to Vfphot1b. The interaction between VfPIP and Vfphot was indicated by a pull-down assay. Northern analysis revealed that the transcription level of VfPIP gene was more abundant in guard cells than in other tissues or cell types. The transiently expressed fusion protein of VfPIP-green fluorescent protein was localized on cortical microtubules in Vicia guard cells. Microtubule-depolymerizing herbicides partially inhibited both blue light-dependent H+ pumping in Vicia guard cell protoplasts and stomatal opening in the Vicia epidermis. From these results, we conclude that VfPIP may act as a downstream component of phototropin (Vfphot1a) in blue light signaling in guard cells. The possible role of VfPIP in blue light signaling of guard cells is discussed.
Blue light induces a variety of physiological responses including phototropism, chloroplast relocation, leaf expansion, and stomatal opening (Briggs and Huala, 1999; Briggs and Christie, 2002). Such responses are mediated by a newly identified class of blue light receptors, phototropins (Huala et al., 1997; Christie et al., 1998; Kagawa et al., 2001; Kinoshita et al., 2001; Sakai et al., 2001). Phototropins (phot1, phot2) are approximately 120-kD proteins that contain two LOV (light, oxygen, voltage) domains at the N terminus and Ser/Thr kinase domains at the carboxyl terminus (Huala et al., 1997). The proteins become activated in response to absorption of blue light by a chromophore FMN (flavin mononucleotide) in the LOV domains, resulting in autophosphorylation of multiple Ser residues (Christie et al., 1998; Salomon et al., 2000, 2003; Sakai et al., 2001; Kinoshita et al., 2003).
Guard cells modulate stomatal apertures in response to various kinds of external stimuli such as hormones, metabolic demands, CO2 concentration, and light (Assmann and Shimazaki, 1999; Schroeder et al., 2001). Recently, a genetic study has demonstrated that the Arabidopsis (Arabidopsis thaliana) phototropins Atphot1 and Atphot2 function as blue light receptors in guard cells (Kinoshita et al., 2001). The opening of stomata is mediated by an accumulation of potassium salt in guard cells, and the potassium accumulation through the voltage-gated potassium channels is driven by an inside-negative, electrical potential across the plasma membrane (Hedrich and Schroeder, 1989; Assmann, 1993; Assmann and Shimazaki, 1999; Schroeder et al., 2001). This electrical potential is created by the activation of plasma membrane H+-ATPase, where the penultimate Thr residue of the C terminus is phosphorylated by blue light with subsequent binding of a 14-3-3 protein to the Thr in guard cells (Assmann et al., 1985; Shimazaki et al., 1986; Kinoshita and Shimazaki, 1999; Emi et al., 2001). However, little is known about signal transduction from phototropins to the H+-ATPase (Schroeder et al., 2001).
Recently, it has been revealed that two proteins interact with phototropins. One is a 14-3-3 protein and the other is NPH3 (nonphototropic hypocotyl 3). The 14-3-3 protein interacts with various target proteins dependent on the phosphorylation of their consensus motifs and has been demonstrated to bind to Vfphots upon their phosphorylations (Kinoshita et al., 2003). NPH3, which contains two protein-protein interaction domains, i.e. a BTB (broad complex, tramtrack, and bric a brac)/POZ (pox virus and zinc finger) domain (Albagli et al., 1995; Aravind and Koonin, 1999) and a coiled-coil domain (Lupas, 1996), has been revealed to interact with Atphot1 in vitro and has been shown to be involved in blue light-induced phototropic bending (Motchoulski and Liscum, 1999). RPT2 (root phototropism2), which shows high homology to NPH3, has been revealed to play a role in blue light-dependent root phototropism (Sakai et al., 2000). More recently, RPT2 has been demonstrated to transduce blue light signals into stomatal opening and phototropism (Inada et al., 2004). However, the proteins 14-3-3, NPH3, and RPT2 do not possess any enzymatic domains, suggesting that these may function as regulators of phototropins or scaffold proteins working together with the enzymatic components of photoactivated phosphorelay. Furthermore, it has been reported that phototropin localizes on the plasma membrane but that it does not contain a membrane localization domain, which suggests that other proteins are present that cause phototropin to localize on the plasma membrane. Thus, it is likely that the other phototropin interaction proteins are present in guard cells.
In this study, we aimed to find protein components that receive signals from phototropins in guard cells, and employed a yeast two-hybrid screening using a Vicia faba guard-cell cDNA library as prey. Since we have already demonstrated the presence of Vicia faba phototropin 1a (Vfphot1a) and Vfphot1b in guard cells of V. faba (Kinoshita et al., 2003), we here used Vfphot1a as bait for the screening. We isolated an interactive protein with Vfphot1a and identified the protein, which has some similarity to a dynein light chain in animal cells. The functional relevance of phototropin signaling to this protein was also investigated.
RESULTS
Isolation of a Protein That Interacts with Vfphot1a in Yeast
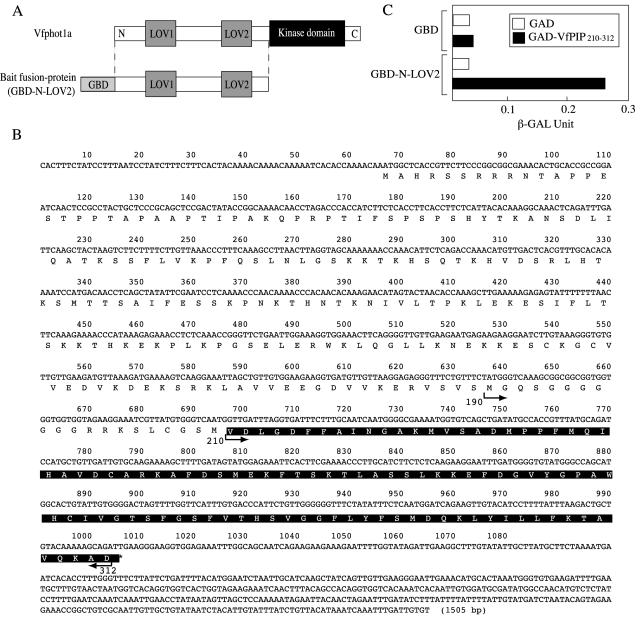
To isolate proteins that interact with Vfphot1a, the N-terminal region of Vfphot1a, which was devoid of a kinase domain, was used as bait (Gal4 DNA-binding domain [GBD]-N-LOV2) in a yeast two-hybrid system to screen a guard-cell cDNA library from V. faba (Fig. 1A). We screened 3×106 colonies and obtained eight positive clones. All of the positive clones possessed a sequence similar to that of the dynein light chain of animal cells in the C terminus but had a long extension in the N terminus that was lacked in the dynein light chain (Fig. 1B). The sequence similarity between this dynein light chain like protein and human dynein light chain was 52.4% in the C terminus. We named this protein the Vicia faba phototropin1a interacting protein (VfPIP). Since the isolated cDNA clone seemed to contain a part of VfPIP gene, 5′ rapid amplification of the cDNA ends was used to obtain the full-length cDNA. The cDNA contained an open reading frame of 939 bp, encoding a deduced polypeptide of 312 amino acid residues with a predicted molecular mass of 34.3 kD (Fig. 1B). The molecular masses of dynein light chain found in animals were usually about 10 kD, but VfPIP had a molecular mass of 34.3 kD because of an added N-terminal region of 24 kD. All of the positive clones obtained by the yeast two-hybrid system encoded the C-terminal region of Val-210 to Asp-312, which possesses a sequence similar to that of dynein light chain.
Figure 1.
Interaction of VfPIP with Vfphot1a in yeast. A, Schematic representation of the Vfphot1a (top) and the fusion protein of GBD with N-terminal region of Vfphot1a including both LOV1 and LOV2 (N-LOV2; bottom). The fusion proteins were used as bait in the yeast two-hybrid screening. The locations of the LOV domains and kinase domain are indicated on the Vfphot1a molecule. B, Nucleotide and deduced amino acid sequence of VfPIP. Residues on a black background indicate the region isolated by yeast two-hybrid screening. C, Quantitative determination of interaction between VfPIP210–312 and Vfphot1a. These two constructs were retransduced into yeast and β-galactosidase activity was measured by yeast two-hybrid assay. GBD and GBD-LOV2, as defined in A; GAD, Gal4 activation domain; VfPIP210–312, the C-terminal region of VfPIP shown with a black background in Figure 1C.
We retransduced these positive clones into yeast and measured the β-galactosidase activity to estimate the interaction between VfPIP210–312 and Vfphot1a (Fig. 1C). The results indicated that N-LOV2 fused to GBD interacted with VfPIP210–312 fused to Gal4 activation domain (GAD) but did not interact with GAD alone. Furthermore, GBD did not interact with VfPIP210–312 fused to GAD.
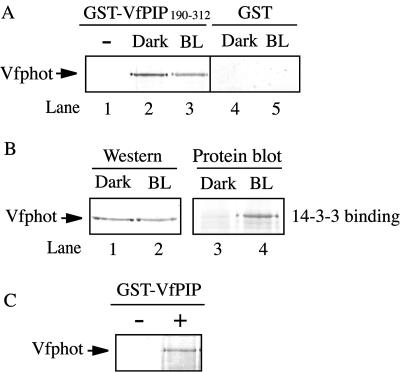
To examine the interaction between Vfphot and VfPIP in vitro, we expressed the glutathione S-transferase (GST)-fused C terminus of VfPIP190–312 (Met-190 to Asp-312) in Escherichia coli and isolated the microsomal fraction from etiolated seedlings of V. faba. The microsomal fraction was used as a source of Vfphot. GST-VfPIP190–312 was incubated with this microsome fraction, and then GST-VfPIP190–312 was pulled down by glutathione-sepharose beads. Proteins bound to the beads were released and the released proteins were subjected to western-blot analysis using antibodies against Vfphot. The results revealed that Vfphot protein was obtained when GST-VfPIP190–312 was present (Fig. 2A, lane 2) but was not found by the GST moiety alone (Fig. 2A, lane 4). In the absence of microsomal fractions, Vfphot was not found (Fig. 2A, lane 1). These results indicate that Vfphot interacts with VfPIP190–312 in vitro. Since it has been demonstrated that phototropin is autophosphorylated in response to blue light, it is possible that the interaction between Vfphot and VfPIP is dependent on phosphorylation. To investigate this, a microsomal fraction was isolated from seedlings that had been preilluminated with blue light for 1 min and was used for a pull-down assay. However, no significant difference in the interaction of Vfphot with VfPIP190–312 was observed (Fig. 2A, lanes 2 and 3). We confirmed the occurrence of autophosphorylation through the binding of 14-3-3 protein to Vfphot by this blue light treatment (Fig. 2B, lanes 3 and 4). The 14-3-3 protein bound to Vfphot only when the seedlings had been illuminated with blue light. Western analysis revealed that the amount of Vfphot used in this experiment was the same (Fig. 2B, lanes 1 and 2). We also confirmed that equal amounts of GST-VfPIP190–312 were used for the pull-down assay by amido black staining (data not shown). Furthermore, full-length VfPIP interacted with Vfphot because Vfphot was efficiently pulled down by the GST-VfPIP (Fig. 2C) in the presence of microsome.
Figure 2.
Assay of interaction between Vfphot by GST-VfPIP190–312. A, Pull-down assay of the Vfphot by GST-VfPIP190–312. Etiolated seedlings of V. faba were kept in the dark (Dark) or illuminated with blue light (BL) for 1 min. Then, microsomal membranes were isolated from the seedlings and the membranes were incubated with GST-VfPIP190–312 (lanes 1–3) or GST (lanes 4 and 5) and pulled down. Vfphot pulled down by GST-VfPIP190–312 was determined by western-blot analysis. VfPIP indicate VfPIP. B, Determination of 14-3-3 protein binding to Vfphot. Dark and BL indicate that the microsome fractions isolated from the seedlings that had been kept in the dark or illuminated with blue light, respectively, as mentioned above. Determination of Vfphot in the membranes was done by western blot (lanes 1 and 2). Blue light-dependent 14-3-3 protein binding to Vfphot in the microsome was done using GST-vf14- 3-3a as a probe (lanes 3 and 4), and was detected by anti-GST antibody. C, Pull-down assay of the Vfphot by GST-fused full length VfPIP. GST-VfPIP was incubated with (+) or without (−) microsome fractions.
Specific Interaction between VfPIP and Vfphot1a Homologs
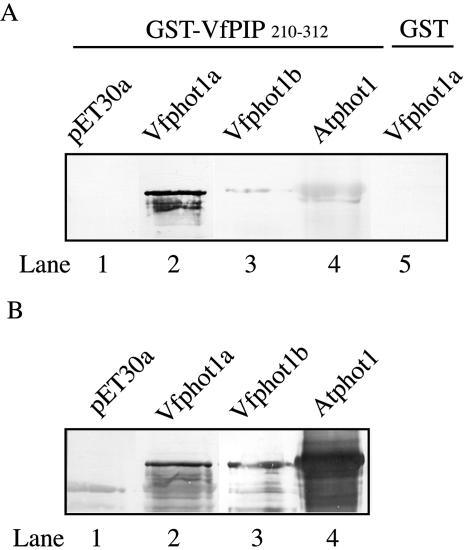
Since Vfphot1a and Vfphot1b migrated similarly on the SDS-PAGE (Kinoshita et al., 2003), we could not determine whether either or both of the phototropins interacted with VfPIP. To determine the binding ability of the phototropins (Vfphot1a and Vfphot1b) to VfPIP, protein-blot analysis was conducted using recombinant His6-tagged-Vfphot1a and Vfphot1b. These recombinant proteins were separated on SDS-PAGE and blotted onto a nitrocellulose membrane. Then, the GST-fused C terminus of VfPIP (GST-VfPIP210–312) was applied as a probe. As shown in Figure 3A, GST-VfPIP210–312 bound to Vfphot1a but did not bind to Vfphot1b or the empty vector (Fig. 3A, lanes 1–3, respectively). We also determined whether the VfPIP interacted with Atphot (Arabidopsis thaliana phototropin). The GST-VfPIP210–312 did not bind to Atphot with high affinity (Fig. 3A, lane 4). GST alone did not bind to Vfphot1a (lane 5). These results indicate that VfPIP210–312 specifically interacts with Vfphot1a. We raised antibodies against Vfphot1a and b, and Atphot1, respectively. Expression of His6-tagged recombinant phototropins was confirmed by western analysis using these antibodies (Fig. 3B, lanes 1–4).
Figure 3.
Specific binding of VfPIP210–312 to Vfphot1a. A, Binding was determined in vitro by protein-blot analysis using GST-VfPIP210–312 as a probe (lanes 1–4) for various phototropins and was detected with anti-GST antibody. His6-tagged- Vfphot1a and Vfphot1b were expressed in E. coli, and His6-tagged Atphot1 was expressed in insect cells. All these recombinant proteins were subjected to SDS-PAGE and blotted onto nitrocellulose membranes. As a control, analysis was done for His6-tagged Vfphot1a using GST alone (lane 5). VfPIP indicates V. faba phototropin 1a interacting protein. B, Western analyses for various phototropin species were done to confirm the presence of these phototropins using the respective antibodies (lanes 1–3, anti-Vfphot antibodies; lane 4, anti-Atphot1 antibodies).
Interaction Site of VfPIP in Vfphot1a
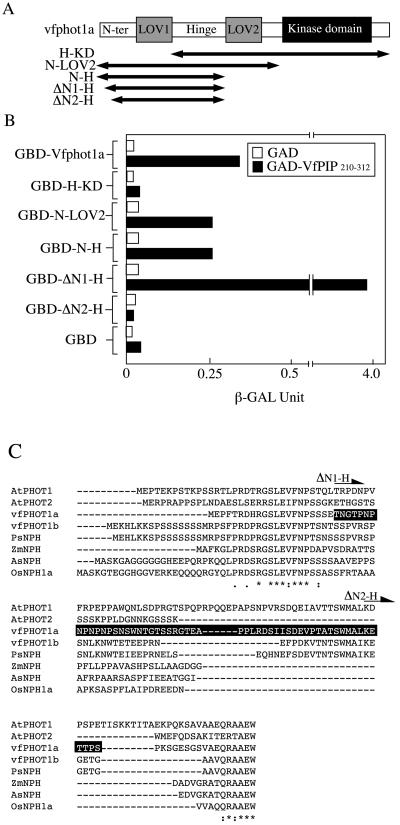
A quantitative interaction assay using a yeast two-hybrid system was employed to determine the binding site of VfPIP in Vfphot1a. A series of Vfphot1a deletion constructs were generated by cloning of Vfphot1a fragments into pAS2-1 bait vectors (Fig. 4A). These constructs were cotransformed into yeast together with VfPIP190–312 that was cloned into the pAD-GAL4-2.1 prey vector. The interaction between bait and prey in yeast results in activation of the lacZ reporter gene and is assayed by measuring β-galactosidase activity. Transformation of the hinge-LOV2-kinase domain (H-KD) of Vfphot1a into yeast resulted in no actual β-galactosidase activity, indicating that VfPIP does not interact with this region of Vfphot1a (Fig. 4B). By contrast, transformation of the N-terminal region of Vfphot1a, which contained the N terminus to the LOV2 domain (N-LOV2) or to the hinge region (N-H), resulted in high β-galactosidase activities (Fig. 4B), indicating that VfPIP interacted with the N-terminal region of Vfphot1a. When ΔN1-H, in which the N terminus of the Thr-22 residue is deleted, was transduced into yeast, the amount of β-galactosidase activity was much higher than that induced by full-length Vfphot1a. However, transformation of ΔN2-H, on which the N terminus of the Pro-77 residue is deleted, did not yield any significant activity (Fig. 4B). These results indicate that VfPIP interacts with the N-terminal region between Thr-22 and Pro-77 in Vfphot1a (Fig. 4C).
Figure 4.
Interaction of VfPIP with Vfphot1a on the N-terminal region. A, Schematic representation of the divided Vfphot1a region used as bait. B, Interactions of VfPIP210–312 with the Vfphot1a N-terminal region were determined by quantitative yeast two-hybrid assay. GBD, H-KD, N-LOV2, N-H, ΔN1-H, and ΔN2-H are as defined in A; GAD and VfPIP210–312 are as defined in Figure 1. VfPIP indicates V. faba phototropin 1a interacting protein. C, Alignment of phototropin amino acid sequences. Residues on a black background indicate interaction region in the VfPIP. The arrowhead shows the start point of ΔN1-H and ΔN2-H, respectively. Psphot1, Pisum sativum phototropin; Zmphot1 and 2, Zea mays phototropin1 and 2, respectively; Osphot1, Oryza sativa phototropin1.
Expression of Gene Products of VfPIP in Several Tissues
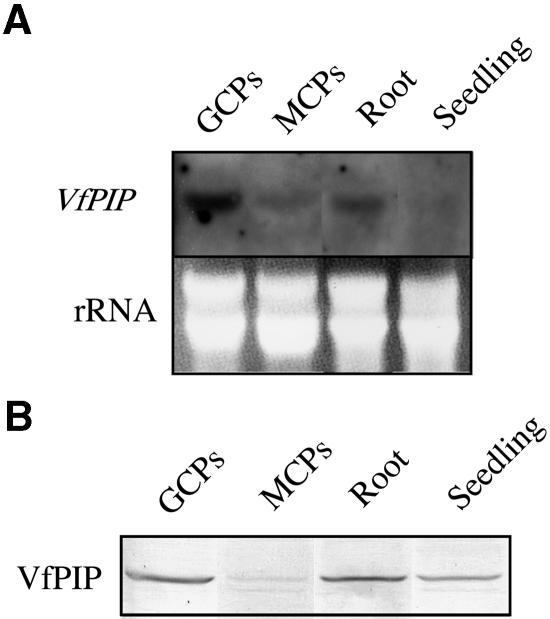
The transcription levels of VfPIP were investigated in guard cell protoplasts, mesophyll cell protoplasts, root, and etiolated seedlings of V. faba (Fig. 5A). A digoxigenin (DIG)-labeled probe for VfPIP gene transcript hybridized to a band of mRNA of 1,500 bp in each lane. This length corresponded to that of the full-length mRNA encoding VfPIP. The transcription level of VfPIP gene was the highest in guard cell protoplasts among other tissues including mesophyll cell protoplast, roots, and etiolated seedlings. Transcript of VfPIP gene was more abundant in roots than in mesophyll cell protoplasts and seedlings.
Figure 5.
Northern and western-blot analyses of VfPIP. A, Northern-blot analysis of the VfPIP gene in guard cell protoplasts (GCPs), mesophyll cell protoplasts (MCPs), roots, and etiolated seedlings. Each lane contained an equal amount (25 μg) of total RNA isolated from guard cell protoplasts, mesophyll cell protoplasts, roots, and etiolated seedlings. These hybridized with DIG-labeled specific probes of VfPIP gene. Staining of gel with ethidium bromide is shown as a loading control (rRNA). Experiments were repeated three times on different occasions and gave similar results. B, Western-blot analysis using antibodies raised against the C-terminal region of VfPIP as a probe. Each lane contained an equal amount (100 μg) of protein isolated from guard cell protoplasts, mesophyll cell protoplasts, roots, and etiolated seedlings.
Expression of VfPIP protein was investigated in these tissues of V. faba by western analysis using antibodies raised against VfPIP190–312. As shown in Figure 5B, a 35.5-kD band was found, and this molecular mass corresponded to that of VfPIP deduced from the amino acid sequence (34.3 kD). The expression level of VfPIP protein was highest in guard cell protoplasts and lowest in mesophyll cell protoplasts. VfPIP was also found in seedlings, although the transcript level was very low.
Subcellular Localization of VfPIP
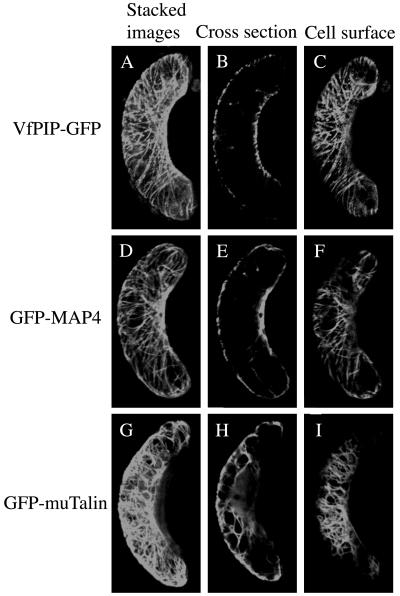
Dynein light chain is a member of the cytoplasmic dynein complex, which is associated with microtubules in animal cells. However, all of the dynein subunits except D are absent in the Arabidopsis genome database. We investigated the distribution of VfPIP in guard cells using VfPIP-green fluorescent protein (GFP) fusion proteins in a transient expression assay. The construct of VfPIP-GFP was introduced into guard cells of Vicia leaf epidermis by particle bombardment to allow the expression of fusion protein. In the transformant, guard cells displayed a marked filamentous fluorescence in the vicinity of the plasma membrane (Fig. 6, A–C). Because this fluorescence profile resembled the distribution of cytoskeletons including the microtubule and actin filament, the localization of VfPIP-GFP was compared with those of the microtubule-associated protein MAP, the microtubule binding domain of MAP4, and the actin-associated protein, mouse Talin. These cytoskeleton-associated proteins were expressed transiently as GFP fusion proteins. GFP-MAP4 revealed the filamentous fluorescence close to the plasma membrane and slightly in cytoplasm (Fig. 6, D–F). By contrast, GFP-muTalin showed a strong mesh-like fluorescence close to the plasma membrane, and the fluorescence was also found inside the cells, probably in the cytoplasm (Fig. 6, G–I).
Figure 6.
Transient expression of VfPIP-GFP, GFP-MAP4, and GFP-muTalin in Vicia guard cells. Fusion proteins of VfPIP-GFP, GFP-MAP4, and GFP-muTalin were expressed transiently by particle bombardment. Guard cells expressing VfPIP-GFP, GFP-MAP4, and GFP-muTalin were examined through GFP fluorescence by a confocal laser microscope. A to C, VfPIP-GFP; G to F, GFP-MAP4; H to J, GFP-muTalin; A, D, and G, stacked images; B, E, and H, cross section; C, H, and I, cell surface.
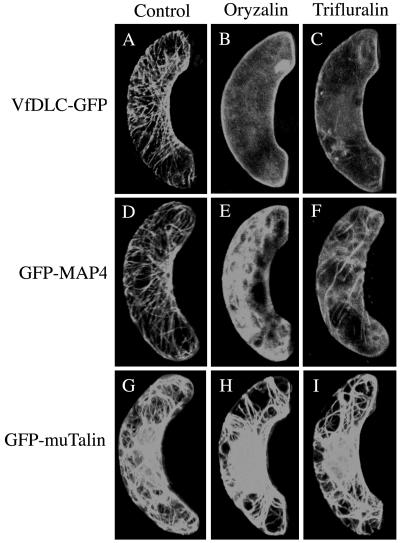
The fluorescence profile of VfPIP-GFP resembled that of GFP-MAP4, and it is most likely that VfPIP-GFP is localized on the microtubules. To demonstrate this, the transformant of Vicia leaf epidermis that expressed GFP fusion proteins was treated with depolymerizing compounds of microtubules. The treatments by oryzalin and trifluralin almost completely destroyed the filamentous structure of VfPIP and released the VfPIP-GFP to the cytoplasm, and stimulated the translocation of VfPIP-GFP to the plasma membrane. The results suggest that VfPIP localized on the cortical microtubules and was released from the microtubules after destruction of their structures by the compounds. In accord with the above results, the treatments with oryzalin and trifluralin destroyed the filamentous structure of GFP-MAP4, and some of the GFP-MAP4 was stimulated to move to the cytoplasm (Fig. 7, D–F). By contrast, neither treatment destroyed the mesh-like fluorescent structure of GFP-muTalin (Fig. 7, G–I), suggesting that the compounds used here specifically affect the microtubule structure but not the actin filaments.
Figure 7.
Effects of microtubule-depolymerizing herbicides on localization of VfPIP-GFP, GFP-MAP4, and GFP-muTalin in Vicia guard cells. Guard cells expressing VfPIP-GFP, GFP-MAP4, and GFP-muTalin were treated with 10 μm trifluralin or 10 μm oryzalin for 2 h in the dark and then measured the GFP fluorescence under a confocal laser microscope. A to C, VfPIP-GFP; D to F, GFP-MAP4; G to I, GFP-muTalin; A, D, and G, 0.1% DMSO; B, E, and H, oryzalin; C, F, and I, trifluralin.
Effects of Microtubule-Depolymerizing Compounds on Blue Light-Dependent Stomatal Opening and H+ Pumping
VfPIP localized on cortical microtubules and interacted with Vfphot1a, and thus it is expected that some function of Vfphot1a is mediated by the microtubules. Since phototropins in V. faba mediate stomatal opening in response to blue light (Kinoshita et al., 2003), we tested the effects of microtubule-depolymerizing compounds on blue light-dependent stomatal opening. Epidermal peels of Vicia leaves were preincubated with each compound for 2 h in the dark before the illumination. Stomata opened in response to red light at 150 μmol m−2 s−1 and opened wider in response to blue light at 15 μmol m−2 s−1 superimposed on the red light (Table I). The latter opening is specific to blue light (Schwartz and Zeiger, 1984). Red light-induced stomatal opening was not inhibited by the compounds. However, blue light-dependent stomatal opening was inhibited by 10 μm trifluralin to 49.2%, 10 μm oryzalin to 38.8%, and 20 μm propyzamide to 79.6%, respectively (Table I).
Table I.
Effects of microtubule-depolymerizing compounds on blue light-dependent stomatal opening in the Vicia leaf epidermis
Vicia epidermal peels were treated with microtubule-depolymerizing compounds and illuminated with red light at 150 μmol m−2 s−1 or blue light at 15 μmol m−2 s−1 under the red light background at150 μmol m−2 s−1. A total of 10 μm trifluralin, 10 μm oryzalin, and 20 μm propyzamide were used in the experiment. All these compounds were solubilized in dimethyl sulfoxide (DMSO) and applied at the final concentration of 0.1% DMSO. As a control, the measurement was done at a final concentration of 0.1% DMSO. Blue light-specific opening was given by the following equation: Blue light-specific opening = Blue light-dependent opening minus red light-dependent opening.
| Stomatal Aperture (μm) ± se (n = 40)
|
||||
|---|---|---|---|---|
| Dark | Red Light | Red + Blue Light | Blue Light-Specific Opening | |
| Control | 3.43 ± 1.06 | 6.47 ± 1.34 | 10.19 ± 0.70 | 3.72 (100%) |
| Trifluralin | 6.37 ± 1.47 | 8.20 ± 1.10 | 1.83 (49.2%) | |
| Control | 3.79 ± 1.03 | 5.66 ± 1.11 | 9.54 ± 0.79 | 3.88 (100%) |
| Oryzalin | 5.89 ± 1.01 | 7.40 ± 1.16 | 1.51 (38.9%) | |
| Control | 3.43 ± 1.06 | 6.47 ± 1.34 | 10.19 ± 0.70 | 3.72 (100%) |
| Propyzamide | 6.26 ± 1.40 | 9.22 ± 1.11 | 2.96 (79.6%) | |
Since inhibition occurs in the opening response specific to blue light, it is expected that these compounds affect blue light-dependent H+ pumping, which in turn drives stomatal opening. To test this, isolated guard cell protoplasts from Vicia were treated with these compounds and blue light-dependent H+ pumping was measured (Table II). As shown in Table II, the magnitudes of blue light-dependent H+ pumping were decreased by 10 μm trifluralin and 10 μm oryzalin to 62.0% and 54.1%, respectively. The maximum rates of blue light-dependent H+ pumping were similarly inhibited by these compounds. Inhibitions by trifluralin and oryzalin were concentration-dependent. However, the inhibitory action of propyzamide on H+ pumping was not obvious, and these results were in accord with those of stomatal opening shown above. From these results, it is likely that the cortical microtubules may play some important roles in mediating light signals from Vfphot1a to the plasma membrane H+-ATPase. However, we note that, in all cases, the effects of compounds are somewhat stronger in the opening responses in epidermal peels than in H+ pumping in guard cell protoplasts.
Table II.
Effects of microtubule-depolymerizing compounds on blue light-dependent H+ pumping in Vicia guard cell protoplasts
Trifluralin, oryzalin, and propyzamide were added to guard cell protoplasts at the indicated concentrations 30 min before illumination with a pulse of blue light. FC was added at 10 μm. Experiments repeated three times on different occasions gave similar inhibitions.
| Concentration | Magnitude | Rate | Rate (+FC) | |
|---|---|---|---|---|
| μm | nmol H+μg protein−1 | nmol H+ h−1μg protein−1 | ||
| Trifluralin | 0 | 0.192 (100) | 2.39 (100) | 3.87 (100) |
| 1 | 0.166 (86.5) | 2.18 (91.2) | 3.60 (93.0) | |
| 10 | 0.119 (62.0) | 1.63 (68.2) | 3.57 (92.2) | |
| Oryzalin | 0 | 0.129 (100) | 1.38 (100) | 3.08 (100) |
| 1 | 0.120 (93.0) | 1.33 (96.4) | 3.08 (100) | |
| 10 | 0.070 (54.1) | 0.72 (52.2) | 3.07 (99.7) | |
| Propyzamide | 0 | 0.130 (100) | 1.70 (100) | 2.01 (100) |
| 20 | 0.122 (93.8) | 1.60 (94.1) | 2.27 (112.9) | |
| 100 | 0.114 (87.7) | 1.33 (78.2) | 2.61 (129.9) |
It is possible that these microtubule-depolymerizing compounds directly affect the activity of plasma membrane H+-ATPase. To check this possibility, we determined the rate of fusicoccin (FC)-dependent H+ pumping in the presence of trifluralin and oryzalin at the same concentrations. FC directly activates the plasma membrane H+-ATPase by stimulation of binding of the 14-3-3 protein to the C terminus. However, the rates of FC-dependent H+-pumping were not inhibited by these compounds, indicating that the H+-ATPase activity was not affected by the compounds. The rate and magnitude of FC-dependent H+ pumping even increased in the presence of 20 μm propyzamide (Table II).
DISCUSSION
Phototropins have been identified as blue light receptors involved in phototropism, chloroplast relocation, leaf extension, and stomatal opening (Briggs and Christie, 2002). Over the past few years, a considerable number of studies have been made on this new class of proteins. Since phototropins induce a wide range of responses, the downstream signaling from phototropins must also diverge. However, the mechanisms by which phototropins transduce the light signal into specific responses are largely unknown. Such a specific response will occur when a phototropin interacts with a specific protein in individual tissues. Only a few studies have so far succeeded in identifying such proteins. NPH3 has been isolated and has been demonstrated to interact with Atphot1, and that NPH3 has been shown to be involved in phototropin-dependent phototropism by a genetic study (Motchoulski and Liscum, 1999). A 14-3-3 protein was shown to bind to phototropin when phototropin was autophosphorylated by blue light in Vicia guard cells, as well as in both leaves and etiolated seedlings (Kinoshita et al., 2003). The binding is fast and reversible. However, the physiological meaning of this finding remains unclear. More recently, RPT2, which is a member of the NPH3 family, was also suggested to interact with Atphot1 in vivo and to mediate Atphot1-induced responses of both phototropism and stomatal opening, but not to affect chloroplast relocation (Inada et al., 2004). There is no evidence that the proteins NPH3, RPT2, or 14-3-3 are substrates for phototropin kinase. Furthermore, it has been reported that phototropin localizes to the plasma membrane (Sakamoto and Briggs, 2002), although phototropins are soluble proteins, and this suggests that it is the presence of a protein that causes phototropin to localize to the plasma membrane. In this study, to isolate new phototropin-interactive proteins, a yeast two-hybrid screening was conducted using the N terminus region of vfphot1a as bait and the Vicia cDNA library from guard cells as prey. We hoped to isolate a guard cell-specific protein by this method. As a result, we obtained a dynein light-chain like protein, VfPIP, as a protein interacting with Vfphot.
Dyneins are molecular motors that translocate to microtubules. The dyneins are comprised of subunits ranging in mass from 9 to over 500 kD. The largest of these subunits, the dynein heavy chains, are involved in force production, whereas the others are defined as accessory subunits based on their association with the dynein complex (King, 2000). The functions of these accessory subunits, i.e. the intermediate, light intermediate, and light chains, are not known, although the intermediate chains have been implicated in targeting dynein molecules to cellular binding sites. Dynein light chains, the smallest dynein subunits, are thought to be associated with the intermediate chains in both axonemal and cytoplasmic dyneins (King and Patel-King, 1995). The dynein light chains also have been shown to interact with unrelated proteins in animal and fungi, such as rhodopsin, neuronal nitric oxide synthase, Bim, IκBα, nuclear respiratory factor-1, and so on, suggesting they may have multiple functions (Jaffrey and Snyder, 1996; Puthalakath et al., 1999; Tai et al., 1999; Herzig et al., 2000).
Dynein heavy chains genes are ubiquitously present in fungi and animals. However, dynein heavy chains genes are absent from the Arabidopsis genome (Lawrence et al., 2001). This indicates that dynein complexes are absent from Arabidopsis. On the other hand, six isoforms of dynein light chain-like protein genes were found in the Arabidopsis genome, and the function of these proteins is unknown.
It has been reported that dynein light chains have been found not only in microtubules but also in the cytoplasm and nucleus (Crépieux et al., 1997; Tai et al., 1998). However, there are no data about the localization of VfPIP. To investigate the localization of VfPIP in Vicia guard cells, we transformed guard cells of Vicia leaf with VfPIP-GFP under the control of the cauliflower mosaic virus (CaMV) 35S promoter by a particle delivery system and expressed VfPIP-GFP transiently. The expression profile in guard cells exhibited a filamentous structure, and VfPIP-GFP localized close to the plasma membrane, with the fluorescence profile resembling that of cortical microtubules (Fig. 6). We thus specifically disrupted the microtubule structures of transformed cells with microtubule-depolymerizing herbicides and found that the filamentous structure of VfPIP-GFP disappeared (Fig. 7). By contrast, the structure of actin filaments was not affected by this treatment. These results strongly suggest that VfPIP localizes to the microtubules. However, we note that no binding motif to the microtubule was found in VfPIP. The VfPIP may indirectly bind to the microtubule through some unidentified proteins.
It has been reported that Atphot1 uniformly localizes on the plasma membrane in guard cells of Arabidopsis and a fraction of Atphot1 dissociates from the plasma membrane in response to blue light (Sakamoto and Briggs, 2002). This Atphot1 localization was distinct from VfPIP localization shown here. It is possible that some portion of Vfphot is dissociated from the plasma membrane and becomes associated with VfPIP after the illumination of blue light.
VfPIP is most likely to be associated directly or indirectly with cortical microtubules in guard cells. Clarifying the functional role of VfPIP in guard cells is the best way to investigate the stomatal response of disruption mutants of VfPIP. By screening the genomic database of Arabidopsis, we found five isoforms of dynein light chains, and the amino acid sequence of one of the isoforms highly resembled that of VfPIP in its C terminus, with 80% identity in the C terminus. We found that Arabidopsis dynein light chain, which has the highest homology to VfPIP, also binds to Atphot1 by protein-blot analysis (data not shown). However, the dynein light chain of Arabidopsis lacked the N terminus found in VfPIP, and a protein homologous to dynein light chain with the N terminus was not found in the database. Two T-DNA insertional lines of Arabidopsis in coding regions of dynein light chain genes were obtained and stomatal openings in the two mutant plants were tested in response to blue light. The stomatal responses of the two mutant lines could not be distinguished from those of wild-type plants (data not shown). This was likely due to functional redundancy in these isoforms of dynein light chains in Arabidopsis.
Since VfPIP seems to localize to the cortical microtubules, the function of VfPIP may be mediated by the microtubules. To investigate this, we treated the epidermis and guard cell protoplasts with microtubule-depolymerizing compounds such as trifluralin, oryzalin, and propyzamide, and measured blue light-dependent-stomatal opening and H+ pumping. As shown in Tables I and II, microtubule disruption by the compounds inhibited both stomatal opening and H+ pumping, and the effect was consistently more severe in opening responses than in H+ pumping. This was probably because the microtubule may not only mediate light signals from phototropins to the plasma membrane H+-ATPase, but may also stimulate stomatal opening by appropriately organizing the microtubule structure.
As shown in Figure 7, however, microtubule-depolymerizing compounds almost completely destroyed the structure of the microtubules, but inhibition of blue light-dependent H+ pumping by these compounds was up to 50%. If all of the blue light signals from phototropins were transmitted to the plasma membrane H+-ATPase via microtubules, the magnitude of inhibition would have been greater than 50%. This discrepancy can be interpreted as follows. Since only Vfphot1a interacts with VfPIP, and since Vfphot1a seems to act redundantly with Vfphot1b in guard cells, a significant portion of the light signals from Vfphot1b may have been transmitted to the plasma membrane H+-ATPase irrespective of the microtubule structure. This may have decreased the sensitivity of blue light-dependent responses to the microtubule-depolymerizing compounds (Tables I and II). Alternatively, the effect of the microtubule-depolymerizing compounds on blue light-dependent H+ pumping may have been indirect.
Taking these results together, we conclude that VfPIP may act as a signal molecule that transduces the light signals from phototropins to cortical microtubules, thereby supporting stomatal opening. It is also possible that the microtubule mediates the light signal to the plasma membrane H+-ATPase for activation via phosphorylation in stomatal guard cells. In accord with these observations, Marcus et al. (2001) indicated that the microtubules of V. faba were involved in events that were upstream of the ion fluxes leading to stomatal opening in response to light. They suggested that some signal molecule associated with microtubules acted as a regulator of ion transport and thereby affected the stomatal opening. The VfPIP identified in this study could be a candidate for such a regulator. Furthermore, Fukuda et al. (1998) demonstrated that the cortical microtubules of V. faba organize in a radial array structure in the daytime with increase in stomatal aperture, and the structure is destroyed from evening to night. Very recently, Lahav et al. (2004) reported that guard-cell microtubules organized in parallel, straight, and dense bundles in response to light, which favors stomatal opening. The effect was specific to blue light, and red light had no effect. They also suggested that microtubule-associated proteins might mediate microtubule reorganization and reorientation via blue light receptors of phototropins. These observations suggest that VfPIP is most likely to mediate organizing microtubules appropriately via the actions of phototropins in guard cells from Vicia.
It has been suggested that, in response to environmental stimuli, guard cells control their cytoskeletons as well as their intracellular ion concentrations (Kim et al., 1995; Eun and Lee, 1997; Assmann and Baskin, 1998; Fukuda et al., 1998; Hwang and Lee, 2001; Marcus et al., 2001). The involvement of actin filaments in stomatal movements has been suggested based on studies showing rapid reorganization of actin in guard cells and inhibitory effects of actin antagonists on stomatal movements (Kim et al., 1995; Eun and Lee, 1997) and on potassium channel activities (Hwang et al., 1997). By contrast, the involvement of microtubules in stomatal movements has also been suggested based on results showing the inhibition of stomatal movement by microtubule-depolymerizing herbicides (Marcus et al., 2001) and a correlation between stomatal movement and the reorganization of microtubules during circadian rhythm (Fukuda et al., 1998). Our results support the notion that microtubules have functional importance in stomatal movement.
MATERIALS AND METHODS
Plant Materials
Plants of Vicia faba L. cv Ryosai Issun were cultured hydroponically for 4 to 7 weeks in a greenhouse (Shimazaki et al., 1992). Fully expanded leaves were harvested from the second or third internode. Etiolated seedlings of V. faba were cultivated for 1 week in the dark at 22°C and 50% to 60% humidity after cold treatment of seeds for 2 d.
Plants of Arabidopsis (Arabidopsis thaliana) were grown in a chamber with a 14-h fluorescent-light/10-h-dark cycle at 24°C and 50% to 60% humidity. Etiolated seedlings of Arabidopsis were grown on an agarose plate containing a 0.5 × Murashige and Skoog salt base, 0.05% MES-KOH, pH 5.7, and 0.8% agarose for 3 d in the dark at 24°C in 50% to 60% humidity after cold treatment of seeds for 4 d.
Protoplast Preparation
Guard cell protoplasts were isolated enzymatically as described previously (Kinoshita and Shimazaki, 1999). Mesophyll protoplasts were prepared according to a method described previously (Shimazaki et al., 1982). Isolated guard cell protoplasts and mesophyll cell protoplasts were stored in 0.4 m mannitol and 1 mm CaCl2, and in 0.6 m mannitol and 1 mm CaCl2, respectively, on ice until use in the dark. Amounts of protein were determined by the method of Bradford (1976).
Yeast Two-Hybrid Screening and Quantitative β-Galactosidase Assays
The cDNA library was prepared using a HybriZAP-2.1 Two-Hybrid cDNA Gigapack Cloning kit and HybriZAP-2.1 Two-Hybrid cDNA Synthesis kit (Stratagene, La Jolla, CA). Total RNA was extracted from V. faba guard cell protoplasts using ISOGEN (Nippon Gene, Tokyo). mRNA was isolated from total RNA using a QuickPrep Micro mRNA Purification kit (Amersham Biosciences, Tokyo). Construction of bait plasmid and yeast two-hybrid screening were performed using a MATCHMAKER Two-Hybrid System 2 (CLONTECH, Palo Alto, CA) according to the manufacturer's instructions.
For measurement of β-galactosidase activity, yeast was grown overnight in synthetic dropout selection medium, diluted to A600 of 0.15 in YPD medium, and grown for an additional 5 h at 30°C in the dark. The yeast cells were washed once in Z buffer (60 mm Na2HPO4, 40 mm NaH2PO4, 10 mm KCl, 1 mm MgSO4, pH 7.0), resuspended in one-fifth volumes of Z buffer, and frozen in liquid N2. After thawing the frozen cells, β-mercaptoethanol and o-nitrophenyl-β-d-galactopyranoside were added at final concentrations of 0.19% and 0.64 mg mL−1, respectively, and extracts were incubated to continue the β-galactosidase reaction at 30°C. The reactions were stopped by adding Na2CO3 to a final concentration of 0.3 m to the extracts. A420 was determined spectrophotometrically, and β-galactosidase activity was indicated in Miller units.
Full-Length VfPIP cDNA Isolation
Two primers, CACTATGCAATGCCATGCTGGTCC and GCCTCTATCCTCGCCGCCGTATC, were used for the 5′ RACE. The 5′ RACE was performed using a GeneRacer kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. PCR products were cloned into a pCR4-TOPO vector (Invitrogen).
Expression and Purification of VfPIP in Escherichia coli
The recombinant VfPIP was expressed and purified from E. coli cells (JM109) as follows. The coding sequence of VfPIP was amplified by PCR using synthetic oligonucleotides that contained an EcoRI site at the 5′ ends and cloned in-frame with GST into the pGEX-4T-1 plasmid vector (Amersham Biosciences). Expression and purification were performed using Bulk and RediPack GST purification modules (Amersham Biosciences) according to the manufacturer's instructions.
Atphot1 Expression in Insect Cells
Recombinant Atphot was expressed in insect cells. The coding sequence of Atphot1 was inserted into the EcoRI site of the pAcHLT-A (BD Biosciences Pharmingen, Palo Alto, CA) and transfected into Sf9 (Spodoptera frugiperda) insect cells using a BaculoGold Transfection kit (BD Biosciences Pharmingen) according to the supplier's instructions. Recombinant baculovirus was used to infect Sf9 insect cells. The expression of recombinant His6-tagged Atphot1 was measured as described previously (Christie et al., 1998).
Pull-Down Assay
To isolate the microsome fraction, V. faba etiolated seedlings were homogenized in a buffer containing 50 mm MOPS-KOH, pH 7.5, 2.5 mm EDTA, 100 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 20 μm leupeptin, and 5 mm dithiothreitol (DTT). The homogenate was centrifuged at 10,000g for 5 min. The supernatant was centrifuged again at 100,000g for 60 min. The microsomal membrane fraction was obtained as a pellet. The pellet was resuspended in Tris-buffered saline (TBS) buffer (20 mm Tris-HCl, pH 7.4, 140 mm NaCl) containing 0.1% (w/v) Tween 20, 1 mm phenylmethylsulfonyl fluoride, 20 μm leupeptin, and 1 mm DTT. Glutathione-sepharose beads that bound to GST-fused VfPIP were suspended in TBS buffer. Five microliters of glutathione-sepharose beads was mixed with 200 μL of 300 μg microsome solution, and the mixture was incubated at 4°C for 4 h. Proteins bound to the beads through VfPIP were released from the beads followed by boiling in Laemmli's buffer. The released proteins were separated by 12.5% SDS-PAGE and transferred onto nitrocellulose membranes (Hybond-C; Amersham Biosciences) using Trans-blot (Bio-Rad, Hercules, CA).
Northern Hybridization
RNA was isolated from guard cell protoplasts, mesophyll cell protoplasts, leaves, and roots of V. faba with ISOGEN (Nippon Gene, Tokyo). DIG-labeled probes from 482 bp to 1,314 bp of VfPIP gene were obtained by PCR using a PCR DIG-labeling mix (Roche, Tokyo) and used as probe. Northern hybridization was performed using a Digoxigenin Luminescent Detection Kit (Roche) according to the manufacturer's instructions. Signals were detected using CDP-Star (Roche).
Generation of Polyclonal Antibodies
The C-terminal region of VfPIP was used as an antigen and was expressed in E. coli as a recombinant protein. The 366-bp DNA fragment encoding Met-190 to Asp-312 of VfPIP was amplified by PCR. The resulting amplified DNA fragment was cloned into the pGEX2T (Amersham Biosciences) and used to transform E. coli. This polypeptide was expressed as a fusion protein with GST and was purified using glutathione-sepharose 4B. The C-terminal region of VfPIP was obtained by digestion with thrombin. Purified polypeptide was used to immunize the rabbits.
Western-Blot Analysis
Phototropins were detected immunologically with antibodies according to the method of Gallagher et al. (1992) with slight modifications. Proteins were subjected to SDS-PAGE and transferred onto nitrocellulose membranes (Hybond-C; Amersham Biosciences) using Trans-blot (Bio-Rad). The membranes were incubated for 30 min at room temperature in blocking medium containing 20 mm Tris-HCl, pH 7.4, 140 mm NaCl, 0.05% (w/v) Tween 20, and 5% (w/v) fatty acid free milk, and then reacted with polyclonal antibodies at 5,000-fold dilution overnight at 4°C. The membranes were washed three times with T-TBS containing 20 mm Tris-HCl, pH 7.4, 140 mm NaCl, and 0.05% Tween 20, then reacted with alkaline phosphatase-conjugated goat anti rabbit IgG antibody (Bio-Rad) at 3,000-fold dilution for 1 h at room temperature. The alkaline phosphatase reaction was developed by 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium.
Protein-Blot Analysis
Protein-blot analysis (far western blotting) was performed as described previously (Kinoshita and Shimazaki, 1999) with slight modifications. Proteins were subjected to SDS-PAGE and transferred onto nitrocellulose membranes using Trans-blot (Bio-Rad). The membranes were incubated at room temperature in 25 mm HEPES-KOH, pH 7.7, 75 mm KCl, 0.1 mm EDTA, 1 mm DTT, 0.04% Tween 20, and 2% fatty acid free milk for 1 h, then reacted with 0.1 μm GST-VfPIP fusion protein overnight at 4°C in the same medium. After washing the membranes with T-TBS three times, GST-VfPIP on the membranes was reacted with anti-GST antibodies (Amersham Biosciences) at 5,000-fold dilution in the blocking medium. The membranes were again washed with T-TBS three times, then reacted with anti-goat IgG antibodies conjugated to alkaline phosphatase (Sigma, Tokyo) at 5,000-fold dilution for 2 h in the blocking medium. The color developing was performed as described above.
Transient Expression Assays by Particle Bombardment
VfPIP-GFP was constructed using full-length VfPIP and GFP under the control of the CaMV 35S promoter. GFP-MAP4 was constructed using the microtubule binding domain of human MAP4 (amino acids 670–1,088) and GFP under the control of the CaMV 35S promoter. GFP-talin was constructed using mouse talin (amino acids 2,345–2,541) and GFP under the control of the CaMV 35S promoter (Olson et al., 1995; Kost et al., 1998).
Plasmid DNA was precipitated onto 1-μm gold particles according to the manufacturer's instructions. The abaxial side of Vicia leaves was shot at a pressure of 1,350 psi (1 psi = 6.89 kPa) at a distance of 4 cm from the macrocarrier holder by a Biolistic PDS-1000/He particle delivery system (Bio-Rad). Approximately 1 μg DNA was used per shot.
Transfected leaves were stored in the dark overnight at room temperature, and the epidermis was peeled. The epidermis was incubated with microtubule-depolymerizing compounds (10 μm trifluralin and 10 μm oryzalin) in incubation medium (5 mm MES-BisTris-propane, 0.1 mm CaCl2, and 50 mm KCl, pH 6.5) for 2 h in the dark and then examined with a confocal laser-scanning microscope (Digital Eclipse C1; Nikon, Tokyo).
Measurement of Stomatal Opening
The epidermis was obtained from a fully expanded Vicia leaf and was placed in incubation medium (5 mm MES-BisTris-propane, 0.1 mm CaCl2, and 50 mm KCl, pH 6.5) in the dark for 1 h. The leaf epidermis was incubated with a microtubule-depolymerizing herbicide (10 μm trifluralin, 10 μm oryzalin, and 20 μm propyzamide) in incubation buffer for 2 h in the dark and illuminated with red light at 150 μmol m−−2 s−1 or with blue light at 15 μmol m−2 s−1 superimposed on the background red light for 2 h.
Measurement of Blue Light-Dependent H+ Pumping
Blue light-dependent H+ pumping by guard cell protoplasts from V. faba was measured with a glass pH electrode (Beckman 39532) using a dual-beam protocol described previously (Shimazaki et al., 1992). The reaction mixture (1.0 mL) consisted of 0.125 mm MES-NaOH, pH 6.0, 1 mm CaCl2, 0.4 m mannitol, 10 mm KCl, and guard cell protoplasts (50 μg of protein) unless otherwise stated. The light intensity was 600 μmol m−2 s−1 for background red light and 100 μmol m−2 s−1 for blue light.
Light Source
Red light was obtained from a tungsten lamp (Sylvania EXR 150 W) by passing the light through a red glass filter (Corning 2–61, >610 nm, Corning, New York) and blue light was obtained from a tungsten lamp (Sylvania EXR 300W) through a blue glass filter (Corning 5–60, peak 420 nm, half band width 45 nm). Photon flux density was determined with a quantum meter (LI-COR model 185A; Lincoln, NE).
Gene Sequence Determination
All sequences were determined using an ABI PRISM 3100 Sequence Kit and ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). Nucleotide and amino acid sequences were analyzed using the GENETYX software system (Software Development, Tokyo).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AB215106.
Acknowledgments
We thank Professor M. Ito, Graduate School of Bioresource and Bioenvironmental Science, Kyushu University for using confocal laser-scanning microscope.
This work was supported by the Ministry of Science, Sports, and Culture of Japan (grant no. 13139202 to K.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.052639.
References
- Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D (1995) The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ 6: 1193–1198 [PubMed] [Google Scholar]
- Aravind L, Koonin EV (1999) Fold prediction and evolutionary analysis of the POZ domain: structural and evolutionary relationship with the potassium channel tetramerization domain. J Mol Biol 285: 1353–1361 [DOI] [PubMed] [Google Scholar]
- Assmann SM (1993) Signal transduction in guard cells. Annu Rev Cell Biol 9: 345–375 [DOI] [PubMed] [Google Scholar]
- Assmann SM, Baskin TI (1998) The function of guard cells does not require an intact array of cortical microtubules. J Exp Bot 49: 163–170 [Google Scholar]
- Assmann SM, Shimazaki K (1999) The multisensory guard cell. Stomatal responses to blue light and abscisic acid. Plant Physiol 119: 809–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann SM, Simoncini L, Schroeder JI (1985) Blue light activates electrogenic ion pumping in guard cell protoplasts of Vicia faba L. Nature 318: 285–287 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Briggs WR, Christie JM (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7: 204–210 [DOI] [PubMed] [Google Scholar]
- Briggs WR, Huala E (1999) Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol 15: 33–62 [DOI] [PubMed] [Google Scholar]
- Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR (1998) Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science 282: 1698–1701 [DOI] [PubMed] [Google Scholar]
- Crépieux P, Kwon H, Leclerc N, Spencer W, Richard S, Lin R, Hiscott J (1997) I kappaB alpha physically interacts with a cytoskeleton-associated protein through its signal response domain. Mol Cell Biol 17: 7375–7385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emi T, Kinoshita T, Shimazaki K (2001) Specific binding of vf14-3-3a isoform to the plasma membrane H+-ATPase in response to blue light and fusicoccin in guard cells of broad bean. Plant Physiol 125: 1115–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun S-O, Lee Y (1997) Actin filaments of guard cells are reorganized in response to light and abscisic acid. Plant Physiol 115: 1491–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Hasezawa S, Asai N, Nakajima N, Kondo N (1998) Dynamic organization of microtubules in guard cells of Vicia faba L. with diurnal cycle. Plant Cell Physiol 39: 80–86 [DOI] [PubMed] [Google Scholar]
- Gallagher S, Winston SE, Hurrell JGR (1992) Immunoblotting and immunodetection. In FM Ausubel, R Brent, RE Kingston, DD Moore, JG Seidman, JA Smith, K Struhl, eds, Current Protocols in Molecular Biology. Greene Publishing and Wiley-Interscience, New York, pp 10.8.1–10.8.16
- Hedrich R, Schroeder JI (1989) The physiology of ion channels and electrogenic pumps in higher plants. Annu Rev Plant Physiol 40: 539–569 [Google Scholar]
- Herzig RP, Andersson U, Scarpulla RC (2000) Dynein light chain interacts with NRF-1 and EWG, structurally and functionally related transcription factors from humans and Drosophila. J Cell Sci 113: 4263–4273 [DOI] [PubMed] [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR (1997) Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278: 2120–2123 [DOI] [PubMed] [Google Scholar]
- Hwang J-U, Lee Y (2001) Abscisic acid-induced actin reorganization in guard cells of dayflower is mediated by cytosolic calcium levels and by protein kinase and protein phosphatase activities. Plant Physiol 125: 2120–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J-U, Suh S, Yi H, Kim J, Lee Y (1997) Actin filaments modulates both stomatal opening and inward K+-channel activities in guard cells of Vicia faba L. Plant Physiol 115: 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada S, Ohgishi M, Mayama T, Okada K, Sakai T (2004) RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin1 in Arabidopsis thaliana. Plant Cell 16: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH (1996) PIN: an associated protein inhibitor of neuronal nitric oxide synthase. Science 274: 774–777 [DOI] [PubMed] [Google Scholar]
- Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M (2001) Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science 291: 2138–2141 [DOI] [PubMed] [Google Scholar]
- Kim M, Hepler PK, Eun S-O, Ha KS, Lee Y (1995) Actin filaments in mature guard cells are radially distributed and involved in stomatal movement. Plant Physiol 109: 1077–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM (2000) The dynein microtubule motor. Biochim Biophys Acta 1496: 60–75 [DOI] [PubMed] [Google Scholar]
- King SM, Patel-King RS (1995) The Mr=8,000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J Biol Chem 270: 11445–11452 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K (2001) phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414: 656–660 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Emi T, Tominaga M, Sakamoto K, Shigenaga A, Doi M, Shimazaki K (2003) Blue-light-and phosphorylation-dependent binding of a 14-3-3 protein to phototropins in stomatal guard cells of broad bean. Plant Physiol 133: 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K (1999) Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J 18: 5548–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Lahav M, Abu-Abied M, Belausov E, Schwartz A, Sadot E (2004) Microtubules of guard cells are light sensitive. Plant Cell Physiol 45: 573–582 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Morris NR, Meagher RB, Dawe RK (2001) Dyneins have run their course in plant lineage. Traffic 2: 362–363 [DOI] [PubMed] [Google Scholar]
- Lupas A (1996) Coiled coils: new structures and new functions. Trends Biochem Sci 21: 375–382 [PubMed] [Google Scholar]
- Marcus AI, Moore RC, Cyr RJ (2001) The role of microtubules in guard cell function. Plant Physiol 125: 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motchoulski A, Liscum E (1999) Arabidopsis NPH3: a NPH1 photoreceptor-interacting protein essential for phototropism. Science 286: 961–964 [DOI] [PubMed] [Google Scholar]
- Olson KR, McIntosh JR, Olmsted JB (1995) Analysis of MAP4 function in living cells using green fluorescent protein (GFP) chimeras. J Cell Biol 130: 636–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, Huang DCS, O'Reilly LA, King SM, Strasser A (1999) The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell 3: 287–296 [DOI] [PubMed] [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K (2001) Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci USA 98: 6969–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Wada T, Ishiguro S, Okada K (2000) RPT2: a signal transducer of the phototropic response in Arabidopsis. Plant Cell 12: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Briggs WR (2002) Cellular and subcellular localization of phototropin1. Plant Cell 14: 1723–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR (2000) Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 39: 9401–9410 [DOI] [PubMed] [Google Scholar]
- Salomon M, Knieb E, von Zeppelin T, Rüdiger W (2003) Mapping of low- and high-fluence autophosphorylation sites in phototropin 1. Biochemistry 42: 4217–4225 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D (2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52: 627–658 [DOI] [PubMed] [Google Scholar]
- Schwartz A, Zeiger E (1984) Metabolic energy for stomatal opening. Roles of photophosphorylation and oxidative phosphorylation. Planta 161: 129–136 [DOI] [PubMed] [Google Scholar]
- Shimazaki K, Gotow K, Kondo N (1982) Photosynthetic properties of guard cell protoplasts from Vicia faba L. Plant Cell Physiol 23: 871–879 [Google Scholar]
- Shimazaki K, Iino M, Zeiger E (1986) Blue light-dependent proton extrusion by guard-cell protoplasts of Vicia faba. Nature 319: 324–326 [Google Scholar]
- Shimazaki K, Kinoshita T, Nishimura M (1992) Involvement of calmodulin and calmodulin-dependent myosin light chain kinase in blue light-dependent H+ pumping by guard cell protoplasts from Vicia faba L. Plant Physiol 99: 1416–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai AW, Chuang JZ, Bode C, Wolfrum U, Sung CH (1999) Rhodopsin's carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell 97: 877–887 [DOI] [PubMed] [Google Scholar]
- Tai AW, Chuang JZ, Sung CH (1998) Localization of Tctex-1, a cytoplasmic dynein light chain, to the Golgi apparatus and evidence for dynein complex heterogeneity. J Biol Chem 273: 19639–19649 [DOI] [PubMed] [Google Scholar]