Abstract
Cyclin-dependent kinases (CDKs) are the main regulators of cell cycle progression in eukaryotes. The role and regulation of canonical CDKs, such as the yeast (Saccharomyces cerevisiae) Cdc2 or plant CDKA, have been extensively characterized. However, the function of the plant-specific CDKB is not as well understood. Besides being involved in cell cycle control, Arabidopsis (Arabidopsis thaliana) CDKB would integrate developmental processes to cell cycle progression. We investigated the role of CDKB in Ostreococcus (Ostreococcus tauri), a unicellular green algae with a minimal set of cell cycle genes. In this primitive alga, at the basis of the green lineage, CDKB has integrated two levels of regulations: It is regulated by Tyr phosphorylation like cdc2/CDKA and at the level of synthesis-like B-type CDKs. Furthermore, Ostreococcus CDKB/cyclin B accounts for the main peak of mitotic activity, and CDKB is able to rescue a yeast cdc28ts mutant. By contrast, Ostreococcus CDKA is not regulated by Tyr phosphorylation, and it exhibits a low and steady-state activity from DNA replication to exit of mitosis. This suggests that from a major role in the control of mitosis in green algae, CDKB has evolved in higher plants to assume other functions outside the cell cycle.
Control of cell cycle progression in eukaryotes relies largely on universally conserved heterodimeric kinases belonging to the cyclin-dependent kinase (CDK) family. These kinases phosphorylate, at the key G1/S and G2/M transitions, a variety of substrate required for DNA replication and mitosis, respectively (Murray, 2004). Cyclins are regulatory subunits enabling periodic activation of CDKs. Whereas animals possess several CDKs, in both fission yeast (Schizosaccharomyces pombe) and budding yeast (Saccharomyces cerevisiae) a single CDK, Cdc2/Cdc28, in association to different cyclins, controls cell cycle progression (Arellano and Moreno, 1997; Mendenhall and Hodge, 1998; Endicott et al., 1999; John et al., 2001). Rapid activation of Cdc2 at mitosis entry is achieved by dephosphorylation of T14 and Y15 residues (Lew and Kornbluth, 1996). CDK/cyclin complexes are also targets of survey mechanisms, which stop or delay cell cycle progression upon failing, such as incomplete DNA replication, DNA damage, or defect of the mitotic spindle (Paulovich et al., 1997; Rhind and Russell, 1998; Lew and Burke, 2003; Lukas et al., 2004; Stark and Taylor, 2004). Inhibitory phosphorylation of T14 and Y15 is largely involved in CDK inhibition upon activation of the DNA integrity checkpoint (Murakami and Nurse, 2000; Donzelli and Draetta, 2003).
Higher plants have a considerable number of core cell cycle genes. Arabidopsis (Arabidopsis thaliana) contains nine cell cycle-related CDK genes and about 30 cyclin genes (Vandepoele et al., 2002). Plant cell cycle genes have been classified on the basis of their sequence homology to their animal counterparts and globally perform similar roles: A-type cyclins are involved in S-phase progression and mitosis, and B-type cyclins are mitotic (Breyne and Zabeau, 2001; Potuschak and Doerner, 2001). In plants, CDKs harboring the consensus PSTAIRE hallmark are referred as to A-type CDK, whereas non-PSTAIRE CDKs encompass a class of CDK unique to plants, the B-type CDKs (Joubes et al., 2000). Overexpression of a dominant negative form of the Arabidopsis CDKA;1 in tobacco plants (Nicotiana tabacum) results in an overall reduction of cell division rate, thus yielding smaller plants. However, the G1/G2 ratio remains unaltered, demonstrating that CDKA;1 is essential at both G1/S and G2/M transition of the cell cycle (Hemerly et al., 1995). The mitotic activity of CDKA;1 and the localization of CDKA;1 with mitotic microtubuli structures, such as the preprophase band and metaphase spindle, further indicates a mitotic role for A-type CDKs (Weingartner et al., 2001; Menges and Murray, 2002). Tyr phosphorylation of A-type CDK was unambiguously detected and shown to down-regulate CDKA under cytokinin deprivation or osmotic stress (Zhang et al., 1996; Schuppler et al., 1998). Furthermore, a CDK inhibitory kinase Wee1 and a divergent antagonist Cdc25-like phosphatase are present in plants (Sun et al., 1999; Sorrell et al., 2002; Landrieu et al., 2004). However, the role of CDKA phosphorylation in plant cell cycle control is still not clear. Arabidopsis plants overexpressing a nonphosphorylable CDKA;1 (Y15A) do not exhibit any obvious phenotype, but expression of fission yeast Cdc25 in tobacco plant accelerates cell division (Bell et al., 1993; McKibbin et al., 1998; Wyrzykowska et al., 2002).
The role of CDKB in cell cycle control is not as well understood. Together, CDKB localization at the preprophase band, the metaphase plate, and its mitotic activity suggest a role of B-type CDKs in the control of mitosis (Magyar et al., 1997; Mészáros et al., 2000; Dewitte and Murray, 2003; Lee et al., 2003). Furthermore, in rice (Oryza sativa), the mitotic CyclinB2;2 was shown to activate CDKB2;1, and both proteins colocalize on chromosomes at metaphase, suggesting that this complex may be involved in the control of mitosis (Lee et al., 2003). Interestingly, the inhibitory phosphorylation sites are also conserved in B-type CDKs, but biochemical evidence of CDKB phosphorylation is still lacking. The role of CDKB in integrating developmental pathways has been clearly demonstrated. Arabidopsis plants overexpressing a dominant negative CDKB1;1 (CDKB1;1 N161) display cells with a higher 4C/2C ratio in specific tissues. This was correlated to dramatic consequences on stomata development (Porceddu et al., 2001; Boudolf et al., 2004a). In the leaves of CDKB1;1N161 plants, cells prematurely exit mitotic cycle and enter endoreduplication cycles associated with leaf development (Boudolf et al., 2004b). Thus, CDKB1;1 would be implicated in both the control of cell cycle progression and the integration of developmental pathways.
We have used a simple cellular system to investigate the control of cell cycle progression by CDK/cyclin partners. Ostreococcus (Ostreococcus tauri) is a marine photosynthetic picoeucaryote, which like plants belongs to the green lineage (Chrétiennot-Dinet et al., 1995). This 1-micron-long unicellular algae has a minimal cellular organization, encompassing one of each organella, and a compact genome (12.6 Mb). Annotation of cell cycle genes revealed an extremely limited set of core cell cycle genes with great similarity to plant cell cycle genes (Robbens et al., 2005). In particular CDKA, CDKB, Cyclin D, Cyclin A, and Cyclin B genes are all present as a single copy. The main CDK inhibitory kinase Wee1 and its antagonist phophatase Cdc25 were also identified. The latter shows a high similarity to animal Cdc25 (in contrast to its plant ortholog) and has been reported to be functional in activating starfish Cdc2 (Khadaroo et al., 2004; Landrieu et al., 2004). In our culture conditions, Ostreococcus cells divide in a binary mode and do not differentiate. When subjected to 12:12 light/dark cycles the cells divide at the end of day. We took advantage of this simple system to investigate the role of CDK/cyclin partners, and more particularly of CDKB, in the control of cell cycle progression. Unexpectedly, CDKB rather than CDKA has the main features of a mitotic CDK, i.e. in association with cyclin B, it is responsible for the peak of mitotic activity of CDKs and is regulated by Tyr phosphorylation. Furthermore, CDKB is able to rescue a budding yeast cdc28ts mutant.
RESULTS
Biochemical Purification of CDKA and CDKB in Ostreococcus
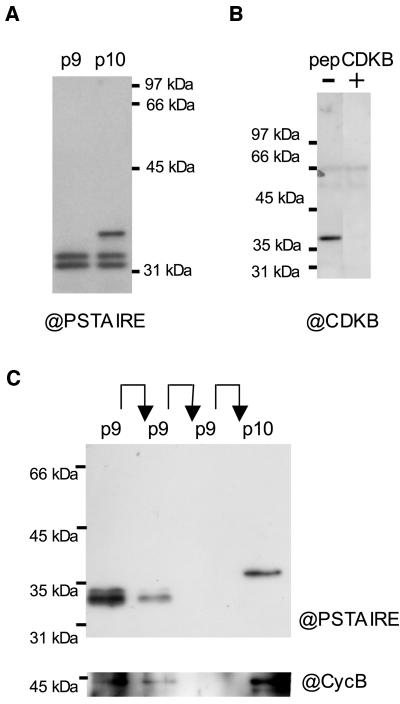
We first assayed the affinity of plant p10CKSAt1 and human p9CKShs1, referred to as p10 and p9, respectively, toward Ostreococcus CDKs (Fig. 1A). An antibody directed against the hallmark PSTAIRE motif of CDKs was used for western blot (Fig. 1A). Two bands of 34 to 35 kD were detected after p9 chromatography, and an additional band of approximately 37 kD was detected after p10 chromatography. The signal intensity of the two 34- to 35-kD bands was similar after p9 or p10 chromatography, suggesting that the corresponding proteins had a similar affinity for p9 and p10. A specific anti-CDKB antibody also detected the purified protein at 37 kD, which is the expected molecular mass of CDKB, indicating that this protein corresponded to CDKB. The specificity of the antibody was tested by competition with the antigenic peptide (CYFDSLDKSQF), which completely prevented binding of the antibody to the 37-kD protein but had no effect on binding of the anti-PSTAIRE antibody (Fig. 1B; data not shown). A single protein at 37 kD was recognized by the anti-CDKB antibody in protein extracts from yeast expressing Ostreococcus CDKB, confirming further the specificity of the antibody (Fig. 7B). It is very likely that the CDKB PSTALRE motif can bind the anti-PSTAIRE antibody since it differs only by a substituted Leu residue (instead of Ile). The genome analysis revealed that Ostreococcus has a single CDKA, suggesting strongly that the 34- to 35-kD bands correspond to two isoforms of CDKA.
Figure 1.
Purification of Ostreococcus CDKs. A, Detection of Ostreococcus CDKs, with a monoclonal anti-PSTAIRE antibody, after affinity purification on CKS from either human p9 or Arabidopsis p10. B, Specificity of the anti-CDKB antibody. CDKs were purified on p10 and detected with the anti-CDKB antibody in the presence or absence of the competing antigenic peptide. C, Sequential purification of Ostreococcus CDKs by affinity chromatography. The 34- to 35-kD bands (CDKA) were depleted from an HU extract, after two rounds of affinity chromatography on p9 as detected with the anti-PSTAIRE (@PSTAIRE) antibody. Only the 37-kD CDK (CDKB) was detected on p10 beads after depletion. A band at approximately 45 kD was detected mainly in association with CDKB (p10) using an anti-cyclin B (@CycB) antibody.
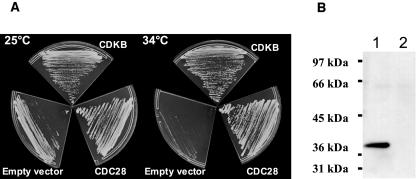
Figure 7.
A, Complementation studies of the budding yeast cdc28-4ts mutant with Ostreococcus CDKA and CDKB. cdc28-4ts mutants were transformed with either CDKB, CDC28 positive control, or no DNA (empty vector). Transformed cells were grown at permissive (25°C) or restrictive (34°C) temperature. B, Immunodetection with the anti-CDKB antibody of affinity-purified proteins on p10 from yeast cdc28-4ts mutant expressing CDKB (lane 1) or untransformed cdc28-4ts control cells (lane2) at permissive temperature.
To isolate CDKB, we took advantage of its differential affinity toward p9 and p10. The two lowest bands were depleted from the protein extract by two successive chromatography procedures on p9, and the last flow-through was loaded on p10 (Fig. 1C).
A signal was detected with an antibody raised against the C-terminal sequence of Ostreococcus cyclin B, at the expected molecular mass of cyclin B (42 kD), and was stronger in the p10 fraction, although it was also detected in the p9 fraction (Fig. 1B, bottom).
CDKB Is Synthesized after the G1/S Transition and Is Phosphorylated upon Activation of the DNA Replication Checkpoint
Natural synchronization of Ostreococcus cells can be achieved by light/dark cycles. However, the shortness of the S/G2/M phases (3 h; Fig. 4A) compared to the G1 phase leads to an overlap of the different cell cycle phases in naturally synchronized cell populations (S/G2/M phases over 10 h), which complicates cell cycle studies.
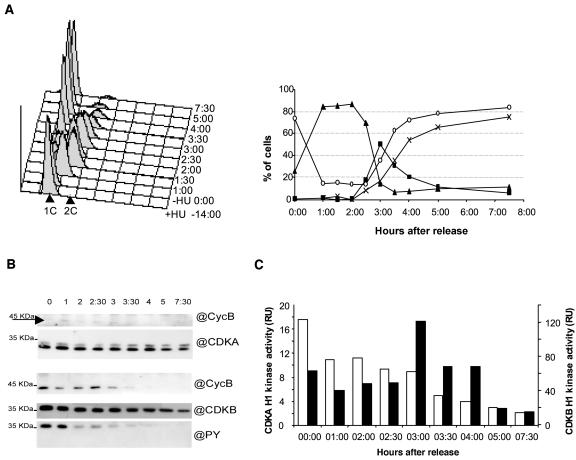
Figure 4.
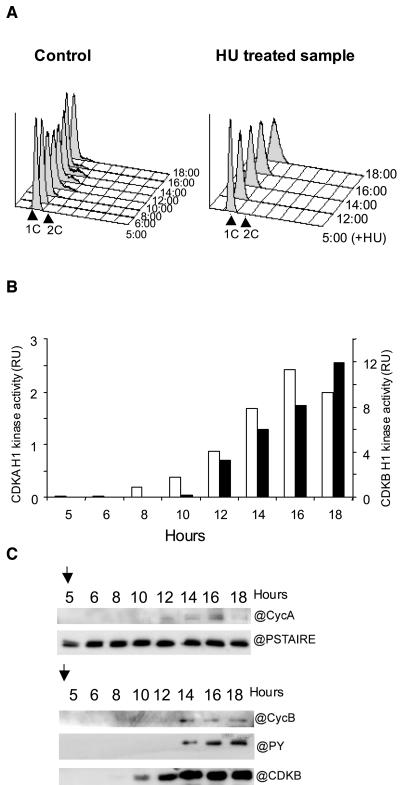
Expression, phosphorylation status, cyclin partners, and activities of CDKA and CDKB at different stages of cell cycle progression in cells released from an HU arrest. A, Time course analysis of DNA content of HU synchronized cells by flow cytometry. Cells were incubated with HU for 14 h and this drug was removed at time 0:00. Overlayed histographs (left section) and relative frequencies of prereplicative nuclei (G1, white circle), replicative nuclei (S, triangle), replicated nuclei (G2M, square) are as estimated by the Modfit software (right section). B, Levels, phosphorylation status, and cyclin partners of CDKA (p9 fraction) and CDKB (p10 fraction). C, Corresponding H1 kinase activities of CDKA (white; left axis) and CDKB (black; right axis).
Therefore, to gain further insight into the biochemistry of CDK/cyclin complexes, we used various cell cycle inhibitors to induce specific cell cycle arrests (Fig. 2A). The CDK inhibitor olomoucine, the DNA replication inhibitor aphidicolin, and the DNA synthesis inhibitor hydroxyurea (HU) were added in late G1 of light/dark-grown cultures (3–5 h after light on) to block early progression in the cell cycle and obtain cells with DNA content corresponding to a G1/S transition and an early S phase, respectively (Fig. 2A). Olomoucine, when added in G1, was expected to completely arrest the cell cycle before the G1/S transition with CDKs in a G1/S status. This arrest will be referred to as a G1s arrest. By contrast, aphidicolin and HU were expected to induce the DNA integrity checkpoint and, as such, to arrest the cells with CDKs in a premitotic status, which mimics a G2/M state, which will be referred to as an S-G2 arrest. Finally, cells were released from a replication arrest by HU and then blocked either at the G2/M transition by olomoucine, referred to as G2m, or in mitosis by the microtubule polymerization inhibitor, propyzamide, referred to as Mp (Planchais et al., 2000). In these cases, CDKs are expected to reflect G2/M and M phases, respectively. Two control stages were used as references, i.e. the late G1 (5 h after light on) and the M/G1 (synchronized cells at exit of mitosis).
Figure 2.
Expression and phosphorylation status of CDKs at different stages of cell cycle progression. Cells were arrested at different stages of the cell cycle using specific inhibitors. Olomoucine (G1s)- and HU (S-G2)-treated cells were collected 14 h after addition of the drugs. G2m and MP phase samples were obtained from artificially synchronized cells, were blocked with olomoucine or propyzamide, and were collected at the time of control mitosis exit (M/G1). Control G1 cells were collected at the time of drug addition. A, Relative frequencies of cells in G1 (white), S (gray), and G2/M (black), as detected by flow cytometry, are shown. B, CDKs were affinity purified on p10 and detected with anti-CDKB, anti-PSTAIRE, or anti-PY antibodies.
The presence and Tyr phosphorylation status of CDKs were first investigated. CDKs were purified using their affinity for p10. As shown in Figure 2B, CDKA was equally present at all these stages, as detected with an anti-PSTAIRE antibody, whereas CDKB was absent in natural G1 cells and in cells arrested before S phase by olomoucine. When the cells were treated with olomoucine, the relative intensities of the two bands corresponding to CDKA changed, the upper band being much stronger. It is unlikely that these two bands reflected different states of phosphorylation of CDKA since the relative intensities of these two bands was not related to changes in CDKA activity during cell cycle progression or upon checkpoint activation (Fig. 4). Olomoucine was reported to compete for ATP in the catalytic pocket of CDK and could therefore induce conformational changes of the protein, which may account for the changes in electrophoretic migration. Surprisingly, CDKB, but not CDKA, was phosphorylated on Tyr, as detected with a monoclonal anti-phosphotyrosine (PY) antibody, when the DNA integrity checkpoint was activated by either aphidicolin or HU. This phosphorylation was also detected in premitotic cells arrested at the G2/M by olomoucine (G2m), but it was absent from cells exiting mitosis or cells blocked in mitosis by propyzamide. Together, these results indicate that CDKB is cell cycle regulated and its synthesis is strictly dependent on the progression of the cells through the G1/S transition.
Cyclin Partners and Activities of CDK/Cyclin Complexes
The same approach, as described in Figure 2, was used to investigate the cyclin partners of CDKs at various cell cycle stages (Fig. 3A). CDKA and CDKB were purified sequentially using their differential affinity for p9 and p10, respectively. Using a specific antibody directed against the C-terminal region of cyclin A, we were able to detect a doublet in the CDKA fraction bound to p9 but not in the CDKB fraction (data not shown). The molecular mass (42 kD) was in agreement with the predicted molecular mass. This doublet probably corresponded to two isoforms of cyclin A and was detected in cells arrested after the G1/S transition, i.e. in HU- and propyzamide-treated cells, but it was not detectable at mitosis exit. Cyclin B was detected in both CDKA (p9) and CDKB (p10) fractions (Fig. 3A). Like cyclin A, cyclin B was found only in cells arrested after the G1/S transition and not in cells exiting mitosis.
Figure 3.
Expression, cyclin partners, and activities of CDKA and CDKB at different stages of cell cycle progression. CDKA and CDKB were sequentially affinity purified (on p9 and p10, respectively) from cells arrested at different stages of cell cycle progression. A, Specific antibodies were used to detect cyclin A, cyclin B, CDKA, and CDKB bound to p9 and p10. B, Corresponding H1 kinase activities of CDKA (white) and CDKB (black) are reported (top). All activities were normalized relative to the HU sample activity (100).
In parallel, the histone H1 kinase activities of CDKA and CDKB were monitored (Fig. 3B). CDKA exhibited a barely detectable activity at the G1/S (G1s) transition compared to the activity found in HU- or aphidicolin-treated cells. CDKA activities from cells blocked in mitosis (Mp) were similar to those of cells in which DNA replication was inhibited (S-G2) or arrested at the G2/M transition (G2m) with olomoucine.
By contrast, CDKB kinase activity was much higher in mitotic cells (Mp) than in cells arrested before mitosis (S-G2 or G2m). These results suggest that CDKB but not CDKA is activated in mitosis and that only CDKB is the target of the DNA replication checkpoint.
In summary, CDKA is present at all stages of the cell cycle and is associated, after the G1/S transition, to cyclin A and to a lesser extent to cyclin B. By contrast, CDKB is synthesized after the G1/S, exclusively associated to cyclin B, and activated in mitosis.
The CDKB/Cyclin B Complex Is Tyr Dephosphorylated and Activated at Mitosis
We next determined the level and activity of CDK/cyclin complexes in cells artificially synchronized by a release from an HU block (Fig. 4). Upon release, the cells progressed synchronously through DNA replication. Over 50% of the cells progressed simultaneously through G2 and M phases compared to less than 15% in naturally synchronous cells (Fig. 4A). The replication of DNA lasted for about 3 h from the release, whereas the cells progressed through G2 and M within a maximum of 1 h (four independent experiments). Mitosis was shorter than 30 min since, by 3 h after the HU release (AR), 50% of the cell was estimated to be in G2-M and only 30 min later 50% of the cells were back in G1 (1C content; Fig. 4A).
In HU-treated cells, CDKB was phosphorylated on Tyr and associated with cyclin B as previously shown. It is interesting to note that cyclin B was barely detectable in CDKA/cyclin-purified complexes, although the exposure time of this western blot was much longer (Fig. 4B, top) than for cyclin B associated to CDKB (Fig. 4B, bottom). While CDKA was fairly constant throughout mitosis, CDKB decreased progressively but significantly from 4 h AR. Cyclin B dropped by 3.5 h AR, attesting that most of the cells were progressing through mitosis. At 5 h AR, cyclin B was no longer detectable. The Tyr phosphorylation of CDKB progressively decreased from 3 h AR and was correlated to CDKB activation (Fig. 4, B and C). By contrast, CDKA did not appear to be activated at mitosis but rather had a constant activity throughout the DNA replication period, which decreased from 3 h AR. No phosphorylation could be detected on CDKA at any time (data not shown). Note that in this experiment, like in most, CDKA H1 kinase activities were about one order of magnitude lower than those of CDKB.
In Vitro Dephosphorylation of CDKB by Cdc25 Results in Its Activation
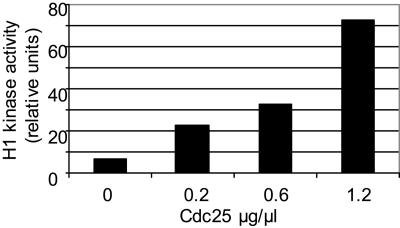
To test whether CDKB Tyr phosphorylation was inhibitory of its kinase activity, we performed an in vitro dephosphorylation assay using the homologous CDK-specific phosphatase Cdc25 (Fig. 5; Khadaroo et al., 2004). CDKs purified from HU-treated cells were incubated with increasing concentrations of Cdc25, and their H1 kinase activity was monitored after dephosphorylation. CDK incubation with Cdc25 resulted in a dose-dependent activation of CDKB kinase activity. This result indicates that the Tyr phosphorylation on CDKB is inhibitory and suggests that the Cdc25 phosphatase has a role in activating CDKB at the G2/M transition.
Figure 5.
In vitro activation of Ostreococcus CDKs by Cdc25. CDKs from HU-treated cells were purified on p10, incubated with increasing amount of Cdc25, and then assayed for H1 kinase activity.
CDKA Activity Precedes CDKB Activity during Cell Cycle Progression
We next investigated the time course of CDK/cyclin appearance during DNA checkpoint activation, which mimics the accumulation of CDKs at the G2/M transition during cell cycle progression (Fig. 6). HU was added at the beginning of the light period (i.e. in G1) and left until the control cells had divided (Fig. 6A, left). The DNA content histographs reflect the partial synchronization of the control cells that entered S phase by 8 h of light (control, left), whereas the HU-treated cells accumulate with a prereplicative DNA content, as attested by the progressive enlargement of the 1C peak (Fig. 6A, right). In HU-treated cutltures, CDKA could be detected at all times, but CDKB was detected only from 8 h after dusk (Fig. 6C). CDKA activity was detectable 8 h after the beginning of the day (3 h after HU addition) and reached a steady-state level by 14 h, whereas CDKB activity was detected only 4 h later and reached a maximal level at 18 h (Fig. 6B). At 14 h, CDKB was seen phosphorylated on Tyr and associated with cyclin B (Fig. 6C). Most of the CDK activity was associated with CDKB. The main cyclin partner of CDKA, cyclin A, was also detected earlier in the CDKA fraction (at 10–12 h) than cyclin B in the CDKB fraction (at 14 h). These results indicate that CDKA operates before CDKB during the cell cycle progression and that CDKB/cyclin B is responsible for most of the kinase activity at the G2/M transition.
Figure 6.
Time course of CDK accumulation from G1 to an S-G2 arrest. HU was added in G1 (5 h after light on) and cells were harvested every 2 h for 13 h. A, Flow cytometry analysis of DNA content of the control population from 5 h until 18 h after light on (left) and in HU-treated cells (right). B, H1 kinase activity of CDKA (white, left axis) and CDKB (black, right axis) in HU-treated cells. C, Time course analysis and phosphorylation status of CDKA, CDKB, cyclin A, and cyclin B in the CDKA (top) and the CDKB (bottom) fractions upon accumulation from G1 to S-G2 arrest in HU-treated cells.
CDKB Complements a Budding Yeast cdc28ts Mutant
The CDKB cDNA was used to complement the budding yeast cdc28-4ts mutant (Fig. 7A). At permissive temperature (25°C), all cells were able to grow. By contrast, only cells transformed with CDKB or CDC28 gave rise to colonies at restrictive temperature (34°C). The expression of Ostreococcus CDKB in yeast was checked using the specific anti-CDKB antibody (Fig. 7B). That CDKB rescues the yeast cdc28-4ts mutant indicates that, despite the lack of a consensus PSTAIRE motive, CDKB is able to bind yeast cyclins and to directly control cell cycle progression.
DISCUSSION
Ostreococcus CDKB, a B1-Type CDK?
Despite a divergent PSTALRE cyclin-binding motif, Ostreococcus CDKB has been classified as a B-type CDK on the basis of sequence analysis (Robbens et al., 2005). In this work, we show that this protein displays a characteristic feature of B-type CDK in that, unlike CDKA, its level fluctuates during the cell cycle. CDKB was absent from naturally synchronized cells in G1 and also from cells blocked at the G1/S transition by olomoucine, a well-known CDK-specific and reversible inhibitor that has been successfully used in other cell cycle studies (Planchais et al., 1997; Corellou et al., 2001). On the contrary, CDKB synthesis was not inhibited when the replication of DNA was impaired, i.e. when the cells were arrested after the G1/S transition. Moreover, CDKB could be detected, although at a very low level, as early as 8 h after the beginning of the light period (Fig. 6B), i.e. when the control cells were entering S phase. Together, these results strongly suggest that CDKB synthesis depends on progression of the cells through the G1/S transition. Through the inhibition of G1 CDK(s), olomoucine is likely to inhibit a downstream pathway like the CDK/Retinoblastoma protein (Rb)-dependent transcription. In plants, phosphorylation of Rb by CycD/CDKA triggers the transcription of genes directly involved in DNA replication like Cdc6 but also of cell cycle genes like CDKB1 (Vlieghe et al., 2003). By contrast, inhibition of DNA replication allows the G1/S transition to occur and consequently the Rb-dependent transcription. In summary, Ostreococcus CDKB resembles more a B1- than a B2-type CDK in that, like CDKB1, it is synthesized in S phase rather than in G2/M like CDKB2. In agreement with these experimental data, BLASTP analysis also assigns the CDKB of Ostreococcus to the CDKB1 class (data not shown).
Ostreococcus CDKB: A B-Type CDK with A-Type Features
Plant A-type and B-type CDKs are associated to cyclin B by the time of mitosis; however, they can be distinguished based on their biochemical properties (Dewitte and Murray, 2003). In Arabidopsis and BY-2 cells, the mitotic activity of CDKA is 5 to 10 times higher than CDKB activity (H. Stals, personal communication; Porceddu et al., 2001). Like animals or fission yeast PSTAIRE CDKs (such as Cdc2/Cdc28, CDK1, or CDK2), plant A-type CDKs are regulated by Tyr phosphorylation (Zhang et al., 1996; Schuppler et al., 1998; Meszaros et al., 2000). Although not well documented in plants, this inhibitory phosphorylation down-regulates PSTAIRE CDKs during cell cycle progression and/or upon activation of the DNA integrity checkpoint like in animals or fission yeast. Furthermore, most of the G2/M H1 kinase activity of CDKs is associated with PSTAIRE CDKs. Artificially synchronized cells of Ostreococcus exhibited a steady-state level of CDKA activity until mitosis, whereas CDKB activity remained low until DNA replication was achieved. By contrast, CDKB peaked at the time of mitosis when it was Tyr dephosphorylated. Moreover, cell arrested in mitosis exhibited a high CDKB activity compared to the premitotic activity, whereas CDKA activity did not significantly change. In all experiments, CDKBs display an activity 5 to 12 times higher than CDKA activity, the larger difference being in mitosis. In cells accumulating at the G2/M checkpoint upon HU treatment (S-G2), CDKB was phosphorylated on Tyr as soon as cyclin B was detected. In parallel, CDKB activity progressively increased from 12 to 18 h. This kinetic suggests that cyclin B/CDKB association most probably occurs in G2 phase of the cell cycle, conferring a basal activity to the phosphorylated complex. By contrast, CDKA activity reached a steady-state level by 12 h, suggesting that by that time all cells had reached the stage corresponding to this maximal activity. In this respect, the profile of Ostreococcus CDKA activity differs from that of Arabidopsis-synchronized cells, which is biphasic with a high peak in mitosis (Menges and Murray, 2002). By contrast, Ostreococcus CDKB activity peaks in mitosis and displays a high H1 kinase activity, which is correlated to Tyr dephosphorylation.
Together, our results suggest that CDKB rather than CDKA plays the main mitotic role, which would be related to the role of animal or yeast Cdc2 or plant CDKA. By contrast, CDKA is more likely to be involved in the control of S phase progression since its activity is maximal during that stage. However, we cannot conclude about the role of CDKA in mitosis since (1) CDKA was associated with cyclin B, which is the universal mitotic cyclin, and (2) CDKA activity was still high when the cells were progressing through mitosis as indicated by the cyclin B decrease. Further work is needed to precisely determine the role of CDKA in the control of mitosis in Ostreococcus.
A Unique Regulation of CDKB in Ostreococcus?
Depending on the organism, inhibitory phosphorylation on Tyr is more or less preponderantly used by the DNA replication checkpoint (Lew and Kornbluth, 1996). Whereas this phosphorylation is essential for the proper function of the checkpoint in fission yeast, it represents only an additional level of regulation in the budding yeast or human as mutants expressing a nonphosphorylable PSTAIRE CDK retain the ability to arrest the cell cycle when DNA replication is inhibited. In plants, the only indication about Tyr regulation during normal cell cycle progression comes from alfalfa (Medicago sativa), where Tyr phosphorylation of a protein in the range of size of A-type CDK was seen to fluctuate inversely to H1 kinase in crude extracts (Mészáros et al., 2000; Wyrzykowska et al., 2002). This putative regulation of CDK by the classical T14 Y15 phosphorylation is further suggested by overexpression experiments of the fission yeast Cdc25 in plants, which leads to an increased rate of cell division (Bell et al., 1993; McKibbin et al., 1998). Tyr phosphorylation of B-type CDK was not detected during normal cell cycle progression or upon activation of the DNA replication checkpoint in Arabidopsis cell suspensions (F. Corellou, personal communication). In Ostreococcus, CDKB but not CDKA was phosphorylated on Tyr when DNA replication was inhibited with either HU or aphidicolin, and it was dephosphorylated in mitotic cells. In vivo dephosphorylation of CDKB correlated with an increase in activity, and the in vitro dephosphorylation assay using the specific and homologous Cdc25 phosphatase confirmed the inhibitory role of this phosphorylation. Our results indicate that, like for the animal mitotic Cdc2, dephosphorylation of CDKB rapidly activates the complex and presumably triggers mitotic events. That this regulatory mechanism also operates during cell cycle progression and not only upon checkpoint activation is suggested by the phosphorylated state of CDKB in cells arrested at mitosis entry (G2m arrest by olomoucine) with a 2C DNA content.
It is interesting to note that the CDKB level did not significantly change between cells arrested at most cell cycle stages (S-G2, G2m, Mp). By contrast, the rice CDKB (Cdc2Os3) was greatly reduced after HU treatment (Umeda et al., 1999), whereas the amount of CDKB1 increased in BY-2 cells and decreased in alfalfa cells upon activation of the DNA replication checkpoint (Magyar et al., 1997). Thus, unlike in higher plants, the turnover of CDKB appears not to be affected by inhibition of DNA replication in Ostreococcus. Regulation of Ostreococcus CDKB upon activation of the DNA replication checkpoint involves an additional level of posttranslational regulation by Tyr phosphorylation, which may account for the absence of regulation at the level of protein synthesis. Similarly, upon HU treatment, CDKB appeared to be continuously synthesized and Tyr phosphorylated (Fig. 6). The accumulation of CDKB/cyclin B may to some extent outcompete its inactivation by Tyr phosphorylation, leading to an overall increase in CDKB activity. This might be a checkpoint adaptation. In any case, dephosphorylation of CDKB by Cdc25 leads to a strong activation of CDKB, consistent with an inhibitory role of Tyr phosphorylation.
Role and Evolution of Plant CDKBs
While the plant-specific B-type CDKs are regulated at the synthesis level, the classical Cdc2-related PSTAIRE CDK, also called CDKA in plants, are down-regulated by Tyr phosphorylation. Ostreococcus CDKB appears to be an unusual CDK since it has integrated two distinct levels of regulation, which are encountered in the plant and in the animal/fungus kingdom, respectively. Protein phosphorylation rather than protein synthesis regulation is used in cell cycle checkpoints and more generally in biological process when rapidity, flexibility, and reversibility are required. By contrast, higher plant CDKB, which is involved in more durable or less reversible responses such as endoreduplication and differentiation, is regulated at the level of protein synthesis. In Ostreococcus, the regulation of CDKB mitotic activity by Tyr phosphorylation together with its major contribution to the mitotic peak of CDK activity strongly suggests that it has an essential role in cell cycle regulation per se. This is further confirmed by the complementation of the budding yeast cdc28-4ts mutant. To which extent or in which context the regulation of CDKB synthesis is important to Ostreococcus cell cycle control (as well as for other organisms) remains to be elucidated. However, that both types of regulations exist for a single CDK points to the importance of this molecule in integrating different types of signals and, thus, cellular responses.
A putative picture of cell cycle control can be drawn from our result: CDKA activity increases after the G1/S transition (since it is barely detectable in G1s-arrested cells) and remains at a steady-state level until the G2/M transition. CDKA/cyclin A complexes are formed in S phase and could drive the progression of DNA replication. A fraction of CDKA would be associated with cyclin B in G2 and could trigger mitosis entry. Cyclin B/CDKB complexes are formed during G2 and are maintained at a basal level of activity until mitosis, where dephosphorylation rapidly activates CDKB, thus triggering mitosis progression. The degradation of cyclin B would trigger mitosis exit.
Ostreococcus belongs to the Prasinophycae, a primitive group of green algae at the base of the green lineage. Ostreococcus contains cell cycle proteins of both animal type, such as Cdc25, or plant type, such as CDKB. That in this ancestor of land plants CDKB is the main mitotic CDK suggests that CDKB evolved in land plants to control specific processes linked to multicellularity such as endoreduplication, whereas CDKA took over the main control of cell cycle progression. Future studies in Ostreococcus should shed light on the respective functions of CDKA and CDKB in this simple model system and possibly also in higher plants.
MATERIALS AND METHODS
Culture and Drugs
Ostreococcus tauri strain, isolated from the Thau lagoon (Courties et al., 1994), was cultivated in filtered sterile seawater supplemented with Keller enrichment medium (Sigma-Aldrich, Lyon, France). Kanamycine (100 μg/mL), neomycine (40 μg/mL), and penicillin (50 μg/mL) were added in our synchronization experiments to avoid bacterial contamination. They did not interfere with Ostreococcus growth. Cultures were grown under constant gentle agitation at 20°C and subjected to 12:12 light/dark cycles under blue-light filter moonlight blue (Texen, Toulouse, France). In exponential phase (below 107 cell/mL), about 90% of the cells divided between 11 and 18 h after light on. Olomoucine, aphidicolin, and HU (Sigma) were stored as 1,000× stock solution and propyzamide (ChemService, West Chester, PA) as a 10,000× stock solution in appropriate solvent. To induce cell cycle arrests before the replication of DNA, olomoucine (80 μm), aphidicolin (20 μm), or HU (1 mm) were added 5 h after light on, i.e. in natural late-G1 phase, and white light was used to boost cell cycle progression. Artificial cell synchronization was achieved as follows: cells were arrested at the beginning of S phase by a 14-h incubation with 1 mm HU, then HU was washed out by two rounds of centrifugation and resuspension of the cells in fresh medium (8,000g, 4°C, 10 min). Pluronic (0.1% final concentration; Sigma) was added to the cells before the first centrifugation to avoid hydrodynamic damage. Under these conditions, cells placed under constant white illumination divide within 5 h after the last wash. G2/M and M arrest were performed by adding, respectively, olomoucine (80 μm) or propyzamide (9 μm) 1 h after the release from HU until the control cells had achieved mitosis.
Flow Cytometry Analysis
One-milliliter cell sample was fixed with 0.25% glutaraldehyde (Sigma) for 15 min at room temperature and then stored at 4°C for 1 d. For longer preservation, samples were frozen in liquid nitrogen and stored at −80°C. Flow cytometry analysis was performed on a FACScan flow cytometer (FACScalibur; Becton-Dickinson, San Jose, CA). Cells were counted from the appropriate gate (FL3-H versus SSC-H) as described previously (Courties et al., 1994). For analysis of the DNA content, whole fixed cells were stained with SYBR green I (3,000× dilution of the commercial solution; Molecular Probes, Eugene, OR) for 30 min, and 20,000 cells per sample were analyzed using the CellQuest software (Marie et al., 1996). Cell cycle analysis was performed with the Modfit software (Verity Software House, Tophsam, ME), and data yielding the best fit were used for graphical representation.
Western-Blot Analysis
Cells were harvested by centrifugation in conical bottles (10,000g, 4°C, 10 min), after addition of pluronic (0.1%) to the medium. Pellets were frozen in liquid nitrogen and stored at −80°C until extraction. Protein extraction was performed at 4°C. A total of 0.8 mL of extraction buffer was added (60 mm β-glycerophosphate, 15 mm p-nitrophenylphosphate, 1 mm phenylphosphate disodium salt, 25 mm MOPS, pH 7.2, 15 mm EGTA, 15 mm MgCl2, 2 mm dithiothreitol, 1 mm NaF, 1 mm Na3VO4, 10 μg/mL leupeptin, 10 μg/mL aprotinin, 10 μg/mL soybean [Glycine max] trypsin inhibitor, 100 μm benzamidine, 100 μL/mL protease inhibitor cocktail P 9599 [Sigma], 0.5% Nonidet P-40) to the frozen pellet. Samples were homogenized with a micropestle, and extraction was performed by several cycles of freeze and thaw. Cell debris and starch were discarded by centrifugation (14,000g, 4°C, 10 min). Protein concentration was determined by the Bradford method (Sigma), and the same amount of protein was incubated with 10 to 20 μL of p9CKShs1or p10CKSAt1 sepharose beads on a rotary shaker at 4°C for 1 h. Beads were spun down at 2,000g for 1 min and washed three times in bead buffer. For sequential chromatography, the supernatant of the bead (flow through) was incubated on p9CKShs1 for 1 h, beads were spun down, and the supernatant was loaded again on p10CKSAt1. Western-blot analysis was performed as follows: 20 μL of 4× Laemmli buffer was added to p9CKShs1 and/or p10CKSAt1 beads and proteins were eluted by heating at 90°C for 10 min. Proteins eluted from p9CKShs1 were resolved on a 10% or 12% SDS-polyacrylamide denaturing gel and liquid-transferred onto a nylon membrane (PVDF; Amersham Life Sciences, Buckinghamshire, UK) for ECL or ECL plus detection (Amersham Life Sciences). The membranes were stained with Ponceau-Red to check the homogeneity of the transfer, blocked in Tris-buffered saline containing 5% milk powder or 3% bovine serum albumin for PY antibody for 1 h and then incubated with the appropriate antibody. The monoclonal anti-PY coupled to HRP PY-99 (Santa Cruz Biotechnology, Santa Cruz, CA) was used at a 1/10,000 dilution for ECL, the monoclonal PSTAIRE antibody (Sigma) at a 1/5,000 dilution for ECL. CDKB antiserum was raised in rabbit against the C-terminal sequence CYFDSLDKSQF (Covalab, Lyon, France). Cyclin A and cyclin B antisera were produced in rabbit using the C-terminal peptides (EUROGENTEC; cyclin A CPNEQNIHREV, cyclin B CTLPVPHDLRL) and further purified (Covalab). Anti-CDKB was used at a 1/2,000 dilution and anti-cyclin antibodies at a 1/1,000 dilution for ECL-plus. The membranes were washed three to six times in Tris-buffered saline containing 0.1% Tween 20, and the bound antibody was detected with a goat anti-mouse IgG (1/5,000) or goat anti-rabbit (1/10,000) coupled to horseradish peroxidase (Sigma) and then visualized by enhanced chemiluminescence. The competition experiment was performed by preincubating the anti-CDKB antibody with the N-terminal peptide of CDKB (CYFDSLDKSQF) for 10 min at room temperature (molar ratio 1:100).
In Vitro Phosphorylation Assays
The activity of purified CDKs was assayed as their histone H1 kinase activity measured at 30°C for 30 min using [γ32P]ATP, as previously reported (Corellou et al., 2001). Quantification of radioactive histone H1 was performed using a /STORM phosphorimager with the Image QuanT software (Molecular Dynamics, Sunnyvale, CA).
Dephosphorylation of Purified CDK by GST-Cdc25
The GST-Cdc25 fusion protein was overproduced in Escherichia coli and purified as previously described (Khadaroo et al., 2004). Dephosphorylation was performed for 2 h at 30°C, in 50 mm Tris-HCl, pH 8, 50 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol.
Yeast Complementation Studies
The CDKB cDNA from O. tauri was amplified by PCR with the following primers: GGA TCC TCT AGA CAT ATG GAG AAC TAC GAG AAG GTG G and TAC GTA CCC GGG TCA GCG ACC GAT GTG TTC C. The cDNAs covering the entire coding region were cloned into the p416TEF vector (Mumberg et al., 1995) by use of XbaI-SmaI and XbaI-SnaBI restriction enzyme sites. For transformation in Saccharomyces cerevisiae, the strain carrying the mutation cdc28-4 (Surana et al., 1991) was transformed with p416TEF-CDKB, p416/CDC28, or p416. The transformants were selected at 25°C, then colonies were plated on selective medium at permissive (25°C) or restrictive (34°C) temperature.
Acknowledgments
Anti-CDKB and anti-cyclin A and B were a kind gift from Hervé Moreau. Thanks also to Wolfgang Zachariae for the yeast cdc28-4ts mutant and to Dominique Marie and Claude Courties for technical advices in flow cytometry.
This work was supported by the Centre National de la Recherche Scientifique (Young Investigator ATIP Fellowship to F.Y.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.059626.
References
- Arellano M, Moreno S (1997) Regulation of CDK/cyclin complexes during the cell cycle. Int J Biochem Cell Biol 29: 559–573 [DOI] [PubMed] [Google Scholar]
- Bell MH, Halford NG, Ormrod JC, Francis D (1993) Tobacco plants transformed with cdc25, a mitotic inducer gene from fission yeast. Plant Mol Biol 23: 445–451 [DOI] [PubMed] [Google Scholar]
- Boudolf V, Barroco R, Engler Jde A, Verkest A, Beeckman T, Naudts M, Inze D, De Veylder L (2004. a) B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GT, Magyar Z, Acosta JA, Maes S, Van Der Schueren E, Inze D, De Veylder L (2004. b) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyne P, Zabeau M (2001) Genome-wide expression analysis of plant cell cycle modulated genes. Curr Opin Plant Biol 4: 136–142 [DOI] [PubMed] [Google Scholar]
- Chrétiennot-Dinet MJ, Courties C, Vaquer A, Neveux J, Claustre H, Lautier J, Machado MC (1995) A new marine picoeukaryote: Ostreococcus tauri gen. et sp. Nov. (Chlorophyta, Prasinophyceae). Phycologia 4: 285–292 [Google Scholar]
- Corellou F, Brownlee C, Detivaud L, Kloareg B, Bouget FY (2001) Cell cycle in the fucus zygote parallels a somatic cell cycle but displays a unique translational regulation of cyclin-dependent kinases. Plant Cell 13: 585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courties C, Vaquer A, Troussellier M, Lautier J, Chrétiennot-Dinet MJ, Neveux J, Machado MC, Claustre H (1994) Smallest eukaryotic organism. Nature 370: 255 [Google Scholar]
- Dewitte W, Murray JA (2003) The plant cell cycle. Annu Rev Plant Biol 54: 235–264 [DOI] [PubMed] [Google Scholar]
- Donzelli M, Draetta GF (2003) Regulating mammalian checkpoints through Cdc25 inactivation. EMBO Rep 4: 671–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott JA, Noble ME, Tucker JA (1999) Cyclin-dependent kinases: inhibition and substrate recognition. Curr Opin Struct Biol 9: 738–744 [DOI] [PubMed] [Google Scholar]
- Hemerly A, Engler J, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P (1995) Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J 14: 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John PC, Mews M, Moore R (2001) Cyclin/Cdk complexes: their involvement in cell cycle progression and mitotic division. Protoplasma 216: 119–142 [DOI] [PubMed] [Google Scholar]
- Joubes J, Chevalier C, Dudits D, Heberle-Bors E, Inze D, Umeda M, Renaudi JP (2000) CDK-related protein kinases in plants. Plant Mol Biol 43: 607–620 [DOI] [PubMed] [Google Scholar]
- Khadaroo B, Robbens S, Ferraz C, Derelle E, Eychenie S, Cooke R, Peaucellier G, Delseny M, Demaille J, Van De Peer Y, et al (2004) The first green lineage cdc25 dual-specificity phosphatase. Cell Cycle 3: 513–518 [PubMed] [Google Scholar]
- Landrieu I, da Costa M, De Veylder L, Dewitte F, Vandepoele K, Hassan S, Wieruszeski JM, Faure JD, Van Montagu M, Inze D, et al (2004) A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 13380–13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Das A, Yamaguchi M, Hashimoto J, Tsutsumi N, Uchimiya H, Umeda M (2003) Cell cycle function of a rice B2-type cyclin interacting with a B-type cyclin-dependent kinase. Plant J 34: 417–425 [DOI] [PubMed] [Google Scholar]
- Lew DJ, Burke DJ (2003) The spindle assembly and spindle position checkpoints. Annu Rev Genet 37: 251–282 [DOI] [PubMed] [Google Scholar]
- Lew DJ, Kornbluth S (1996) Regulatory roles of cyclin dependent kinase phosphorylation in cell cycle control. Curr Opin Cell Biol 8: 795–804 [DOI] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J (2004) Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair (Amst) 3: 997–1007 [DOI] [PubMed] [Google Scholar]
- Magyar Z, Meszaros T, Miskolczi P, Deak M, Feher A, Brown S, Kondorosi E, Athanasiadis A, Pongor S, Bilgin M, et al (1997) Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 9: 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie D, Vaulot D, Partensky F (1996) Application of the novel nucleic acid dyes YOYO-1, YO-PRO-1, and PicoGreen for flow cytometric analysis of marine prokaryotes. Appl Environ Microbiol 62: 1649–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKibbin RS, Halford NG, Francis D (1998) Expression of fission yeast cdc25 alters the frequency of lateral root formation in transgenic tobacco. Plant Mol Biol 36: 601–612 [DOI] [PubMed] [Google Scholar]
- Mendenhall MD, Hodge AE (1998) Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62: 1191–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Murray JA (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Meszaros T, Miskolczi P, Ayaydin F, Pettko-Szandtner A, Peres A, Magyar Z, Horvath GV, Bako L, Feher A, Dudits D (2000) Multiple cyclin-dependent kinase complexes and phosphatases control G2/M progression in alfalfa cells. Plant Mol Biol 43: 595–605 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Murakami H, Nurse P (2000) DNA replication and damage checkpoints and meiotic cell cycle controls in the fission and budding yeasts. Biochem J 349: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW (2004) Recycling the cell cycle: cyclins revisited. Cell 116: 221–234 [DOI] [PubMed] [Google Scholar]
- Paulovich AG, Toczyski DP, Hartwell LH (1997) When checkpoints fail. Cell 88: 315–321 [DOI] [PubMed] [Google Scholar]
- Planchais S, Glab N, Inze D, Bergounioux C (2000) Chemical inhibitors: a tool for plant cell cycle studies. FEBS Lett 476: 78–83 [DOI] [PubMed] [Google Scholar]
- Planchais S, Glab N, Trehin C, Perennes C, Bureau JM, Meijer L, Bergounioux C (1997) Roscovitine, a novel cyclin-dependent kinase inhibitor, characterizes restriction point and G2/M transition in tobacco BY-2 cell suspension. Plant J 12: 191–202 [DOI] [PubMed] [Google Scholar]
- Porceddu A, Stals H, Reichheld JP, Segers G, De Veylder L, Barroco RP, Casteels P, Van Montagu M, Inze D, Mironov V (2001) A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J Biol Chem 276: 36354–36360 [DOI] [PubMed] [Google Scholar]
- Potuschak T, Doerner P (2001) Cell cycle controls: genome-wide analysis in Arabidopsis. Curr Opin Plant Biol 4: 501–506 [DOI] [PubMed] [Google Scholar]
- Rhind N, Russell P (1998) Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol 10: 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbens S, Khadaroo B, Camasses A, Derelle E, Ferraz C, Inze D, Van de Peer Y, Moreau H (2005) Genome-wide analysis of core cell cycle genes in the unicellular green alga Ostreococcus tauri. Mol Biol Evol 22: 589–597 [DOI] [PubMed] [Google Scholar]
- Schuppler U, He PH, John PC, Munns R (1998) Effect of water stress on cell division and cell-division-cycle 2-like cell-cycle kinase activity in wheat leaves. Plant Physiol 117: 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell DA, Marchbank A, McMahon K, Dickinson JR, Rogers HJ, Francis D (2002) A WEE1 homologue from Arabidopsis thaliana. Planta 215: 518–522 [DOI] [PubMed] [Google Scholar]
- Stark GR, Taylor WR (2004) Analyzing the G2/M checkpoint. Methods Mol Biol 280: 51–82 [DOI] [PubMed] [Google Scholar]
- Sun Y, Dilkes BP, Zhang C, Dante RA, Carneiro NP, Lowe KS, Jung R, Gordon-Kamm WJ, Larkins BA (1999) Characterization of maize (Zea mays L.) Wee1 and its activity in developing endosperm. Proc Natl Acad Sci USA 96: 4180–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher AB, Nasmyth K (1991) The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell 65: 145–161 [DOI] [PubMed] [Google Scholar]
- Umeda M, Umeda-Hara C, Yamaguchi M, Hashimoto J, Uchimiya H (1999) Differential expression of genes for cyclin-dependent protein kinases in rice plants. Plant Physiol 119: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K, Vuylsteke M, Florquin K, Rombauts S, Maes S, Ormenese S, Van Hummelen P, Van de Peer Y, Inzé D, De Veylder L (2003) Microarray analysis of E2Fa-DPa-overexpressing plants uncovers a cross-talking genetic network between DNA replication and nitrogen assimilation. J Cell Sci 116: 4249–4259 [DOI] [PubMed] [Google Scholar]
- Weingartner M, Binarova P, Drykova D, Schweighofer A, David JP, Heberle-Bors E, Doonan J, Bogre L (2001) Dynamic recruitment of Cdc2 to specific microtubule structures during mitosis. Plant Cell 13: 1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrzykowska J, Pien S, Shen WH, Fleming AJ (2002) Manipulation of leaf shape by modulation of cell division. Development 129: 957–964 [DOI] [PubMed] [Google Scholar]
- Zhang K, Letham DS, John PC (1996) Cytokinin controls the cell cycle at mitosis by stimulating the tyrosine dephosphorylation and activation of p34cdc2-like H1 histone kinase. Planta 200: 2–12 [DOI] [PubMed] [Google Scholar]