Abstract
Arabidopsis MAP kinase 4 (MPK4) functions as a regulator of pathogen defense responses, because it is required for both repression of salicylic acid (SA)-dependent resistance and for activation of jasmonate (JA)-dependent defense gene expression. To understand MPK4 signaling mechanisms, we used yeast two-hybrid screening to identify the MPK4 substrate MKS1. Analyses of transgenic plants and genome-wide transcript profiling indicated that MKS1 is required for full SA-dependent resistance in mpk4 mutants, and that overexpression of MKS1 in wild-type plants is sufficient to activate SA-dependent resistance, but does not interfere with induction of a defense gene by JA. Further yeast two-hybrid screening revealed that MKS1 interacts with the WRKY transcription factors WRKY25 and WRKY33. WRKY25 and WRKY33 were shown to be in vitro substrates of MPK4, and a wrky33 knockout mutant was found to exhibit increased expression of the SA-related defense gene PR1. MKS1 may therefore contribute to MPK4-regulated defense activation by coupling the kinase to specific WRKY transcription factors.
Keywords: disease resistance, phosphoserine, protein kinase
Introduction
Plant disease resistance is induced via host recognition of pathogen elicitors, which may be either general to a large class of pathogens or specific to a race of pathogen. Bacterial flagellin is an example of a general elicitor that induces resistance through interaction with a plasma membrane-localized receptor kinase (Gomez-Gomez and Boller, 2000; Zipfel et al, 2004). This mode of pathogen recognition resembles pathways of innate immunity induction upon perception of pathogen-associated molecular patterns in insects and vertebrates. In addition, plants use a second recognition mode to perceive specific factors produced by the pathogen to promote infection. While normally acting as virulence factors, these race-specific elicitors activate resistance in plant hosts harboring specific, matching resistance (R) genes. Race-specific elicitors are therefore referred to as avirulence (Avr) factors. Recognition of Avr factors by R proteins may occur predominantly via ‘surveillance' of host proteins manipulated by Avr activity rather than by direct R–Avr interaction, although examples of both scenarios have been reported (Dangl and Jones, 2001). At the infection site, responses to R–Avr interactions include Ca2+ influx (Scheel, 1998), production of reactive oxygen species (Delledonne et al, 1998) and expression of pathogenesis-related PR genes, some of which have antimicrobial activity (Narasimhan et al, 2005). Several protein kinases are implicated in local resistance signaling (Romeis et al, 2001), and signal transduction components may be shared between resistance responses induced by general elicitors and Avr factors. For example, the MAP kinases WIPK and SIPK are activated upon the Cf-9/Avr9 R–Avr interaction in tobacco, and their Arabidopsis orthologs MPK3/6 function in flagellin-induced resistance activation as well as in resistance mediated by specific R genes (Romeis et al, 1999; Asai et al, 2002; Menke et al, 2004).
Upon R protein activation, immunity may also be promoted in uninfected tissues by the induction of systemic acquired resistance (SAR) dependent upon salicylic acid (SA). Thus, impairment of SA biosynthesis by mutation of the isochorismate synthase ICS1/SID2 or the putative transporter EDS5 abrogates activation of SAR (Nawrath and Métraux, 1999; Wildermuth et al, 2001). In response to some infections, the interacting proteins of unknown biochemical function EDS1 and PAD4 are requisite, positive regulators of SA biosynthesis (Feys et al, 2001). As SA itself may not be a systemic signal (Forouhar et al, 2005), genes such as DIR1 (Maldonado et al, 2002) and CDR1 (Xia et al, 2004) are necessary for the production or transmission of the signal(s) required for SAR.
A well-characterized component in SA signaling is the BTB/ankyrin repeat protein NPR1. Following SA application, monomeric NPR1 released by reduction from cytosolic, disulfide-bound oligomers translocates to the nucleus, where it interacts with and induces the binding of TGA transcription factors to PR promoters (Johnson et al, 2003; Mou et al, 2003). BTB proteins function as substrate receptors for cullin-3-based ubiquitin E3 ligases in plants and animals (Furukawa et al, 2003; Dieterle et al, 2005), but no direct substrates of NPR1-mediated degradation have been identified as yet. Despite the importance of NPR1, NPR1-independent pathways exist (Clarke et al, 1998; Petersen et al, 2000). These may involve the WHY1 DNA-binding protein (Desveaux et al, 2004) and WRKY transcription factors that bind to W-boxes in PR genes (Eulgem et al, 2000; Maleck et al, 2000; Petersen et al, 2000).
In addition to the SA-dependent defense system, defense pathways mediated by ethylene (ET) and jasmonates (JA) exist. Such pathways provide resistance to herbivores and to pathogens distinct from those that induce SA-dependent defenses (Turner et al, 2002; Wang et al, 2002). While overlapping gene sets are induced by SA, JA and ET (Schenk et al, 2000), SA and ET/JA responses exhibit clear antagonisms (Norman-Setterblad et al, 2000; Glazebrook et al, 2003), and the PDF1.2 defensin is a prototype ET/JA target gene negatively regulated by SA. The significance of the antagonistic relationship between these two plant defense pathways is unknown, and its molecular basis also remains poorly characterized. For example, genetic studies in Arabidopsis have placed the positive regulators of SA signaling NPR1 and WRKY70 in the same pathway negatively regulating PDF1.2 expression (Spoel et al, 2003; Li et al, 2004).
We have previously shown that knockout of Arabidopsis MAP kinase 4 (mpk4) leads to accumulation of SA and activation of SA-dependent resistance, as well as a block of PDF1.2 induction by JA (Petersen et al, 2000), and hypersusceptibility to herbivore feeding and infection by an ET/JA pathway-inducing fungus (E Andreasson and P Brodersen, unpublished data, 2004). This suggests that MPK4 functions in the control of pathogen defense pathway antagonism. Together with MPK6, MPK4 is also activated by a range of abiotic stresses and, since the MAP kinase kinase MKK2 is required for both MPK4 activation and appropriate abiotic stress adaptation, MPK4 may also have a role in such stress signaling (Huang et al, 2000; Ichimura et al, 2000; Teige et al, 2004). Similar complexities of MAP kinase function have been described in yeast and animal cells. A key to understanding such complexity is the identification and characterization of kinase substrates. For example, mating in Saccharomyces cerevisiae is induced by activation of the MAP kinases Fus3 and Kss1, while specific induction of filamentous growth involves Kss1 activation. Fus3 activation ensures specificity of the mating response because it phosphorylates the Tec1 transcription factor required for expression of filamentation genes, thereby targeting Tec1 for degradation (Bao et al, 2004).
Arabidopsis contains 20 MAP kinases, like MPK4 of the ERK type, and only five typical MPK phosphatases (MAPK Group, 2002). Therefore, numerous plant MAP kinase pathway specificity determinants can be expected. To date, the only known MAP kinase substrates are two isoforms of ET biosynthetic enzymes that are phosphorylated and activated by MPK6, explaining the effect of MPK6 activation on ET accumulation (Liu and Zhang, 2004). Here, we report the identification and characterization of an MPK4 substrate, designated MAP kinase 4 substrate 1 (MKS1). MKS1 acts downstream of MPK4 in the SA-dependent pathway, but does not appear to be involved in the ET/JA pathway. MKS1 interacts with the transcription factors WRKY 25 and WRKY33, and may thereby couple the MAP kinase to regulation of specific transcription factors.
Results
MPK4 interacts in yeast and in vitro with MKS1
To identify intermediates in the signaling pathway(s) mediated by MPK4, we isolated MPK4 interacting proteins by yeast two-hybrid (Y2H) cDNA library screening with full-length MPK4 as bait. Initial immunoblotting with anti-MPK4 antibody (Huang et al, 2000) showed that the GAL4BD-MPK4 bait construct in plasmid pGBKT7 was expressed in the yeast strain PJ69-4A, and Y2H assays showed that this construct, alone or transformed with the empty pACT prey vector, did not activate expression of selectable markers. A total of 2 × 107 clones were then screened representing 6.6-fold redundancy of the Clontech library used. Four clones encoding full-length MKS1 protein were identified by prototrophy for adenine and histidine. The interaction between MPK4 and MKS1 was confirmed by selectable marker and β-galactosidase reporter activation upon cotransformation of MPK4 and MKS1 in bait and prey vectors, and vice versa. Directed Y2H assays indicated that this interaction was specific, as MKS1 did not interact with MAP kinases closely (MPK5) or more distantly (MPK17) related to MPK4 (MAPK Group, 2002), or with MPK3 or MPK6 involved in innate immunity responses (Asai et al, 2002). The MKS1 and MPK4 interaction was confirmed by in vitro binding assays. This showed that recombinant, N-terminal 6xHis-tagged MKS1 from Escherichia coli could pull down labeled MPK4, but not a control human lamin (Figure 1A).
Figure 1.

MKS1 interactions and phosphorylation in vitro. (A) MKS1 interactions with MPK4, W25 and W33 used N-terminal 6xHis-tagged MKS1 (His-MKS1) purified from E. coli. 35S-methionine-labeled MPK4, W25 or W33 was incubated without (−) or with (+) nickel-agarose-coupled His-MKS1. Human lamin (Clontech) was a negative control. (B) Left: Phosphorylation of full-length recombinant MKS1, C-terminal MKS1 truncations (C1–C3) and myelin basic protein (MBP) as positive control. HA-tagged MPK4 (HA-MPK4) was immunoprecipitated (IP) from complemented mpk4 transgenic plants. Right: In vitro kinase assays with mutated full-length MKS1 (MKS1 S30A) and mutated C3 (C3 S30A). (C) Phosphorylation of full-length MKS1 and increasing molar ratios of a 22-amino-acid, MKS1-derived peptide, Pep22 (left; Supplementary Figure S1) and the 22-amino-acid flagellin elicitor Flg22 (right).
MKS1 homologs are found in diverse plant species
MKS1 is a 222-amino-acid protein (predicted mass of 23 851 and isoelectric point of 6.0) that lacks predicted subcellular targeting sequences. MKS1 is a member of an apparently plant-specific family including proteins from monocots and dicots (Figure 2). Family members share a conserved VQ motif of unknown function (Protein Family PF05678; Figure 2, domain II), while those most similar to MKS1 include a less conserved, N-terminal region (domain I). In contrast, the C-terminal regions are divergent although they contain numerous Ser-Pro residues that are potential MAP kinase phosphorylation sites (Sharrocks et al, 2000; Liu and Zhang, 2004). MKS1 contains 12 Ser-Pro sites, one each in domains I and II and 10 in the C-terminal region.
Figure 2.
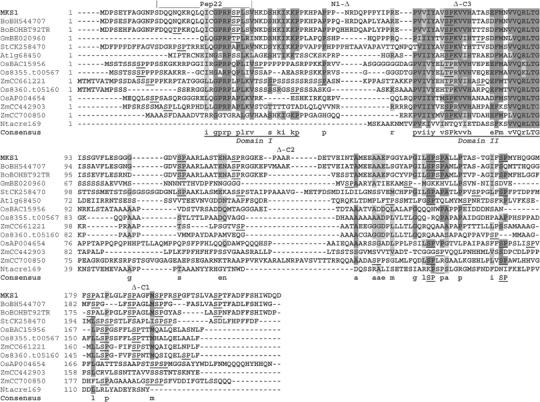
MKS1 and homologs. DNA and amino-acid sequences of MKS1 (At3g18690) were used to query databases at www.ncbi.nlm.nih.gov/BLAST for proteins similar to MKS1. Protein sequences of selected accessions were aligned at clustalw.genome.jp and identical/similar residues highlighted at www.ch.embnet.org/software/BOX_form.html. Putative MAP kinase phosphorylation sites (S/TP) are underlined. The sequence of the Pep22 peptide is indicated by an overbar. The ends of an N-terminally truncated (N1) and three C-terminally truncated (C1–3) MKS1 versions described in the text are noted above the MKS1 sequence. Putative domains I and II are underlined in the consensus. Species abbreviations are as follows: At: Arabidopsis thaliana; Bo: Brassica oleracea; Gm: Glycine max; St: Solanum tuberosum; Os: Oryza sativa; Nt: Nicotiana tabacum; Zm: Zea mays. Similar plant proteins not aligned here include BM340911, CAD40925, CC613160, CC635639, Al390921, AL138658, T46022, AP004654, AP003260, AC143340 and Arabidopsis At1g21326, At2g41180, At2g44340, At2g42140 and At3g56710 (Sig1; Morikawa et al, 2002).
Two family members have been partially characterized: Sib1, a nuclear-encoded, chloroplast targeted protein that interacts with plastid-encoded, plastid RNA polymerase σ-factor Sig1 (Morikawa et al, 2002), and tomato ACRE169 whose mRNA is rapidly induced following activation of Cf-9-dependent resistance by the race-specific elicitor Avr9 (Durrant et al, 2000). This indicates that MKS1 family members may be involved in transcriptional regulation and in responses to pathogens.
MPK4 phosphorylates MKS1 in vitro
We have previously shown that the expression of a wild-type MPK4 gene including a C-terminal, 3 × HA epitope tag complements the mpk4 loss-of-function mutant and produces immunoprecipitable MPK4 that phosphorylates the standard MAP kinase substrate myelin basic protein in vitro (Petersen et al, 2000). We therefore used this system to demonstrate that immunoprecipitated, HA-tagged MPK4 from plants phosphorylated recombinant His-tagged MKS1 and three C-terminal MKS1 truncations purified from E. coli (C1–3; Figures 1B and 2). This indicates that MKS1 is an in vitro substrate of MPK4, and that MKS1 Ser30 and/or Ser72, both contained in the C3 truncation, may be phosphorylation sites. This was confirmed by demonstrating that although MPK4 phosphorylated full-length MKS1 carrying a Ser30Ala substitution, phosphorylation of the C3 truncation carrying this substitution was severely reduced (Figure 1B).
MPK4 phosphorylation of full-length MKS1 was competed at a 10 molar ratio, and abolished at a 100 molar ratio, by a synthetic 22-residue peptide derived from MKS1 (pep22; Figures 1C and 2). This peptide (SDQQNQKRQLQICGPRPSPLSV) was synthesized because it included Ser30 and because the sequence upstream is reminiscent of MAP kinase substrate docking domains (Sharrocks et al, 2000). In contrast, the unrelated, 22-residue Flg22 peptide (Gomez-Gomez and Boller, 2000; Asai et al, 2002) did not affect MKS1 phosphorylation. This indicates that MKS1-derived pep22 specifically interacts with MPK4. Furthermore, the C1–3 MKS1 truncations interacted with MPK4 in Y2H experiments, while an N-terminal truncation, N1, failed to interact (Figure 2). These results indicate that MPK4 interacts with the N-terminal region of MKS1 including pep22 and domain I, and that this interaction is necessary for MPK4 phosphorylation of MKS1.
MKS1 interacts with MPK4 in vivo
A selected monoclonal antibody raised in mice against MKS1 pep22 was used to detect recombinant MKS1 extracted from E. coli and endogenous MKS1 from plants, both with the expected mobility in SDS–PAGE (Figure 3A). Monoclonal anti-pep22 antibodies also co-immunoprecipitated from plants MKS1 with MPK4 immunodetected by the HA tag (Figure 3B). In addition, the levels of phosphorylated MKS1 detected with a phosphoserine/phosphothreonine-specific antibody were markedly higher in wild-type plants than in mpk4 mutants (Figure 3C). These results indicate that MKS1 and MPK4 interact in vivo, and that MPK4 is the major kinase activity that phosphorylates MKS1 under default conditions.
Figure 3.
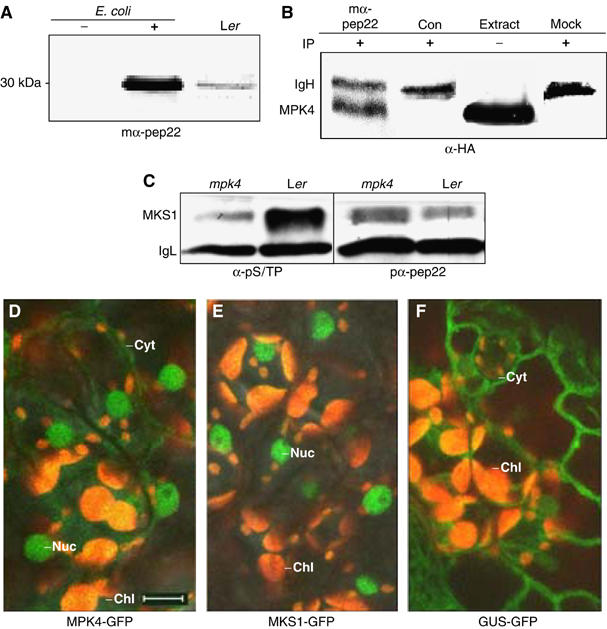
MKS1 interaction, phosphorylation and localization in vivo. (A) Immunodetection of MKS1 in E. coli extracts before (−) and after (+) IPTG induction, and from extracts of leaves (Ler) by a monoclonal anti-Pep22 antibody (mα-pep22) recognizing a protein of the predicted size of MKS1 (∼28 kDa). (B) Immunodetection of HA-MPK4 by anti-HA antibody (α-HA) in immunoprecipitates from plant extracts with mα-pep22 (lane 1), negative control monoclonal antibody (Con; lane 2) or in mock immunoprecipitation lacking extract (lane 4). MPK4 from plant extracts is shown (raw extract; lane 3). MPK4 and immunoglobin heavy chain (IgH) are indicated. (C) Immunodetection of MKS1 following immunoprecipitation with mα-Pep22 from Ler or mpk4 extracts. MKS1 is detected with an anti-phosphoserine/phosphothreonine antibody (α-pS/TP) and polyclonal anti-Pep22 antibody (pα-pep22). MKS1 and immunoglobin light chain (IgL) are indicated. In planta localization in mesophyll cells of (D) MPK4-GFP, (E) MKS1-GFP and (F) GUS-GFP fusion proteins. Cyt: cytoplasm; Nuc: nuclei; Chl: chloroplast (orange autofluorescent); — 10 μm size bar.
Transgenic plants expressing gene fusions between the green fluorescent protein (GFP) and MPK4 or MKS1 were produced to examine their subcellular localization. The MPK4-GFP gene fusion fully complemented the mpk4 mutant phenotype, indicating that it provided functional MPK4 activity. Western analysis with a GFP antibody detected a single band of the expected size of the MPK4-GFP fusion protein (data not shown), indicating that the intact fusion, and not a cleavage product liberating MPK4, was responsible for complementation. The MPK4-GFP fusion produced weak but readily detectable GFP fluorescence in cytoplasmic strands and strong fluorescence in nuclei (Figure 3D). Similarly, the MKS1-GFP fusion under control of the cauliflower mosaic virus (CaMV) 35S promoter or 1.9 kbp of MKS1 5′ upstream sequence produced strong fluorescence only detectable in nuclei (Figure 3E). The MKS1-GFP fusion was apparently functional, as its overexpression via the 35S promoter produced the same phenotype as overexpression of MKS1 alone (see below, data not shown). In contrast, a control GUS-GFP fusion from pCAMBIA3300, under control of the 35S CaMV promoter, produced strong fluorescence in cytoplasmic strands (Figure 3F). These results indicate that MKS1 is primarily localized in nuclei and that the presence of both MKS1 and MPK4 in nuclei is consistent with their ability to interact in vivo.
Altered MKS1 expression affects defense responses and mpk4 phenotypes
MKS1 function was addressed in transgenic plants that overexpressed immunodetectable MKS1 from the constitutive CaMV 35S promoter (35S-MKS1), or that underexpressed MKS1 by RNA interference (MKS1-RNAi). Compared to wild type, 35S-MKS1 plants were semi-dwarfed (Figure 4A and B), accumulated increased levels of PR1 mRNA (Figure 4C) and of SA (∼13 500 ng SAG/g fresh weight leaf tissue in 35S-MKS1 compared to ∼3500 ng in wild type), and were more resistant to the virulent biotrophic pathogen Pseudomonas syringae pv. tomato DC3000 (Figure 4D). Analysis of independent 35S-MKS1 lines indicated that such characteristics of heightened defense signaling were correlated with MKS1 expression (data not shown). All of the 35S-MKS1 lines examined were fully fertile and the expression of the MKS1 transgene was stable over at least four generations. In contrast, MKS1-RNAi plants did not exhibit observable growth phenotypes (Figure 4A). These results provide functional links between MKS1 and MPK4, as mpk4 mutants exhibit dwarfism and constitutive defense signaling (Petersen et al, 2000).
Figure 4.
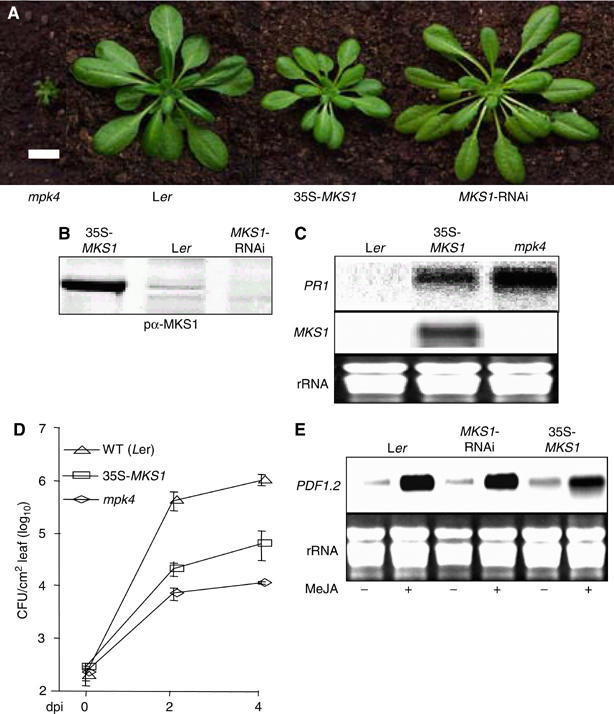
Effects of MKS1 over- and underexpression. (A) Phenotypes of mpk4, wild type (Ler) and transgenic (Col) overexpressing MKS1 (35S-MKS1) or underexpressing MKS1 (MKS1-RNAi). The size bar is 2 cm. (B) Immunodetection with pα-Pep22 antibody of MKS1 in extracts from 35S-MKS1, wild type (Ler) and MKS1-RNAi. (C) RNA blot detection of PR1 mRNA in 35S-MKS1, mpk4 and wild-type Ler. (D) Growth of P. syringae pv. tomato DC3000 in mpk4, 35S-MKS1 and Ler. (E) Detection of PDF1.2 mRNA in wild type (Col), MKS1-RNAi and 35S-MKS1 following MeJA (+) and mock (−) treatments.
As noted above, PDF1.2 defensin gene induction by JA is blocked in mpk4, suggesting that MPK4 mediates JA-dependent responses to necrotrophic pathogens (Pieterse and van Loon, 1999). In contrast, MKS1 under- or overexpression had little, if any effect, on PDF1.2 mRNA levels (Figure 4E), indicating that MKS1 is not involved in JA-responsive gene expression downstream of MPK4.
To position genetically MKS1 in MPK4 signaling, mpk4/MKS1-RNAi mutant plants were constructed. These plants exhibited partial suppression of mpk4 dwarfism and a clear reduction in PR1 mRNA levels (Figure 5A), again indicating that MKS1 acts downstream of MPK4. Furthermore, mpk4/MKS1-RNAi plants exhibited decreased resistance to P. syringae DC3000 compared to mpk4 (Figure 5B). This partial suppression of the mpk4 phenotype by MKS1-RNAi indicates that components other than MKS1 perform functions downstream of MPK4.
Figure 5.
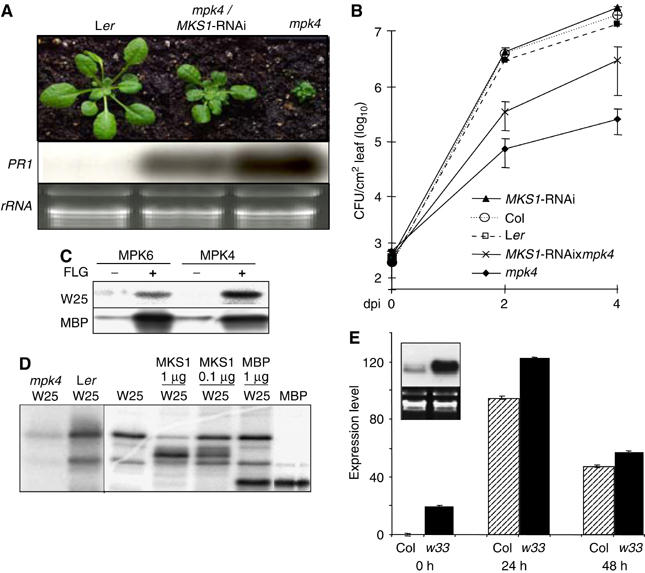
Suppression of mpk4 by MKS1-RNAi, WRKY phosphorylation and w33 phenotype. (A) Top: Phenotypes of wild type (Ler), mpk4 carrying MKS1-RNAi (mpk4/MKS1-RNAi) and mpk4. Bottom: Detection of PR1 mRNA in wild type (Ler), mpk4/MKS1-RNAi and mpk4 versus rRNA loading control. (B) Growth of P. syringae pv. tomato DC3000 in wild types Ler and Col, mpk4, MKS1-RNAi and mpk4/MKS1-RNAi. (C) Phosphorylation of full-length W25 by immunoprecipitation of MPK6 or MPK4 from Arabidopsis cells without (−) or with (+) treatment with flagellin. (D) Phosphorylation of N-terminal region of W25 with MPK4 immunoprecipitation from mpk4 or Ler (left) and of W25, MKS1 and MBP with immunoprecipitation of MPK4-HA (right). (E) RNA blot (inset) and real-time PCR detection of PR1 mRNA in Col and w33 before (0 h and inset) or after infiltration with DC3000.
Gene expression profiling of 35S-MKS1 and mpk4
Comparative transcript profiling revealed that of 22 810 genes probed, 800 (3.5%) exhibited significantly differential expression patterns between wild-type, mpk4 and 35S-MKS1 plants (Supplementary Figure S1, clusters A–G). mRNAs of PR genes including PR1 and PR2 and the SA biosynthetic enzyme ICS1 (encoded by SID2) were increased in both mpk4 and 35S-MKS1 plants compared to wild type (Supplementary Figure S1, cluster A). This suggests that MPK4 and MKS1 participate in regulating the expression of these defense-related genes. Analyses of 5′ upstream regions of putative target genes revealed significant occurrences of two sequence motifs TAGACT and TGACTT, the latter a putative WRKY binding site or W-box (Lebel et al, 1998; Eulgem et al, 2000; Petersen et al, 2000). This suggests that WRKY factors participate in regulating genes downstream of MPK4 and MKS1.
To further analyze the overlap between genes affected by the mpk4 mutation and the 35S-MKS1 transgene, the two were tested individually against wild type using t-tests. Because the t-test has substantially less power than the ANOVA, only the 800 most significant genes in the ANOVA test were considered. This revealed that mpk4 and 35S-MKS1 significantly affected 350 and 156 genes, respectively, of which 76 genes were affected by both mpk4 and 35S-MKS1. Considering the total number of genes measured (n=22 810), the overlap of 76 genes is highly significant (P=6e−99). This supports the finding that MPK4 depends on MKS1 in the regulation of a set of genes. Of the 76 genes affected by both mpk4 and 35S-MKS1, only one gene was affected in the opposite direction, further supporting a relation between MPK4 and MKS1 in regulating a subset of genes. However, this analyses also confirmed that mpk4 affects the expression of other genes independently of 35S-MKS1, as can be seen in clusters B, E and F that contain 112, 169 and 173 genes, respectively. Among these are a number of genes that are implicated in defense and stress responses: WRKYs 30, 32, 46, 48, 60 and 75, basic chitinase (PR3) and EDS1. This and the partial suppression of the mpk4 phenotype by MKS1-RNAi (Figure 5A) indicate that other effects of MPK4 are mediated by effectors other than MKS1. Of particular interest are genes overexpressed in both mpk4 and 35S-MKS1 (Figure 6A, cluster A), as well as genes underexpressed only in the mpk4 background (Figure 6B, cluster B). This latter group includes the JA-inducible PDF1.2 gene, which was more than 10-fold underexpressed in the mpk4 background compared to both 35S-MKS1 and wild type. This confirms the RNA blot results (Figure 4E), and indicates that MKS1 function downstream of MPK4 does not affect defense responses mediated by JA.
Figure 6.
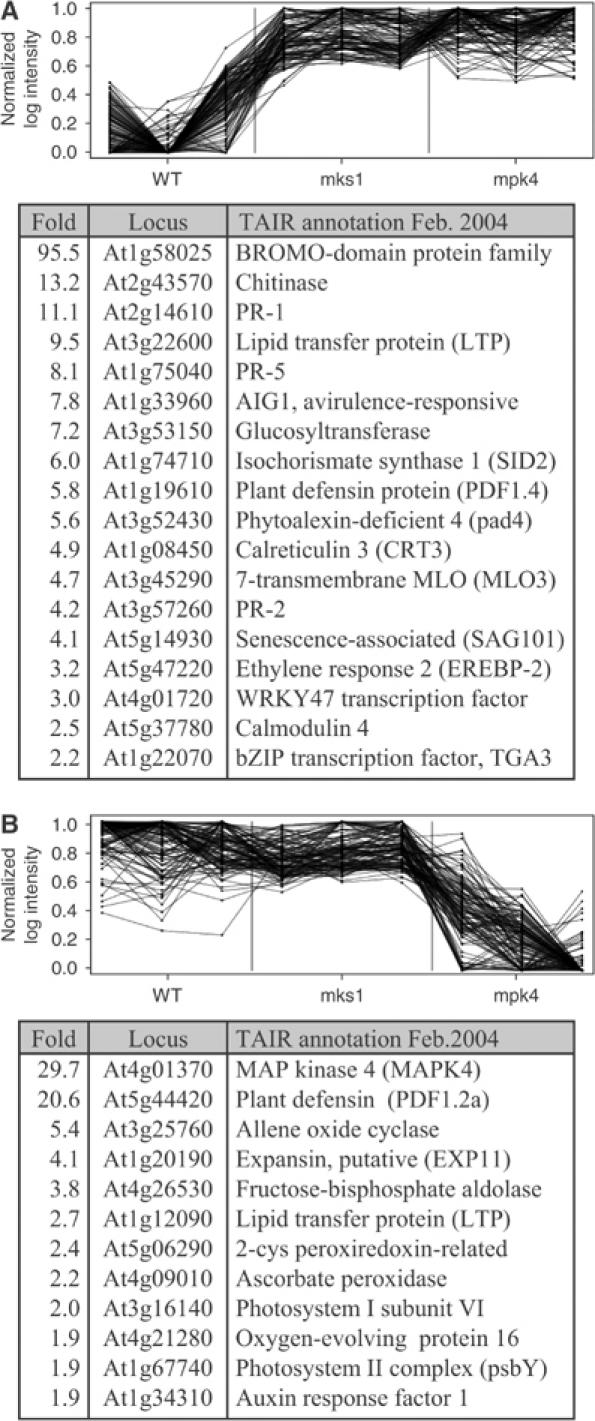
Expression profiles of genes differentially expressed between wild-type, mpk4 and 35S-mks1 plants. Two clusters A and B of significantly differentially expressed genes are shown. (A) Top: Expression profiles of genes in cluster A through triplicates of the three genotypes (x-axis) versus normalized log expression index values. Vertical lines mark the borders between the genotypes. Bottom: List of selected genes from cluster A. (B) as in (A) but for cluster B genes. See Supplementary Figure S1 for a comprehensive list of clustered genes.
MKS1 interacts in yeast and in vitro with WRKY25 and WRKY33
To further investigate MKS1 function, we conducted a second Y2H library screen to identify additional MKS1 interacting proteins. Of 6 × 107 clones screened, four encoding WRKY25 (W25) and 11 encoding WRKY33 (W33) were identified and confirmed by β-galactosidase assays. The interactions of MKS1 with W25 and W33 were also confirmed in vitro using His-tagged MKS1 (Figure 1A). These interactions were apparently specific, as Y2H assays with MKS1 as bait did not permit the growth of yeast expressing the closest homolog W26, or yeast expressing W29 involved in flagellin responses (Asai et al, 2002). Furthermore, W33 was found to interact in yeast with the truncated MKS1 versions N1, C1 and C2, but not with C3 (Figure 2). This suggests that MKS1 domain II including the VQ motif is involved in the interaction between MKS1 and W33.
W25 and W33 contain N-terminal D-domains with Ser-Pro residues that may be MAP kinase phosphorylation sites (Eulgem et al, 2000; Liu and Zhang, 2004). Although we did not detect interactions between MPK4 and W25 or W33 in directed Y2H assays, recombinant W25 and W33 were phosphorylated by MPK4 in vitro. Kinase assays with MPK6 or MPK4 immunoprecipitated from plant cells showed that full-length W25 was preferentially phosphorylated by MPK4 (Figure 5C). In addition, the anti-MPK4 antibody preferentially precipitated, from wild type versus mpk4, MPK4 that phosphorylated the N-terminal regions of W25 and W33 (Figure 5D, left panel and data not shown). Interestingly, MKS1, but not the standard MAP kinase substrate MBP, inhibited W25 or W33 phosphorylation by MPK4 (Figure 5D, right). These results suggest that MKS1 may be involved in modulating the phosphorylation of the WRKYs. Two additional lines of evidence indicate a role for W33 in regulating gene expression downstream of MPK4. First, as for mpk4 and 35S-MKS1, plants of an available insertion line with a T-DNA in the first intron of the W33 gene accumulated significantly more PR1 mRNA under normal conditions and following infection with P. syringae DC3000 (Figure 5E). Second, these w33 mutants were significantly smaller than wild type when grown under short-day (8 h) conditions. Of the 30 plants examined, the total leaf area (n=80 each) of w33 plants was 45% (±6%) and the area of the two largest leaves was 34% (±11%) of wild type. Such reduced growth phenotypes are typical of a number of constitutive defense mutants (Clarke et al, 1998; Xia et al, 2004).
Discussion
Using Y2H screening with MPK4 as bait, we identified the MPK4 interacting protein MKS1. In vitro and in vivo interaction and phosphorylation assays, together with the low level of phosphorylated MKS1 in the mpk4 mutant, confirmed that MKS1 is an MPK4 substrate. In addition, analyses of transgenic plants showed that over- or underexpression of MKS1 oppositely affect pathogen resistance and PR gene expression. For example, compared to wild type, plants that overexpressed MKS1 from a constitutive promoter (35S-MKS1) were smaller, accumulated increased total SA and exhibited increased resistance to the virulent pathogen P. syringae DC3000, as well as increased expression of PR genes. This indicates that plants that accumulate excess levels of unphosphorylated MKS1 exhibit similar phenotypes to plants lacking MPK4 kinase activity, and is consistent with a model in which default phosphorylation of MKS1 represses SA signaling. These data, combined with partial suppression of SA-dependent defense activation in mpk4 mutants by reduced expression of MKS1 via MKS1-RNAi, indicate that MKS1 is required downstream of MPK4 to effect defense responses regulated by the SA pathway.
These results were confirmed and extended by comparative gene expression profiling of mpk4 and 35S-MKS1 plants. While this revealed a significant overlap in genes differentially regulated in mpk4 and 35S-MKS1 compared to wild type, it showed that loss of MPK4 function affected the expression of other genes independently of MKS1. This is consistent with the expectation that MPK4 phosphorylates substrates other than MKS1. For example, other MPK4 substrates affecting JA signaling may exist because loss of MPK4 activity reduces basal and JA/ET-inducible PDF1.2 mRNA accumulation (Petersen et al, 2000; P Brodersen, unpublished data, 2004), while PDF1.2 mRNA levels are not significantly affected by MKS1 over- or underexpression. This indicates that MKS1 overexpression produces plants with increased resistance to biotrophic pathogens, while maintaining a functional JA defense pathway.
Additional evidence for a role of MKS1 in defense signaling comes from our identification by Y2H screening of the WRKY transcription factors W25 and W33 as specific MKS1 interactors. Of the more than 70 Arabidopsis WRKY factors, W33 is most similar to W25 and to parsley WRKY1 involved in responses to pathogen elicitors (Turck et al, 2004). As shown here, W25 and W33 can be phosphorylated by MPK4 in vitro, presumably in what has been referred to as their D-domains (Eulgem et al, 2000), which contain the only Ser-Pro sites in the proteins:
W25 67YLDSPLLLSSSHSLISPTTGT84
W33 47SISSPSLVSPSTCFSPSLFLDSPAFVSSSANVLASPTTGA84.
The several Ser-Pro sites in such domains suggest that the WRKYs may be multiply phosphorylated, as we have shown here for MKS1 and as has been recently demonstrated for the ET biosynthetic enzyme ACS6 by MPK6 (Liu and Zhang, 2004).
Preliminary functional characterization of W33 is provided by our demonstration that a w33 reduced or loss-of-function mutant shares at least two characteristics with mpk4 and 35S-MKS1 plants. First, they are significantly smaller than wild type, as seen for other constitutive defense mutants (Clarke et al, 1998; Xia et al, 2004). Second, w33 accumulates heightened basal levels of PR-1 mRNA. Although these phenotypes are tightly linked with the T-DNA insert in GABI KAT line 324B11, complementation of this allele with wild-type W33 sequences and analyses of other reduced or loss-of-function w33 alleles are required to further assess W33 function. Given the potential redundancy of W25 and W33 function and the complexity of WRKY auto- and inter-regulation (Turck et al, 2004), analyses of w25/w33 double mutants and of W25 and W33 target genes are required to substantiate their function(s) downstream of MPK4 and MKS1.
MKS1 is a member of a novel family of plant proteins that contain a conserved VQ motif in what we here refer to as domain II. MKS1 and other members also contain an N-terminal region, here termed domain I, which includes Ser30 that is phosphorylated by MPK4 in vitro. We have shown that this is not the only site of in vitro phosphorylation, indicating that MKS1 may be phosphorylated by MPK4 and other MAP kinases in vivo on one or more of 12 Ser-Pro sites. Nonetheless, the MKS1 N-terminal region, including domain I, is important for interaction with MPK4, as shown by directed Y2H assays and reduced MKS1 phosphorylation in the presence of pep22 derived from domain I. The reduced levels of phosphorylated MKS1 in mpk4 plants also argue for the specificity of in vivo phosphorylation. In contrast, the Y2H assays with MKS1 truncations indicated that domain II is required for WRKY interaction with MKS1. This suggests that the VQ motif may represent the core of a protein–protein interaction domain, and is consistent with the interaction between another VQ motif protein with an RNA polymerase σ-factor (Morikawa et al, 2002). Such interactions may explain the nuclear localization of MKS1, which lacks predicted nuclear localization signals.
MKS1 may function as an MPK4 adaptor or coupling protein that affects the activities of WRKY factors and perhaps other proteins. Such activities may be determinants of the specificity of MAP kinase pathways (Baker et al, 2001), and suppression of the mpk4 phenotype by MKS1-RNAi indicates this role for MKS1. Consistent with MKS1 nuclear localization and the interaction of different MKS1 domains with MPK4 and the WRKYs, these proteins may be part of transcription or chromatin remodeling complexes, as described in yeast (de Nadal et al, 2004; Edmunds and Mahadevan, 2004). Models of an MPK4 pathway must account for the ability of MPK4 and W33 to default repress, and of MKS1 to mediate defense signaling. Our observations are consistent with a binary interaction model in which MPK4–MKS1 complexes are largely distinct from MKS–WRKY complexes. If so, MKS1 phosphorylation by MPK4 could be modulated by sequestration of MKS1 in complexes with WRKYs or other proteins. Such a model would explain our inability to detect interactions between MPK4 and W25 or W33 in yeast, although we cannot exclude transient interactions between MPK4 and the WRKYs, as noted for other MAP kinase substrates (Manning and Cantley, 2002). Alternatively, MKS1 forms a ternary or larger complex with MPK4 and W25 and/or W33 that regulates their activities. In this case, MKS1 overexpression may effect the inhibition of MPK4 activity toward W25 and W33 seen here in vitro. Overall, the results presented here indicate that plants have a mechanism for repression of defense signaling, and that controlled overexpression of MKS1 provides a potential tool for sustainable agriculture by inhibiting this signaling.
Materials and methods
Yeast two-hybrid assays and cloning
MKS1 (NM112755) is encoded by At3g18690, WRKY 25 (AAL13040) by At2g30250 and WRKY33 (AAM34736) by At2g38470. Y2H screening was performed with full-length MPK4 and MKS1 cDNA baits and the Arabidopsis MATCHMAKER library according to the manufacturer (Clontech).
Molecular biology and biochemistry
In vitro interaction assays were performed with His-tagged MKS1 purified from E. coli and 35S-methionine-labeled MPK4, WRKY33, WRKY25 and control lamin generated using a T7 polymerase-coupled reticulocyte lysate system (Promega). A 10 μl portion of 35S-labeled protein was mixed with 200 μl 1% BSA in binding buffer (BB: 50 mM KPO4, pH 7.5, 150 mM KCl, 1 mM MgCl2, 2 μg/ml leupeptin, 1 mM AEBSF, 2 μg/ml antipain, 2 μg/ml aprotinin), incubated on ice for 15 min and then centrifuged for 10 min at 4°C. The supernatant was added to 1 μg His-MKS1 protein bound to Ni beads in 200 μl 1% BSA in BB and incubated for 2 h at 4°C. Beads were washed three times with 1 ml wash buffer (50 mM KPO4, pH 7.5, 150 mM KCl, 1 mM MgCl2, 10% glycerol, 5% Triton X-100). Purification of active HA-tagged MPK4 was as described previously (Petersen et al, 2000). Phosphorylation assays were performed by combining MPK4, substrate protein (1 μg unless specified), 3 μCi [32P-γ]ATP and kinase buffer (200 μM ATP, 80 mM Tris–HCl, pH 7.5, 8 mM EGTA, 120 mM MgCl2, 4 mM Na3VO4, 4 mM DTT). Samples were incubated for 60 min at 30°C. The N-terminal deletions used were Met1–Gln220 for W25 and Met1–Asn220 for W33. For detection of MKS1, polyclonal sera and monoclonal (HYB 330-01, Danish Serum Institute, Copenhagen) antibody were used. A 1 mg portion of extract protein was used for co-immunoprecipitation experiments, as described previously (Feys et al, 2001). pS/TP was detected with 2321L antibody (Cell Signaling). Antibodies against MPK4 and MPK6 and flagellin treatments of Arabidopsis cells (Ler) have been described previously (Nühse et al, 2000). Free and glycosylated SA were analyzed, as described previously (Newman et al, 2001). GFP expression in mesophyll cells of young leaves of stably transformed lines was visualized with a Zeiss LSM 510 microscope. RNA was prepared for gel blot analysis, as described previously (Petersen et al, 2000). Probe templates were amplified by PCR from cDNAs or genomic DNA with primer sequences from PR1 (At2g14610), PDF1.2 (At5g44420) and MKS1. For RT–PCR and Q-PCR analysis, RNA samples were incubated with 1 U of DNase according to the manufacturer (Promega, Madison, WI). RT reactions were carried out with 1 μg of RNA and 0.5 μg of random hexamer primer at 42°C with 0.1 U of RT (Promega) and 2 U of RNasin (Promega) for 1 h in 20 μl reactions. Product aliquots were used as template for RT–PCR and Q-PCR analysis. Q-PCR was performed using the SYBR Green protocol (Applied Biosystems, Foster City, CA) with 10 pmol of each primer and 0.5 μl aliquot of RT reaction product in a 25 μl reaction. A standard curve was made by determining Ct (threshold cycle) values for a dilution series of the RT reaction product for each primer pair. Using this standard curve, the relative quantification for each reaction was calculated from its Ct value due to a linear relationship between Ct value and log. Q-PCR reactions were in triplicate and averaged for each line individually. PR1 expression was standardized relative to 18S rRNA expression for each data point. The relative quantity for the control sample (Col) at t=0 was arbitrarily set to 0. Expression levels of all other samples were expressed relative to this control sample. Primers were as follows: PR1: forward 5′-gtgggttagcgagaaggcta-3′, reverse 5′-actttggcacatccgagtct-3′; 18S: forward 5′-cggctaccacatccaaggaa, reverse 5′-gctgcaattaccgcggct-3′.
Genetics and transgenics
The CaMV 35S-MKS1 transgene was stably introduced into ecotype Ler with pCAMBIA1301, and MKS1-RNAi construct was introduced into ecotype Col via plasmids SLJ1382B1 (A Ludwig and JDG Jones, The Sainsbury Laboratory, Norwich) and pCAMBIA3300. The W33 insertional mutant was GABI KAT line 324B11 (Rosso et al, 2003). RT–PCR was used to show that it did not accumulate detectable W33 mRNA. The MKS1-GFP gene fusion under control of 1.9 kb 5′ upstream MKS1 sequence was made, as previously described for producing the MPK4-GFP fusion (Petersen et al, 2000). Bacterial resistance assays were performed as described previously (Parker et al, 1996; Petersen et al, 2000). Plants were infiltrated with 1 × 105 colony forming units per ml (CFU/ml). Values represent average and standard deviations of CFU extracted from leaf disks 0, 2 and 4 days after inoculation in three independent samplings. Plants were grown under 8 h light and 16 h darkness at 22°C for up to 5 weeks.
Microarray hybridization and analysis
Total RNA was isolated from three replicates of wild type, mpk4 and 35S-MKS1 grown in a chamber with 16 h light (21°C) and 8 h dark (16°C). RNA was amplified according to the standard Affymetrix protocol and hybridized to the Affymetrix ATH1 oligonucleotide microarray (accession # E-MEXP-173, ArrayExpress database, EBI). Raw intensity data were normalized using R implementation of qspline (Workman et al, 2002; Gautier et al, 2004). An implementation in the statistical language R of the logit-t method (Lemon et al, 2003) applying one-way ANOVA was used to calculate statistical significance for differential gene expression. Likewise, logit-t was applied for the comparison between wild type and both mpk4 and S35-MKS1. Genes with P-value less than 0.01 were considered significant. Hypergeometric statistic was used to access the significance of the overlap of genes significantly affected by mpk4 and S35-MKS1. Gene expression index values were calculated using the perfect match only implementation method (Li and Wong, 2001). Gene expression profiles for significantly differentially expressed genes were clustered by k-means. For promoter analysis, 800 bp upstream regions from all genes profiled in the microarray experiments were extracted. All patterns from 5 to 12 bp in length were tested for significant over- or under-representation between upstream regions of genes included in one of the seven clusters (Supplementary Figure S2) and all 22 810 genes. Hypergeometric statistics was applied, as described previously (Jensen and Knudsen, 2000).
Supplementary Material
Supplementary Figures
Acknowledgments
We thank Suksawad Vongvisuttikun and Ying Kaaring for technical assistance. This work was supported by grants to EA and JM from the Danish Research Councils and European Union, and by funds from the Danish Biotechnology Instrument Center for microarray analyses.
References
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Baker DA, Mille-Baker B, Wainwright SM, Ish-Horowicz D, Dibb NJ (2001) Mae mediates MAP kinase phosphorylation of Ets transcription factors in Drosophila. Nature 411: 330–334 [DOI] [PubMed] [Google Scholar]
- Bao MZ, Schwartz MA, Cantin GT, Yates JR III, Madhani HD (2004) Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell 119: 991–1000 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Liu Y, Klessig DF, Dong X (1998) Uncoupling PR gene expression from NPR1 and bacterial resistance, characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10: 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- De Nadal E, Zapater M, Alepuz PM, Sumoy L, Mas G, Posas F (2004) The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427: 370–374 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- Desveaux D, Subramaniam R, Després C, Mess J-N, Lévesque C, Fobert PR, Dangl JL, Brisson N (2004) A ‘Whirly' transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev Cell 6: 229–240 [DOI] [PubMed] [Google Scholar]
- Dieterle M, Thomann A, Renou JP, Parmentier Y, Cognat V, Lemonnier G, Muller R, Shen WH, Kretsch T, Genschik P (2005) Molecular and functional characterization of Arabidopsis Cullin 3A. Plant J 41: 386–399 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JD (2000) cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12: 963–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC (2004) MAP kinases as structural adaptors and enzymatic activators in transcription complexes. J Cell Sci 117: 3715–3723 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton P, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouhar F, Yang Y, Kumar D, Chen Y, Fridman E, Park SW, Chiang Y, Acton TB, Montelione GT, Pichersky E, Klessig DF, Tong L (2005) Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc Natl Acad Sci USA 102: 1773–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M, He YJ, Borchers C, Xiong Y (2003) Targeting of protein ubiquitination by BTBCullin 3-Roc1 ubiquitin ligases. Nat Cell Biol 5: 1001–1007 [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Chen W, Estes B, Chang H-S, Nawrath C, Métraux J-P, Zhu T, Katagiri F (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J 34: 217–228 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li H, Gupta R, Morris PC, Luan S, Kieber JJ (2000) ATMPK4, an Arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol 122: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000) Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24: 655–665 [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Knudsen S (2000) Automatic discovery of regulatory patterns in promoter regions based on whole cell expression data and functional annotation. Bioinformatics 16: 326–333 [DOI] [PubMed] [Google Scholar]
- Johnson C, Boden E, Arias J (2003) Salicylic acid and NPR1 induce the recruitment of transactivating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15: 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E (1998) Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J 16: 223–233 [DOI] [PubMed] [Google Scholar]
- Lemon WJ, Liyanarachchi S, You M (2003) A high performance test of differential gene expression for oligonucleotide arrays. Genome Biol 4: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2: R0032.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK (2002) A putative lipid transfer protein involved in systemic resistance signaling in Arabidopsis. Nature 419: 399–403 [DOI] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26: 403–410 [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC (2002) Hitting the target: emerging technologies in the search for kinase substrates. Sci STKE 162: pe49. [DOI] [PubMed] [Google Scholar]
- MAPK Group (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Menke LH, van Pelt JA, Pieterse CMJ, Klessig DF (2004) Silencing of mitrogen-activated protein kinase MPK6 comprises disease resistance in Arabidopsis. Plant Cell 16: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa K, Shiina T, Murakami S, Toyoshima Y (2002) Novel nuclear-encoded proteins interacting with a plastid sigma factor, Sig1, in Arabidopsis thaliana. FEBS Lett 514: 300–304 [DOI] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944 [DOI] [PubMed] [Google Scholar]
- Narasimhan ML, Coca MA, Jin J, Yamauchi T, Ito Y, Kadowaki T, Kim KK, Pardo JM, Damsz B, Hasegawa PM, Yun DJ, Bressan RA (2005) Osmotin is a homolog of mammalian adiponectin and controls apoptosis in yeast through a homolog of mammalian adiponectin receptor. Mol Cell 17: 171–180 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, von Roepenack-Lahaye E, Parr A, Daniels MJ, Dow JM (2001) Induction of hydroxycinnamoyl-tyramine conjugates in pepper by Xanthomonas campestris, a plant defense response activated by hrp gene-dependent and hrp gene-independent mechanisms. Mol Plant–Microbe Interact 14: 785–792 [DOI] [PubMed] [Google Scholar]
- Norman-Setterblad C, Vidal S, Palva ET (2000) Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora. Mol Plant–Microbe Interact 13: 430–438 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Boller T, Peck SC (2000) A plasma membrane syntaxin is phosphorylated in response to the bacterial elicitor flagellin. J Biol Chem 275: 7521–7526 [DOI] [PubMed] [Google Scholar]
- Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, Sharma SB, Klessig DF, Martienssen R, Mattsson O, Jensen AB, Mundy J (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, van Loon LC (1999) Salicylic acid-independent plant defence pathways. Trends Plant Sci 4: 52–58 [DOI] [PubMed] [Google Scholar]
- Romeis T, Ludwig AA, Martin R, Jones JD (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20: 5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Zhang S, Klessig DF, Hirt H, Jones JD (1999) Rapid Avr9- and Cf-9-dependent activation of MAP kinases in tobacco cell cultures and leaves: convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11: 273–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Scheel D (1998) Resistance response physiology and signal transduction. Curr Opin Plant Biol 1: 305–310 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97: 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks AD, Yang SH, Galanis A (2000) Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem Sci 25: 448–453 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, van Pelt JA, Mueller MJ, Buchala AJ, Métraux J-P, Brown R, Kazan K, van Loon LC, Dong X, Pieterse CMJ (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, Doczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 2: 141–152 [DOI] [PubMed] [Google Scholar]
- Turck F, Zhou A, Somssich IE (2004) Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense-related gene PcPR1-1 in Parsley. Plant Cell 16: 2573–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A (2002) The jasmonate signal pathway. Plant Cell (Suppl): S153–S164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL-C, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell (Suppl): S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielsen HB, Saxild HH, Nielsen C, Brunak S, Knudsen S (2002) A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol 3: research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Suzuki H, Borevitz J, Blount J, Guo Z, Patel K, Dixon RA, Lamb C (2004) An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J 23: 980–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures


