Abstract
Mitogen-activated protein kinase (MAPK) cascades are activated in plants during responses to pathogens or to pathogen-derived elicitors and mediate intracellular stress responses. Here, we show that a rice (Oryza sativa) MAPK, OsMAPK6, was posttranslationally activated in a cell culture by a sphingolipid elicitor. Suppression of OsMAPK6 expression by RNA interference resulted in a strong reduction of pathogen-induced Phe ammonia-lyase mRNA, whereas the mRNA level of another rice MAPK, OsMAPK5a, was highly increased. Silencing of a small GTPase, OsRac1, by RNA interference or loss-of-function mutation (d1) of the heterotrimeric G-protein α-subunit gene resulted in a strong reduction of the OsMAPK6 protein levels and of kinase activation by a sphingolipid elicitor. Furthermore, coimmunoprecipitation experiments with OsRac1 and OsMAPK6 proteins showed that OsMAPK6 is closely associated with the active form of OsRac1, but not with inactive forms of OsRac1. These results indicate that these two G-proteins regulate an elicitor-inducible MAPK in rice at the protein level.
Recently, several protein kinases with high sequence similarity to mammalian mitogen-activated protein kinases (MAPKs) have been identified in plants (Hirt, 1997; Mizoguchi et al., 1997; Tena et al., 2001; Zhang and Klessig, 2001; Ichimura et al., 2002). A growing body of evidence indicates that plant MAPKs play an important role in signal transduction related to biotic and abiotic stresses. For example, mechanical stress, drought, and cold activate MsMMK4 in alfalfa (Medicago sativa; Jonak et al., 1996; Bogre et al., 1997) and AtMPK4 and AtMPK6 in Arabidopsis (Arabidopsis thaliana; Ichimura et al., 2000). Furthermore, the salicylic acid-induced protein kinase (SIPK) and the wound-induced protein kinase (WIPK) in tobacco (Nicotiana tabacum) are activated by wounding, fungal elicitors, infection with avirulent pathogens, and salicylic acid (Seo et al., 1995; Zhang and Klessig, 1997; Zhang et al., 1998; Zhang and Klessig, 1998a, 1998b; Romeis et al., 1999). Wounding and infection with an avirulent pathogen also activate LeMPK3 in tomato (Mayrose et al., 2004), and treatment with an elicitor activates Arabidopsis AtMPK6 (Nuhse et al., 2000; Asai et al., 2002) and parsley (Petroselinum crispum) PcMPK6 (Kroj et al., 2003), both orthologs of tobacco NtSIPK.
Several MAPKKs and MAPKKKs upstream of well-characterized MAPKs have been identified, suggesting that MAPK cascades also operate in plant defense signaling responses. Among these, the constitutively active MAPKK NtMEK2 activates NtSIPK and NtWIPK, which is followed by induced hypersensitive response-like cell death and defense gene expression (Yang et al., 2001). Parsley PcMKK5 is an activator of PcMPK6, PcMPK3a, and PcMPK3b and is required for the activation of the pathogenesis-related (PR) gene promoter following elicitor treatment (Lee et al., 2004). In Arabidopsis, AtMKK1 is the upstream MAPKK for AtMPK4 (Ichimura et al., 2002), which, however, turned out to be a negative regulator of defense response (Petersen et al., 2000). Finally, a complete MAPK cascade (comprising MEKK1, MKK4/MKK5, and MPK3/MPK6) has been proposed in Arabidopsis (Asai et al., 2002). It is located downstream of the flagellin receptor kinase FLS2, and its signaling cascade components lead to the activation of WRKY22/WRKY29 transcription factors.
Most of the recently reported plant MAPKs have been isolated and characterized from dicot plant species, such as Arabidopsis, tobacco, and alfalfa. In rice (Oryza sativa), an economically important monocot, several MAPKs have been investigated (He et al., 1999; Agrawal et al., 2003; Cheong et al., 2003; Xiong and Yang, 2003). A 60-kD MAPK (OsBWMK1) has been reported to be activated in rice leaves by rice blast fungus (Magnaporthe grisea) infection, elicitor treatment, and wounding (He et al., 1999; Cheong et al., 2003). OsBWMK1, which is localized in the nucleus in Arabidopsis protoplasts, phosphorylates transcription factor OsEREBP1 in vitro and positively regulates PR genes in tobacco plants (Cheong et al., 2003). Another group has reported that a multiple stress-responsive MAPK (OsMAPK5a) inversely modulates abiotic stress and disease resistance (Xiong and Yang, 2003). Rice plant lines overexpressing OsMAPK5a exhibited increased OsMAPK5a kinase activity and increased tolerance to drought, salt, and cold stresses, whereas OsMAPK5a-silenced plants had a significant reduction in abiotic stress tolerance, but enhanced resistance to fungal and bacterial pathogens.
We have previously reported that the rice small GTPase OsRac1 plays an essential role in disease resistance of rice (Kawasaki et al., 1999; Ono et al., 2001; Wong et al., 2004). The constitutively active form of OsRac1 induces production of reactive oxygen species (ROS), cell death, and phytoalexin production, as well as expression of defense-related genes leading to resistance to infection by rice blast fungus and bacterial blight. Further study with a sphingolipid elicitor (SE) isolated from a rice blast fungus has demonstrated that the heterotrimeric G-protein is located upstream of OsRac1 in the SE signaling pathway and is important for resistance against rice blast and bacterial blight infection (Suharsono et al., 2002).
In mammals and yeast (Saccharomyces cerevisiae), a number of reports have shown a close relationship among MAPKs, G-proteins, and small GTPases in the transduction of external stimuli into intracellular responses (Dohlman and Thorner, 2001; Lowes et al., 2002; Kurose, 2003; Ramezani-Rad, 2003; Ory and Morrison, 2004), but no such association has been reported so far in plants.
In this study, we have characterized a rice MAPK termed OsMAPK6 that was activated in suspension cell cultures by a SE. Silencing of this MAPK caused a reduction of pathogen-induced Phe ammonia-lyase (PAL) and OsBWMK1 mRNAs and an increase in the mRNA of another MAPK of rice, OsMAPK5a. Suppression of OsRac1 by RNA interference (RNAi) or loss-of-function mutation of the heterotrimeric G-protein α-subunit (Gα) gene resulted in a strong reduction of both OsMAPK6 protein levels and kinase activation by a SE. Furthermore, coimmunoprecipitation experiments showed that OsMAPK6 and OsRac1 form a protein complex and that OsMAPK6 is only associated with the active form of OsRac1, suggesting a close functional link between these two proteins.
RESULTS
A Rice MAPK Is Activated in Cell Culture by a SE
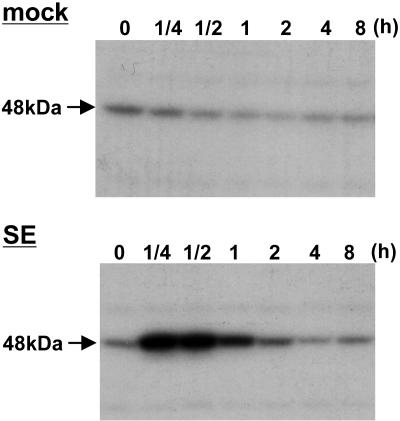
Previous studies have shown that a SE, purified from the rice blast fungus activates defense responses in rice plants and suspension cell cultures (Koga et al., 1998; Suharsono et al., 2002; Umemura et al., 2002). Therefore, to better characterize MAPK signaling in rice defense responses, we performed an in-gel kinase activity assay with myelin basic protein (MBP) as a substrate by using the crude soluble protein extract of SE-treated cells. The assays revealed the activation of a MAPK with an apparent molecular mass of 48 to 49 kD (Fig. 1). A sustained activation was observed from 15 min to 2 h, and the activity then decreased to a basal level after 4 h of elicitor treatment. Because the SE-induced 48- to 49-kD MAPK activity was strongly reduced in OsMAPK6-silenced cell cultures, the activity most likely corresponds to OsMAPK6 (see below).
Figure 1.
Induction of MAPK activity in a suspension cell culture by a SE. Total soluble proteins extracted from cell cultures treated with mock or 5 μg mL−1 SE were separated on a 10% SDS-PAGE gel containing 0.1 mg mL−1 MBP. After renaturation, the gel was incubated in 15 mL of a reaction buffer containing 25 μCi (approximately 3,000 Ci mmol−1) of 5′-[γ-32P]ATP, dried after washing, and exposed to x-ray film.
Isolation of a MAPK Gene Homologous to Arabidopsis MPK6 and Tobacco SIPK
To isolate a MAPK gene whose protein corresponds to the kinase activity detected in SE-treated cell cultures, we isolated a rice ortholog of AtMPK6 because this MAPK is required for resistance to pathogens in Arabidopsis (Asai et al., 2002; Menke et al., 2004; Zipfel et al., 2004), and its homolog in tobacco, NtSIPK, is involved in disease resistance and the hypersensitive response (Zhang and Klessig, 2001). We found an ortholog of AtMPK6 in rice and named it OsMAPK6. The OsMAPK6 gene contains six exons, is located on chromosome 6 of rice (bacterial artificial chromosome AP004239), and is present in a single copy in the rice genome (data not shown).
The OsMAPK6 gene encodes a protein of 398 amino acid residues with an estimated molecular mass of 44.9 kD. OsMAPK6 contains the 11 subdomains that are conserved among all MAPK families and possesses a dual phosphorylation motif TEY (Thr-225/Tyr-227; Fig. 2A). OsMAPK6 is highly similar to NtSIPK (Zhang and Klessig, 1997; 85% identity) and AtMPK6 (Mizoguchi et al., 1993; 83% identity). However, the N-terminal domains of these MAPKs are poorly conserved. Phylogenetic analysis of representative plant MAPKs based on whole-sequence alignment (Fig. 2B) places OsMAPK6 in the A2 subgroup (Ichimura et al., 2002) together with AtMPK6, NtSIPK, and PcMPK6. Most of the members of the A1 and A2 subgroups are activated by various biotic and abiotic stresses (Zhang and Klessig, 2001).
Figure 2.
Comparison of OsMAPK6, NtSIPK, AtMPK6, and representative MAPKs from other higher plants. A, Alignment of the deduced amino acid sequences of OsMAPK6 with NtSIPK and AtMPK6. Identical amino acid residues are shaded, and the TEY motif, with normally phosphorylated Thr (T) and Tyr (Y) for the activation of MAPKs, is boxed. The C-terminal region used to raise the antibody is indicated by a dotted line. B, Phylogenetic analysis of representative plant MAPKs involved in plant defense signaling. Five of the eight subgroups of plant MAPKs (Ichimura et al., 2002) are represented in this tree obtained by the neighbor-joining method. The confidence values of a bootstrap test with 1,000 replications were greater than 95%, with one exception of 72% at the end branch between PcMPK6 and NtSIPK (data not shown). The numbers in brackets after the name of MAPKs indicate the GenBank accession numbers of the corresponding genes. Subgroups are indicated on the right. The scale represents 0.1 substitutions per site.
Activation of OsMAPK6 by a SE Is Posttranslational
We raised an antibody against the C-terminal region of OsMAPK6 (amino acids 274–398), which exhibits lower similarity to other rice MAPKs. The antibody could recognize a purified recombinant protein containing a His tag (His-OsMAPK6), as well as a major band with an apparent molecular mass of 48 kD in a crude protein extract (Fig. 3, A and B). This size is 3 kD greater than that estimated from the deduced amino acid sequence of OsMAPK6. A similar discrepancy between predicted size and migration in SDS-PAGE gels was also observed for NtSIPK (Zhang and Klessig, 1997; Zhang et al., 1998) and AtMPK6 (Ichimura et al., 2000; Nuhse et al., 2000). The 48-kD MAPK activity detected in the SE-treated cell cultures corresponded well in size to that of OsMAPK6. Immunodepletion experiments showed that our antibody precipitated the SE-inducible kinase activity, indicating that the antibody indeed reacted with the observed kinase protein (Fig. 3C). After SE treatment, we used our antibody to analyze the levels of the OsMAPK6 protein. The OsMAPK6 protein levels remained constant and did not increase in response to the elicitor (Fig. 3D). The mRNA levels of OsMAPK6 remained unchanged throughout the elicitor treatment, with a slight decrease at 24 h, indicating that OsMAPK6 mRNA was not induced by the elicitor (Fig. 3E). Compared with the rapid induction of the kinase activity shown in Figure 1, these results indicated that OsMAPK6 is posttranslationally activated by a SE in a rice cell culture.
Figure 3.
The activation of OsMAPK6 by a SE is posttranslational. A and B, Specificity of the anti-OsMAPK6 antibody. Eight micrograms of total protein extract (T) from rice suspension cells (cv Kinmaze) and 250 (A) or 10 (B) ng of affinity-purified fusion protein His-OsMAPK6 (H) purified from a bacterial culture were separated on 10% SDS-PAGE and stained with Coomassie Brilliant Blue or detected with the anti-OsMAPK6 antibody. The mobilities of the protein ladder are indicated on the left. C, The kinase activity was removed by immunodepletion with anti-OsMAPK6 antibody from total proteins purified from rice cells at 15 min after treatment with 5 μg mL−1 SE. D, Rice cell cultures treated with 5 μg mL−1 SE were analyzed by immunoblotting using the specific antibody against OsMAPK6 or anti-α-tubulin antibody. Total soluble proteins were extracted, and 8 μg of each sample were separated by 10% SDS-PAGE, electrotransferred onto a polyvinylidene difluoride membrane, and probed with the anti-OsMAPK6 antibody or anti-α-tubulin antibody. E, Rice suspension cell cultures were treated with 5 μg mL−1 SE. Total RNAs were isolated from suspension cell samples at the indicated times and used for RNA gel-blot analysis. The membrane was probed with a [32P]-labeled gene-specific probe. Equal loading was monitored by ethidium bromide staining of rRNA (bottom).
Silencing of OsMAPK6 Activity by RNAi Causes Alterations of mRNA Levels of PAL and Two Other MAPK Genes
To investigate the function of OsMAPK6 in SE signaling, we generated transgenic rice cell cultures and plants in which the OsMAPK6 mRNA level was specifically reduced by RNAi. The construct was made by inserting a 422-bp region (covering a portion from the C-terminal encoding region and 3′-untranslated region) in inverse orientation, separated by a β-glucuronidase sequence linker and under the control of the maize Ubq1 promoter (Miki and Shimamoto, 2004). Four RNAi cell lines (nos. 3, 20, 31, and 32) were analyzed for OsMAPK6 silencing by northern blot. In RNAi line 3, OsMAPK6 mRNA was almost completely abolished, whereas lines 20, 31, and 32 showed various levels of silencing (Fig. 4A). In addition to mRNA levels, the RNAi lines showed strong reduction of OsMAPK6 protein levels (Fig. 4B). In particular, OsMAPK6 was almost undetectable in RNAi line 3, correlating with its very low level of transcripts. When the four RNAi lines were further tested for kinase activation in response to SE (Fig. 4C), they all showed almost no activation with similar levels, regardless of the variation in their mRNA and protein levels. These results confirmed that the SE-inducible MAPK activity is encoded by OsMAPK6.
Figure 4.
Effect of OsMAPK6-RNAi on the expression of other genes. A to C, Effect of OsMAPK6-RNAi on the protein level and kinase activity in a cell culture. A, Total RNAs were isolated from cv Kinmaze (wild-type) cells and from cell lines with a silenced OsMAPK6 gene (OsMAPK6-RNAi) and analyzed by RNA gel blot. Membranes were probed with a [32P]-labeled OsMAPK6 gene-specific probe. B, Total soluble proteins were extracted from the same cell lines and analyzed by immunoblotting with the anti-OsMAPK6 and anti-α-tubulin antibodies. C, The same cell lines were treated with 5 μg mL−1 SE, sampled at the indicated times, and assayed for in-gel kinase activity. D, Effect of OsMAPK6-RNAi on mRNA levels of OsMAPK5a, OsBWMK1, and PAL genes. Wild-type cv Kinmaze and OsMAPK6-RNAi line 3 (Ri 3) cells were incubated with 5 μg mL−1 SE, sampled at the indicated times for RNA extraction, and analyzed by northern blot.
In response to elicitors, plant cells exhibit a rapid induction of a specific set of genes, including MAPKs, PR genes, chitinase, PAL, and lipoxygenase (Peng et al., 1994; Tanaka et al., 2003). We examined the mRNA levels of three genes, OsMAPK5a, OsBWMK1, and PAL, which showed significant changes in their expression between the wild-type and the RNAi lines (Fig. 4D). We observed a strong, but transient, expression of OsMAPK5a mRNA in the silenced lines, while in the wild type, very low levels of OsMAPK5a mRNA were detected in each of the time points analyzed. These results indicate that the suppression of OsMAPK6 resulted in a significantly increased response of OsMAPK5a to a SE, suggesting the presence of antagonistic effects of the two MAPKs at the RNA level. Alternatively, the enhanced OsMAPK5a mRNA level may compensate for the loss of OsMAPK6. On the other hand, OsBWMK1, whose expression was slightly reduced in the RNAi lines at the steady state, was induced in the OsMAPK6-RNAi line by a SE, but its mRNA was decreased at 9 h compared to the wild type. PAL mRNA was strongly suppressed in the OsMAPK6 RNAi line over the course of the elicitor treatment. One of the rice phytoalexins, Sakuranetin, is synthesized via the phenylpropanoid pathway (Kodama et al., 1992). Together, these results indicate that OsMAPK6 regulates the mRNA levels of various genes that are involved in biotic and abiotic stresses.
OsRac1 and Heterotrimeric G-Protein Regulate OsMAPK6 at the Protein Level
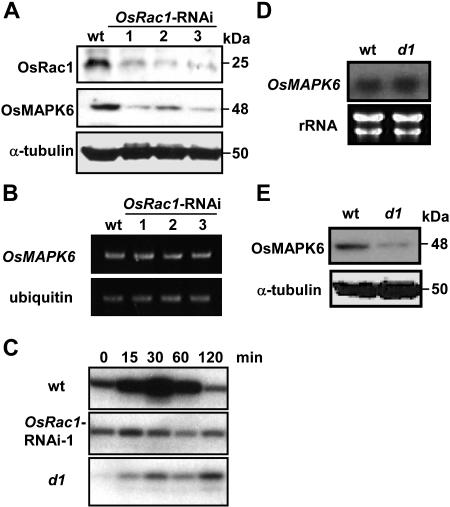
In mammals and yeast, accumulating evidence indicates a close relationship between MAPKs and GTPases, such as Ras and heterotrimeric G-proteins, in the transduction of external stimuli into intracellular responses (Dohlman and Thorner, 2001; Lowes et al., 2002; Kurose, 2003; Ramezani-Rad, 2003; Ory and Morrison, 2004), but no such evidence has been reported so far in plants. We have previously demonstrated that defense responses to a SE and blast fungus infection require both OsRac1 and Gα, the latter acting upstream of OsRac1 (Ono et al., 2001; Suharsono et al., 2002). To investigate a possible link between OsMAPK6 and these GTP-binding proteins, we first analyzed transgenic cell lines in which OsRac1 was specifically silenced by RNAi (Miki and Shimamoto, 2004) for protein level (Fig. 5A). Wild-type cv Kinmaze and three independent RNAi lines representative of six lines were used in our experiment. Immunoblot detection of the OsRac1 protein showed a strong decrease in the transgenic lines. Interestingly, a significant reduction in the OsMAPK6 protein level was observed in these cell lines (Fig. 5A). However, its mRNA levels were not reduced (Fig. 5B), suggesting that OsRac1 regulates OsMAPK6 accumulation by affecting its stability. We further tested whether the OsMAPK6 protein level directly affected its kinase activation upon elicitor treatment (Fig. 5C). In the OsRac1-RNAi line, the MAPK activity was not induced by an elicitor, indicating that a threshold OsMAPK6 level is required for the kinase activity response (Fig. 5C). Alternatively, lack of OsMAPK6 kinase activity could be explained by a requirement for OsRac1.
Figure 5.
Effects of OsRac1-RNAi and heterotrimeric G-protein Gα mutation on OsMAPK6 protein, gene expression, and kinase activity. A, Total soluble proteins were extracted from cv Kinmaze (wild-type) cells and from cell lines with a silenced OsRac1 gene (OsRac1-RNAi) and analyzed by immunoblotting with the anti-OsRac1 antibody (top), anti-OsMAPK6 antibody (middle), or anti-α-tubulin antibody (bottom) as the loading control. B, Total RNAs were isolated from OsRac1-RNAi lines and analyzed by RT-PCR using specific primers of OsMAPK6 and ubiquitin. C, The OsRac1-RNAi line (line 1) and d1 mutant were treated with elicitor 5 μg mL−1 SE, sampled at the indicated times, and assayed for in-gel kinase activity. D, Total RNAs were isolated from wild-type cells and the heterotrimeric protein Gα-subunit null mutant (d1) and analyzed by RNA gel blot. Membranes were probed with a [32P]-labeled OsMAPK6 gene-specific probe. E, Total soluble proteins were extracted from wild-type cells and the heterotrimeric protein Gα-subunit null mutant (d1) and analyzed by immunoblotting with the anti-OsMAPK6 antibody and anti-α-tubulin antibody as the loading control.
We next analyzed OsMAPK6 mRNA, protein, and kinase activity in cell cultures containing the Gα mutation (d1; Fujisawa et al., 1999; Suharsono et al., 2002). The OsMAPK6 protein level was strongly reduced in the d1 cells (Fig. 5E), while the mRNA levels remained unchanged (Fig. 5D), indicating that Gα is required for the maintenance of the OsMAPK6 protein level, as was shown for OsRac1. The observed reduction of OsMAPK6 protein is the likely cause of the lack of kinase activation in the d1 cells (Fig. 5C). Our results indicate that OsRac1 and heterotrimeric G-protein regulate OsMAPK6 at the protein level.
OsMAPK6 and OsRac1 Proteins Are in the Same Protein Complex
Because we found a close functional link between OsRac1 and OsMAPK6, we decided to examine whether they are in the same protein complex by coimmunoprecipitation experiments. For these experiments, we used transgenic rice cell cultures that express constitutively active myc-OsRac1 (CA), dominant-negative myc-OsRac1 (DN), and myc-CA-OsRac1, in which the C-terminal Cys residue of OsRac1 was exchanged by Ser (CS) to block the plasma membrane localization of myc-CA-OsRac1 (Ono et al., 2001). Western-blot analysis of proteins immunoprecipitated with the anti-myc antibody revealed that the OsMAPK6 protein was coimmunoprecipitated with myc-CA-OsRac1, but not with DN or CS mutant OsRac1 proteins (Fig. 6A). In the coimmunoprecipitation experiments, we always found two OsRac1 bands and reasons for this observation remain to be analyzed. The control experiments showed that the input protein levels of CA-, DN-, and CS-OsRac1 were comparable (Fig. 6B). Assays of the OsMAPK6 kinase activities in these OsRac1 mutant cell cultures indicated that SE activated the kinase activity in wild-type and CA cell cultures and that the activity remained slightly longer in CA than in wild type (Fig. 6C). In DN and CS mutants, however, the kinase activity was suppressed (Fig. 6C). These results indicate that OsMAPK6 protein makes a complex only with the active form of OsRac1, but not with the inactive OsRac1 or the active OsRac1, which was not able to bind to the plasma membrane. Since OsRac1 and OsMAPK6 did not bind directly in the yeast two-hybrid assays (L. Chen and K. Shimamoto, unpublished data), it is likely that they form a protein complex with other proteins.
Figure 6.
OsMAPK6 and active OsRac1 are in the same protein complex. A and B, Protein extracts obtained from wild-type rice cells and transgenic rice cell cultures expressing myc-tagged OsRac1 proteins were immunoprecipitated with the anti-myc antibody (Santa Cruz Biotechnology). The immunoprecipitated proteins were subjected to western-blot analysis with the anti-OsMAPK6 (A) or anti-OsRac1 (B) antibodies. IgH corresponds to the IgG heavy chain. WT, Wild-type cv Kinmaze; CA, myc-constitutively active OsRac1; DN, myc-dominant negative OsRac1; CS, myc-CA-OsRac1 having a mutation of a C-terminal Cys residue. This mutation blocks plasma membrane localization of OsRac1. All these OsRac1 mutants have been described previously (Kawasaki et al., 1999; Ono et al., 2001). C, Rice cell cultures of wild type, CA, DN, and CS were treated with a SE and analyzed for the OsMAPK6 kinase activity.
In contrast to OsRac1-RNAi lines in which levels of OsMAPK6 proteins were strongly reduced (Fig. 5A), no reduction of OsMAPK6 protein was observed in DN or CS mutants (Fig. 6A). However, OsMAPK6 protein was not associated with the mutant OsRac1 proteins. These results indicate that, in DN and CS mutants, OsMAPK6 protein was still present, but not in the OsRac1 complex, and that the kinase activity was suppressed, suggesting the requirement of the close association of OsMAPK6 protein with OsRac1 for the kinase activity. Another explanation could be that these OsRac1mutants may activate other proteins that have inhibitory effects on OsMAPK6 activity.
DISCUSSION
Defense signaling and SE signaling in rice involve two GTPases: OsRac1, a molecular switch for defense responses, hypersensitive cell death, and ROS production (Kawasaki et al., 1999; Ono et al., 2001; Wong et al., 2004) and Gα, located upstream of OsRac1 in rice defense signaling (Suharsono et al., 2002). We report that, in plants, a MAPK is regulated by OsRac1 and Gα at the protein level. This conclusion is based on the observations that OsMAPK6 protein levels were strongly reduced in OsRac1-silenced cells and in the d1 mutant (Gα mutant) and that SE-induced OsMAPK6 activation is greatly reduced in these mutant cells. These results suggest that the two GTP-binding proteins are required for the accumulation of the OsMAPK6 protein and, possibly, for its activation as well. Furthermore, our finding that OsMAPK6 and OsRac1 proteins are in the same protein complex supports this conclusion.
The analysis of OsRac1-RNAi lines and DN- and CS-OsRac1 mutants suggests that there might be more than one mechanism for regulation of OsMAPK6 kinase activity by OsRac1. In OsRac1-RNAi lines, OsMAPK6 protein levels were strongly reduced. In DN and CS mutants, OsMAPK6 protein levels were not reduced and it was not associated with the OsRac1 complex. Therefore, understanding molecular mechanisms for regulation of OsMAPK6 kinase activity by OsRac1 is important in future study.
Our previous study showed that Gα functions upstream of OsRac1 in the SE signaling pathway, leading to the induction of ROS production and defense gene expression (Suharsono et al., 2002). Therefore, a MAPK cascade may be similarly activated by these two G-proteins along with other pathways. The mechanism of how Gα activates OsRac1 in these signaling pathways remains to be studied.
It is known that, in mammals and yeast, Ras-like GTPases are involved in upstream signaling for MAPK cascade activation (Dohlman and Thorner, 2001; Lowes et al., 2002; Kurose, 2003; Ramezani-Rad, 2003; Ory and Morrison, 2004). These Ras-MAPK or G-protein-MAPK cascades occur in response to various stimuli, such as hormones or environmental stresses. The signals are either transduced into the cascade components by direct protein interactions or require additional intermediate regulating factors (Dohlman and Thorner, 2001; Ramezani-Rad, 2003; Ory and Morrison, 2004).
In the strongest OsMAPK6-silenced line (line 3), we observed the induction of OsMAPK5a mRNA (Fig. 4D). Silencing of NtSIPK in tobacco and AtMPK6 in Arabidopsis (both orthologous to OsMAPK6) was shown to increase the activation of NtWIPK and AtMPK3 (both orthologous to OsMAPK5a) under ozone stress and after wounding, respectively (Samuel and Ellis, 2002; Menke et al., 2004). In a similar way, the enhanced OsMAPK5a mRNA level may compensate for the loss of OsMAPK6.
OsMAPK6 silencing reduced the level of PAL mRNA. The PAL gene has been shown to be involved in defense signaling in tobacco and rice by its rapid induction in response to pathogens or an elicitor (Zhu et al., 1995; Shadle et al., 2003; Tanaka et al., 2003). This enzyme represents the entry point of the phenylpropanoid pathway for the synthesis of antimicrobial compounds, such as phytoalexins (Dixon, 2001), or of the lignin polymer, which can act as an inducible physical barrier against pathogen ingress (Mitchell et al., 1999). A rice phytoalexin, Sakuranetin, is synthesized through the phenylpropanoid pathway (Kodama et al., 1992).
Recently, AtMPK6 was shown to be a component of a complete MAPK cascade, leading to resistance in Arabidopsis (Asai et al., 2002). Menke et al. (2004) reported that the silencing of AtMPK6 compromises resistance in Arabidopsis against both avirulent and virulent pathogens. Therefore, OsMAPK6-silenced plants were tested for resistance to avirulent and virulent isolates of rice blast. In both cases, with more than 20 independent RNAi plants tested, we could not detect significant differences in the resistance or susceptibility of the silenced plants (data not shown). As is the case for AtMPK6 in Arabidopsis, OsMAPK6 may be required for only some of the specific pathogens. Therefore, other rice pathogens need to be tested with OsMAPK6-silencing plants to further understand how this MAPK functions in disease resistance in rice.
MATERIALS AND METHODS
Cell Cultures, Chemical Treatment, and Pathogen Inoculation
Rice (Oryza sativa) suspension cultures expressing dominant-negative OsRac1 and the heterotrimeric G-protein α-subunit mutant dwarf1 (d1) have been described previously (Kawasaki et al., 1999; Ono et al., 2001; Suharsono et al., 2002). A SE was purified as described previously (Koga et al., 1998). For the SE assay and chemical treatment, suspension cell cultures were subcultured for 3 d in a fresh medium before being divided into microplate wells (0.15–0.2 g cells, 1 mL fresh medium per 22-mm-diameter well) and maintained in suspension for 16 h at 30°C. A SE or chemicals was added to the cells, and incubation was continued for various lengths of time after treatment.
Isolation and Sequence Analysis of OsMAPK6
OsMAPK6 cDNA was isolated by reverse transcription (RT)-PCR. Total RNA was extracted from approximately 0.15 g of a suspension cell culture from cv Kinmaze, as previously described (Lieberherr et al., 2003). For the RT reaction, cDNAs were synthesized at 37°C for 1 h in a final volume of 20 μL containing 1 μg total RNA, 2.5 μm oligo(dT) primers, 500 μm each deoxynucleotide triphosphate (dNTP), and 50 units Moloney-murine leukemia virus reverse transcriptase (Promega, Madison, WI). DNA was then amplified by PCR in a reaction volume of 20 μL containing the DNA template (1 μL RT reaction), 200 μm dNTP, 1 unit of platinum Pfx DNA polymerase, buffer conditions according to the manufacturer's instructions (Invitrogen, Carlsbad, CA), and 0.5 μm of each primer: FW1, 5′-AAATCTCGCGACGAATTCCGC-3′ and RV1, 5′-GACCACGTCTTACAACATGGAGCATTA-3′. After a 2-min denaturation step at 94°C, PCR was performed at 94°C, 58°C (0.5 min each), and 68°C (2 min) for 30 cycles, with a termination step of 10 min at 68°C. The PCR products were separated on a 1% agarose gel, and a band of approximately 1.7 kb was excised, gel extracted, and cloned into pCR-Blunt II-TOPO (Invitrogen). Three clones were sequenced in both directions to obtain the complete sequences of the cDNAs.
Sequence analysis was performed using BLAST (Altschul et al., 1990) for a similarity search in the rice database, and amino acid sequence alignment was performed with GeneWorks software (version 2.5.1; Intelligenetics, Mountain View, CA). PHYLIP (Phylogeny Inference Package; http://evolution.genetics.washington.edu/phylip.html) was used to analyze protein phylogeny by the neighbor-joining method. The phylogenetic tree was displayed with TreeExplorer (http://evolgen.biol.metro-u.ac.jp/TE/TE_man.html).
Plasmid Constructs and Rice Transformation
Agrobacterium-mediated transformation of rice calli was performed as described previously (Hiei et al., 1994). Plants were regenerated from transformed calli by selecting for hygromycin resistance. The OsMAPK6-RNAi (dsRNAi) was made by cloning an amplified fragment of 422 bp, in inverse orientation and separated by a β-glucuronidase sequence linker, into the p2K1+ vector under the control of the maize Ubq1 promoter (Miki and Shimamoto, 2004). The 422-bp OsMAPK6 DNA fragment for the RNAi construct was amplified from a cDNA region covering the C-terminal encoding region (29 bp upstream of the stop codon) and the 3′-untranslated region (393 bp downstream of the stop codon) using standard PCR methods and the following primers: RiFW1, 5′-GCCTTGCGTTCAACCCTGATTACC-3′ and RiRV1, 5′-GACCACGTCTTACAACATGGAGCATTA-3′.
RNA Analyses
Total RNA was isolated from about 0.15 g ground rice tissue, and RNA gel-blot analysis was performed as described previously (Lieberherr et al., 2003). Gene-specific DNA probes were obtained by standard PCR amplification methods using the following primers: OsMAPK6, FWRT 5′-AATGGAGCTCATCGGAACGCC-3′ and RTRV 5′-CAAATCCGAATCCGGCCATGGAC-3′; OsMAPK5a, F24 5′-TTAGGTTGGTCAATTCGGC-3′ and R496 5′-TGGCAGTGCTCTTCTGACAG-3′; OsBWMK1, FW1 5′-CTTCCTCTATCCAAGTGGGGTTGATC-3′ and RV1 5′-ATCGTTGTGCACTAGGAGTGCATC-3′; PAL, F 5′-CTACCTTCTCCTCCGGCTC-3′ and R 5′-TGTTGAGCAGCTTGGTGATG-3′.
All the DNA probes were electrophoresed in agarose gel, excised, gel extracted, and purified before labeling.
Production of Recombinant OsMAPK6, Anti-OsMAPK6, and Anti-OsRac1 Antibodies
A sequence from the 3′ end of OsMAPK6 (from codon V274 to the stop codon) was amplified by PCR using primers containing BamHI/XhoI restriction sites (abBFW1, 5′-GGATCCGTCCATCAATTACGTCTACTAATGGAG-3′; abXRV1, 5′-CTCGAGCACCAGCTACTGGTAATCAGGGTTG-3′) and cloned into pCR-Blunt II-TOPO (Invitrogen) for further in-frame subcloning into a pGEX-4T-1 vector (Amersham Biosciences, Little Chalfont, UK) using BamHI/XhoI sites. Glutathione S-transferase fusion recombinant protein was produced in Escherichia coli BL21 (DE3) RIL cells (Stratagene, La Jolla, CA). Overnight cultures of the transformant E. coli cells were diluted 1:100 in a fresh Luria-Bertani medium supplemented with 100 μg mL−1 carbenicillin and grown to A600 = 0.8 at 37°C; then expression of the recombinant proteins was induced by addition of 1 mm isopropyl β-d-thiogalactoside and followed by growth at 37°C for a further 4 h. The bacteria were collected by centrifugation, frozen, and sent to MBL (Nagoya, Japan) for recombinant protein purification and rabbit polyclonal antiserum production.
To construct His-OsMAPK6, the OsMAPK6 sequence was amplified using primers including XhoI and BamHI restriction sites (15bXFW1, 5′-CTCGAGCGCGATCCAAATCCGAATCCG-3′; 15bBRV1, 5′-GGATCCAACACCAGCTACTGGTAATCAGGGTTG-3′), cloned into pCR-Blunt II-TOPO, and further subcloned into the pET-15b vector (Novagen, Madison, WI) using XhoI/BamHI sites. The recombinant protein was produced as described above and purified with HiTrap chelating horseradish peroxidase (1 mL) according to the manufacturer's instructions (Amersham Biosciences).
To produce an anti-OsRac1-specific antibody, the OsRac1 gene was cloned into pGEX-4T-1 (Amersham Biosciences), and the recombinant protein was produced in E. coli. After purification, the glutathione S-transferase tag was removed at the thrombin cleavage site and the OsRac1 protein was used to produce the antibody in rabbit (services provided by MBL).
Antiserum (10 mL) was incubated at 25°C for more than 4 h with a membrane containing bound pET32a-Trx-His-S-OsRac1 (Novagen) recombinant protein from E. coli. The membrane was washed five times with a Tris-buffered saline buffer (137 mm NaCl, 2.68 mm KCl, 25 mm Tris, pH 7.4) for 5 min each time. The OsRac1-specific antibody was eluted with 2 mL 0.1 m Gly and 0.15 m HCl (pH 2.5–3.0) and neutralized with 450 μL of 0.5 m HEPES, pH 8.5. Similar procedures, with a membrane saturated with bound His-OsMAPK6, were used to purify the anti-OsMAPK6 antibody.
Protein Extraction, Immunoblot Analysis, In-Gel Kinase Assay, and Immunodepletion
About 0.15 g frozen tissue were ground in liquid nitrogen and homogenized in an extraction buffer (100 mm HEPES, pH 7.5, 5 mm EDTA, 5 mm EGTA, 10 mm Na3VO4, 10 mm NaF, 50 mm β-glycerophosphate, Roche complete [without EDTA], 1 tablet per 50 mL buffer, 10 mm dithiothreitol [DTT]). After centrifugation at 18,000g for 10 min at 4°C, the supernatant was transferred to a new tube and stored at −40°C. The protein concentration was determined using the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin as standard. The protein samples were used both for immunoblot analysis and for the in-gel kinase activity assay.
For immunoblot detection, equal amounts of protein extracts (8 μg) were separated by 10% SDS-PAGE and electrotransferred for 1 h onto an Immobilon-P membrane (Millipore, Billerica, MA) in a transfer buffer (25 mm Tris, 192 mm Gly, 20% MeOH). The membrane was blocked for 1 h in phosphate-buffered saline plus Tween (PBS-T; 137 mm NaCl, 8.1 mm Na2HPO4, 2.68 mm KCl, 1.47 mm KH2PO4, 0.1% Tween 20) containing 5% skim milk powder, washed three times with PBS-T, incubated for 1 h with anti-OsMAPK6 (diluted 1:500), anti-OsRac1 (diluted 1:200), or anti-α-tubulin (diluted 1:2,000) antibodies in PBS-T containing 0.5% skim milk, and washed five times with PBS-T. The membrane was then incubated for 1 h in PBS-T containing 0.5% skim milk with anti-rabbit IgG conjugated to horseradish peroxidase (diluted 1:5,000; Amersham Biosciences) and then washed as described above. Detection was performed using ECL western-blot detection reagents (Amersham Biosciences) according to the manufacturer's instructions.
For the in-gel kinase activity assay, protein samples (8 μg) were separated on 10% SDS-PAGE gels containing 0.1 mg mL−1 MBP. The subsequent washing, renaturation, and kinase activity steps were performed as described by Zhang et al. (1998), except for the washing steps after electrophoresis to remove SDS. The gel was washed three times (30 min each) at room temperature in a buffer containing 25 mm Tris, pH 7.5, 0.5 mm DTT, 0.1 mm Na3VO4, and 5 mm NaF.
For the immunodepletion assay, 1 mg of protein extracts was incubated with anti-OsMAPK6 antibody for 4 h, and then precipitated with protein A Sepharose (Amersham Biosciences). After washing with a buffer (100 mm HEPES, pH 7.5, 5 mm EDTA, 5 mm EGTA, 10 mm Na3VO4, 10 mm NaF, 50 mm β-glycerophosphate, 10 mm DTT), the beads were eluted with an SDS sample buffer and subjected to western-blot analysis with anti-MAPK6 antibody.
Coimmunoprecipitation Assays
Rice cells (cv Kinmaze) expressing myc-tagged OsRac1 protein were generated by Agrobacterium-mediated transformation of a rice callus. All myc-tagged OsRac1 constructs were driven by the maize Ubq1 promoter. Three mutants of OsRac1 (CA, DN, and CS) have been previously described (Kawasaki et al., 1999; Ono et al., 2001).
Total proteins from rice cells expressing myc-tagged OsRac1 proteins were extracted in an extraction buffer (137 mm NaCl, 8.1 mm Na2HPO4, 1.47 mm KH2PO4, pH7.0, 10% Suc, and protease inhibitor (Roche, Mannheim, Germany). Cell debris was removed by centrifugation at 12,000g for 25 min. The supernatants were mixed with the anti-myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 8 h at 4°C and 3 h at room temperature, then precipitated with protein A Sepharose beads (Amersham Biosciences). After washing with a buffer (137 mm NaCl, 8.1 mm Na2HPO4, 1.47 mm KH2PO4, pH7.0, 10% Suc, 150 mm NaCl, and 0.5% Triton X-100), the beads were eluted with an SDS sample buffer and subjected to western-blot analysis with anti-MAPK6 antibody or anti-OsRac1 antibody.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AB183398.
Supplementary Material
Acknowledgments
We thank Dr. Hirohiko Hirochika for unpublished data on OsMAPK6 cDNA and protein and Dr. Ian Smith for reading the manuscript. We thank the members of the Laboratory of Plant Molecular Genetics at Nara Institute of Science and Technology (NAIST) for technical assistance, comments, and participation in discussions.
This work was supported by the Japanese Society for the Promotion of Science (JSPS; postdoctoral fellowship no. P01701 to D.L.), the Research for the Future Program of the JSPS (grant no. JSPS–RFTF 00L01604), and the Ministry of Agriculture, Forestry, and Fisheries of Japan, Rice Genome Project.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.057414.
References
- Agrawal GK, Iwahashi H, Rakwal R (2003) Rice MAPKs. Biochem Biophys Res Commun 302: 171–180 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bogre L, Ligterink W, Meskiene I, Barker PJ, Heberle-Bors E, Huskisson NS, Hirt H (1997) Wounding induces the rapid and transient activation of a specific MAP kinase pathway. Plant Cell 9: 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Moon BC, Kim JK, Kim CY, Kim MC, Kim IH, Park CY, Kim JC, Park BO, Koo SC, et al (2003) BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol 132: 1961–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA (2001) Natural products and plant disease resistance. Nature 411: 843–847 [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Thorner JW (2001) Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem 70: 703–754 [DOI] [PubMed] [Google Scholar]
- Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T, Asahi T, Iwasaki Y (1999) Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci USA 96: 7575–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Fong SH, Yang D, Wang GL (1999) BWMK1, a novel MAP kinase induced by fungal infection and mechanical wounding in rice. Mol Plant Microbe Interact 12: 1064–1073 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hirt H (1997) Multiple roles of MAP kinases in plant signal transduction. Trends Plant Sci 2: 11–15 [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000) Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24: 655–665 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, Sheen J, Henri Y, Champion A, Kreis M, Zhang SQ, Hirt H, Wilson C, et al (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Jonak C, Kiegerl S, Ligterink W, Barker PJ, Huskisson NS, Hirt H (1996) Stress signaling in plants: a mitogen-activated protein kinase pathway is activated by cold and drought. Proc Natl Acad Sci USA 93: 11274–11279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K (1999) The small GTP-binding protein rac is a regulator of cell death in plants. Proc Natl Acad Sci USA 96: 10922–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama O, Miyakawa J, Akatsuka T, Kiyosawa S (1992) Sakuranetin, a flavanone phytoalexin from ultraviolet-irradiated rice leaves. Phytochemistry 31: 3807–3809 [Google Scholar]
- Koga J, Yamauchi T, Shimura M, Ogawa N, Oshima K, Umemura K, Kikuchi M, Ogasawara N (1998) Cerebrosides A and C, sphingolipid elicitors of hypersensitive cell death and phytoalexin accumulation in rice plants. J Biol Chem 273: 31985–31991 [DOI] [PubMed] [Google Scholar]
- Kroj T, Rudd JJ, Nurnberger T, Gabler Y, Lee J, Scheel D (2003) Mitogen-activated protein kinases play an essential role in oxidative burst-independent expression of pathogenesis-related genes in parsley. J Biol Chem 278: 2256–2264 [DOI] [PubMed] [Google Scholar]
- Kurose H (2003) Galpha12 and Galpha13 as key regulatory mediator in signal transduction. Life Sci 74: 155–161 [DOI] [PubMed] [Google Scholar]
- Lee J, Rudd JJ, Macioszek VK, Scheel D (2004) Dynamic changes in the localization of MAPK cascade components controlling pathogenesis-related (PR) gene expression during innate immunity in parsley. J Biol Chem 279: 22440–22448 [DOI] [PubMed] [Google Scholar]
- Lieberherr D, Wagner U, Dubuis PH, Metraux JP, Mauch F (2003) The rapid induction of glutathione S-transferases AtGSTF2 and AtGSTF6 by avirulent Pseudomonas syringae is the result of combined salicylic acid and ethylene signaling. Plant Cell Physiol 44: 750–757 [DOI] [PubMed] [Google Scholar]
- Lowes VL, Ip NY, Wong YH (2002) Integration of signals from receptor tyrosine kinases and G protein-coupled receptors. Neurosignals 11: 5–19 [DOI] [PubMed] [Google Scholar]
- Mayrose M, Bonshtien A, Sessa G (2004) LeMPK3 is a mitogen-activated protein kinase with dual specificity induced during tomato defense and wounding responses. J Biol Chem 279: 14819–14827 [DOI] [PubMed] [Google Scholar]
- Menke FL, van Pelt JA, Pieterse CM, Klessig DF (2004) Silencing of the mitogen-activated protein kinase MPK6 compromises disease resistance in Arabidopsis. Plant Cell 16: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Shimamoto K (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Mitchell HJ, Hall SA, Stratford R, Hall JL, Barber MS (1999) Differential induction of cinnamyl alcohol dehydrogenase during defensive lignification in wheat (Triticum aestivum L.): characterisation of the major inducible form. Planta 208: 31–37 [Google Scholar]
- Mizoguchi T, Hayashida N, Yamaguchi-Shinozaki K, Kamada H, Shinozaki K (1993) ATMPKs: a gene family of plant MAP kinases in Arabidopsis thaliana. FEBS Lett 336: 440–444 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Ichimura K, Shinozaki K (1997) Environmental stress response in plants: the role of mitogen-activated protein kinases. Trends Biotechnol 15: 15–19 [DOI] [PubMed] [Google Scholar]
- Nuhse TS, Peck SC, Hirt H, Boller T (2000) Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J Biol Chem 275: 7521–7526 [DOI] [PubMed] [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 98: 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory S, Morrison DK (2004) Signal transduction: implications for Ras-dependent ERK signaling. Curr Biol 14: R277–R278 [DOI] [PubMed] [Google Scholar]
- Peng YL, Shirano Y, Ohta H, Hibino T, Tanaka K, Shibata D (1994) A novel lipoxygenase from rice. Primary structure and specific expression upon incompatible infection with rice blast fungus. J Biol Chem 269: 3755–3761 [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Ramezani-Rad M (2003) The role of adaptor protein Ste50-dependent regulation of the MAPKKK Ste11 in multiple signalling pathways of yeast. Curr Genet 43: 161–170 [DOI] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Zhang S, Klessig DF, Hirt H, Jones JD (1999) Rapid Avr9- and Cf-9-dependent activation of MAP kinases in tobacco cell cultures and leaves: convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11: 273–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Ellis BE (2002) Double jeopardy: both overexpression and suppression of a redox-activated plant mitogen-activated protein kinase render tobacco plants ozone sensitive. Plant Cell 14: 2059–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Okamoto M, Seto H, Ishizuka K, Sano H, Ohashi Y (1995) Tobacco MAP kinase: a possible mediator in wound signal transduction pathways. Science 270: 1988–1992 [DOI] [PubMed] [Google Scholar]
- Shadle GL, Wesley SV, Korth KL, Chen F, Lamb C, Dixon RA (2003) Phenylpropanoid compounds and disease resistance in transgenic tobacco with altered expression of L-phenylalanine ammonia-lyase. Phytochemistry 64: 153–161 [DOI] [PubMed] [Google Scholar]
- Suharsono U, Fujisawa Y, Kawasaki T, Iwasaki Y, Satoh H, Shimamoto K (2002) The heterotrimeric G protein alpha subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 99: 13307–13312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Che FS, Watanabe N, Fujiwara S, Takayama S, Isogai A (2003) Flagellin from an incompatible strain of Acidovorax avenae mediates H2O2 generation accompanying hypersensitive cell death and expression of PAL, Cht-1, and PBZ1, but not of Lox in rice. Mol Plant Microbe Interact 16: 422–428 [DOI] [PubMed] [Google Scholar]
- Tena G, Asai T, Chiu WL, Sheen J (2001) Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol 4: 392–400 [DOI] [PubMed] [Google Scholar]
- Umemura K, Ogawa N, Koga J, Iwata M, Usami H (2002) Elicitor activity of cerebroside, a sphingolipid elicitor, in cell suspension cultures of rice. Plant Cell Physiol 43: 778–784 [DOI] [PubMed] [Google Scholar]
- Wong HL, Sakamoto T, Kawasaki T, Umemura K, Shimamoto K (2004) Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol 135: 1447–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KY, Liu Y, Zhang S (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Du H, Klessig DF (1998) Activation of the tobacco SIP kinase by both a cell wall-derived carbohydrate elicitor and purified proteinaceous elicitins from Phytophthora spp. Plant Cell 10: 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (1997) Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9: 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (1998. a) Resistance gene N-mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc Natl Acad Sci USA 95: 7433–7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (1998. b) The tobacco wounding-activated mitogen-activated protein kinase is encoded by SIPK. Proc Natl Acad Sci USA 95: 7225–7230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6: 520–527 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Dabi T, Beeche A, Yamamoto R, Lawton MA, Lamb C (1995) Cloning and properties of a rice gene encoding phenylalanine ammonia-lyase. Plant Mol Biol 29: 535–550 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.