Abstract
The breast cancer tumor suppressor BRCA2-interacting protein, DSS1, and its homologs are critical for DNA recombination in eukaryotic cells. We found that Dss1p, along with Mlo3p and Uap56p, Schizosaccharomyces pombe homologs of two messenger RNA (mRNA) export factors of the NXF–NXT pathway, is required for mRNA export in S. pombe. Previously, we showed that the nuclear pore-associated Rae1p is an essential mRNA export factor in S. pombe. Here, we show that Dss1p and Uap56p function by linking mRNA adapter Mlo3p to Rae1p for targeting mRNA–protein complex (mRNP) to the proteins of the nuclear pore complex (NPC). Dss1p preferentially recruits to genes in vivo and interacts with –FG (phenylalanine glycine) nucleoporins in vivo and in vitro. Thus, Dss1p may function at multiple steps of mRNA export, from mRNP biogenesis to their targeting and translocation through the NPC.
Keywords: Dss1p, Mlo3p, mRNA export, Uap56p
Introduction
Studies from various laboratories indicate that messenger RNA (mRNA) is exported by homologs of the heterodimeric NXF–NXT class of non-beta-type carrier proteins (Zenklusen and Stutz, 2001; Cullen, 2003; Stutz and Izaurralde, 2003; Thakurta et al, 2003; Erkmann and Kutay, 2004). Although the mechanism of mRNA export remains unclear, many soluble export factors and critical protein–protein interactions in the NXF–NXT-dependent mRNA export pathway have been discovered (Zenklusen and Stutz, 2001; Lei and Silver, 2002a; Cullen, 2003; Vinciguerra and Stutz, 2004). In Saccharomyces cerevisiae, mRNA export involves recruitment of the splicing factor Sub2p onto activated genes and transcripts (Zenklusen et al, 2002). Subsequently, Sub2p or metazoan UAP56 recruits an RNA annealing factor S. cerevisiae Yra1p/ALY that functions as an adapter between the mRNA and the NXF–NXT heterodimer (Luo et al, 2001; Strasser and Hurt, 2001). Yra1p is an essential mRNA export factor in S. cerevisiae (Strasser and Hurt, 2000). ALY was shown to link splicing of pre-mRNA to export of mRNA in metazoan cells (Zhou et al, 2000). However, suppression of ALY gene expression by RNAi in Drosophila did not produce any mRNA export defect, suggesting that its functions are dispensable in that organism for exporting mRNA (Gatfield and Izaurralde, 2002).
Sub2p/UAP56 belongs to the ATP-dependent DExD-box RNA helicase family whose members are critical in the splicing of pre-mRNA and in the export of both intron-containing and intron-less mRNAs (Jensen et al, 2001; Kistler and Guthrie, 2001; Libri et al, 2001; Luo et al, 2001; Strasser and Hurt, 2001; Zhang and Green, 2001). The encoding gene is essential in S. cerevisiae, Caenorhabditis elegans and Drosophila (Gatfield et al, 2001; Jensen et al, 2001; Strasser and Hurt, 2001; MacMorris et al, 2003). Schizosaccharomyces pombe uap56 was previously isolated as a suppressor of the cold-sensitive S. cerevisiae Δnam8 strain (Libri et al, 2001).
S. cerevisiae Mex67p and Sub2p bind at the N- and C- terminal regions of Yra1p. In a competitive binding assay, purified Mex67p–Mtr2p displaced bound Sub2p from Yra1p (Strasser and Hurt, 2001). It was suggested that this displacement event allows Mex67p to interact with Yra1p within the mRNPs, targeting them to the nuclear pores (Strasser and Hurt, 2001). Whether in S. pombe, Uap56p and Mlo3p, the homologs of Yra1p (Javerzat et al, 1996), function in mRNA export is not known. However, in S. pombe, Mex67p–Nxt1p heterodimer is not required for normal mRNA export (Yoon et al, 2000; Thakurta et al, 2004). Instead, nuclear pore-associated Rae1p (S. cerevisiae Gle2p, human RAE1 or Chironomus tentans ct-RAE1) (Murphy et al, 1996; Bharathi et al, 1997; Kraemer and Blobel, 1997; Sabri and Visa, 2000) functions as an essential mRNA export factor (Brown et al, 1995). rae1-167 ts mutant rapidly accumulates poly(A)+ RNA in the nucleus when the cells are shifted to nonpermissive temperatures (Brown et al, 1995). Rae1p homologs are linked directly to mRNA export by their ability to interact with nuclear pore proteins such as Nup98p (Bailer et al, 1998; Blevins et al, 2003), to associate with poly(A)+ RNA in vivo (Kraemer and Blobel, 1997), and to shuttle between the nucleus and the cytoplasm (Pritchard et al, 1999). S. pombe, Xenopus and human Rae1p homologs were shown to interact with their NXF homologs in vivo and/or in vitro (Bachi et al, 2000; Yoon et al, 2000; Blevins et al, 2003). The Chironomous, RAE1 was shown to associate with Balbiani ring mRNP particles at the nuclear pore complex (NPC) but not during transcription (Sabri and Visa, 2000), suggesting that it may function at a late step in mRNA export.
Previously, we described Elf1p as an mRNA export factor in S. pombe (Kozak et al, 2002). Elf1p is an ATPase, and its ability to hydrolyze ATP is essential for its mRNA export function. By using a synthetic lethal genetic screen using an ATPase-deficient elf1-1 mutant S. pombe strain, we identified Dss1p as a novel mRNA export factor. We show that Mlo3p and Uap56p, two key components of NXF–NXT pathway, are also required for normal mRNA export in S. pombe. In addition, we show that Dss1p and Uap56p function by linking mRNA adapter Mlo3p to Rae1p for targeting mRNA–protein complex to the proteins of the NPC. In humans, DSS1 functions with breast cancer tumor suppressor BRCA2 in DNA recombination (Yang et al, 2002). Human DSS1 and S. cerevisiae Sem1p copurify with19S proteasomes and Sem1p was found associated with double-strand breaks along with proteasomes (Krogan et al, 2004). The identification of Dss1p as an integral component of the mRNA export machinery extends its biological role beyond recombination.
Results
Dss1p is required for mRNA export in S. pombe
A synthetic lethal genetic screen based on elf1-1 mutant strain led to the isolation of several ESL (elf synthetic lethal) strains including ESL49 harboring elf1-1 and a putative esl1-49 mutation. To keep the strain viable, the ESL49 cells expressed the wild-type elf1 gene from the thiamine (vitamin B1)-repressible nmt81 promoter in pREP81X (Maundrell, 1993). ESL49 mutant grew normally in the absence of B1 but did not grow when the expression of elf1 was inhibited by the addition of B1 (Figure 1A). Additionally, poly(A)+ RNA accumulated in the double mutant even when the cells were grown in the absence of B1 (Figure 1Ba and b). Complementation by an S. pombe genomic library identified a plasmid carrying the dss1 gene that was able to rescue growth in +B1 condition (Figure 1A) as well as the mRNA export defects of ESL49 strain under both +B1 (Figure 1Bc) and −B1 conditions (not shown). DNA sequences corresponding to dss1 were able to marker rescue the synthetic lethal growth and mRNA export defect phenotypes of ESL49 cells (data not shown). These results demonstrate that dss1 gene carried the esl1-49 mutation and that dss1 was not a multicopy suppressor of ESL49 synthetic lethality.
Figure 1.
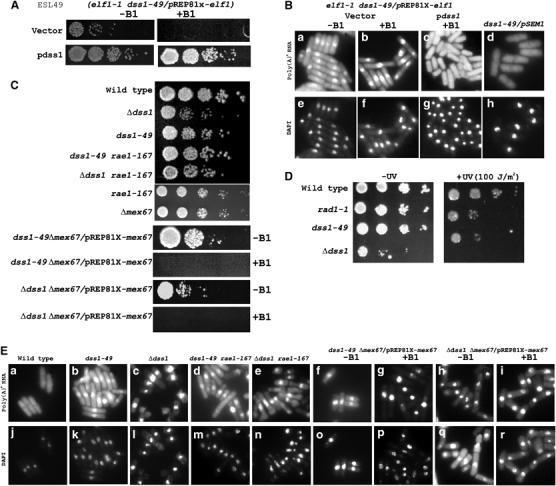
Growth, UV sensitivity and poly(A)+ RNA accumulation in S. pombe mRNA export mutants. (A) Synthetic lethal growth and complementation of ESL49 (elf1-1 dss1-49/pREP81X-elf1) by dss1 in the presence (+B1) or absence (−B1) of thiamine (see Materials and methods for details). (B) Nuclear accumulation of poly(A)+ RNA in ESL49 (elf1-1 dss1-49/pREP81X-elf1) −B1 and +B1 with vector (a, b). (c) ESL49 complemented with a genomic multicopy plasmid expressing dss1 in +B1 condition. (d) dss1-49 complemented by pSEM1. (e–h) Corresponding DAPI-stained cells to visualize DNA. (C) Growth of dss1 and dss1-49 and double mutant strains of dss1 or dss1-49 with rae1-167 or Δmex67, respectively, with or without B1 on EMM plates. (D) UV sensitivity of dss1-49 and Δdss1 strains. For comparison, wild-type and rad1-1 (Rowley et al, 1992) strains were used. See Materials and methods for details. (E) mRNA export of strains in (C), as indicated (a–i). (j–r) DAPI-stained DNA.
The dss1 gene (SPAC3G6.02) encodes a 71-amino-acid acidic protein with conserved homologs from yeast to human (Marston et al, 1999). DNA sequence analyses of the dss1 locus in ESL49 cells revealed that a tryptophan residue at amino acid 26 was changed to a termination codon. Human DSS1 binds the DNA-binding region of BRCA2 and forms a co-complex whose structure was recently solved (Yang et al, 2002). Furthermore, amino-acid residues in DSS1 and BRCA2 that directly interact with each other were found mutated in some breast cancer patients (Yang et al, 2002).
An S. pombe dss1 null strain was previously shown to be viable but had poor growth properties (Marston et al, 1999). Independently, we made a null strain that also was viable but grew poorly compared to the wild-type strain (Figure 1C). In contrast, the dss1-49 strain grew only a little slower than the wild-type strain at 30°C (Figure 1C). To test the level of mRNA export in the dss1-49 and Δdss1 strains, poly(A)+ RNA was visualized by in situ hybridization using an oligo-dT probe. dss1-49 cells had a modest level of poly(A)+ RNA accumulation in the nucleus, but in Δdss1 cells, the accumulation was extensive compared to the wild-type cells (Figure 1Ea–c). These results demonstrate that the loss of Dss1p impairs mRNA export in S. pombe cells, while Dss1-49p inhibits mRNA export without significantly affecting growth. To find out whether mRNA export function by Dss1p is conserved, we tested the ability of S. cerevisiae Sem1p to complement the mRNA export defect of dss1-49 strain. Expression of Sem1p in S. pombe was sufficient to rescue the mRNA export defect of dss1-49 mutation, suggesting that the S. cerevisiae protein could functionally substitute for Dss1p (Figure 1Bd). Taken together, Dss1p functions as a conserved mRNA export factor.
Since human DSS1 functions with BRCA2, and both human and S. cerevisiae homolog of Dss1p may be involved in DNA double-strand break repair (Yang et al, 2002; Krogan et al, 2004), we wanted to know whether dss1-49 and Δdss1 strains have sensitivity to UV ray, indicator for a likely role in recombination. As a UV-sensitive strain, we used rad1-1 mutant (Rowley et al, 1992). We found that both dss1-49 and Δdss1 strains were very sensitive to moderate dose of UV (100 J/m2) (Figure 1D). Thus Dss1p, like its human and S. cerevisiae counterpart, may function in recombination repair.
Dss1p functions with Rae1p in mRNA export
We tested genetic interactions of dss1-49 or Δdss1 alleles with rae1-167 and Δmex67 gene mutations in S. pombe. We found that dss1-49 rae1-167 or Δdss1 rae1-167 double mutants were viable (Figure 1C). The amount of poly(A)+ RNA accumulation in the nucleus in the double mutant cells resembled that in the dss1-49 or Δdss1 mutant cells (Figure 1Ed and e). However, dss1-49 Δmex67 and Δdss1 Δmex67 double mutants were not viable. The double mutant cells carrying a plasmid expressing mex67 from the nmt81 promoter grew in the absence of B1, but not, when the expression of mex67 was inhibited in the presence of B1 (Figure 1C). Thus, both dss1-49 and Δdss1 alleles were synthetically lethal with the Δmex67 mutation. Furthermore, compared to dss1-49 strain, the dss1-49 Δmex67 cells accumulated high levels of poly(A)+ RNA in the nucleus under the conditions of synthetic lethality (Figure 1E, compare panels f and h with g and i). For Δdss1 Δmex67 cells, the accumulation of poly(A)+ RNA in the nucleus was significantly more than the dss1-49 Δmex67 cells. Since Δdss1 already possessed high levels of poly(A)+ RNA in the nucleus, whether additional accumulation had taken place in the double mutant cells could not be determined. The observed increase of poly(A)+ RNA accumulation in dss1-49 Δmex67 cells under the conditions of synthetic lethal growth could be explained if the mutations simultaneously blocked both Mex67p- and Rae1p-mediated mRNA export. The simplest interpretation of the above results is that Dss1p is a component of the essential Rae1p-mediated mRNA export process.
S. pombe Mlo3p is functionally linked to Dss1p and Rae1p in mRNA export
We wanted to know if there is a functional relationship between Dss1p and Mlo3p, a putative RNA annealing protein whose homolog Yra1p/ALY directly functions with the NXF–NXT heterodimer in mRNA export. mlo3 was originally identified as one of the genes whose overexpression led to a failure to segregate chromosomes properly in mitosis and caused lethality in S. pombe (Javerzat et al, 1996). We found that mlo3 gene was not essential for growth of S. pombe cells although the null strain grew very slowly compared to a wild-type strain (Figure 2A). We next tested the extent of poly(A)+ RNA localization in Δmlo3 cells. There was substantial accumulation of poly(A)+ RNA in the nucleus of Δmlo3 cells, suggesting that Mlo3p is important for mRNA export in S. pombe (Figure 2Ba). When the Δmlo3 mutation was combined with Δdss1 or dss1-49 mutations, the double mutants were viable (Figure 2A). Δmlo3 was not lethal with rae1-167 mutation; in fact the Δmlo3 rae1-167 double mutant cells grew slightly better than Δmlo3 cells (Figure 2A). In contrast, the Δmlo3Δmex67 double mutant was not viable. For growth, Δmlo3 Δmex67 double mutant cells carried a plasmid (pREP81X-mlo3) expressing mlo3 from a thiamine-repressible nmt81 promoter. The double mutant cells were able to grow in the absence of B1, but growth was inhibited in the presence of B1 when the expression of mlo3 was turned off (Figure 2A). We tested the extent of poly(A)+ RNA localization in nonlethal double mutant strains Δmlo3 Δdss1, Δmlo3 dss1-49, Δmlo3 rae1-167 and in the synthetic lethal Δmlo3 Δmex67/pREP81X-mlo3 strain under nonlethal and synthetically lethal growth conditions. The Δmlo3 Δmex67/pREP81X-mlo3 cells had no mRNA export defect when grown in the absence of B1 (Figure 2Be). There was a significant amount of poly(A)+ RNA in the nucleus of Δmlo3 Δmex67 double mutants compared to Δmlo3 strain when the cells were grown under the synthetic lethal conditions (+B1) (Figure 2B, compare panel f with a). In contrast, there was no additive increase in the amount of poly(A)+ RNA in the nucleus of the double mutants Δmlo3 dss1-49, Δmlo3 Δdss1 and Δmlo3 rae1-167 (Figure 2B, compare panels b–d with panel a). These results are consistent with an epistatic relationship among mlo3, dss1 and rae1 gene mutations in mRNA export. Yra1p/ALY functions with NXF–NXT, and in S. cerevisiae, it is essential for mRNA export and cell viability. The nonlethality of Δmlo3 rae1-167 cells indicates that Mex67p may be able to function in the absence of Mlo3p, presumably by directly interacting with RNA and/or by utilizing as yet unidentified adapter protein(s).
Figure 2.
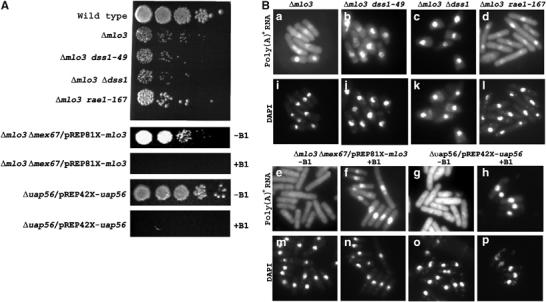
Growth and poly(A)+ RNA accumulation in S. pombe mRNA export mutants. (A) Growth of Δmlo3, its double mutants and Δuap56/pREP42X-uap56 strain, as indicated. (B) In situ hybridization of poly(A)+ RNA of strains shown in (A), grown in the presence (20 h) or absence of B1, as indicated (a–h). Corresponding lower panels (i–p) show DAPI-stained DNA.
uap56 is required for mRNA export in S. pombe
We found that uap56 was an essential gene for growth in S. pombe (Figure 2A). For viability, a haploid null strain carried a plasmid (pREP42X-uap56) containing a wild-type copy of uap56 gene under the control of the nmt41 promoter. In situ hybridization experiments showed no nuclear accumulation of poly(A)+ RNA in Δuap56/pREP42X-uap56 cells grown in the absence of B1 (Figure 2Bg). When uap56 gene expression was turned off by the addition of B1, the cells showed extensive accumulation of poly(A)+ RNA in the nucleus (Figure 2Bh). These results show that Uap56p is essential in S. pombe for normal mRNA export.
We wanted to find out the cellular localization of Dss1p, Mlo3p and Uap56p in S. pombe cells. Both Mlo3p-GFP and Uap56p-GFP localized to the nucleus while Dss1p-GFP was found both in the cytoplasm and in the nucleus including a fraction stably associated with the nuclear pore (see Supplementary data for details).
Dss1p or Uap56p links Mlo3p to Rae1p
Physical interactions among Mlo3p, Dss1p, Uap56p and Rae1p were tested by using purified proteins made in bacteria. A Mal-Mlo3p fusion protein was unable to bind purified His-Rae1p (Figure 3A, lane 5). However, under the same experimental condition, Mal-Mlo3p was able to bind a GST-Dss1p fusion (Figure 3A, lane 4) but not GST alone (not shown). Moreover, Maltose-binding protein alone did not bind GST-Dss1p (data not shown). Thus, Mal-Mlo3p could directly interact with GST-Dss1p. When GST-Dss1p and His-Rae1p were added to the binding reaction mixture together, Mal-Mlo3p was able to retain both proteins in the bound fraction (Figure 3A, lane 6). These results demonstrate that Rae1p could be present in a ternary complex with Mlo3p dependent on Dss1p. We show later that GST-Dss1p can directly interact with Rae1p and Mlo3p independent of each other. Thus, in the ternary complex, Dss1p most likely acts as a linker between Rae1p and Mlo3p by simultaneously interacting with them.
Figure 3.
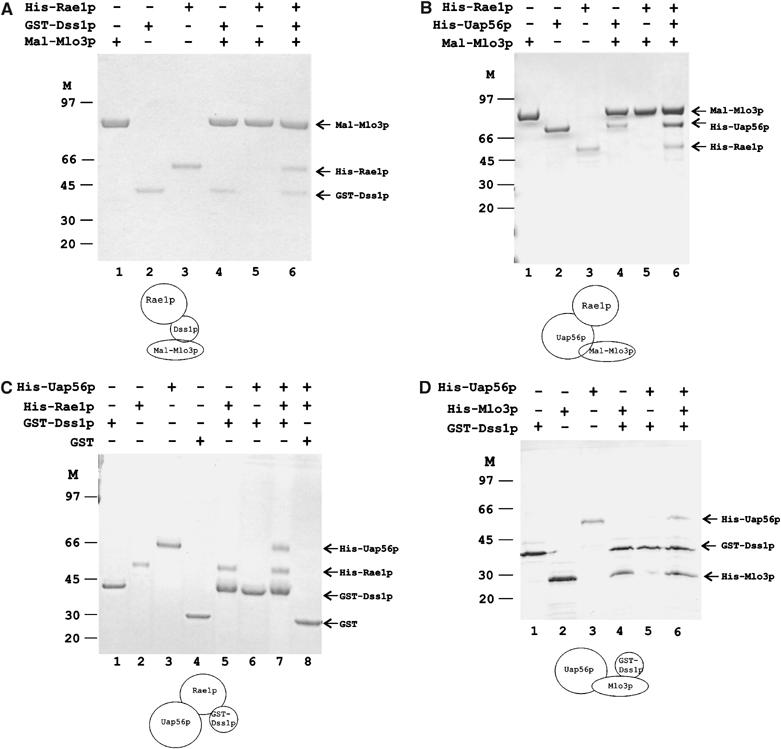
In vitro protein complex formation by Mlo3p, Dss1p, Uap56p and Rae1p. Presence or absence of a protein in a binding reaction is indicated by (+) or (−) on top of the respective lanes. Individual protein bands are identified with arrowheads. Molecular weight markers are shown. A cartoon below each gel depicts hypothetical ternary complexes. (A) GST-Dss1p links Mal-Mlo3p and His-Rae1p. Inputs of Mal-Mlo3p, GST-Dss1p and His-Rae1p are shown (lanes 1–3). Mal-Mlo3p binds GST-Dss1p (lane 4) but not His-Rae1p (lane 5). Mal-Mlo3p pulls down His-Rae1p in the presence of GST-Dss1p (lane 6). (B) His-Uap56p links Mal-Mlo3p to His-Rae1p. Inputs are shown (lanes 1–3). Mal-Mlo3p binds His-Uap56p (lane 4) but not His-Rae1p as in (A) (lane 5), but pulls down His-Rae1p in the presence of his-Uap56p (lane 6). (C) His-Rae1p binds His-Uap56p and GST-Dss1p simultaneously. Inputs including GST are shown (lanes 1–4). GST-Dss1p binds His-Rae1p (lane 5) but not His-Uap56p (lane 6). GST-Dss1p brings down His-Uap56p in the presence of His-Rae1p (lane 7). GST alone fails to bind either His-Rae1p or His-Uap56p (lane 8). (D) His-Mlo3p binds GST-Dss1p and His-Uap56p simultaneously. Lanes 1–3: Input of individual proteins. GST-Dss1p binds His-Mlo3p directly (lane 4) but not His-Uap56 (lane 5). GST-Dss1p forms a complex with His-Uap56p via His-Mlo3p (lane 6).
Next, we wanted to confirm direct interaction between Mlo3p and Uap56p. As expected, purified His-Uap56p was able to directly bind Mal-Mlo3p, but as above, Mal-Mlo3p did not interact with His-Rae1p (Figure 3B, lanes 4 and 5). By comparison with the Mlo3p–Dss1p–Rae1p interaction, the formation of an Mlo3p–Uap56p complex raised the possibility of whether Uap56p could also physically link Mlo3p to Rae1p. When His-Uap56p was added to the binding reaction of Mal-Mlo3p and His-Rae1p, Mal-Mlo3p fusion protein bound His-Rae1p along with His-Uap56p (Figure 3B, lane 6) indicating the formation of a ternary complex (Mal-Mlo3–His-Uap56p–His-Rae1p). Thus, Uap56p also could connect Mlo3p with Rae1p. These results immediately suggest that Rae1p can directly interact with Uap56p. Taken together, Dss1p and Uap56p may act as linker proteins between Mlo3p and Rae1p in the formation of specific protein complexes in vitro. In addition, if they act similarly in vivo, then the formation of these complexes could have clear implications about Rae1p function in the nuclear mRNP targeting (see Discussion).
Uap56p and Dss1p can simultaneously bind Mlo3p or Rae1p but not each other
The above results led us to investigate the nature of interactions of the linker proteins with Mlo3p or Rae1p. We separately examined the interactions of Rae1p or Mlo3p with Dss1p and Uap56p, respectively. A direct interaction between Dss1p and Rae1p was predicted from the previous binding experiment (Figure 3A, lane 6). Indeed GST-Dss1p fusion protein was able to bind His-Rae1p (Figure 3C, lane 5). Under the same condition, however, GST-Dss1p did not bind His-Uap56p (Figure 3C, lane 6). Thus the linker proteins do not interact with each other directly. When both proteins were added to the binding reaction mixture, we found that GST-Dss1p could pull down His-Uap56p along with His-Rae1p (Figure 3C, lane 7). GST alone was unable to bind any of the proteins in a control binding reaction (lane 8). Taken together, these results demonstrate that Rae1p could simultaneously bind Dss1p and Uap56p.
Next, we set out to test the formation of the complementary complex among Mlo3p, Dss1p and Uap56p. GST-Dss1p was immobilized on agarose beads and mixed with purified His-Mlo3p or His-Uap56p. As shown before, GST-Dss1p did not interact with His-Uap56p but bound His-Mlo3p efficiently (Figure 3D, lanes 4 and 5). In a mixture of three proteins, both His-Mlo3p and His-Uap56p were captured in the bound fraction by GST-Dss1p, demonstrating the formation of a ternary complex (Figure 3D, lane 6). Taken together, these results indicate that the linker proteins can coexist in a complex with Mlo3p or Rae1p. Further, the four proteins may be able to form an interlinked structure in which Dss1p and Uap56p link Mlo3p with Rae1p within a single complex.
Dss1p binds Mlo3p at Nvr and Cvr sequences
In order to understand the spatial relationship of Dss1p and Uap56p binding on Mlo3p, we tested which regions of Mlo3p are involved in its interaction with Dss1p. By alignment with the amino-acid sequence of Yra1p, we divided Mlo3p into an N-terminal (1–55 aa) domain, a middle RNA binding domain (RBD; 56–134 aa) and a C-terminal (134–199 aa) region (Figure 4A). Further, the N-terminal region could be subdivided into a conserved N1 (1–15 aa) segment and a variable Nvr (16–55 aa) region known to be required for interacting with Mex67p and RNA (Zenklusen et al, 2001). Similarly, the C-terminal sequences could be subdivided into variable Cvr (134–155 aa) and conserved C2 (156–199 aa) regions.
Figure 4.
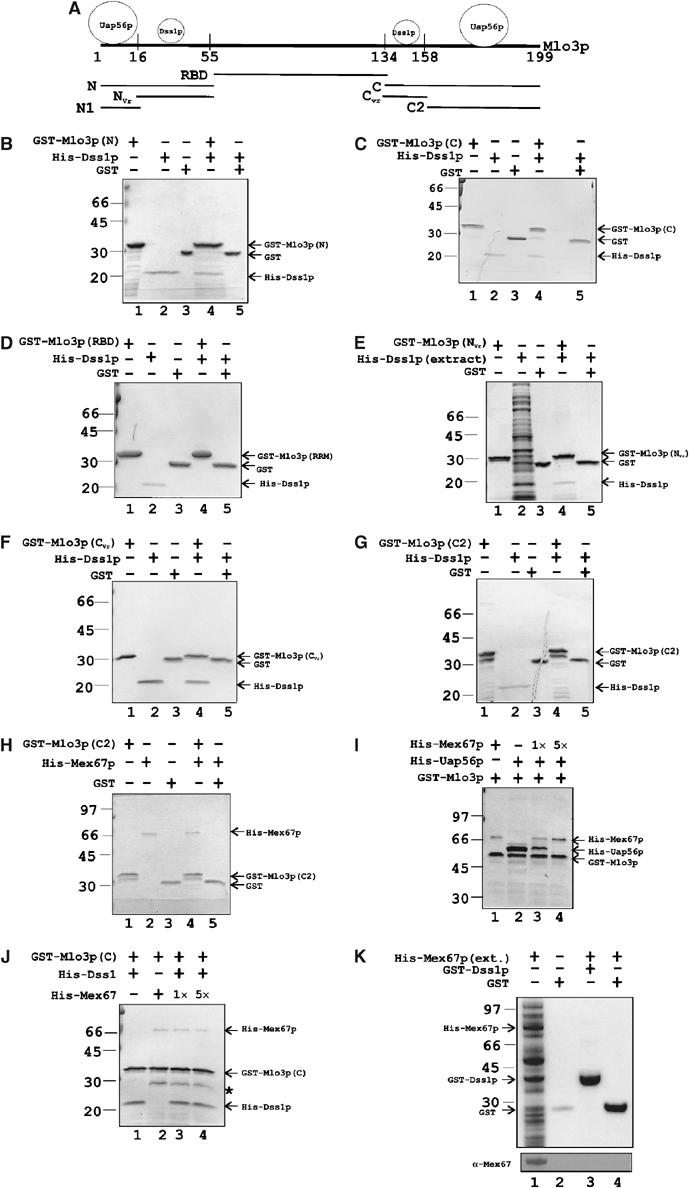
GST-Mlo3p interaction with His-Dss1p, His-Mex67p and His-Uap56p. (A) Schematic diagram of amino-acid residue positions on Mlo3p indicating various regions by comparison with Yra1p primary amino-acid sequence (Zenklusen et al, 2001). Approximate locations of Dss1p and Uap56p binding sites on Mlo3p are shown on top of Mlo3p sequence. GST fusions of Mlo3p are indicated. N: N-terminal sequence (1–55 aa); NVr: variable sequence (16–54 aa); N1: conserved sequence (1–15 aa); RBD: RNA binding domain (56–134 aa); C: C-terminal region (135–199 aa); CVr: variable sequence (135–157 aa); C2: conserved region (158–199 aa). (B–G) Binding of His-Dss1p by GST fusions of Mlo3p as indicated in each gel. Lanes 1–3: Input. Lanes 4 and 5: Binding and negative control. Individual bands are identified with arrowheads and molecular weight markers are shown on the left-hand side. (H) Binding of His-Mex67p by GST-Mlo3p-(C2). Lanes 1–3: Input. Lane 4: GST-Mlo3p-(C2) binds His-Mex67p. Lane 5: GST control. (I) Displacement of prebound His-Uap56p from GST-Mlo3p by His-Mex67p. Lane 1: Binding of GST-Mlo3p to His-Mex67p. Lane 2: Binding of His-Uap56p to GST-Mlo3p. The effect of the addition of 1 × (lane 3) or 5 × (lane 4) molar amounts of His-Mex67p to prebound fractions of GST-Mlo3p and His-Uap56 (lane 2) is shown. (J) Noncompetitive binding of His-Mex67p and His-Dss1p on GST-Mlo3p-C. GST-Mlo3p-C binds His-Dss1p (lane 1) or His-Mex67p (lane 2) or both (lane 3). Addition of five-fold excess Mex67p for the dissociation of His-Dss1p from GST-Mlo3p (lane 4) is shown. (K) GST-Dss1p does not bind His-Mex67p. Extract containing His-Mex67p was added to GST-Dss1p (lane 3) or GST alone (lane 4). Bottom panel: Western blot of a duplicate gel probed with anti-Mex67p antibody. In (J), * indicates a copurifying contaminant.
We found that GST fusions of both the N- and the C-terminal regions of Mlo3p, but not GST alone, could bind His-Dss1p directly (Figure 4B and C, lanes 4 and 5). In contrast, a GST fusion of the Mlo3p RBD was incapable of binding His-Dss1p (Figure 4D, lane 4). In order to further dissect what sequences within the N- and the C-terminal regions are responsible for interacting with Dss1p, we tested GST fusion of the Nvr and the Cvr region of Mlo3p for their ability to bind His-Dss1p. Both fragments were able to bind His-Dss1p (Figure 4E and F, lane 4). When a GST fusion of C2 or N1 was used, they both failed to bind Dss1p (Figure 4G, lane 4, and data not shown). In the context of the full-length protein, whether Nvr and Cvr bind Dss1p independently or synergistically is not known.
Mex67p can displace Uap56p from Mlo3p
We found that, like S. cerevisiae Yra1p, a GST fusion of Mlo3p-C2 (156–199 aa) could directly bind His-Mex67p while GST alone could not (Figure 4H, compare lane 4 with 5). Since C2 does not contain the Cvr region, this result shows that Mex67p could bind Mlo3p outside the variable region. Similarly, a GST fusion of Mlo3p-N could also interact with His-Mex67p (data not shown). Further, GST fusion of both C2 and N could separately bind His-Uap56p (data not shown). These results confirm that S. pombe Mlo3p behaves similar to Yra1p or ALY in its ability to interact with Mex67p and Uap56p proteins.
It is known from previous studies that Mex67p could displace bound Sub2p from Yra1p (Strasser and Hurt, 2001). It was suggested that binding of Mex67p or Sub2p on Yra1p is exclusive in nature. To test whether the S. pombe proteins exhibit similar properties, a competition experiment was set up. First, the efficiency of His-Mex67p–GST-Mlo3p interaction was tested. When an equimolar quantity of His-Mex67 was mixed to bead-bound GST-Mlo3p, His-Mex67p was retained inefficiently but reproducibly (Figure 4I, lane 1). For testing the competitive binding of His-Uap56p and His-Mex67p to GST-Mlo3p, a large molar excess of His-Uap56p was added to bead-bound GST-Mlo3p to saturate the binding by Uap56p molecules. After incubating the binding reaction for 1 h, the reaction was split into three equal parts. The first part was untreated. To the second part, equimolar His-Mex67p was added. To the third part, five-fold excess of His-Mex67p was added. After further incubation, the binding reactions were washed and bead-bound fractions were loaded onto SDS–polyacrylamide gel in loading dye. When excess His-Uap56p was added, over-stoichiometric quantity of the protein was bound to GST-Mlo3p (Figure 4I, lane 2). Since both N and C2 regions can bind His-Uap56p, our interpretation of this result is that more than one molecule of Uap56p is bound to GST-Mlo3p under the conditions of this experiment. In the challenge experiment, when low (1 × ) quantity of His-Mex67p was added, GST-Mlo3p interacted with both His-Uap56p and His-Mex67p, even though His-Mex67p bound rather inefficiently. The binding reaction may represent a mixture of binary complexes such as GST-Mlo3p–His-Mex67p and GST-Mlo3p–His-Uap56p. Alternatively, the binding may represent a ternary complex of His-Uap56p–GST-Mlo3p–His-Mex67p since His-Uap56p and His-Mex67p did not interact with each other directly (data not shown). When excess His-Mex67p was added, the entire amount of His-Uap56p bound to GST-Mlo3p was lost with a concomitant increase in the amount of His-Mex67p. These results concur with previous findings with S. cerevisiae or metazoan proteins and demonstrate that S. pombe Mex67p and Uap56p bind Mlo3p via the same region and that Mex67p could displace Uap56p from Mlo3p. However, at comparable protein concentration, these results are consistent with the same molecule of Mlo3p simultaneously binding both Uap56p and Mex67p presumably via the two ends. A similar result was obtained previously in the binding studies of Yra1p, Sub2p and Mex67p (Figure 5C, lane 2 of Strasser and Hurt, 2001).
Figure 5.
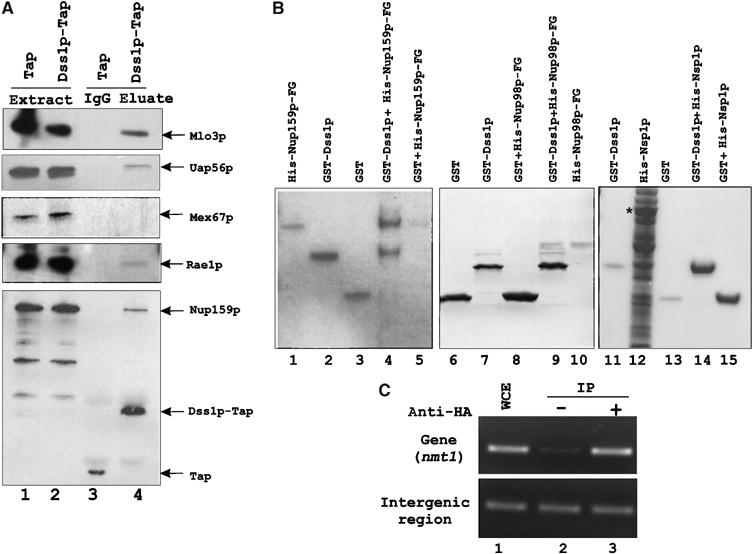
Association of mRNA export factors with Dss1p-Tap in S. pombe extracts. (A) Input extracts from strains expressing Tap or Dss1p-Tap fusion (lanes 1 and 2). Tap- or Dss1p-Tap-associated proteins were captured on IgG-Sepharose beads and were separated on 4–12% NuPAGE, transferred onto a PVDF membrane and detected by Western blot using polyclonal antibodies as indicated (lanes 3 and 4). (B) Binding of GST-Dss1p to –FG-containing sequences of S. pombe His-Nup159p (481–850 aa) or His-Nup98p (141–612 aa). Lanes 1–5: His-Nup159p binds GST-Dss1p (lane 4), but not GST (lane 5); input (lanes 1–3). Lanes 6–10: His-Nup98p is bound by GST-Dss1p (lane 9) but not by GST alone (lane 8); input (lanes 6, 7 and 9). His-Nsp1p is not bound by GST-Dss1p (lane 14) or GST alone (lane 15); input (lanes 11–13). * denotes full-length Nsp1p band. (C) ChIP analysis by PCR of S. pombe nmt1 gene and an intergenic region of chromosome III. Lane 1: WCE, whole-cell extract. Lane 2: Mock immunoprecipitation without antibody. Lane 3: Immunoprecipitation with HA antibody. For PCR, 1:50 dilution of WCE and 1:2.5 dilution of immunoprecipitation samples were used.
Mex67p and Dss1p binding on Mlo3p is noncompetitive
In an analogous competitive experiment, we tested whether binding of Dss1p to Mlo3p could withstand a challenge from His-Mex67p or His-Uap56p. For this purpose, we chose GST fusion of Mlo3p-C to which His-Dss1p and His-Mex67p bind via Cvr and C2 regions, respectively. As expected, GST-Mlo3p-C bound His-Dss1p and His-Mex67p (Figure 4J, lanes 1 and 2). When an equimolar amount of His-Mex67 was added to bead-bound GST-Mlo3p-C–His-Dss1p, both proteins were retained (Figure 4J, lane 3). But importantly, the amount of His-Dss1p bound to GST-Mlo3p-C remained unchanged upon challenge from His-Mex67p. Similarly, a five-fold excess His-Mex67p did not reduce the quantity of His-Dss1p bound to GST-Mlo3p-C (Figure 4J, lane 4). Increased His-Mex67p did not significantly increase its own binding either, presumably reflecting inefficient binding by the protein. Taken together, these results suggest that the binding of Dss1p and Mex67p (or Uap56p) occurs independently on GST-Mlo3p-C and that the binding of His-Mex67p may not significantly alter the structure of GST-Mlo3p-C to affect its interaction with His-Dss1p. Finally, we also tested for a possible direct interaction between GST-Dss1p and His-Mex67p. GST-Dss1p was unable to bind His-Mex67p (Figure 4K, lane 3). Western analysis with anti-Mex67p antibody confirmed the lack of direct binding between the two proteins (Figure 4K, lower panel).
Rae1p, Uap56p, Mlo3p and Nup159p but not Mex67p associate with Tap-tagged Dss1p in S. pombe extract
We wanted to know whether the observed interactions in vitro represent bona fide functional interactions in vivo. An S. pombe strain was constructed in which the genomic dss1 gene was replaced by a dss1-tap gene fusion. As a negative control for the experiment, Tap was expressed in wild-type S. pombe cells from a plasmid (pNTAP41) carrying the nmt41 promoter. Extracts containing Dss1p-Tap or Tap alone were incubated with IgG-Sepharose beads, and following cleavage of the tag with TEV-protease, bound protein fractions were analyzed by Western blot analyses. Dss1p-Tap was able to interact with cellular proteins that were recognized by polyclonal antibodies against Mlo3p, Uap56p, Rae1p and Nup159p (Figure 5A, lane 4). In contrast, in the control experiments, Tap alone did not associate with these proteins. Interestingly, Mex67p and Nsp1p (data not shown) did not interact with Dss1p-Tap. Nup98p was also tested but the protein degraded very rapidly during the preparation of the extract (data not shown). These results are consistent with the observed direct interaction between Dss1p and Mlo3p or between Dss1p and Rae1p and an indirect interaction of Dss1p with Uap56p via Mlo3p or Rae1p. The absence of Mex67p in the pull-down is also in agreement with its lack of direct binding to Dss1p in vitro. However, in contrast to its interaction with Mlo3p–Dss1p complex in vitro, Mex67p may not be able to associate with functional, Mlo3p-containing complexes of Dss1p in vivo. The ability of Dss1p-Tap to associate with Nup159p indicates that Dss1p may interact with the NPC proteins directly or indirectly. Recently, Tap-tagged Sem1p and human DSS1 were shown to copurify a number of proteasomal subunits (Krogan et al, 2004). But interestingly, no mRNA export factor was found associated with either protein. It is possible that under the purification conditions, only a minor fraction of Sem1p/DSS1 is associated with mRNA export factors. Alternatively, DSS1/Sem1p may be part of a minor mRNP complex in these organisms.
Dss1p binds –FG-containing regions of Nup159p and Nup98p but not Nsp1p
In vivo pull-down results led us to test whether Dss1p could interact directly with Nup159p. We used an –FG (phenylalanine glycine)-containing region of S. pombe Nup159p (481–850 aa) that directly binds Mex67p sequences (Thakurta et al, 2004). His-Nup159p could successfully bind GST-Ds1p while it could not bind GST alone (Figure 5B, lanes 4 and 5). We then tested –FG-containing region of Nup98p (141–612 aa) for its ability to interact with Dss1p. GST-Dss1p could bind the Nup98p fragment (lane 9) but GST alone did not bind the protein (lane 10). However, under the same experimental conditions, GST-Dss1p could not bind Nsp1p (lane 14), a ubiquitous –FG-containing protein (homolog of vertebrate p62). We also found that GST-Dss1p did not bind S. pombe Npp106p (data not shown). Thus, in addition to Mlo3p and Rae1p, Dss1p appears to be able to directly interact with the –FG-containing regions of two NPC components whose functions are critical in mRNA export.
Dss1p preferentially recruits to genes
In vivo association of Dss1p with Mlo3p prompted us to ask whether the protein recruited early to transcriptionally active genes. Using an HA-tagged dss1 strain, we analyzed by chromatin immunoprecipitation (ChIP) the amount of DNA from a transcribed region and an intergenic region that associated with Dss1p. There was a significant amplification of DNA from the transcribed region of nmt1 gene as compared to the mock immunoprecipitation experiment (Figure 5C, upper panel, compare lane 3 with 2). In contrast, similar amount of DNA amplified from an intergenic region by the HA antibody or by the mock immunoprecipitation (Figure 5C, lower panel, compare lane 3 with 2). These results suggest that Dss1p recruited preferentially to genes, and presumably to transcripts, over untranscribed intergenic regions of the chromosome.
Discussion
In this study, we identified Dss1p as a novel mRNA export factor in S. pombe. We show that it associates with genes and together with Uap56p links mRNA adapter Mlo3p to Rae1p for targeting the mRNP complex to the NPC and likely participates in the translocation of the cargo through the NPC. Homologs of Uap56p and Mlo3p are known components of mRNA export in other organisms, but this is the first demonstration of their direct role in mRNA export in S. pombe. Finally, we describe how Rae1p could function in mRNA export by associating with proteins of the mRNP complex.
An mRNP-targeting complex in S. pombe
Previous studies indicated a direct role of Rae1p homologs in the mRNA export process but they did not demonstrate a direct interaction of Rae1p homologs with the soluble mRNP machinery. Our results are consistent with the proposal that Rae1p facilitates the docking of mRNP to the nuclear pore via Nup98p. Further, in vitro binding results imply that by combining two ternary interactions (Rae1p–Uap56p–Mlo3p and Rae1p–Dss1p–Mlo3p), a single complex may form in vivo for targeting mRNPs to the NPC.
How is mRNP targeted in S. pombe? For a comparative analysis, current understanding of the NXF–NXT-mediated mRNP targeting in S. cerevisiae/human and the Rae1p-mediated process in S. pombe is presented in Figure 6. In the NXF–NXT pathway, Sub2p/Uap56p recruits to elongating transcripts by interacting with the THO complex (Zenklusen et al, 2002). Sub2p/UAP56 then recruits Yra1p/Aly by directly interacting with the protein. Subsequently, NXF–NXT heterodimer displaces Sub2p/UAP56 and directly binds Yra1p/Aly and links the mRNP by interacting with –FG sequences of Nup98p at the NPC. The initial events may be similar in S. pombe, although the recruitment of Uap56p may be mechanistically different, as components of the THO complex appear to be absent in the fission yeast. Next, Uap56p could recruit Mlo3p–Dss1p together onto the transcripts. Alternatively, Dss1p could be recruited only after Mlo3p is recruited. In contrast to being displaced by NXF–NXT, Dss1p and Uap56p remain associated with Mlo3p and directly link it to Rae1p on the nuclear pore. Rae1p is likely bound to Nup98p via the latter's Rae1p binding (GLEBS) domain.
Figure 6.
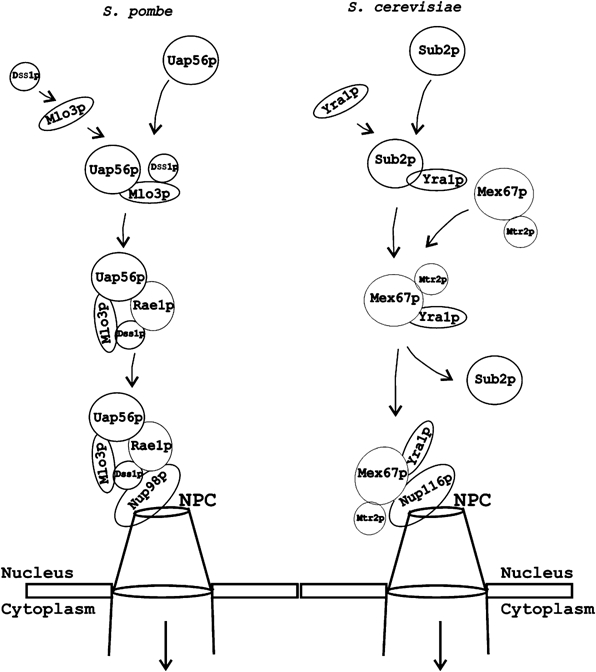
mRNP targeting in S. pombe and S. cerevisiae. In S. pombe, the progression of mRNP formation starts following recruitment of Uap56p, Mlo3p and Dss1p. Direct interaction of Uap56p and Dss1p with Rae1p connects Mlo3p to the NPC (mRNA cargo not shown). Presumptive interactions between Dss1p inside the complex and –FG sequences of Nup98p are shown based on their direct interaction in vitro. At the nuclear pore, Rae1p associates with Nup98p (our unpublished results). Additional proteins may be involved in the process in vivo (not shown). For comparison, the major mRNA export pathway in S. cerevisiae via NXF–NXT homolog Mex67p–Mtr2p is shown. Unlike in S. cerevisiae, NXF–NXT pathway plays a minor role in S. pombe (see text for details).
In S. pombe, the functions of Mex67p–Nxt1p are critical in the presence of a mutant Rae1p. How does Mex67p–Nxt1p function in S. pombe cells? One possibility is that in the mutant Rae1p background, Uap56p is displaced by Mex67p–Nxt1p to target the mRNP. Another possibility is that Mex67p–Nxt1p associates with the Uap56p–Mlo3p–Dss1p complex without displacing Uap56p. If so, it will be expected to associate with the mRNP at the NPC.
Role of Uap56p and Dss1p in mRNP targeting
Both Rae1p and NXF–NXT appear to function with Mlo3p/Yra1p/ALY. However, there are clear differences in their mechanisms and requirements for mRNP targeting to the NPC. In the Rae1p complex, Dss1p and Uap56p simultaneously link Mlo3p (hence the mRNP) to Rae1p in the nucleoplasm or at the NPC. Thus, Uap56p and Dss1p remain associated with Mlo3p when the mRNPs are connected to Rae1p. This requires Uap56p to play a critical role beyond the initial recruitment of Mlo3p. In S. cerevisiae, Sub2p affects recruitment of Yra1p on intron-containing genes but not on intron-less genes (Lei and Silver, 2002b). However, Sub2p is required for the export of both kinds of mRNA (Strasser and Hurt, 2001). Thus, it must have a role in addition to Yra1p recruitment in mRNA export. A direct interaction of Sub2p with Gle2p may provide such a role for linking some mRNAs to Gle2p for export.
Dss1p binds Mlo3p directly and the latter binds both Mex67p and Uap56p. We expected that Mex67p would associate with Dss1p complex in vivo. However, Uap56p, but not Mex67p, was found associated with Dss1p complex in the S. pombe extract. Since Mlo3p binds Uap56p and Mex67p in an exclusive manner (Figure 4I), one possible reason for this result could be a much higher cellular concentration of Uap56p as compared to Mex67p in S. pombe cells. Notwithstanding the cause, the presence of Uap56p within the Dss1p complex clearly suggests that it remains associated with Mlo3p when the mRNP complex interacts with Rae1p. In further support of this possibility, in C. tentans, Uap56p homolog HEL was shown to remain associated with mRNPs until they reach the NPC (Kiesler et al, 2002).
The multicopy expression of dss1 could not rescue the mRNA export defect associated with Δuap56/pREP42X-uap56 strain; conversely, the multicopy expression of uap56 could not rescue the mRNA export defect of Δdss1 strain (data not shown). Even though each protein could form a link between Mlo3p and Rae1p in vitro, the partial complexes could not adequately function in mRNA export in vivo. Thus, linking Mlo3p and Rae1p is not sufficient for exporting mRNA. One possibility is that both Dss1p and Uap56p are necessary for the formation of the proper targeting complex. Another, not mutually exclusive, possibility is that targeting of some mRNPs specifically requires Dss1p or Uap56p.
Significance of Dss1p–FG interaction
Dss1p may participate at many steps in the mRNA export process. Its recruitment to genes indicates a possible early role. A second likely role is in the formation of the targeting complex. A third functional role of Dss1p was suggested from its direct interaction with the –FG-containing fragments of Nup159p and Nup98p. In this regard, it could function like Mex67p–Nxt1p. However, multicopy expression of dss1 did not rescue the mRNA export defect either of rae1-167 at nonpermissive temperature or that of Δuap56/pREP42X-uap56 in +B1 conditions (data not shown). Thus, Dss1p and Mex67p are not functional homologs. Dss1p–FG interactions likely assume functional importance at a step beyond the initial targeting of mRNP to the pore. Dss1p localization in the nucleus, cytoplasm and the NPC implies that the protein likely shuttles. Human RAE1 was shown to shuttle (Pritchard et al, 1999). The S. pombe Rae1p could also be dynamic in nature. It is possible that Rae1p and Dss1p function beyond the mRNP targeting step, by participating together in the translocation of the cargo through the nuclear pore.
Materials and methods
Strains and culture
Basic genetic and cell culture techniques used have been described previously (Moreno et al, 1991; Alfa et al, 1993). Strains are described in Table I. The null strains for Δuap56, Δdss1 and Δmlo3 were constructed by standard gene disruption techniques. uap56 null strain was constructed in a diploid strain in the presence of a pREP42X-uap56 plasmid and was confirmed by Southern blot analyses.
Table 1.
S. pombe strains used in this study
| Strain | Genotype(s) | Source |
|---|---|---|
| Wild type | h− leu1-32 ura4D18 | Yoon et al (2000) |
| Wild type | h+ leu1-32 ura4D18 | Yoon et al (2000) |
| rae1-167 | h− leu1-32 ura4D18 rae1-167 | Yoon et al (2000) |
| rae1-167 | h+ leu1-32 ura4D18 rae1-167 | Yoon et al (2000) |
| elf1-1 | h− leu1-32 ura4D18 elf1-1 | Kozak et al (2002) |
| Δmex67 | h− leu1-32 ura4D18 mex67∷kan | Yoon et al (2000) |
| elf1-1 esl1-49 | h− leu1-32 ura4D18 elf1-1 dss1-49/pRep81X-elf1 | This study |
| dss1-49 rae1-167 | h− leu1-32 ura4D18 dss1-49 rae1-167 | This study |
| Δdss1 rae1-167 | h− leu1-32 ura4D18 Δdss1∷kan rae1-167 | This study |
| Δdss1 Δmex67 | h+leu1-32 ura4D18 Δdss1∷kan mex67∷kan/pRep81X-mex67 | This study |
| Δmlo3 | h− leu1-32 ura4D18 Δmlo3∷kan | This study |
| dss1-49 Δmlo3 | h− leu1-32 ura4D18 dss1-49 rae1-167 | This study |
| Δmlo3rae1-167 | h+ leu1-32 ura4D18 Δmlo3∷kan rae1-167 | This study |
| Δmex67Δmlo3 | h− leu1-32 ura4D18 Δmlo3∷kan mex67∷kan/pRep81X-mlo3 | This study |
| Δuap56 | h− leu1-32 ura4D18 uap56∷kan pRep42X-uap56 | This study |
Spot assay for growth, UV sensitivity and in situ hybridization
Growth and in situ hybridization methods were described before (Amberg et al, 1992; Yoon et al, 2000; Thakurta et al, 2004). For UV sensitivity, growing cultures (OD=0.5) were serially diluted and plated on YEA medium and exposed to various doses of UV (75–150 J/m2). Growth was monitored after 3 days at 30°C.
Construction of plasmids
dss1 genomic clone was isolated from a genomic library. cDNA clone of dss1 was made by RT–PCR from total RNA made from wild-type S. pombe cells. The gene for uap56 was amplified from the genome and inserted into pREP42X vector (Maundrell, 1993). C-terminal GFP fusions of S. pombe genes were constructed in a Ura+ promoter-less vector into which respective genes including its promoter were placed with their 3′-termini fused to DNA sequences expressing GFP. The construction of plasmids for bacterial expression of the N-terminal GST fusion proteins was made in pGEX5X-3 or pGEX4T-3 (Amersham). His-tagged proteins were made by inserting cDNA or genomic DNA of yeast into pET11b vector (Novagen, CA). For Mal fusion of Mlo3p, its coding cDNA was inserted into pMal2X (New England Biolabs, Beverly, MA). For various GST-Mlo3p deletions, appropriate cDNA fragments representing various domains were expressed similar to the full-length protein. Other plasmids were described before (Yoon et al, 2000; Kozak et al, 2002).
Purification and binding of bacterial proteins
For His-tagged protein purification, a Talon column was used; for GST fusion, a GSH-Sepharose bead was used; and Mal-Mlo3 was purified on amylose resin. Proteins were eluted in 500 mM imidazole (His proteins), 20 mM glutathione (GST fusions) and 10 mM Maltose (Mal-Mlo3 fusion). Eluted proteins were dialyzed into the universal binding buffer (Kunzler and Hurt, 1998) and stored at −70°C in aliquots. Binding reactions were performed in the universal binding buffer in Talon matrix (Clontech), GSH-Sepharose beads or amylose resins with 5–10 μg of purified proteins. Bound proteins were separated on 4–12% NuPAGE gels, and stained by Simply Blue (Invitrogen). For Western blot analyses, 10% of the reaction was run on 4–12% NuPAGE (Invitrogen, CA) gels and transferred to a PVDF membrane.
Antibodies and Western blot detection method
Polyclonal antibodies to S. pombe Mlo3p and Uap56p were made by expressing full-length proteins in bacteria and injecting them into rabbits. Nup159 (450–830 aa) of S. pombe was expressed from pET11b vector for raising antibody. Rae1p and Mex67p antibodies were described before (Bharathi et al, 1997; Yoon et al, 2000). GST fusion proteins were detected by using a monoclonal anti-GST antibody (BABCO). Standard chemiluminescent detection methods (NEN, MA) were used to detect proteins.
Immunoprecipitation
An S. pombe strain expressing Dss1-Tap fusion from its chromosomal locus was constructed based on published method (Gould et al, 2004). For control experiments, cells transformed with pNTAP41 were used. Purification of Tap-tagged proteins from S. pombe cells was based on published methods except for the following modification. Extracts were made in buffer containing 0.5% Triton and 250 mM KI and proteins bound to IgG-Sepharose beads. The complexes were cleaved from the beads using TEV-protease and the bound proteins analyzed by gel electrophoresis.
We performed ChIP by following published methods (Takahashi et al, 2000) using a monoclonal antibody to HA epitope. Gene-specific PCR primers were used to amplify the 3′ end of nmt1 gene. A pair of primers corresponding to untranscribed portion of S. pombe chromosome III was used for the intergenic region. Sequences of primers are available on request.
Supplementary Material
Supplementary Information
Supplementary Figure
Acknowledgments
We thank Dr A Ashworth and Dr K Gould for providing us with strains and plasmids, and Drs Alain Mir, J Landry and D Chattoraj for critically reading the manuscript.
References
- Alfa C, Fantes P, Hyams J, Mcleod M, Warbrick E (1993) Experiments with Fission Yeast. Cold Spring Harbor, NY: Cold Spring Harbor Press [Google Scholar]
- Amberg DC, Goldstein AL, Cole CN (1992) Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev 6: 1173–1189 [DOI] [PubMed] [Google Scholar]
- Bachi A, Braun IC, Rodrigues JP, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, Izaurralde E (2000) The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6: 136–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailer SM, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M, Hurt E (1998) Nup116p and Nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor Gle2p. EMBO J 17: 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathi A, Ghosh A, Whalen WA, Yoon JH, Pu R, Dasso M, Dhar R (1997) The human RAE1 gene is a functional homologue of Schizosaccharomyces pombe rae1 gene involved in nuclear export of poly(A)+ RNA. Gene 198: 251–258 [DOI] [PubMed] [Google Scholar]
- Blevins M, Smith A, Phillips E, Powers M (2003) Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J Biol Chem 278: 20979–20988 [DOI] [PubMed] [Google Scholar]
- Brown JA, Bharathi A, Ghosh A, Whalen W, Fitzgerald E, Dhar R (1995) A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J Biol Chem 270: 7411–7419 [DOI] [PubMed] [Google Scholar]
- Cullen BR (2003) Nuclear RNA export. J Cell Sci 116: 587–597 [DOI] [PubMed] [Google Scholar]
- Erkmann JA, Kutay U (2004) Nuclear export of mRNA: from the site of transcription to the cytoplasm. Exp Cell Res 296: 12–20 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E (2002) REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J Cell Biol 159: 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Le Hir H, Schmitt C, Braun IC, Kocher T, Wilm M, Izaurralde E (2001) The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol 11: 1716–1721 [DOI] [PubMed] [Google Scholar]
- Gould KL, Ren L, Feoktistova AS, Jennings JL, Link AJ (2004) Tandem affinity purification and identification of protein complex components. Methods 33: 239–244 [DOI] [PubMed] [Google Scholar]
- Javerzat JP, Cranston G, Allshire RC (1996) Fission yeast genes which disrupt mitotic chromosome segregation when over expressed. Nucleic Acids Res 24: 4676–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Boulay J, Rosbash M, Libri D (2001) The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr Biol 11: 1711–1715 [DOI] [PubMed] [Google Scholar]
- Kiesler E, Miralles F, Visa N (2002) HEL/UAP56 binds cotranscriptionally to the Balbiani ring pre-mRNA in an intron-independent manner and accompanies the BR mRNP to the nuclear pore. Curr Biol 12: 859–862 [DOI] [PubMed] [Google Scholar]
- Kistler AL, Guthrie C (2001) Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for Sub2, an essential spliceosomal ATPase. Genes Dev 15: 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak L, Gopal G, Yoon JH, Sauna ZE, Ambudkar SV, Thakurta AG, Dhar R (2002) Elf1p, a member of the ABC class of ATPases, functions as an mRNA export factor in Schizosacchromyces pombe. J Biol Chem 277: 33580–33589 [DOI] [PubMed] [Google Scholar]
- Kraemer D, Blobel G (1997) mRNA binding protein mrnp41 localizes to both nucleus and cytoplasm. Proc Natl Acad Sci USA 94: 9119–9124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, Greenblatt JF (2004) Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell 16: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Kunzler M, Hurt EC (1998) Cse1p functions as the nuclear export receptor for importin alpha in yeast. FEBS Lett 433: 185–190 [DOI] [PubMed] [Google Scholar]
- Lei EP, Silver PA (2002a) Protein and RNA export from the nucleus. Dev Cell 2: 261–272 [DOI] [PubMed] [Google Scholar]
- Lei EP, Silver PA (2002b) Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes Dev 16: 2761–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Graziani N, Saguez C, Boulay J (2001) Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev 15: 36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R (2001) Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413: 644–647 [DOI] [PubMed] [Google Scholar]
- MacMorris M, Brocker C, Blumenthal T (2003) UAP56 levels affect viability and mRNA export in Caenorhabditis elegans. RNA 9: 847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A (1999) Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol 19: 4633–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K (1993) Thiamine repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127–130 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR (1996) GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell 7: 1921–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CE, Fornerod M, Kasper LH, van Deursen JM (1999) RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol 145: 237–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley R, Subramani S, Young PG (1992) Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J 11: 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri N, Visa N (2000) The Ct-RAE1 protein interacts with Balbiani ring RNP particles at the nuclear pore. RNA 6: 1597–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E (2000) Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J 19: 410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E (2001) Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413: 648–652 [DOI] [PubMed] [Google Scholar]
- Stutz F, Izaurralde E (2003) The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol 13: 319–327 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Saitoh S, Yanagida M (2000) Application of the chromatin immunoprecipitation method to identify in vivo protein–DNA associations in fission yeast. Sci STKE 56: PL1. [DOI] [PubMed] [Google Scholar]
- Thakurta AG, Gopal G, Dhar R (2003) Explaining mRNA export in Schizosacharomyces pombe. Res Dev Genet 3: 121–128 [Google Scholar]
- Thakurta AG, Gopal G, Yoon JH, Saha T, Dhar R (2004) Conserved nuclear export sequences in Schizosaccharomyces pombe Mex67 and human TAP function in mRNA export by direct nuclear pore interactions. J Biol Chem 279: 17434–17442 [DOI] [PubMed] [Google Scholar]
- Vinciguerra P, Stutz F (2004) mRNA export: an assembly line from genes to nuclear pores. Curr Opin Cell Biol 16: 285–292 [DOI] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP (2002) BRCA2 function in DNA binding and recombination from a BRCA2–DSS1–ssDNA structure. Science 297: 1837–1848 [DOI] [PubMed] [Google Scholar]
- Yoon JH, Love DC, Guhathakurta A, Hanover JA, Dhar R (2000) Mex67p of Schizosaccharomyces pombe interacts with Rae1p in mediating mRNA export. Mol Cell Biol 20: 8767–8782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Stutz F (2001) Nuclear export of mRNA. FEBS Lett 498: 150–156 [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Strahm Y, Stutz F (2001) The yeast hnRNP-like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol Cell Biol 21: 4219–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Wyss JC, Stutz F (2002) Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol 22: 8241–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Green MR (2001) Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes Dev 15: 30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Strasser K, Katahira J, Hurt E, Reed R (2000) The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401–405 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figure


