Abstract
α-l-Arabinofuranosidase (α-l-arafase) was purified from fruit of Japanese pear (Pyrus pyrifolia). The enzyme solubilized from the cell wall by NaCl and Triton X-100 had the homogeneity of a single 62-kD polypeptide on SDS-PAGE after purification through the steps of hydroxyapatite, anion-exchange chromatography, and size-exclusion chromatography. A related cDNA clone was isolated (PpARF2). The transcript and related protein were detected solely in the ripening fruit corresponding to the increase of α-l-arafase activity. Transcripts of PpARF2 were not detected in buds, leaves, roots, or shoots of the Japanese pear. The deduced amino acid sequences of PpARF2 had low identity with those of other plants or bacteria. This α-l-arafase belonged to glycoside hydrolase family 3, which includes some β-xylosidases. The purified enzyme hydrolyzed mainly p-nitrophenyl α-l-arabinofuranoside and also reacted bifunctionally with p-nitrophenyl β-d-xylopyranoside. However, it released only arabinose from native cell wall polysaccharides prepared from Japanese pear and from sugar beet arabinan. The enzyme did not release xylose from arabinoxylan and xylan. The only activity of the α-l-arafase presented here was hydrolyzing the arabinosyl residue from native polysaccharides, whereas it showed bifunctional activity against artificial substrates. According to the expression pattern and properties of the enzyme, it is a new member of the glycoside hydrolase family 3 isolated from fruit, and it may be responsible for modification of the cell wall architecture during fruit softening.
The modification of cell wall architecture is involved in plant growth, development, and formation of shape. The cooperative biosynthesis and degradation of several cell wall components are necessary, and numerous cell wall-related enzymes are implicated in these processes. Fruit softening or textural changes are important factors that decide fruit quality, and they are caused by modification of cell wall polysaccharide architecture during fruit ripening. Several cell wall-metabolizing enzymes contribute to the changes in cell wall architecture (Fischer and Bennett, 1991). During fruit ripening, pectic and some hemicellulosic polysaccharides become increasingly soluble and depolymerize with the release of neutral sugar residues from side chains of matrix polysaccharides (Huber and O'Donoghue, 1993; Brummell and Labavitch, 1997; Sakurai and Nevins, 1997; Brummell and Harpster, 2001). It seems that the loss of the neutral sugar residues of the side chains occurs during the early stage of ripening associated with pectin solubilization (Sakurai and Nevins, 1997; Rose et al., 1998). The initial solubilization of pectic polysaccharides during ripening occurs without changes in the degree of polymerization (Dawson et al., 1992; Redgwell et al., 1992).
Side chains of matrix polysaccharides are removed by the actions of several glycosidases (Fry, 1995). The release of neutral sugar residues during fruit ripening was assumed to increase the sensitivity of enzymatic degradation or accessibility of other glycan hydrolases because almost all pectic or hemicellulosic polysaccharide backbones constituting the cell wall are associated with branched side chains, except for homogalacturonan. For example, β-galactosidase II, one of the glycosidases isolated from tomato (Lycopersicon esculentum) fruit, possesses the ability to release galactosyl residues from pectic side chains (Pressey, 1983). Recently, Smith et al. (2002) demonstrated that β-galactosidase II, which is encoded by TBG4, had a significant effect on fruit softening by using antisense suppression of a related gene. Therefore, the release of galactosyl residues from side chains of pectic polysaccharides, at least, is involved in fruit softening in addition to the pectin depolymerization caused by polygalacturonase activity at the later stage of ripening. However, the actual role of other glycosidases in vivo remains to be established.
Besides the release of galactosyl residues, the loss of arabinosyl residues during fruit softening was also observed in many kinds of fruits (Gross, 1984; Gross and Sams, 1984). Terminal arabinosyl residues are widely distributed in pectic and hemicellulosic polysaccharides such as arabinan, arabinogalactan, arabinoxylan, arabinoxyloglucan, and glucuronoarabinoxylan (Beldman et al., 1997; Saha, 2000; Sozzi et al., 2002b). α-l-Arabinofuranosidase (α-l-arafase; α-l-arabinofuranoside arabinofuranohydrolase, EC 3.2.1.55) is an enzyme that is able to hydrolyze nonreducing arabinofuranosyl residues. Yamaki et al. (1979) have reported the loss of arabinosyl residues associated with fruit ripening in Japanese pear (Pyrus pyrifolia). Indeed, α-l-arafase activity increased during ripening of Japanese pear fruit (Tateishi et al., 1996) and its rise was also observed in ripening fruits of apple (Malus domestica; Yoshioka et al., 1995), avocado (Persea americana; Tateishi et al., 2001b), tomato (Sozzi et al., 2002a), persimmon (Diospyros kaki; Xu et al., 2003), and peach (Prunus persica; Brummell et al., 2004a). Recently, Sozzi et al. (2002b) characterized α-l-arafase isoforms from tomato fruit and reported their different hormonal regulation and contribution to fruit development and ripening. One of the α-l-arafase isoforms is regulated by ethylene and releases arabinosyl residues from the pectic fraction. This indicates that the isoform plays an important role in arabinose (Ara) metabolism during ripening of tomato fruit.
To date, we have found α-l-arafase cDNA sequences from higher plants in the DDBJ/EMBL/GenBank databases; however, except for arabinoxylan arabinofuranohydrolase (AXAH; Ferré et al., 2000; Lee et al., 2001) and α-l-arafase (Lee et al., 2003) purified from barley (Hordeum vulgare), they appear to be defined as α-l-arafase on the basis of low sequence homology to bacterial α-l-arafase. There is no evidence that the protein translated from them actually possesses α-l-arafase activity. We have already reported the increase in α-l-arafase activity with the ripening of Japanese pear fruit and the purification of the enzyme (Tateishi et al., 1996). However, it was difficult to further analyze the enzyme properties due to low recovery of the enzyme protein after purification. In this study, we repurified α-l-arafase from ripening Japanese pear fruit, characterized its properties against native cell wall polysaccharides, and, moreover, isolated a related cDNA clone by elucidating the primary structure of the enzyme.
RESULTS
Purification of α-l-Arafase from Cell Walls of Japanese Pear Fruit
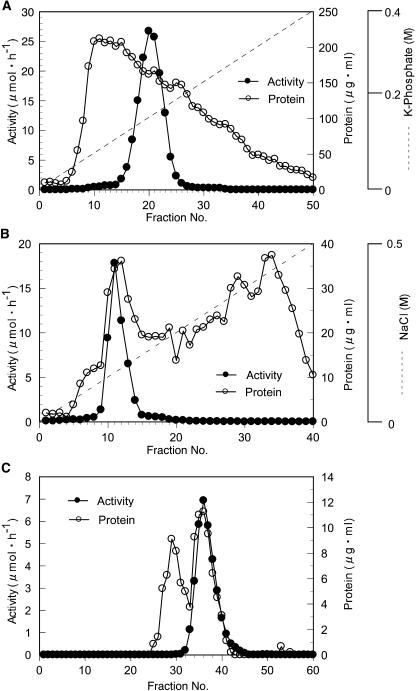
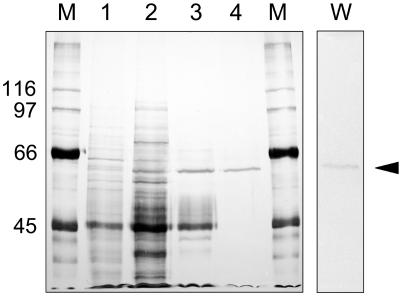
In a previous study, approximately 30% of α-l-arafase activity was solubilized from cell walls using trans-1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid (CDTA) for 38 h (Tateishi et al., 1996). Using the buffer containing NaCl and Triton X-100 for solubilization, 20 times the activity was recovered compared to that solubilized by CDTA (Tateishi and Inoue, 2000). The condition also contributed to stabilization of the enzyme. α-l-Arafase was purified by hydroxyapatite, Q-Sepharose anion-exchange chromatography, and Sephacryl S-200 gel-filtration chromatography (Fig. 1; Table I). Through the purification steps, we could not use ammonium sulfate or ultrafiltration membrane to purify or concentrate the enzyme because the recovery was too low (data not shown). The enzyme was eluted by gel media with a delay, compared to ideal elution position, regardless of the addition of 0.2 m NaCl (data not shown); this is similar to a case previously reported by Sozzi et al. (2002b). After size-exclusion chromatography, α-l-arafase was purified 106-fold compared to the activity of the crude enzyme, and the activity against p-nitrophenyl α-l-arabinofuranoside (pNPA) became 6.95 μmol h−1. The purified α-l-arafase appeared as a single protein with a molecular mass of 62 kD by staining with silver nitrate after SDS-PAGE (Fig. 2). It was larger than that obtained in the previous study (Tateishi et al., 1996). Proteolytic degradation may have occurred during long-term incubation with CDTA. The pH optimum of α-l-arafase against pNPA was 4.5, and the activity was detected at pH 4.0 to 6.5 (data not shown). Cu2+ and Hg2+ were found to be potent inhibitors (data not shown). The exact Km value of α-l-arafase against pNPA was not determined because pNPA could not be dissolved at concentrations higher than approximately 20 mm. All the kinetic parameters assayed were almost the same as those of a previous study (Tateishi et al., 1996).
Figure 1.
Elution profiles of α-l-arafase activity by hydroxyapatite (A), Q-Sepharose (B), and Sephacryl S-200 (C) chromatography. The activity was represented by a black circle (•) and the protein by a white circle (○). Fractions eluted from hydroxyapatite were collected (fraction nos. 18–23; 36 mL), dialyzed, and loaded onto a Q-Sepharose anion-exchange column. Then fraction numbers 10 to 12 (15 mL) were collected, concentrated, and then loaded onto a Sephacryl S-200 gel-filtration chromatograph. Fractions were collected in 2-mL aliquots.
Table I.
Typical purification steps of α-l-arafase from the cell wall of Japanese pear fruit
Solubilized enzyme extract from the cell wall was defined as the crude enzyme.
| Step | Activity | Protein | Specific Activity | Fold | Yield |
|---|---|---|---|---|---|
| μmol h−1 | mg | μmol h−1 mg protein−1 | % | ||
| Crude enzyme | 150.4 | 50.4 | 2.99 | 1.0 | 100 |
| Hydroxyapatite | 121.3 | 5.79 | 20.9 | 7.0 | 80.6 |
| Q-Sepharose FF | 38.58 | 0.49 | 77.9 | 26.1 | 25.6 |
| Sephacryl S-200 | 6.95 | 0.02 | 315 | 105.7 | 4.62 |
Figure 2.
SDS-PAGE of each purification step of α-l-arafase and western-blot analysis. Marker (lane M), crude enzyme (lane 1), hydroxyapatite (lane 2), Q-Sepharose FF (lane 3), and Sephacryl S-200 gel filtration (lane 4). The gel was stained with silver nitrate. Lane W is a western-blot of α-l-arafase protein using anti-Japanese pear α-l-arafase IgG raised from recombinant protein.
Isolation of a PpARF2 cDNA Clone
We determined the amino acid sequences of several tryptic peptide fragments generated from purified α-l-arafase (Fig. 3). Degenerate primers were designed based on the obtained amino acid sequences and PCR was performed using a combination of all primers. The primer set of ARA-Fr39 and ARA-Fr57C gave a single cDNA fragment of 350 bp (data not shown). The deduced amino acid sequence from the clone obtained matched the internal amino acid sequence of purified α-l-arafase. Based on the sequence obtained, the 5′- and 3′-ends of the cDNA fragment were determined using RACE-PCR (see “Materials and Methods”). Finally, we obtained a full-length cDNA clone encoding Japanese pear α-l-arafase (named PpARF2; accession no. AB195230; Fig. 3). The cloned cDNA fragment (2,575 bp) contained an untranslated region at both the 5′- and the 3′-ends, and a presumptive coding sequence of 2,322 nucleotides. The molecular mass calculated from the deduced amino acid sequence was 84 kD, consisting of 774 amino acids (nucleotides 37–2,358), which had a calculated pI of 8.29. The N-terminal amino acid sequence from purified α-l-arafase was Arg-Pro-Pro-Phe-Ala-Cys-Asp-Pro-Arg-Asn, which corresponds to the amino acid sequence of 29 to 38 deduced from PpARF2. This indicates that a signal sequence existing in the N terminus of PpARF2 protein would be cleaved in the mature protein. The sequences of a total of 41 amino acid residues from three different tryptic peptides exactly matched the sequence deduced from PpARF2 (Fig. 3). PpARF2 was subcloned into an expression vector and protein expression was induced by the addition of isopropylthio-β-galactoside. Under our conditions, the induced protein was recovered as an inclusion body without activity (see “Materials and Methods”). Therefore, we dissolved the protein and used it as an antigen to raise an antibody against α-l-arafase. The prepared antibody recognized a single polypeptide in the crude extract and also reacted with purified α-l-arafase after gel-filtration chromatography (Fig. 2).
Figure 3.
Amino acid sequence of α-l-arafase (PpARF 2) from Japanese pear fruit. The shade-boxed amino acids are the sequences of N-terminal amino acids and tryptic peptide fragments generated from purified α-l-arafase. White-boxed amino acid sequences exhibited possible N-glycosylation sites. The cleaved signal peptide is indicated by an underline. The putative catalytic nucleophile (amino acid position 292) and catalytic acid/base (amino acid position 498) are circled.
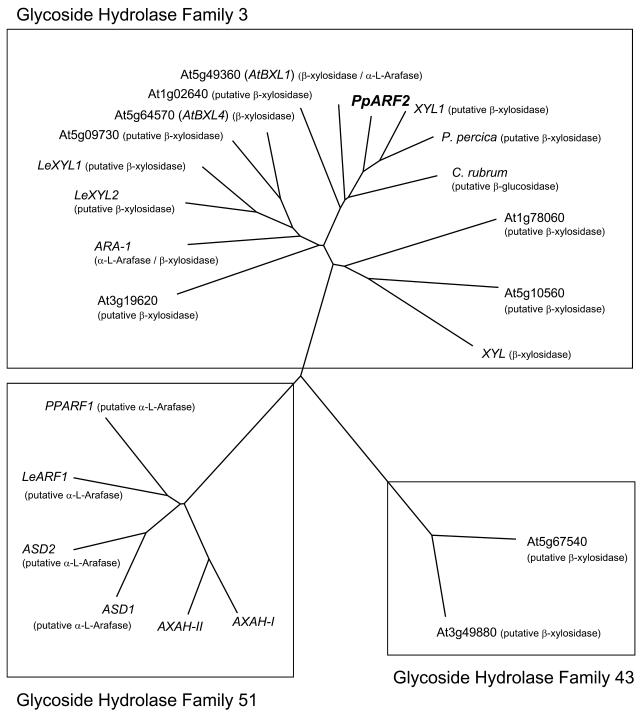
Two N-glycosylation sites were predicted from the deduced amino acid sequence of PpARF2. PSORT (Nakai and Kanehisa, 1991) and TargetP (Emanuelsson et al., 2000) programs predicted that the encoded α-l-arafase would be exported to the apoplast, indicating an enzyme would act on cell wall substrates. Both nucleotide and deduced amino acid sequences showed low identity with those of other plant and bacterial α-l-arafases. However, they showed relatively higher similarity with putative β-glucosidase and β-xylosidase cDNA clones isolated from Chenopodium rubrum and Arabidopsis (Arabidopsis thaliana), respectively, and extremely high identity with a Japanese pear cv Nijisseiki β-xylosidase-like gene expressed during fruit ripening and senescence (Itai et al., 1999). Bioinformatics analysis of deduced amino acid sequences of PpARF2 also revealed that purified α-l-arafase was grouped into the glycoside hydrolase (GH) family 3 (http://afmb.cnrs-mrs.fr/CAZY; Fig. 4), which contains β-glucosidase, β-xylosidase, and α-l-arafase cDNA clones.
Figure 4.
Comparison of deduced amino acid sequence of PpARF2 and several α-l-arafases, β-xylosidases, and β-glucosidase. Using the ClustalW program, amino acid sequences were aligned and an unrooted phylogenetic tree was generated. Their GenBank accession numbers are ASD1 and ASD2 from Arabidopsis (AY243509 and AY243510), PPARF1 from Japanese pear (AB073311), LeARF from tomato (AB073310), AXAH-I and AXAH-II from barley (AF320324 and AF320325), β-glucosidase from C. rubrum (AY320257), β-xylosidase from peach (partial, AF362990), XYL1 from Fragaria × ananassa (partial, AY486104), ARA-1 and XYL from barley (AY029259 and AY029260), and LeXYL1 and LeXYL2 from tomato (AB041811 and AB041812). At1g02640, At5g49360 (AtBXL1), At5g10560, At1g78060, At3g19620, At5g64570 (AtBXL4), and At5g09730 were β-xylosidases from Arabidopsis.
The Activity and Gene Expression of α-l-Arafase with Fruit Development and Ripening
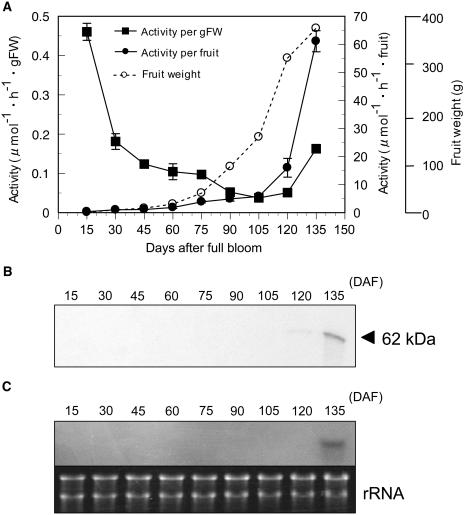
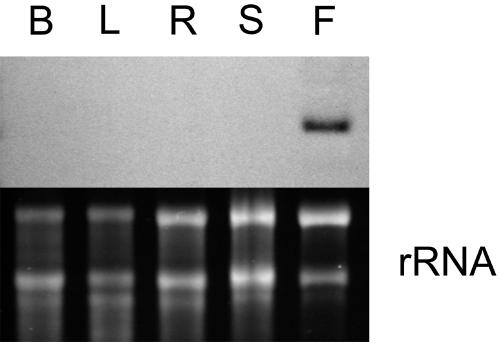
Western-blot analysis revealed that α-l-arafase protein accumulated as ripening progressed in fruit (120 and 135 days after full bloom [DAF]; Fig. 5B). RNA gel-blot analysis also detected accumulation of PpARF2 transcripts only in the ripening stage (Fig. 5C). We also assayed α-l-arafase activity during fruit development and ripening (Fig. 5A). The highest activity on a per gram fresh weight basis was detected in both young fruit (15 DAF) and fully ripened fruit (135 DAF). To exclude the dilution of the activity with fruit enlargement, the fluctuation of activity on a whole-fruit basis is also described in the same figure. The activity increased vigorously with fruit ripening after fruit enlargement (Fig. 5A). Moreover, PpARF2 transcripts were detected solely in ripening fruit, but not in buds, young and mature leaves, and shoots (Fig. 6). The increase of the activity, accumulation of the enzyme protein, and expression of PpARF2 were strongly specific to ripening fruit.
Figure 5.
Changes in α-l-arafase activities, accumulation of protein, and expression of mRNA during fruit development and ripening. A, Activity of α-l-arafase solubilized from the cell wall using pNPA as substrate. The activity on a per gram fresh weight basis is indicated by a black box (▪) and the activity on a whole-fruit basis is indicated by a black circle (•). Fruit growth was indicated by a white circle (○) with a dashed line. The values are the means of three independent experiments. Vertical bars indicate the se. B, Accumulation of α-l-arafase protein. C, Ripening-specific expression of PpARF2. The probe was prepared from mainly the 3′-untranslated region of PpARF2. The hybridization temperature was 68°C and the final washing condition was 0.1× SSC, 0.1% SDS at 68°C. rRNAs stained with ethidium bromide were used as a loading control.
Figure 6.
Fruit-specific expression of PpARF2. RNA samples were prepared from buds (lane B), leaves (lane L), roots (lane R), shoots (lane S), and ripe fruit (lane F). Ethidium bromide-stained gel was used as a loading control (indicated rRNA).
Activities against Native Cell Wall Polysaccharides and Substrates Containing Xylose
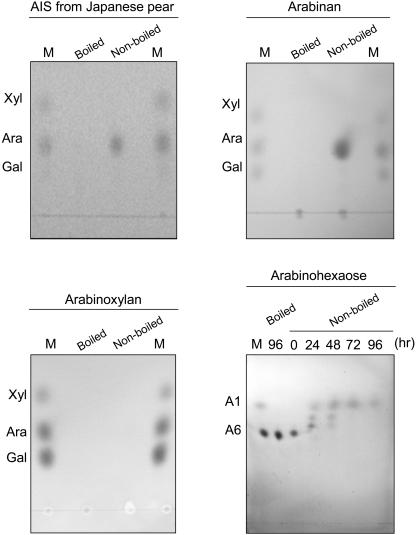
We assayed the activity against several Ara-containing polysaccharides because the activity against an artificial substrate does not necessarily reflect that against polysaccharides in vivo (Fig. 7). α-l-Arafase released only Ara as a consequence of hydrolysis of alcohol-insoluble substances (AIS) prepared from the cell wall of Japanese pear fruit. α-l-Arafase also possessed the activity of hydrolyzing sugar beet arabinan to Ara. However, no Ara could be detected in the reaction mixture incubated up to 72 h with wheat (Triticum aestivum) arabinoxylan. α-l-Arafase also hydrolyzed an arabino-oligosaccharide, arabinohexaose, to Ara within 72 h (Fig. 7). The relative activity against arabinan and/or arabinohexaose seems to be lower than that against pNPA estimated from the Ara spot of thin-layer chromatography.
Figure 7.
Hydrolysis of polysaccharides and arabinohexaose by purified α-l-arafase. Released monosaccharides were separated using thin-layer chromatography. AIS isolated from Japanese pear fruit, arabinan from sugar beet, and arabinoxylan from wheat were used as polysaccharide substrates. Lane M indicates standard sugars.
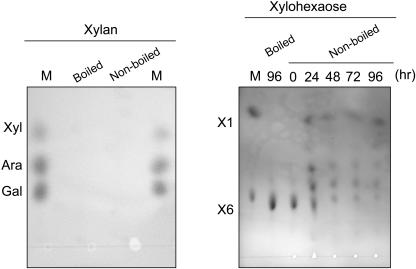
Bioinformatics analysis revealed that purified α-l-arafase is grouped into GH family 3 involving mainly plant β-xylosidases. Therefore, we also assayed the activity against substrates containing xylose (Xyl). α-l-Arafase had not hydrolyzed xylan to Xyl after a long incubation (72 h; Fig. 8), and we also did not detect Xyl in the reaction mixture with Japanese pear AIS and wheat arabinoxylan (Fig. 7). On the other hand, α-l-arafase also hydrolyzed xylohexaose to Xyl; however, some hydrolysis reactions were still in progress after a 96-h incubation (Fig. 8). As an enzymatic property, α-l-arafase possessed hydrolysis activity against p-nitrophenyl β-d-xylopyranoside (pNPX) that was 31% of that against pNPA. It was clear that the rate of hydrolysis of xylohexaose to Xyl was slower than that of arabinohexaose to Ara.
Figure 8.
Hydrolysis activity against Xyl-containing substrate. Xylan from birchwood and xylohexaose were used as substrates. Released monosaccharides were separated using thin-layer chromatography. Lane M indicates standard sugars.
DISCUSSION
We purified an α-l-arafase to the homogeneity of a single polypeptide on SDS-PAGE. We designed degenerate primers based on internal amino acid sequences and then obtained the cDNA clone encoding α-l-arafase (PpARF2; Fig. 4). The sequences of the three independent peptide fragments were actually found in the deduced amino acid sequence of PpARF2. The N-terminal amino acid sequence also exactly matched the deduced amino acid sequence of PpARF2 (below amino acid position 29). Furthermore, the antibody raised against recombinant α-l-arafase from PpARF2 recognized purified α-l-arafase (Fig. 2). Thus, we determined the complete primary structure of α-l-arafase from corresponding cDNA. The calculated molecular mass of PpARF2 was 84 kD, which is considerably larger than that of the α-l-arafase obtained on SDS-PAGE, 62 kD. A single 62-kD polypeptide was detected by immunoblot analysis when the frozen sample was inactivated immediately in SDS-denatured sample buffer. Therefore, it is hard to consider that proteolytic degradation of the protein was caused by isolation. This discrepancy may be due to a process of the C-terminal region of the protein during maturation, besides cleaving of the N-terminal signal peptide. A similar case has been reported in barley α-l-arafase (Lee et al., 2003). A hydrophobic signal peptide is present at the N terminus, and both PSORT and TargetP programs predicted that the protein would be exported to the outside of the plasma membrane. These results imply that α-l-arafase works actively in cell wall degradation.
Based on amino acid sequences of active sites rather than their substrate specificities, α-l-arafases were classified into five GH families (family 3, 43, 51, 54, and 62), and we can find α-l-arafase sequence information for several plants in families 3, 43, and 51 (Fig. 4). Almost all α-l-arafases were found in family 51 and indeed were cloned from fruit of Japanese pear (PpARF1, AB073311), tomato (LeARF; Itai et al., 2003), and Arabidopsis (ASD1 and ASD2; Fulton and Cobbett, 2003). Ferré et al. (2000) and Lee et al. (2001) reported that α-l-arafases belonging to family 51 encoded AXAH protein. According to the substrate specificity, the α-l-arafase purified from Japanese pear should be separated in other α-l-arafase groups. Indeed, Japanese pear α-l-arafase (PpARF2), which was not classified in family 51, could not hydrolyze arabinoxylan (Figs. 4 and 7) and it was isolated from ripening fruit.
Japanese pear α-l-arafase belongs to GH family 3, which includes β-glucosidase and β-xylosidase as well as α-l-arafase. Due to limits in the availability from plants, it has not been elucidated whether the proteins encoded by the clones possessed β-xylosidase or β-glucosidase activity or not, except for β-xylosidases from barley and Arabidopsis (Goujon et al., 2003; Lee et al., 2003; Minic et al., 2004). Recently, Lee et al. (2003) reported that α-l-arafase purified from barley was classified into GH family 3 and that it possessed β-xylosidase activity besides α-l-arafase activity. Moreover, most recently, it was reported that one of the β-xylosidases of Arabidopsis was classified into GH family 3 and also possesses α-l-arafase activity (Goujon et al., 2003; Minic et al., 2004). Therefore, we determined the properties of Japanese pear α-l-arafase as β-xylosidase and β-glucosidase. The purified enzyme did not possess β-glucosidase activity; however, through all the purification steps, elution profiles of β-xylosidase activity corresponded with that of α-l-arafase (data not shown). Purified α-l-arafase also hydrolyzed xylohexaose at a relatively slow rate compared to arabinohexaose (Figs. 7 and 8). This indicates that the enzyme possesses bifunctional activity as α-l-arafase/β-xylosidase as an enzymatic property. Bifunctional activity has been reported in plant α-l-arafases purified from radish (Raphanus sativus) seeds (Hata et al., 1992) and spinach (Spinacia oleracea) leaf (Hirano et al., 1994) besides barley (Lee et al., 2003). It is not clear whether these α-l-arafases belong to GH family 3 or not; however, it may be reasonable that several β-xylosidases grouped family 3 were called α-l-arafase rather than β-xylosidase. It should be noted that the activities of bifunctional α-l-arafase against pNPA are 3-fold higher than those of β-xylosidase activity against pNPX.
α-l-Arafase isolated from Japanese pear actually released Ara and Xyl hydrolyzed bifunctionally from both p-nitrophenyl (pNP) aglycon and oligosaccharides. Surprisingly, only Ara was detected in hydrolysates from AIS prepared from Japanese pear fruit, which contains xylosyl-polysaccharides (Yamaki et al., 1979). Moreover, the α-l-arafase did not release Xyl from xylan (Fig. 8). The case of the barley bifunctional α-l-arafase was different (Lee et al., 2003). The bifunctional β-xylosidase from Arabidopsis released Ara and Xyl from arabinoxylan polysaccharides (Minic et al., 2004), but our α-l-arafase did not. One of the possible reasons is that there are few xylosyl sites, which would be cleaved by α-l-arafase, in native substrates such as AIS or xylan compared to those in pNPX or xylohexaose. However, Japanese pear α-l-arafase did not release Xyl from xylan that had been previously partially hydrolyzed by trifluoroacetic acid (data not shown). According to substrate specificity, we consider that the α-l-arafase we isolated from pear fruit was a distinct type of bifunctional α-l-arafase/β-xylosidase and may release only arabinosyl residues from cell wall polysaccharides in situ. In fact, PpARF2 is expressed highly in ripening fruit (Figs. 5 and 6), whereas barley ARA-1 is expressed in grain, root, and leaf, and AtBXL1 is expressed highly in stem. Moreover, during fruit ripening, including Japanese pear, the loss of arabinosyl residues from the fruit cell wall was generally observed, but loss of xylosyl residues was not. The release of arabinosyl residues from Japanese pear was observed in an acid-soluble hemicellulose fraction where uronic acids were also detected (Yamaki et al., 1979) during the over-ripening stage. α-l-Arafase may act in the hydrolysis of the highly branched side chain of pectic and hemicellulosic polysaccharides and contribute to a part of the fruit-softening process.
In this study, we purified α-l-arafase from the cell wall of ripening pear fruit and determined that its mRNA is expressed highly only in ripening fruit. It was also determined that PpARF2 is a new member of the GH family 3, according to the primary structure of the enzyme and its activity against native substrates. However, we did not elucidate how the loss of arabinosyl residues during fruit ripening affects fruit softening. The existence of arabinosyl residues in wall polysaccharides seems to affect the adhesion of each polysaccharide or cell. In apple fruit, Ara content decreased during the over-ripening stage (Peña and Carpita, 2004) and a decrease of arabinosyl residues from cell wall polysaccharides was observed in the development of mealiness of apple fruit during postharvest storage (Nara et al., 2001). On the other hand, in peach fruit, extensive loss of arabinosyl residues from both loosely and tightly bound matrix glycans was observed in normal ripening fruit, but did not occur in mealy fruit, which showed a decline in the loss of arabinosyl residues containing polysaccharides firmly attached to cellulose (Brummell et al., 2004a, 2004b). A larger amount of tightly bound arabinosyl-containing polysaccharides was observed in softening-suppressed, colorless, nonripening (Cnr) mutant tomato fruit, which has a mealy texture (Thompson et al., 1999; Orfila et al., 2001, 2002). It was also reported that the existence of arabinosyl residues in pectic side chains affected cell-to-cell adhesion in cultured cells (Iwai et al., 2001). Fruit softening is a complex process and several cell wall-metabolizing enzymes contribute to modification of cell wall architecture. Therefore, α-l-arafase, in particular, may play an important role in alteration of fruit texture during softening.
MATERIALS AND METHODS
Plant Material
Japanese pear (Pyrus pyrifolia) cv Housui fruit was harvested from Shinohara Orchard in Yokohama City, Kanagawa Prefecture, Japan. Fruit was picked 15, 30, 45, 60, 75, 90, 105, 120, and 135 DAF. The fruit harvested on 135 DAF was at optimum maturity for eating. Buds, leaves, roots, and shoots were sampled to use in RNA gel-blot analysis. Peeled and cored fruit and the other organs were immediately frozen in liquid nitrogen and then stored at −85°C.
Enzyme Extraction and Purification
The extraction of α-l-arafase was carried out at 4°C, and all buffers contained 10 mm 2-mercaptoethanol, unless stated otherwise. The cell wall-bound enzyme fraction was prepared by the method described previously (Tateishi et al., 1996). To solubilize this enzyme from the cell wall, the cell wall fraction was resuspended in 10 mm borate buffer, pH 9.0, containing 1.0 m NaCl and 0.1% (v/v) Triton X-100, and gently stirred for 1 h. The suspension was centrifuged at 15,000g for 30 min, and the resultant supernatant was defined as the crude enzyme. The crude enzyme was then dialyzed against 10 mm potassium phosphate buffer, pH 8.0, containing 0.1% (v/v) Triton X-100 (buffer A). It was then applied to a hydroxyapatite (Wako Pure Chemical Industries, Osaka) column (2.5 × 6.1 cm) previously equilibrated with buffer A. The column was washed with buffer A and eluted with a linear gradient of 0 to 400 mm potassium phosphate buffer, pH 8.0, containing 0.1% (v/v) Triton X-100. Active fractions were pooled and dialyzed against 10 mm Tris-HCl buffer, pH 8.5, containing 0.1% (v/v) Triton X-100. After dialysis, the dialysate was applied to a Q-Sepharose FF (Amersham-Pharmacia Biotech, Uppsala) column (1.6 × 10 cm) previously equilibrated with 10 mm Tris-HCl buffer, pH 8.5, containing 0.01% (v/v) Triton X-100 (buffer B). The column was washed by the same buffer and the enzyme was eluted with a linear gradient of 0 to 0.5 m NaCl in buffer B. The active fractions were inserted into a dialysis tube and concentrated to 3 mL using polyethylene glycol (average Mr 20,000) surrounding the tube. Finally, the concentrated protein was loaded onto a Sephacryl S-200 (Amersham-Pharmacia Biotech) size-exclusion chromatography column previously equilibrated with buffer B containing 0.2 m NaCl. Assay of α-l-arafase activity using pNPA (Sigma, St. Louis) was performed according to the method described previously (Tateishi et al., 1996).
Protein Determination and SDS-PAGE
The protein concentration of the fractions was determined using the Coomassie Brilliant Blue dye-binding method (Bradford, 1976) with bovine serum albumin as a standard. SDS-PAGE was carried out by the method of Laemmli (1970) on 8.5% separation gel, and the protein bands were stained with silver nitrate (Wray et al., 1981).
Activity against Native Substrate and Oligosaccharides
Cell wall polysaccharides from Japanese pear fruit were prepared according to the method previously described (Tateishi et al., 2001b). Briefly, fruit (105 DAF) was homogenized in ice-cold 80% ethanol, and enzymes included in precipitates were inactivated in Tris-buffered phenol (Huber and O'Donoghue, 1993). The ethanol-insoluble materials were washed by ethanol and reprecipitated by 80% (v/v) ethanol. The resultant cell wall precipitate was freeze dried and defined as AIS. Xylan from birchwood was purchased from Sigma. Sugar beet arabinan, wheat arabinoxylan, arabinofuranohexaose, and xylopyranohexaose were purchased from Megazyme (Bray, Ireland).
Purified α-l-arafase was dialyzed against 5 mm sodium-citrate buffer, pH 4.5, and then concentrated using polyethylene glycol surrounding the dialysis tube. Reaction mixtures consisting of 0.5% (w/v) substrate, 5 mm sodium-citrate buffer (pH 4.5), and purified α-l-arafase, equal to 1 μmol h−1 activity against pNPA were incubated at 37°C for 72 h for native substrates, and 96 h for oligosaccharides. Toluene (20 μL) was added to each reaction mixture to prevent bacterial effects.
The monosaccharide product released from native substrates during enzymatic hydrolysis by α-l-arafase was determined by using thin-layer chromatography on silica plates (silica gel 64 F254; Merck, Rahway, NJ). After incubation, the reaction mixture was centrifuged at 15,000 rpm for 15 min. Then ethanol was added to the supernatants (200 μL) to the final concentration of 80% (v/v), and the mixture was centrifuged at 15,000 rpm for 30 min. The supernatant was collected and evaporated to dryness with a rotary vacuum evaporator, then dissolved again in 5 μL of water. A silica plate was sprayed with 0.2 m NaH2PO4, dried in air for more than 12 h, and dried completely at 110°C. Concentrated samples (0.5 μL) were spotted onto the plate and developed in n-propanol:water (85:15). Sugars were visualized by heating the plate at 95°C after spraying ethanol:sulfuric acid:p-anisaldehyde (95:2.5:2.5) on the plate. For analysis of the products of oligosaccharide substrates, 1.0 μL of the reaction mixture was spotted on a silica plate (no pretreatment) and developed in ethyl acetate:acetic acid:water (2:1:1) for arabinohexaose or in n-propanol:ethanol:water (7:1:2) for xylohexaose.
Determination of Internal Amino Acid Sequence
Protein purified by SDS-PAGE was electroblotted onto a polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, CA) and the N-terminal amino acid residues were sequenced with an ABI automated protein sequencer using Edman degradation chemistry. In addition, purified α-l-arafase was extracted from the gel and digested by trypsin. The peptide fragments generated were separated with reversed-phase HPLC and the sequences of the peptide fragments were also determined.
Cloning of α-l-Arafase cDNA
Total RNA was extracted from ripe Japanese pear fruit using the hot borate extraction method (Wan and Wilkins, 1994); the extraction buffer contained 1% Nonidet P-40, 2% polyvinylpyrrolidone, and 1% sodium deoxycholate (Tateishi et al., 2001a). First-strand cDNA was synthesized from 1 μg total RNA using SuperScript II (Invitrogen, Carlsbad, CA) reverse transcriptase with a NotI-oligo d(T)18 primer (5′-ACCTGGAAGAATTCGCGGCCGCAGGAA(T)18-3′). The degenerate primers were designed based on amino acid sequences from purified Japanese pear α-l-arafase fragments; ARA-Fr39 (5′-CCNGGNCAYCARCARGARTTRGC-3′, upstream) and ARA-Fr57C (5′-CCCATNCCRAANGGRAANCANAC-3′, downstream) were used. The temperature program was 1 cycle of 3 min at 94°C; 30 cycles of 1 min at 94°C, 1 min at 52°C, and 1 min at 72°C; 1 cycle of 20 min at 70°C. PCR products were analyzed on 1% agarose gel and the cDNA fragments were recovered from the gel using QIAEX II (Qiagen, Valencia, CA) followed by cloning into a pGEM-T easy vector (Promega, Madison, WI), according to the manufacturer's instructions. After the sequence was confirmed, the 5′-end of the PpARF2 cDNA was amplified using the 5′-RACE system (version 2.0; Invitrogen); ARA-GSP95C (5′-TCACATCAATAGGGCCGCCT-3′) was used as the gene-specific primer. After incubating oligo-dC with terminal deoxynucleotidyl transferase, the dC-tailed cDNA was purified and amplified by PCR using the Japanese pear-specific nested primer ARA-GSP44C (5′-AAGCCCTGGCCACCCTAGAC-3′) and the abridged anchor primer supplied with the kit. An amplified DNA fragment was cloned into a pGEM-T easy vector and sequenced. The full-length PpARF2 cDNA was synthesized using high-fidelity DNA polymerase (Platinum Pfx DNA polymerase; Invitrogen). The primers, ARA-START2 (5′-GGGAGAAAAATACACAATATTCC-3′, sense) designed from the obtained sequence of 5′-RACE and NotI-oligo d(T)18 (antisense) were used, incubated with a temperature program of 1 cycle of 2 min at 94°C; 30 cycles of 1 min at 94°C, 2 min at 50°C, and 3 min at 68°C, and then incubated at 70°C for 20 min after addition of 1 unit of Taq DNA polymerase to the reaction tube. An approximately 2.6-kb band was recovered from the agarose gel and cloned into a pGEM-T easy vector.
DNA Sequencing and Sequencing Analysis
Sequencing was carried out by the primer walking method with an ALF Express II DNA sequencer (Amersham-Pharmacia Biotech). BLAST programs were used for searching homology against nucleotide sequence databases and deducing amino acids. The targets of the deduced protein were analyzed with PSORT (Nakai and Kanehisa, 1991) and TargetP (Emanuelsson et al., 2000), and the motifs were subjected to the PROSITE program. The ClustalW program was used for alignment of the deduced amino acid sequences and drawing the unrooted phylogenetic tree.
RNA Gel-Blot Analysis
Total RNA (1.5 μg per lane) was separated by electrophoresis on 0.8% formaldehyde denaturing agarose gel and blotted onto a positively charged nylon membrane. The blotted membrane was prehybridized at 68°C for 1 h in a solution containing 5× SSC, 50% formamide, 1% (w/v) blocking reagent (Roche Diagnostics, Mannheim, Germany), 0.1% (v/v) N-lauroylsarcosine, and 0.02% (w/v) SDS. Then overnight hybridization followed in the same solution containing a digoxygenin (DIG)-labeled RNA probe that was prepared from the 3′-untranslated region of a full-length α-l-arafase cDNA clone (nucleotide sequence corresponding to 2,265–2,575) with a DIG RNA-labeling kit (Roche Diagnostics). After hybridization, the membrane was washed twice in 2× SSC containing 0.1% (w/v) SDS for 5 min, followed by two washes in 0.1× SSC containing 0.1% (w/v) SDS at 68°C for 15 min. The hybridized DIG-labeled probe was detected by a DIG nucleic acid detection kit using CDP-Star (Tropix, Bedford, MA).
Expression of PpARF2 and Western Blotting
The DNA fragment excluding signal peptides of PpARF2 was amplified using the primers of SalI ARA103 (5′-ACGCGTCGACATGGCAGTTGTGCATGCTCG-3′, sense) and NotI-oligo d(T)18 (antisense) with Pfx DNA polymerase. The temperature program was 1 cycle of 2 min at 94°C; 30 cycles of 1 min at 94°C, 2 min at 57°C, 3 min at 68°C, and incubation at 70°C for 5 min. The PCR fragment was digested by SalI and NotI and subcloned into a pPROEX expression vector (Gibco-BRL, Cleveland) previously digested by the same endonuclease. The subcloned insert was sequenced to confirm that no nucleotide error had occurred during PCR amplification. The subcloned PpARF2 was cultured and the protein was induced with isopropylthio-β-galactoside, according to the manufacturer's instructions. The induced protein obtained as an inclusion body was solubilized with urea and purified using a HisTrap kit (Amersham-Pharmacia Biotech). The purified protein was injected into rabbit to raise the antibody that was used for immunoblot analysis. Proteins separated in the gel by SDS-PAGE were electroblotted on PVDF membrane, and the membrane was blocked in 1% blocking reagent in Tris-buffered saline plus Tween buffer. The blocked membrane was incubated for 1 h with the generated polyclonal antibody (1,000-fold dilution) in the same buffer. After washing steps, the membrane was incubated with anti-rabbit IgG-alkaline phosphatase-conjugated goat antibody. The signal was visualized using nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate color development after the washing steps.
Sequence data from this article have been deposited with the DDBJ/EMBL/GenBank data libraries under accession number AB195230.
Acknowledgments
The authors are very grateful to Prof. Naoki Sakurai, Hiroshima University, for helpful discussions and advice. We also thank Mr. Minoru Shinohara, Yokohama City, Kanagawa Prefecture, for assisting in the cultivation of Japanese pear.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.056655.
References
- Beldman G, Schols HA, Pitson SM, Searle-van Leeuwen MJF, Voragen AGJ (1997) Arabinans and arabinan degrading enzymes. In RJ Sturgeon, ed, Advances in Macromolecular Carbohydrate Research, Vol 1. Jai Press, Greenwich, CT, pp 1–64
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brummell DA, Cin VD, Crisosto CH, Labavitch JM (2004. a) Cell wall metabolism during maturation, ripening and senescence of peach fruit. J Exp Bot 55: 2029–2039 [DOI] [PubMed] [Google Scholar]
- Brummell DA, Cin VD, Lurie S, Crisosto CH, Labavitch JM (2004. b) Cell wall metabolism during the development of chilling injury in cold-stored peach fruit: association of mealiness with arrested disassembly of cell wall. J Exp Bot 55: 2041–2052 [DOI] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH (2001) Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol Biol 47: 311–340 [PubMed] [Google Scholar]
- Brummell DA, Labavitch JM (1997) Effect of antisense suppression of endopolygalacturonase activity on polyuronide molecular weight in ripening tomato fruit and in fruit homogenates. Plant Physiol 115: 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson DM, Melton LD, Watkins CB (1992) Cell wall changes in nectarines (Prunus persica). Solubilization and depolymerization of pectic and neutral polymers during ripening and in mealy fruit. Plant Physiol 100: 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Ferré H, Broberg A, Duus JØ, Thomsen KK (2000) A novel type of arabinoxylan arabinofuranohydrolase isolated from germinated barley. Eur J Biochem 267: 6633–6641 [DOI] [PubMed] [Google Scholar]
- Fischer RL, Bennett AB (1991) Role of cell wall hydrolases in fruit ripening. Annu Rev Plant Physiol Plant Mol Biol 42: 675–703 [Google Scholar]
- Fry SC (1995) Polysaccharide-modifying enzymes in the plant cell wall. Annu Rev Plant Physiol Plant Mol Biol 46: 497–520 [Google Scholar]
- Fulton LM, Cobbett CS (2003) Two α-l-arabinofuranosidase genes in Arabidopsis thaliana are differentially expressed during vegetative growth and flower development. J Exp Bot 54: 2467–2477 [DOI] [PubMed] [Google Scholar]
- Goujon T, Minic Z, Amrani AEI, Lerouxel O, Aletti E, Lapierre C, Joseleau J-P, Jouanin L (2003) AtBXL1, a novel higher plant (Arabidopsis thaliana) putative beta-xylosidase gene, is involved in secondary cell wall metabolism and plant development. Plant J 33: 677–690 [DOI] [PubMed] [Google Scholar]
- Gross KC (1984) Fractionation and partial characterization of cell walls from normal and non-ripening mutant tomato fruit. Physiol Plant 62: 25–32 [Google Scholar]
- Gross KC, Sams CE (1984) Changes in cell wall neutral sugar composition during fruit ripening: a species survey. Phytochemistry 23: 2457–2461 [Google Scholar]
- Hata K, Tanaka M, Tsumuraya Y, Hashimoto Y (1992) α-l-Arabinofuranosidase from radish (Raphanus sativus L.) seeds. Plant Physiol 100: 388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Tsumuraya Y, Hashimoto Y (1994) Characterization of spinach leaf α-l-arabinofuranosidases and β-galactosidases and their synergistic action on an endogenous arabinogalactan protein. Physiol Plant 92: 286–296 [Google Scholar]
- Huber DJ, O'Donoghue EM (1993) Polyuronides in avocado (Persea americana) and tomato (Lycopersicon esculentum) fruits exhibit markedly different patterns of molecular weight downshifts during ripening. Plant Physiol 102: 473–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itai A, Ishihara K, Bewley JD (2003) Characterization of expression, and cloning, of β-d-xylosidase and α-l-arabinofuranosidase in developing and ripening tomato (Lycopersicon esculentum Mill.) fruit. J Exp Bot 54: 2615–2622 [DOI] [PubMed] [Google Scholar]
- Itai A, Yoshida K, Tanabe K, Tamura F (1999) A β-d-xylosidase-like gene is expressed during fruit ripening in Japanese pear (Pyrus pyrifolia Nakai). J Exp Bot 50: 877–878 [DOI] [PubMed] [Google Scholar]
- Iwai H, Ishii T, Satoh S (2001) Absence of arabinan in the side chains of the pectic polysaccharides strongly associated with cell walls of Nicotiana plumbaginifolia non-organogenic callus with loosely attached constituent cells. Planta 213: 907–915 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lee RC, Burton RA, Hrmova M, Fincher GB (2001) Barley arabinoxylan arabinofuranohydrolases: purification, characterization and determination of primary structures from cDNA clones. Biochem J 356: 181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Hrmova M, Burton RA, Lahnstein J, Fincher GB (2003) Bifunctional family 3 glycoside hydrolases from barley with α-l-arabinofuranosidase and β-d-xylosidase activity. J Biol Chem 278: 5377–5387 [DOI] [PubMed] [Google Scholar]
- Minic Z, Rihouey C, Do CT, Lerouge P, Jouanin L (2004) Purification and characterization of enzymes exhibiting β-d-xylosidase activities in stem tissues of Arabidopsis. Plant Physiol 135: 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M (1991) Expert system for predicting protein localization sites in gram-negative bacteria. Proteins 11: 95–110 [DOI] [PubMed] [Google Scholar]
- Nara K, Kato Y, Motomura Y (2001) Involvement of terminal-arabinose and -galactose pectic compounds in mealiness of apple fruit during storage. Postharvest Biol Tech 22: 141–150 [Google Scholar]
- Orfila C, Huisman MMH, Willats WGT, van Alebeek G-JWM, Schols HA, Seymour GB, Knox JP (2002) Altered cell wall disassembly during ripening of Cnr tomato fruit: implications for cell adhesion and fruit softening. Planta 215: 440–447 [DOI] [PubMed] [Google Scholar]
- Orfila C, Seymour GB, Willats WGT, Huxham IM, Jarvis MC, Dover CJ, Thompson AJ, Knox JP (2001) Altered middle lamella homogalacturonan and disrupted deposition of (1→5)-α-l-arabinan in the pericarp of Cnr, a ripening mutant of tomato. Plant Physiol 126: 210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña MJ, Carpita NC (2004) Loss of highly branched arabinans and debranching of rhamnogalacturonan I accompany loss of firm texture and cell separation during prolonged storage of apple. Plant Physiol 135: 1305–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressey R (1983) β-Galactosidases in ripening tomatoes. Plant Physiol 71: 132–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redgwell RJ, Melton LD, Brasch DJ (1992) Cell wall dissolution in ripening kiwifruit (Actinidia deliciosa). Solubilization of the pectic polymers. Plant Physiol 98: 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Hadfield KA, Labavitch JM, Bennett AB (1998) Temporal sequence of cell wall disassembly in rapidly ripening melon fruit. Plant Physiol 117: 345–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha BC (2000) α-l-Arabinofuranosidases: biochemistry, molecular biology and application in biotechnology. Biotechnol Adv 18: 403–423 [DOI] [PubMed] [Google Scholar]
- Sakurai N, Nevins DJ (1997) Relationship between fruit softening and wall polysaccharides in avocado (Persea americana Mill) mesocarp tissues. Plant Cell Physiol 38: 603–610 [Google Scholar]
- Smith DL, Abbott JA, Gross KC (2002) Down-regulation of tomato β-galactosidase 4 results in decreased fruit softening. Plant Physiol 129: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzi GO, Fraschina AA, Navarro AA, Cascone O, Greve LC, Labavitch JM (2002. a) α-l-Arabinofuranosidase activity during development and ripening of normal and ACC synthase antisense tomato fruit. HortScience 37: 564–566 [Google Scholar]
- Sozzi GO, Greve LC, Prody GA, Labavitch JM (2002. b) Gibberellic acid, synthetic auxins, and ethylene differentially modulate α-l-arabinofuranosidase activities in antisense 1-aminocyclopropane-1-carboxylic acid synthase tomato pericarp discs. Plant Physiol 129: 1330–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi A, Inoue H (2000) Purification and characterization of α-l-arabinofuranosidase from Japanese pear fruit. Acta Hortic 517: 397–403 [Google Scholar]
- Tateishi A, Inoue H, Shiba H, Yamaki S (2001. a) Molecular cloning of β-galactosidase from Japanese pear (Pyrus pyrifolia) and its gene expression with fruit ripening. Plant Cell Physiol 42: 492–498 [DOI] [PubMed] [Google Scholar]
- Tateishi A, Inoue H, Yamaki S (2001. b) Fluctuation of the activities of three β-galactosidase isoforms from avocado (Persea americana) fruit with fruit ripening and different activities against its cell wall polysaccharides. J Jpn Soc Hortic Sci 70: 586–592 [Google Scholar]
- Tateishi A, Kanayama Y, Yamaki S (1996) α-l-Arabinofuranosidase from cell walls of Japanese pear fruits. Phytochemistry 42: 295–299 [Google Scholar]
- Thompson AJ, Tor M, Barry CS, Vrebalov J, Orfila C, Jarvis MC, Giovannoni JJ, Grierson D, Seymour GB (1999) Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiol 120: 383–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C-Y, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223: 7–12 [DOI] [PubMed] [Google Scholar]
- Wray W, Boulikas T, Wray VP, Hancock R (1981) Silver staining of polyacrylamide gels. Anal Biochem 118: 197–203 [DOI] [PubMed] [Google Scholar]
- Xu CG, Nakatsuka A, Kano H, Itamura H (2003) Changes in ethylene production and activities of cell wall degrading enzymes during rapid fruit softening of Japanese persimmon ‘Saijo’. J Jpn Soc Hortic Sci 72: 460–462 [Google Scholar]
- Yamaki S, Machida Y, Kakiuchi N (1979) Changes in cell wall polysaccharides and monosaccharides during development and ripening of Japanese pear fruit. Plant Cell Physiol 20: 311–321 [Google Scholar]
- Yoshioka H, Kashimura K, Kaneko K (1995) β-d-Galactosidase and α-l-arabinofuranosidase activities during the softening of apples. J Jpn Soc Hortic Sci 63: 871–878 [Google Scholar]