Summary
We report the role of dCREB2, the Drosophila homolog of CREB/CREM, in circadian rhythms. dCREB2 activity cycles with a 24 hr rhythm in flies, both in a light:dark cycle and in constant darkness. A mutation in dCREB2 shortens circadian locomotor rhythm in flies and dampens the oscillation of period, a known clock gene. Cycling dCREB2 activity is abolished in a period mutant, indicating that dCREB2 and Period affect each other and suggesting that the two genes participate in the same regulatory feedback loop. We propose that dCREB2 supports cycling of the Period/Timeless oscillator. These findings support CREB’s role in mediating adaptive behavioral responses to a variey of environmental stimuli (stress, growth factors, drug addiction, circadian rhythms, and memory formation) in mammals and long-term memory formation and circadian rhythms in Drosophila.
Introduction
The cAMP response element binding protein (CREB) has been shown to be involved in adaptive behavioral responses to various external stimuli. These responses include stress (Borsook et al., 1994; Tan et al., 1996), growth factor stimulation (reviewed by Segal and Greenberg, 1996), drug addiction and withdrawal (Widnell et al., 1994; Carlezon et al., 1998; reviewed by Self and Nestler, 1995), learning and memory (Bourtchuladze et al., 1994; Yin et al., 1994, 1995a; reviewed by Dubnau and Tully, 1998; Silva et al., 1998), and circadian rhythms (Ginty et al., 1993; Stehle et al., 1993; Foulkes et al., 1996).
Several groups have studied the involvement of CREB in mammalian circadian rhythms. Ginty et al. (1993) showed that CREB is phosphorylated in the suprachiasmatic nucleus (SCN) of the hypothalamus, the location of the mammalian clock, in response to a light pulse delivered during the dark period. The light pulse, administered at a time that resets the clock, causes a dramatic increase in the amount of Ser-133 phosphorylation, implicating CREB in the reset mechanism. Recently, a CREB-responsive reporter was found to be activated in the SCN by a light pulse, as well as to cycle in a circadian rhythm (D. Storm, personal communication; also see Discussion). A circadian response has also been shown for the mouse CREM gene. Transcripts of an isoform of CREM called ICER (inducible cAMP early repressor), were found to cycle in a circadian rhythm in the pineal gland (Stehle et al., 1993; Foulkes et al., 1996), another anatomical component of the mammalian clock system. These experiments suggest that CREB and CREM are regulated by the circadian system but do not show whether or not they play a causative role in establishing circadian rhythms.
In Drosophila, the mechanism of the circadian clock has been worked out in some detail. The two best-characterized circadian genes in Drosophila are period (per) and timeless (tim). per encodes a PAS domain protein (Reddy et al., 1984; Jackson et al., 1986; Citri et al., 1987), while tim has no homology to known genes (Gekakis et al., 1995; Myers et al., 1995). They both encode components of the clock and constitute an autoregulatory, negative feedback loop that controls expression of their own transcripts (Hardin et al., 1990; Zwiebel et al., 1991; Hardin et al., 1992). Flies mutant for either per or tim display aberrant circadian locomotor rhythms (Konopka and Benzer, 1971; Sehgal et al., 1994). Three additional circadian genes have recently been identified in Drosophila, dClock (Allada et al., 1998; Darlington et al., 1998), cycle (Rutila et al., 1998), and double-time (Kloss et al., 1998; Price et al., 1998). These genes all function to promote the oscillation of Per and Tim (see Discussion).
We examined the role of dCREB2 in circadian rhythms using a phenotypic assay in the intact adult fly. We chose to use a transcriptional fusion with luciferase as a reporter molecule for two reasons. First, luciferase can be quantitatively measured in the behaving fly, and this measurement can be confirmed in vitro. Second, dynamic measurements can be made easily because luciferase activity is short lived. Potentially, the use of luciferase allows a more accurate picture of the timing of transcriptional activation. Our analysis of such a reporter shows that dCREB2 activity cycles in a 24 hr rhythm in Drosophila. Using mutations in both dCREB2 and per, our results suggest that dCREB2 participates in the circadian feedback loop, and along with dClock, Cycle, and Double-time functions to promote oscillations of Per and Tim.
Results
CRE Reporter Activity Cycles in a Circadian Rhythm in Drosophila
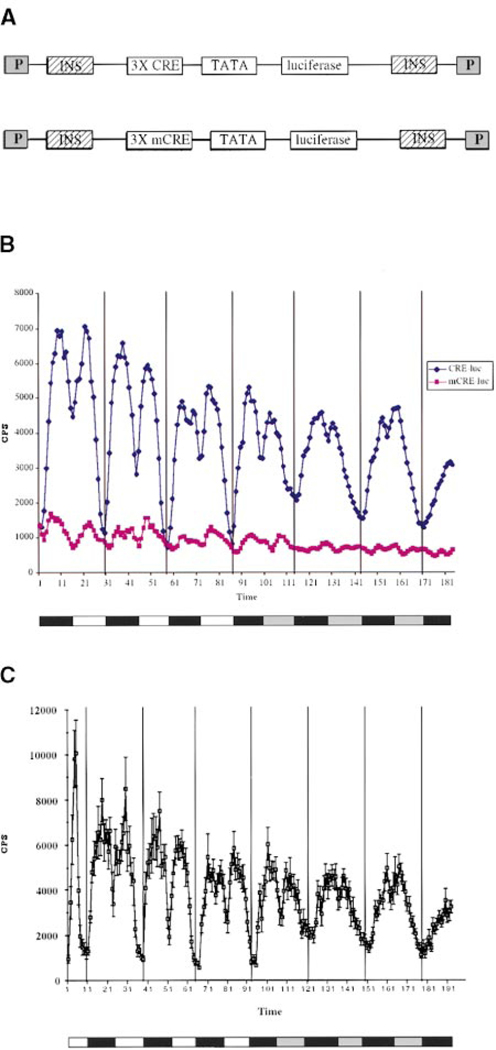
To measure dCREB2 activity in vivo, we constructed transgenic Drosophila lines carrying the luciferase reporter gene driven by an enhancer element comprised of consensus CREB binding sites. Three cAMP response elements (CREs), 5′-TGACGTCA-3′, were placed upstream of the TATA box region of the hsp70 gene promoter, followed by the luciferase reporter gene (Figure 1A; see Experimental Procedures). This sequence was flanked by the scs and scs′ insulator elements (Udvardy et al., 1985; Kellum and Schedl, 1992; Vazquez and Schedl, 1994) to reduce potential positional effects caused by the random insertion site of the transgene (Henikoff, 1994). The construct was cloned into the pCaSpeR transformation vector and injected into Drosophila embryos to generate transgenic lines (Rubin and Spradling, 1982), referred to as CRE–luc lines throughout this paper. A mutant CRE–luc reporter construct (mCRE–luc) was also generated in which the consensus CRE sites were mutated to TGAAATCA (Figure 1A). dCREB2 protein binds this mutant CRE site with at least 20-fold lower affinity in gel shift experiments (Yin et al., 1995b). This construct is otherwise identical to wild-type CRE–luc.
Figure 1.
Cycling of a dCREB2-Responsive Reporter
(A) Constructs used to generate CRE–luc and mCRE–luc transgenic flies. Abbreviations: P, P transposable element inverted repeats; INS, SCS and SCS′ insulator elements; TATA, TATA box sequence from hsp 70 promoter. These constructs were cloned into the pCaSpeR transformation vector.
(B) In vivo cycling of CRE–luc and mCRE–luc reporter expression as measured in a Packard TopCount Luminometer (see Experimental Procedures). All time points represent an average of data points from 30 flies. The bar below the graph indicates light:dark conditions. White box, light period; black box, dark period; gray box, dark period during former light hours (subjective day). Vertical bars in the graph represent lights out and are spaced 24 hr apart.
(C) Similar graph as in (B) but with standard error bars added.
Luciferase activity in the CRE–luc flies was monitored according to the method of Brandes et al. (1996), with some modifications (see Experimental Procedures). Briefly, flies were maintained on a 12 hr light:12 hr dark cycle at 25°C on standard food. The flies were then loaded into 96-well microtiter dishes containing an agar/sucrose solution supplemented with luciferin, the substrate for luciferase. In vivo expression of luciferase activity was measured in a plate-reading luminometer, with the expression level of each fly being measured hourly over a period of 6–10 days.
The expression of luciferase in the wild-type CRE–luc flies oscillates in a 24 hr rhythm (Figure 1B), both in a light:dark cycle (white bars:black bars) and in constant darkness (gray bars:black bars). The main peak of activity occurs just after lights out, with the nadir just before the main peak. Since this rhythmic transcription pattern is sustained in constant darkness, it is regulated by the circadian system, rather than simply being a response to light. In light:dark conditions, a second peak is observed in the middle of the day; however, these two peaks gradually blend together under conditions of constant darkness (Figure 1B). This pattern is very similar to that seen for per activity (Brandes et al., 1996; Stanewsky et al., 1997). The per–luc reporter also exhibits a similar secondary peak under light:dark conditions, even though per RNA peaks only once per cycle (Brandes et al., 1996; Stanewsky et al., 1997). It is likely that the secondary peaks of both reporters, which occur during the day, are due to a light response of luciferase rather than a circadian response. The expression level of the mCRE–luc reporter is drastically reduced relative to the wild-type reporter (Figure 1B), indicating that the CRE sites mediate the high-level expression of the wild-type reporter. A similar experiment with standard error bars is shown in Figure 1C, giving an indication of the variation between flies. Three independent lines of CRE–luc and mCRE–luc were tested and showed nearly identical qualitative patterns of transcription (periodicity, peaks, and troughs), with minor quantitative differences in the levels (data not shown). Thus, this rhythmic transcription is independent from positional effects of the site of transgene insertion. When extracts were made from the CRE–luc transgenic flies and luciferase activity was measured in vitro, a similar rhythmic expression pattern was seen (data not shown). This demonstrates that the cyclic transcription measured in vivo is not due to circadian effects on feeding and substrate availability.
S162 Is a Mutation in the dCREB2 Gene
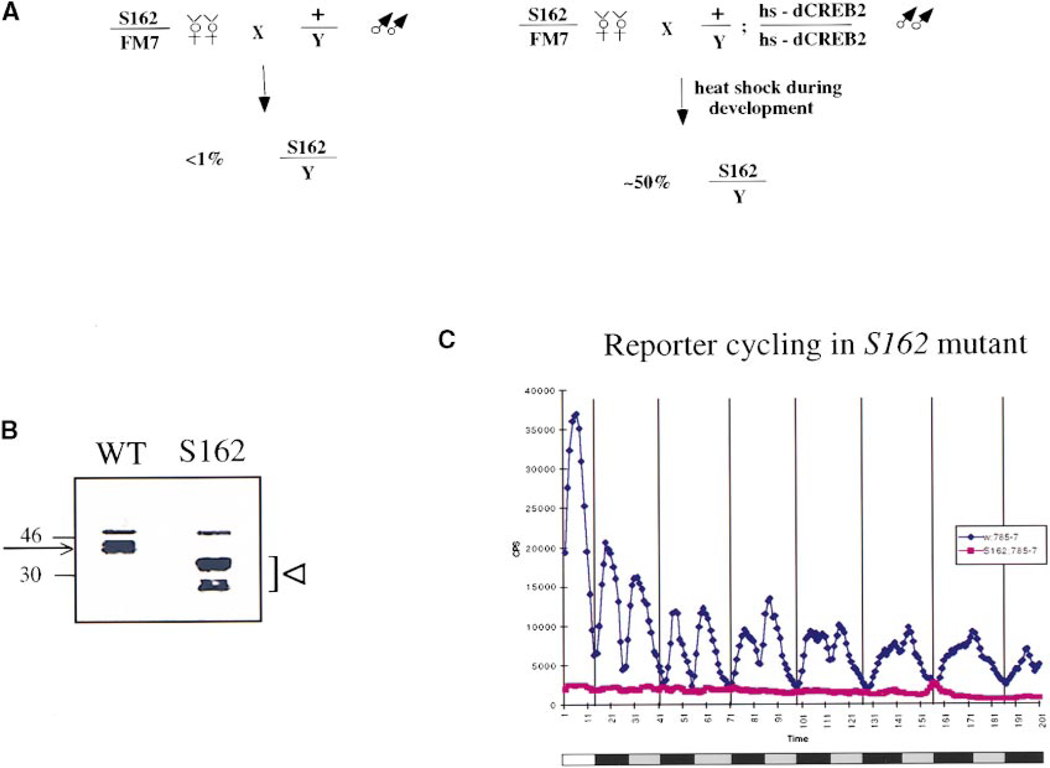
S162 is a mutation that was isolated in a screen for lethal mutations on the Drosophila X chromosome (Eberl et al., 1992). The S162 mutation and the dCREB2 gene both map to the 17A1–5 region (Eberl et al., 1992; Usui et al., 1993). S162 is lethal, although not completely penetrant, with fewer than 0.5% of hemizygous S162 males surviving to adulthood (Figure 2A, left). These escaper males are about three-fourths the size of wild-type flies but are otherwise apparently normal. The lethality can be completely rescued by induction of a dCREB2 transgene, hs-dCREB2–10, during development (Figure 2A, right panel). This suggests that S162 is an allele of dCREB2. To confirm this, we performed Western blot analysis of extracts made from S162 escaper males (Figure 2B). A monoclonal antibody (mAb 27) was used that recognizes a doublet of dCREB2 isoforms, indicated by the arrow. In S162 extracts, these wild-type forms do not exist while several smaller, mutant forms are present, indicated by the arrowhead. Taken together, these three converging lines of data (comapping, transgene rescue, and protein analysis) suggest very strongly that S162 is a mutation in the dCREB2 gene.
Figure 2.
Characterization of the S162 Mutation
(A) Crossing S162/FM7 heterozygous females to wild-type males results in fewer than 0.5% of the male progeny carrying the S162 mutation (left panel). This percentage of S162 males can be increased by crossing S162/FM7 females to transgenic males homozygous for a heat shock–inducible form of dCREB2, hs-dCREB2–10. This construct encodes an isoform of dCREB2 (see Experimental Procedures). The transgene is induced by a daily 60 min heat shock of 37°C during the larval and pupal stages of development. This results in 50% of the male progeny carrying the S162 mutation (right panel).
(B) Western blot analysis of extracts from wild-type males and S162 escaper males. Extracts were prepared from whole flies and were analyzed on a 12% denaturing SDS–polyacrylamide gel followed by Western blot analysis. A mouse monoclonal antibody (mAb 27) raised against full-length dCREB2 was used to probe the membrane. This antibody recognizes two wild-type dCREB2 proteins of about 38–40 kDa, indicated by the arrow. The S162 mutant forms are indicated by an arrowhead. The high molecular weight band observed in both lanes is a cross-reacting band.
(C) Expression of the CRE–luc reporter in S162 mutant flies. S162/FM7 females were mated to males homozygous for the CRE–luc reporter transgene. Escaper males of the genotype S162/Y; CRE–luc/+ were assayed in the luminometer. The traces represent an average of data from 15 flies (S162 mutants) or 30 flies (wild type). See Figure 1 legend for description of the graph.
The CRE–luc Reporter Is dCREB2 Responsive, Demonstrating that dCREB2 Activity Cycles in a Circadian Rhythm
To verify that the CRE–luc reporter reflected dCREB2 activity, we crossed the reporter into the S162 mutant background. Figure 2C shows that the S162 mutation dramatically reduces both the expression levels (amplitude) and cycling pattern (periodicity) of the reporter, demonstrating that the reporter is dCREB2 responsive. Therefore, the cycling of the reporter indicates that dCREB2 activity cycles in a circadian manner.
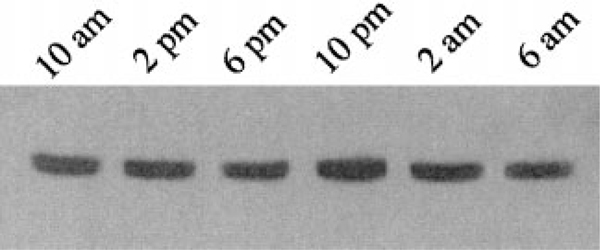
This data shows that the activity of dCREB2 cycles but does not show whether or not the actual protein level cycles. Flies entrained on a 12 hr light:12 hr dark cycle were collected every 4 hr over a 24 hr period, and head extracts were prepared and analyzed by Western blot using the mAb 27 antibody (Figure 3). The levels of dCREB2 protein did not appear to vary over time. Similar samples were also probed with a phospho-specific antibody that recognizes dCREB2 only when it is phosphorylated on Ser-230, which corresponds to Ser-133 in mammalian CREB, a residue whose phosphorylation is required for activation of CREB (Gonzalez and Montminy, 1989). However, the phosphorylation status of Ser-230 did not vary detectably over the circadian cycle (data not shown). Cycling of Ser-133 phosphorylation of CREB has been observed in the SCN of mice (D. Storm, personal communication). In Drosophila, it may be that dCREB2 Ser-230 phosphorylation oscillates, but only in a small subset of cells in the brain. Alternatively, the oscillation of the CRE–luc reporter may be due to the oscillation of a dCREB2 binding partner or a different kinase (see Discussion).
Figure 3.
dCREB2 Protein Levels Do Not Appear to Cycle in a Circadian Rhythm
Western blot analysis of extracts prepared from adult fly heads. Flies were collected every 4 hr over a 24 hr period. Total extracts were prepared from heads, and equal amounts of protein were loaded and analyzed by Western blot as described in Figure 2.
dCREB2 Affects the Circadian Clock
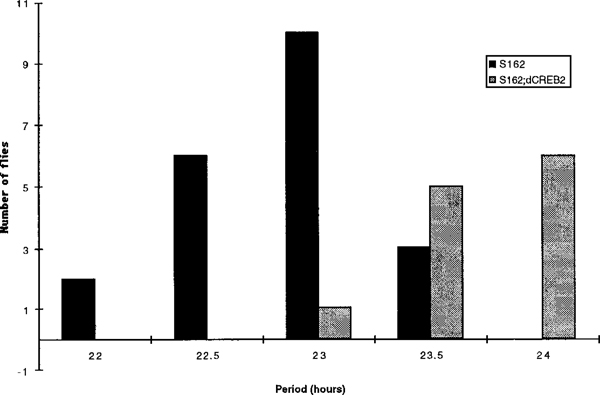
These experiments show that dCREB2 activity is under circadian control, but they do not show whether dCREB2 plays a role in maintaining the rhythms or is just responsive to them. We used the S162 mutation to address this question. To test for behavioral effects of the mutation, S162 escaper males were assayed for circadian locomotor activity (see Experimental Procedures). The flies were tested for 10 days in constant darkness to determine whether they displayed normal circadian fluctuations in activity. Of the 34 S162 mutants tested, 13 (38%) were arrhythmic while the 21 that were rhythmic had a short period averaging 22.8 hr (Figure 4 and Table 1).
Figure 4.
Circadian Locomotor Activity Defect of S162 Mutant Flies and Rescue by hs-dCREB2–10
Flies were analyzed for circadian locomotor activity as described in the Experimental Procedures. Both S162 mutant flies (black bars) and S162 mutants that had been rescued by developmental induction of hs-dCREB2–10 (gray bars) were assayed. See also Table 1.
Table 1.
Period Length of S162 Mutants and S162; hs-dCREB2–10 Rescued Flies
| S162 | S162 ; dCREB2 | |
|---|---|---|
| % Arrhythmic | 38 | 0 |
| Period (hr) | 22.8 | 23.7 |
| Standard error | 0.09 | 0.10 |
| n | 21 | 12 |
Flies that displayed no obvious rhythm after periodogram analysis were considered to be arrhythmic. Period, mean period of rhythmic flies; standard error, standard error of the mean period; n, number of flies for which a period could be calculated.
None of these flies had a wild-type 24 hr rhythm. The high percentage of arrhythmicity is typical of mutations that affect period length (e.g., see Allada et al., 1998; Rutila et al., 1998).
To verify that this behavioral phenotype was specific for the S162 mutation, we rescued the phenotype by induction of the hs-dCREB2–10 transgene. To induce the transgene, larvae and pupae were subjected to a daily 60 min heat pulse of 37°C during development. All of the rescued flies (12/12) were rhythmic, and they all displayed normal circadian locomotor rhythms of 23.5–24 hr (Figure 4 and Table 1). This demonstrates that the short period phenotype is caused by the S162 mutation rather than a second site mutation elsewhere on the chromosome. It also shows the involvement of dCREB2 in the timing of the clock.
S162 Affects per Expression
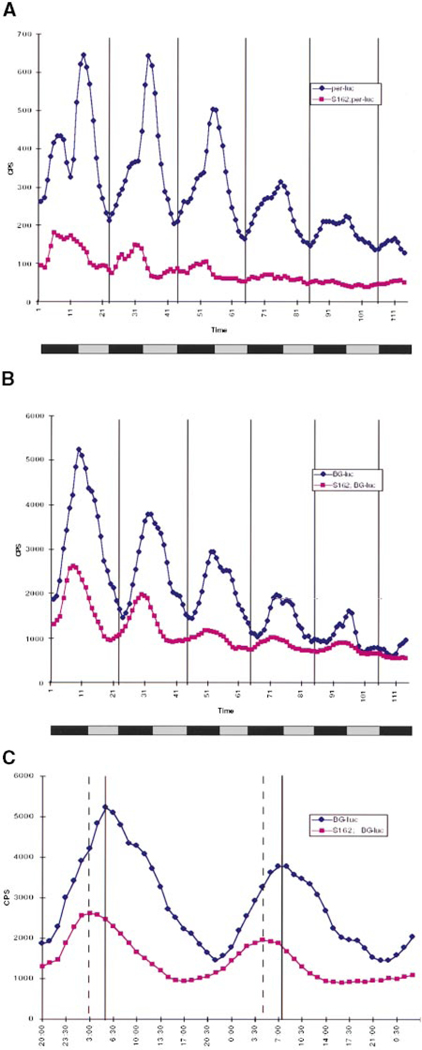
If S162 is indeed acting in the clock, then it should affect the per clock gene. We assayed the effects of S162 on two different per-dependent reporters. The first is a transcriptional fusion with a 4.2 kb fragment of the per promoter upstream of the luciferase reporter gene, referred to as per–luc (Brandes et al., 1996). The second is a translational fusion containing the same promoter fragment, plus the 5′ untranslated region and the first 2.4 kb of the per coding region fused in frame to the luciferase gene, referred to as BG–luc (Stanewsky et al., 1997). When the expression of these reporters was compared, it was found that the BG–luc reporter cycles much more robustly than the per–luc reporter, consistent with the interpretation that there are at least two mechanisms contributing to the cycling of Per: one mediated by the promoter, and the other(s) mediated by sequences in either the per transcript or Per protein itself (Stanewsky et al., 1997). S162 affects the two reporters differently. As shown in Figure 5A, the S162 mutation reduces both the expression level and cycling pattern of the per–luc reporter. However, its effect on the BG–luc reporter is weaker (Figure 5B). In S162 flies, the BG–luc reporter maintains a robust cycling pattern, although its expression level and amplitude are reduced. The peak in the mutant background also occurs in advance of the peak in wild-type flies (Figure 5C), consistent with the short period phenotype of these flies.
Figure 5.
Expression of per–luc and BG–luc Reporters in the S162 Mutant Background
(A) Expression of the per–luc reporter in a wild-type or S162 mutant background. For the wild-type background, +/+ females were crossed to per–luc homozygous males, and per–luc/+ male progeny were tested. For the S162 background, S162/FM7 females were crossed to homozygous per–luc males, and escaper males of the genotype S162/Y; per–luc/+ were tested. Crosses were similar for experiments with BG–luc.
(B) Expression of the BG–luc reporter in a wild-type or S162 mutant background.
(C) Replotting of the first 48 hr of the graph in (B). The solid lines are placed at the peaks of the wild-type curve, and the dashed lines are at the peaks of the S162 curve.
In this experiment, both the wild-type and mutant flies appear to have slightly longer than 24 hr rhythms (Figure 5B; see also Figure 1B). This is probably a result of using the luciferase reporter to monitor rhythms, since the circadian rhythms measured via monitoring luciferase in the luminometer tend to be a bit long. Therefore, the locomotor assay is better for measuring exact period lengths (Figure 4 and Table 1), whereas the luminometer assay is useful for rougher measurements of period length and for comparisons between lines. The luminometer is also more amenable to high-throughput assays. The strong disruption of per–luc expression in S162 shows that per expression is controlled, at least in part, by dCREB2. The smaller effect of S162 on BG–luc, however, suggests that while dCREB2 affects per expression via its promoter, there are compensatory posttranscriptional mechanisms that can partially overcome the transcriptional defects.
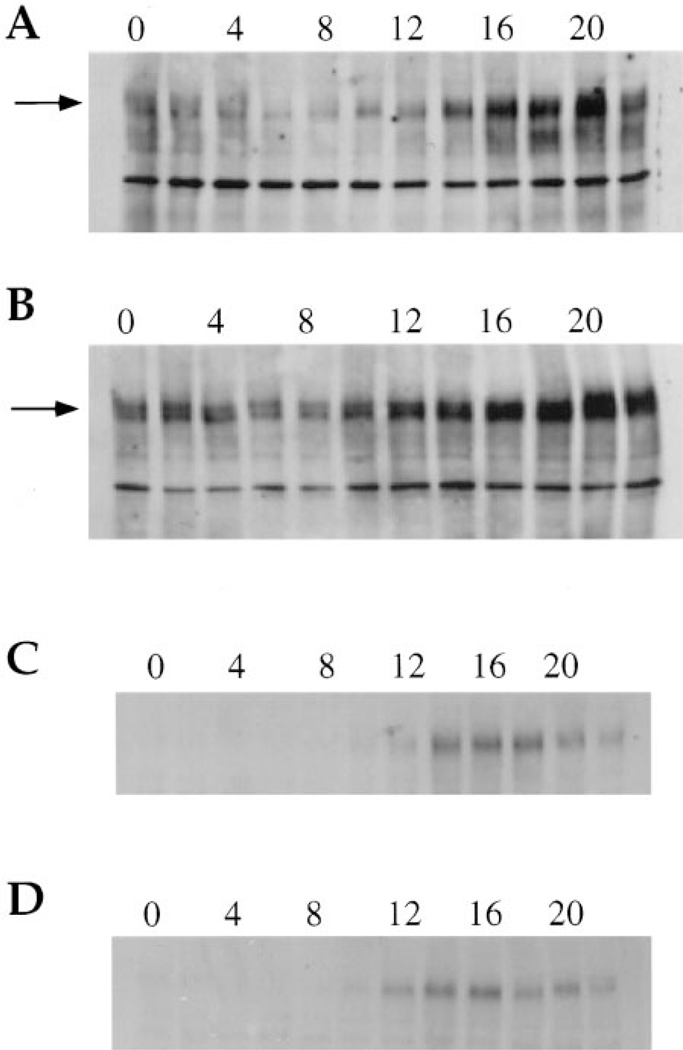
In order to demonstrate a direct effect of the S162 mutation on the clock, we also examined its effect on the Per protein itself. Wild-type and S162 flies were entrained on a 12 hr light:12 hr dark cycle and aliquots were frozen every 2 hr throughout the cycle. Head extracts were prepared and analyzed by Western blot using an antibody directed against Per. Two independently isolated sets of samples were analyzed and generated the same results, although only one is shown. Figure 6A shows the circadian oscillation of Per protein in wild-type flies. Per is present at very low levels at ZT 6 and ZT 8 (Zeitgeber time), increasing to peak levels before lights on, which occurs at ZT 0. A corresponding change in phosphorylation, and protein mobility, accompanies the change in absolute levels, with Per becoming more highly phosphorylated as it accumulates (Edery et al., 1994). This temporal pattern of Per is altered in the S162 mutant background (Figure 6B), where Per is present at more equal levels throughout the circadian cycle. At the peak time, ZT 20, the amount of Per is at least comparable to that in wild-type flies; however, it decreases less at ZT 6 and ZT 8, when Per is virtually absent in wild-type flies. At these trough periods of Per expression, a discrete doublet protein band persists in S162, perhaps representing preservation of certain phosphorylated forms (Edery et al., 1994). There also seems to be a general increase in the amount of Per protein throughout the cycle in the mutant flies. The change in both per–luc expression and Per protein levels in the S162 mutant background demonstrates that Per activity is under the influence of the dCREB2 gene. The effects of S162 on Tim protein were assayed in the same experiment. Figures 6C and 6D show the same blots as in Figures 6A and 6B after they were stripped and reprobed with an antibody that recognizes Tim. The effect of S162 on Tim is much more subtle than the effect on Per. The Tim protein appears to accumulate slightly sooner in the mutant than in the wild type (ZT 12 versus ZT 14); however, its overall oscillation remains fairly normal.
Figure 6.
Per Protein Expression in Wild-Type and S162 Mutant Flies
(A) Head extracts from wild-type flies were analyzed by Western blot using a Per antibody. Flies were collected every 2 hr throughout the circadian cycle and are plotted from ZT 0 (far left lane) to ZT 22 (far right lane). Per protein is present as a series of bands indicated by the arrow.
(B) Head extracts from S162 flies analyzed as in panel (A).
(C and D) Same blots as in (A) and (B) but probed with an antibody to Tim protein.
dCREB2 and per Form a Feedback Loop
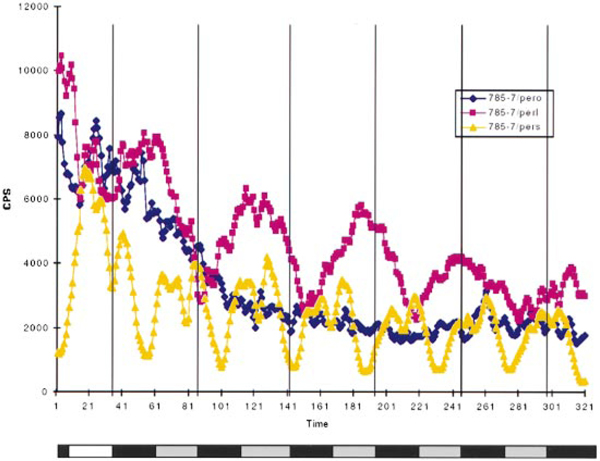
Because S162 affects the per–luc reporter and the Per protein, dCREB2 may be part of the feedback loop controlling rhythmic per expression. If dCREB2 and per are part of the same feedback loop, then per mutants should affect the cycling of the CRE–luc reporter. We crossed CRE–luc into three different per mutant backgrounds: per0, a null mutation resulting in arrhythmic flies; pers (per short), which causes a short (~19 hr) period; and perl (per long), which causes a long (~29 hr) period (Konopka and Benzer, 1971). As shown in Figure 7, CRE–luc expression mirrored the per phenotype in all cases. In the per0 background, the CRE–luc reporter did not cycle; in the perl background, it cycled with a long rhythm; and in the per s background, it cycled with a short rhythm. Therefore, mutations in per affect dCREB2 activity.
Figure 7.
Expression of the CRE–luc Reporter in per Mutant Backgrounds
Females homozygous for one of three per mutations (described in the text) were crossed to males homozygous for the CRE–luc reporter, and male progeny of the genotype per/Y; CRE–luc/+ were assayed in the luminometer. Abbreviations: pero, per0 or per null; perl, perl or per long; pers, pers or per short. The flies had been entrained to a 12 hr light:12 hr dark cycle for 4 days before the start of the experiment. The flies were then switched to constant darkness for the duration of the experiment. Each trace represents the average of data from 40 flies.
Discussion
dCREB2 Activity Cycles in a Circadian Rhythm in Drosophila
Using an in vivo reporter assay, we have shown that dCREB2 activity cycles with a circadian rhythm in Drosophila. This cycling takes place in tissues throughout the fly, because isolated heads, thoraces, and abdomens all cycled when assayed independently (data not shown). This is not unusual for a circadian gene in flies, since the per gene is known to cycle in multiple tissues (Liu et al., 1988; Saez and Young, 1988; Siwicki et al., 1988; Hardin, 1994; Emery et al., 1997; Plautz et al., 1997).
Although the CREM ICER transcript has been shown to cycle in a circadian rhythm in mammals (Stehle et al., 1993), cycling of CREB activity has not previously been observed. Changes in CREB phosphorylation have only been observed after a light pulse (Ginty et al., 1993). Recent analysis of a CRE-mediated reporter transgene in mice, however, indicates that CREB activity does in fact cycle, at least in the SCN (D. Storm, personal communication). Circadian fluctuations in Ser-133 phosphorylation were also observed. We were not able to observe circadian differences in dCREB2 protein levels or Ser-230 phosphorylation in Drosophila, even though the activity of the protein clearly oscillates as assayed by reporter activity. The dClock and Cycle proteins also do not cycle, although their activities are rhythmic (Allada et al., 1998; Rutila et al., 1998; but see Darlington et al., 1998). It is believed that this pattern of transcriptional activity is caused by the cycling of a different binding partner, Per (Allada et al., 1998; Darlington et al., 1998; Gekakis et al., 1998; Rutila et al., 1998). Similarly, dCREB2 may have an unknown binding partner that cycles. For instance, the levels or activity of the dCREB2 cofactor, CBP, may cycle. Alternatively, a kinase that phosphorylates dCREB2 at a residue other than Ser-230, and which affects its activity, may cycle. It is also possible that dCREB2 RNA or protein levels cycle in a small subset of cells in the brain, causing fluctuations that are below the detection levels of our assays.
Per and dCREB2 Affect the Activity of Each Other
Traditionally, circadian genes have been divided into three groups: input genes, clock genes, and output genes. Input genes mediate the effects of light on clock genes. As of yet, no input genes in Drosophila have been identified. However, the target of the input is Timeless degradation (Hunter-Ensor et al., 1996; Lee et al., 1996; Myers et al., 1996; Zeng et al., 1996; Suri et al., 1998; Yang et al., 1998), which does not depend on any previously identified photoreceptive pathway (Suri et al., 1998; Yang et al., 1998). Output genes mediate the effects of the clock on behavioral outputs such as locomotor activity and eclosion rhythms. Two output genes have been identified so far in Drosophila. Flies mutant for protein kinase A (PKA) have defective locomotor rhythms but normal eclosion rhythms (Majercak et al., 1997). Conversely, the lark gene, encoding a novel RNA-binding protein, affects eclosion rhythms but not locomotor rhythms (McNeil et al., 1998).
Clock genes comprise the timekeeping mechanism itself and consist of a feedback loop designed to maintain the oscillation of Per and Tim. Of the known genes involved in circadian rhythms, Per and Tim are the only two that cycle at both the RNA and protein levels (Hardin et al., 1990; Sehgal et al., 1995; Hunter-Ensor et al., 1996; Myers et al., 1996; Zeng et al., 1996) and are known to control their own levels of transcription (Hardin et al., 1990; Zwiebel et al., 1991; Hardin et al., 1992). Other components, including dClock, Cycle, and Double-time, function to maintain strong oscillations of Per and Tim but do not cycle at the transcript or protein level.
If dCREB2 were acting only as an input gene, then it would act upstream of the clock. Therefore, a dCREB2 mutation would affect Per activity but not vice versa. Likewise, if dCREB2 acted as an output gene, then it would act downstream of the clock. Thus, a mutation in per would be expected to influence dCREB2 activity, but a dCREB2 mutation would have no effect on Per activity. Our data shows that dCREB2 acts as if it were both an input and an output gene. The S162 dCREB2 mutation affects Per levels, and the per0 mutation reduces dCREB2 activity, showing that the two genes mutually affect each other’s activity. Two models would be most consistent with this data. One is that dCREB2 plays two separate roles in circadian rhythms: one as an input gene, and one as an output gene. In support of this, the work in mammals showing that CREB is phosphorylated in the SCN in response to a light pulse (Ginty et al., 1993) implies that CREB may act as an input gene. dCREB2 could play separate roles as an input and an output gene. This model, while a formal possibility, is not the simplest model, and in our view is less likely than the one described below.
A more straightforward model is that dCREB2 participates in the clock feedback loop. This would explain why dCREB2 and Per mutually affect each other. In this model, dCREB2 functions to promote transcription of per, although the mechanism by which it acts is not clear. Recent work from several labs has shown that dClock and Cycle activate transcription of per and tim via the E box sites in their promoters (Hao et al., 1997; Darlington et al., 1998; Gekakis et al., 1998). E boxes are bound by basic helix-loop-helix (bHLH) transcription factors (Murre et al., 1989), of which both dClock and Cycle are members (Allada et al., 1998; Darlington et al., 1998; Gekakis et al., 1998; Rutila et al., 1998). Per protein antagonizes this process (Darlington et al., 1998), presumably by forming nonfunctional heterodimers with dClock and/or Cycle, via their PAS domains (Huang et al., 1993). Since dCREB2 is a bZIP protein and lacks a PAS domain, it is not likely that it would interact directly with any of the PAS domain–containing proteins. However, it could bind a different region of the promoter and exert its effects additively.
Hardin and colleagues have identified separate regions of the per promoter that are required for its cycling pattern and wild-type expression levels (Hardin et al., 1992; Hao et al., 1997, 1999). They made transgenic lines carrying reporters under the control of various portions of the per promoter, focusing on the first 4 kb upstream of the transcription start site, which was shown to be sufficient for generating wild-type levels of per expression. A 69 bp fragment from the per promoter containing an E box site was found to be sufficient for rhythmic expression of per and rhythmic behavior in flies (Hao et al., 1999). However, the absolute levels of transcript produced by this fragment could not be tested since it was fused to two different heterologous basal promoter elements, which varied in their effect on expression. Previous work showed that 1.2 kb of the endogenous per promoter (containing this 69 bp fragment) was sufficient for rhythmic expression. However, this fragment yielded only one-fifth of wild-type levels of per transcript, whereas a 4.0 kb fragment of the promoter produced fully wild-type levels (Hardin et al., 1992). Therefore, the region between 1.2 and 4 kb upstream of the transcription start site is required for normal expression levels of per. There are three CREB binding sites, at positions −3210, −2990, and −1335, which lie in this region (P. Hardin, personal communication). The −1335 site, CGACGTCA, differs by only one base from the consensus CRE site (TGACGTCA), while the −3210 and −2990 sites contain the first five bases of the consensus site (TGACG), which are sufficient for CREB-mediated transcription (Sassone-Corsi, 1995). Since the S162 mutation reduces per–luc reporter expression, it is possible that dCREB2 contributes to per expression by acting through these sites. Further experiments will be needed to test this hypothesis and clarify the mechanism by which dCREB2 affects per expression.
We do not know if there are any CRE sites in the tim promoter. However, if dCREB2 affected the transcription of per only, it would still contribute to increased levels of the Per/Tim heterodimer, since Per protein is stabilized by the interaction with Tim (Gekakis et al., 1995). Analysis of the S162 mutation suggests that dCREB2, in addition to affecting per transcription (as measured using the per–luc and BG–luc reporters), is likely to affect other components of the clock. The levels of Per protein appear increased in the mutant background (Figures 6A and 6B), indicating that there is likely to be a compensatory change, reversing the effects on transcription. These other changes probably affect posttranslational processes (phosphorylation, dephosphorylation, and/or degradation).
Our results place dCREB2 in the clock mechanism and suggest that its function is to contribute to high levels of per expression. Other genes that affect per and tim act either at the transcriptional level (dClock and Cycle) or at the posttranslational level, as in the case of Double-time (Kloss et al., 1998; Price et al., 1998), which acts by phosphorylating Per, thereby accelerating its degradation. Our data is most consistent with a model in which dCREB2 also acts to maintain Per/Tim cycling. In this model, it acts on the per promoter, along with dClock and Cycle, to affect transcription of per, thereby modulating levels of the Per/Tim heterodimer. Our data also suggest that dCREB2 affects the levels of the Per protein through indirect posttranscriptional mechanism(s).
How Common Is Rhythmic Transcription?
Although only a small number of genes have been identified that control the clock, many have been identified whose transcription cycles in a circadian manner in response to the clock. Large-scale molecular screens for differentially expressed transcripts between day and night have yielded collections of genes in Drosophila (Van Gelder et al., 1995; Rouyer et al., 1997), Neurospora (Loros et al., 1989), and cyanobacteria (Liu et al., 1995) that are regulated by the circadian clock.
Recently, it has been demonstrated that cyclical transcription can also occur in a cell culture system when the cells are challenged with a high serum shock (Balsalobre et al., 1998). This result suggests that the machinery exists in cells to synchronize and maintain rhythmic transcription. Since the per–luc reporter gene can cycle in most tissues of the adult fly, it is reasonable to speculate that many genes may have rhythmic patterns in their activity, which when assayed in unsynchronized tissue culture cells appear to be independent of the circadian system. Indeed, in the fly it has been shown that many tissues are actually photoreceptive, and the oscillations of per activity in these tissues can be reset by light (Plautz et al., 1997). However, this is not likely to be the case in mammals, where light is unable to reach most tissues. Autonomous cycling of transcription in tissue and tissue culture cells need not imply that these actually function in vivo in an independent manner. Instead, it is more likely that in whole mammals, cells and tissues are capable of maintaining rhythmic transcription, but they are subservient to the central circadian system, which entrains them, insuring synchrony.
In the mammalian SCN, the immediate-early gene c-fos cycles in a circadian rhythm (Geusz et al., 1997). In addition, c-fos, as well as c-jun, junB, and junD, are upregulated in the SCN after a nighttime light pulse (Kornhauser et al., 1990, 1992; Rusak et al., 1992). fos is known to be regulated by CREB in a number of different contexts (Sassone-Corsi, 1995). Recent work in the hippocampus shows that brain-derived neurotrophic factor (BDNF) also cycles in its expression, with a peak during the nighttime (Berchtold et al., submitted). The regulation of BDNF is complex, with multiple promoter elements that determine transcription initiation. However, the only transcript that cycles is the one that is initiated from exon III, which is the promoter regulated by CREB. Therefore, rhythmic transcription mediated by CREB may be quite general.
What Is the Biological Significance of dCREB2’s Participation in the Clock?
Since a mutation in the dCREB2 gene affects the clock, the possibility exists that stimuli that activate CREB could affect circadian rhythmicity. This is clearly true for light pulses that reset the clock when delivered at the appropriate times during the nighttime period and have been shown to induce CREB phosphorylation in mammals (Ginty et al., 1993; D. Storm, personal communication). What about “noncircadian” stimuli? One example of this phenomenon is work by Amir and Stewart (1996) showing that rats can learn an association between air puffs delivered to the eye (conditioned stimulus) and light (unconditioned stimulus). After acquisition, the conditioned stimulus alone, when delivered during the nighttime period, phase shifts the clock. This suggests that there may be much more overlap in the conditioning circuitry and the circadian circuitry than is generally appreciated. Since the molecules within all of these neurons are similar, the molecular pathways may also overlap.
What Is the Biological Significance of the Circadian Clock Affecting CREB Activity?
CREB-mediated transcription occurs in response to stimuli that induce stress, long-term memory formation, and growth factor responses. What is the significance of the fact that this transcription factor also responds to circadian signals, which happen regularly over a 24 hr period? One speculation is that the cyclical pattern in CREB activity means that there are optimal periods during the 24 hr cycle for CREB-responsive physiological processes. One physiological process for which there is evidence that the nighttime period is important is the consolidation of long-term memory formation. Over the years, a large number of experiments, primarily on rodents and humans, suggest a possible involvement of some aspect of nighttime sleep in the consolidation of memory (Wilson and McNaughton, 1994; Karni et al., 1994; reviewed by Sejnowski, 1995; Dotto, 1996). A unifying interpretation of the physiological and behavioral data is that the brain utilizes the sleep period to “replay” plasticity-related events, thereby insuring their total consolidation and maintenance (Wilson and McNaughton, 1994). This replay occurs during sleep, when there is minimal external input into the brain. Our speculation is that the circadian system controls some of the neuronal activity that occurs during the sleep period. This activity in turn leads to activation of CREB-responsive transcription, which may be important in consolidating and maintaining preexisting circuits and memories. Experiments to test these types of ideas are underway.
Experimental Procedures
Reporter Constructs and Transgenic Lines
Standard recombinant DNA techniques were used to assemble the CRE–luc and mCRE–luc constructs. The enhancer-detecting vector pCaSpeR hs43 βgal was modified so that 3X CRE (TGACGTCA) or 3X mCRE (TGAAATCA) sites were placed in front of the hsp70 TATA box, and the lacZ gene was replaced with a modified luciferase gene. The details of the construction are available upon request. The entire cassette was subcloned into a P element vector insulated with the SCS and SCS9 insulator elements (Kellum and Schedl, 1992). The constructs were injected into Drosophila embryos to create transgenic lines as previously described (Rubin and Spradling, 1982). The hs-dCREB2–10 transgenic line was made from the dCREB2-d cDNA (Yin et al., 1995b).
Luciferase Assay
In vivo luciferase expression in Drosophila was measured as previously described (Brandes et al., 1996; Stanewsky et al., 1997), with the following modifications: each well of a black 96-well microtiter dish (Dynatech) was filled with 250 0μl of a 1% agar, 5% sucrose solution, followed by 100 μl of a 1% agar, 5% sucrose, 5 mM luciferin (BioSynth) solution. Flies were anesthetized with CO2 and then placed individually into the wells. The plates were covered with adhesive plastic sheets (Packard, Top-Seal), and air holes were punched over each well with a needle. The plates were returned to a 25°C incubator on a 12 hr light:12 hr dark cycle overnight. The following day, the plates were loaded into a Packard TopCount Microplate Scintillation and Luminescence Counter and maintained either on a 12 hr light:12 hr dark cycle (LD) or constant darkness (DD) according to the experiment. The flies were then cycled through the luminometer so that their luciferase expression was measured roughly every hour for the duration of the experiment. The data were imported into Microsoft Excel using the Import-and-Analysis program (J. Plautz and S. Kay, personal communication). The data was then analyzed using Microsoft Excel. The graphs shown are averages of multiple flies, usually between 20 and 40 per experiment. The averages are subjected to a smoothing curve, whereby each data point represents the average of itself plus the two data points on either side.
Rescue of S162 by Induction of dCREB2
S162/FM7 females were mated to +/Y; hs-dCREB2–10 males. Progeny from this cross were subjected to a daily 60 min heat shock at 37°C throughout the larval and pupal stages by submerging the vials in a water bath. Flies were maintained at 25°C prior to and following the heat shocks.
Locomotor Assays
The circadian locomotor activity of flies was assayed and analyzed as previously described (Hamblen et al., 1986; Sehgal et al., 1992). Periods were calculated by χ2 periodogram analysis.
Western Blotting
For all blots, flies were quickly frozen in liquid nitrogen. For analysis of dCREB2 proteins, fly heads were separated from bodies by sieving, and total head extracts were prepared by powdering the fly tissue in the cold, periodically dipping the tubes (which contained the tissue) in a liquid nitrogen bath. A standard 1X SDS sample buffer was added, and the samples were boiled. Samples were clarified by centrifugation prior to gel loading. SDS–polyacrylamide gel electrophoresis and Western blot analysis were carried out as described (Ausubel et al., 1989). The membranes were probed with a 1:20 dilution of dCREB2 mouse monoclonal antibody (27–118), followed by a goat anti-mouse HRP-conjugated secondary antibody (Bio-Rad), followed by chemiluminescent detection (Amersham). The monoclonal antibody was raised by injecting bacterially expressed, gel-purified, full-length dCREB2-b protein into mice. Monoclonal antibodies were generated from these mice using standard techniques. The Ser-230 phospho-specific antibody will be described elsewhere (J. Horiuchi and J. C. P. Y., unpublished data).
The Per and Tim Western analyses were performed on circadian entrained flies as previously described (Price et al., 1998). The equivalence of protein levels in different samples was established using cross-reacting bands on the Western, direct comparison of protein staining intensity on Coomassie gels, and reprobing of stripped blots with antibodies against other fly antigens.
Acknowledgments
We thank Jeff Plautz and Steve Kay for providing software to import the luminometer data to Microsoft Excel, and Jeff Plautz and Ralf Stanewsky for assistance with the luminometer assay. Mike Young provided the antibody to Timeless, and Ralf Stanewsky provided the Per antibody as well as suggestions for the Per Western blots. We thank Amita Sehgal and Joan Hendricks for valuable discussions and for help with the circadian locomotor assay. Sally Till and Jun Horiuchi participated in the preparation of the dCREB2 Ser-230 phospho-specific antibody, and Elizabeth Wilder and Norbert Perrimon helped with the initial characterization of S162. We also thank John Connolly, Josh Dubnau, Michael Regulski, Eric Drier, and an anonymous reviewer for insightful comments on the manuscript. This work was supported by National Institutes of Health grants 5RO1 NS3557 (J. C. P. Y.) and 1RO1 HL/AR59649 (Joan Hendricks, J. C. P. Y., and Amita Sehgal) and a McKnight Scholar’s Award to J. C. P. Y.
Footnotes
Note Added in Proof
The data referred to throughout as ‘‘D. Storm, personal communication,’’ are now in press: Obrietan, K., Impey, S., Smith, D., Athos, J., and Storm, D. (1999). Circadian regulation of cAMP response element–mediated gene expression in the suprachiasmatic nucleus. J. Biol. Chem., in press.
References
- Allada R, White NE, So WV, Hall JC, and Rosbash M (1998). A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93, 791–804. [DOI] [PubMed] [Google Scholar]
- Amir S, and Stewart J (1996). Resetting of the circadian clock by a conditioned stimulus. Nature 379, 542–545. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Kingston RE, Moore DD, Seidman JG, Smith JA, and Struhl K, eds. (1989). Current Protocols in Molecular Biology, Sections 10.2 and 10.8 (New York: Wiley Interscience; ). [Google Scholar]
- Balsalobre A, Damiola F, and Schibler U (1998). A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937. [DOI] [PubMed] [Google Scholar]
- Borsook D, Konradi C, Falkowski O, Comb M, and Hyman SE (1994). Molecular mechanisms of stress-induced proenkephalin gene regulation: CREB interacts with the proenkephalin gene in the mouse hypothalamus and is phosphorylated in response to hyperosmolar stress. Mol. Endocrinol. 8, 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchuladze R, Frenguelli B, Blendy J, Coffi D, Schutz G, and Silva AJ (1994). Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 79, 59–68. [DOI] [PubMed] [Google Scholar]
- Brandes C, Plautz JD, Stanewsky R, Jamison CF, Straume M, Wood KV, Kay SA, and Hall JC (1996). Novel features of Drosophila period transcription revealed by real-time luciferase reporting. Neuron 16, 687–692. [DOI] [PubMed] [Google Scholar]
- Carlezon WA, Thome J, Olson VG, Lane-Ladd SB, Brodkin ES, Hiroi N, Duman RS, Neve RL, and Nestler EJ (1998). Regulation of cocaine reward by CREB. Science 282, 2272–2275. [DOI] [PubMed] [Google Scholar]
- Citri Y, Colot HV, Jacquier AC, Yu Q, Hall JC, Baltimore D, and Rosbash M (1987). A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature 326, 42–47. [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, and Kay SA (1998). Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim [see comments]. Science 280, 1599–1603. [DOI] [PubMed] [Google Scholar]
- Dotto L (1996). Sleep stages, memory and learning. Can. Med. Assoc. J. 154, 1193–1196. [PMC free article] [PubMed] [Google Scholar]
- Dubnau J, and Tully T (1998). Gene discovery in Drosophila: new insights for learning and memory. Annu. Rev. Neurosci. 21, 407–444. [DOI] [PubMed] [Google Scholar]
- Eberl DF, Perkins LA, Engelstein M, Hilliker AJ, and Perrimon N (1992). Genetic and developmental analysis of polytene section 17 of the X chromosome of Drosophila melanogaster. Genetics 130, 569–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I, Zweibel LJ, Dembinska ME, and Rosbash M (1994). Temporal phosphorylation of the Drosophila period protein. Proc. Natl. Acad. Sci. USA 91, 2260–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery IF, Noveral JM, Jamison CF, and Siwicki KK (1997). Rhythms of Drosophila period gene expression in culture. Proc. Natl. Acad. Sci. USA 94, 4092–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes NS, Duval G, and Sassone-Corsi P (1996). Adaptive inducibility of CREM as transcriptional memory of circadian rhythms. Nature 381, 83–85. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Saez L, Delahaye-Brown AM, Myers MP, Sehgal A, Young MW, and Weitz CJ (1995). Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL [see comments]. Science 270, 811–815. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, and Weitz CJ (1998). Role of the CLOCK protein in the mammalian circadian mechanism [see comments]. Science 280, 1564–1569. [DOI] [PubMed] [Google Scholar]
- Geusz ME, Fletcher C, Block GD, Straume M, Copeland NG, Jenkins NA, Kay SA, and Day RN (1997). Long-term monitoring of circadian rhythms in c-fos gene expression from suprachiasmatic nucleus cultures. Curr. Biol. 7, 758–766. [DOI] [PubMed] [Google Scholar]
- Ginty DD, Kornhauser JM, Thompson MA, Bading H, Mayo KE, Takahashi JS, and Greenberg ME (1993). Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science 260, 238–241. [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, and Montminy MR (1989). Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59, 675–680. [DOI] [PubMed] [Google Scholar]
- Hamblen M, Zehring WA, Kyriacou CP, Reddy P, Yu Q, Wheeler DA, Zwiebel LJ, Konopka RJ, Rosbash M, and Hall JC (1986). Germ-line transformation involving DNA from the period locus in Drosophila melanogaster: overlapping genomic fragments that restore circadian and ultradian rhythmicity to per0 and per2 mutants. J. Neurogenet. 3, 249–291. [DOI] [PubMed] [Google Scholar]
- Hao H, Allen DL, and Hardin PE (1997). A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol. Cell. Biol. 17, 3687–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Glossop NRJ, Lyons L, Qiu J, Morrish B, Cheng Y, Helfrich-Förster C, and Hardin P (1999). The 69 bp circadian regulatory sequence (CRS) mediates per-like developmental, spatial, and circadian expression and behavioral rescue in Drosophila. J. Neurosci. 19, 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE (1994). Analysis of period mRNA cycling in Drosophila head and body tissues indicates that body oscillators behave differently from head oscillators. Mol. Cell. Biol. 14, 7211–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, and Rosbash M (1990). Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343, 536–540. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, and Rosbash M (1992). Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc. Natl. Acad. Sci. USA 89, 11711–11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S (1994). A reconsideration of the mechanism of position effect. Genetics 138, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JH, Edery I, and Rosbash M (1993). PAS is a dimerization domain common to Drosophila Period and several transcription factors. Nature 364, 259–262. [DOI] [PubMed] [Google Scholar]
- Hunter-Ensor M, Ousley A, and Sehgal A (1996). Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell 84, 677–685. [DOI] [PubMed] [Google Scholar]
- Jackson FR, Bargiello TA, Yun SH, and Young MW (1986). Product of per locus of Drosophila shares homology with proteoglycans. Nature 320, 185–188. [DOI] [PubMed] [Google Scholar]
- Karni A, Tanne D, Rubenstein BS, Askenasy JJM, and Sagi D (1994). Dependence on REM sleep of overnight improvement of a perceptual skill. Science 265, 679–682. [DOI] [PubMed] [Google Scholar]
- Kellum R, and Schedl P (1992). A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 12, 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, and Young MW (1998). The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell 94, 97–107. [DOI] [PubMed] [Google Scholar]
- Konopka RJ, and Benzer S (1971). Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 68, 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornhauser JM, Nelson DE, Mayo KE, and Takahashi JS (1990). Photic and circadian regulation of c-fos gene expression in the hamster suprachiasmatic nucleus. Neuron 5, 127–134. [DOI] [PubMed] [Google Scholar]
- Kornhauser JM, Nelson DE, Mayo KE, and Takahashi JS (1992). Regulation of jun-B messenger RNA and AP-1 activity by light and a circadian clock. Science 255, 1581–1584. [DOI] [PubMed] [Google Scholar]
- Lee C, Parikh V, Itsukaichi T, Bae K, and Edery I (1996). Resetting the Drosophila clock by photic regulation of PER and a PER–TIM complex [see comments]. Science 271, 1740–1744. [DOI] [PubMed] [Google Scholar]
- Liu X, Lorenz L, Yu QN, Hall JC, and Rosbash M (1988). Spatial and temporal expression of the period gene in Drosophila melanogaster. Genes Dev. 2, 228–238. [DOI] [PubMed] [Google Scholar]
- Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, Ishiura M, and Kondo T (1995). Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 9, 1469–1478. [DOI] [PubMed] [Google Scholar]
- Loros J, Denome S, and Dunlap J (1989). Molecular cloning of genes under the control of the circadian clock in Neurospora. Science 243, 385–388. [DOI] [PubMed] [Google Scholar]
- Majercak J, Kalderon D, and Edery I (1997). Drosophila melanogaster deficient in protein kinase A manifests behavior-specific arrhythmia but normal clock function. Mol. Cell. Biol. 17, 5915–5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil GP, Zhang X, Genova G, and Jackson FR (1998). A molecular rhythm mediating circadian clock output in Drosophila. Neuron 20, 297–303. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, and Baltimore D (1989). A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56, 777–783. [DOI] [PubMed] [Google Scholar]
- Myers MP, Wager-Smith K, Wesley CS, Young MW, and Sehgal A (1995). Positional cloning and sequence analysis of the Drosophila clock gene, timeless [see comments]. Science 270, 805–808. [DOI] [PubMed] [Google Scholar]
- Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, and Young MW (1996). Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock [see comments]. Science 271, 1736–1740. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, and Kay SA (1997). Independent photoreceptive circadian clocks throughout Drosophila [see comments]. Science 278, 1632–1635. [DOI] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, and Young MW (1998). double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94, 83–95. [DOI] [PubMed] [Google Scholar]
- Reddy P, Zehring WA, Wheeler DA, Pirrotta V, Hadfield C, Hall JC, and Rosbash M (1984). Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 38, 701–710. [DOI] [PubMed] [Google Scholar]
- Rouyer F, Rachidi M, Pikielny C, and Rosbash M (1997). A new gene encoding a putative transcription factor regulated by the Drosophila circadian clock. EMBO J. 16, 3944–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, and Spradling AC (1982). Genetic transformation of Drosophila with transposable element vectors. Science 218, 348–353. [DOI] [PubMed] [Google Scholar]
- Rusak B, McNaughton L, Robertson HA, and Hunt SP (1992). Circadian variation in photic regulation of immediate-early gene mRNAs in rat suprachiasmatic nucleus cells. Brain Res. Mol. Brain Res. 14, 124–130. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, and Hall JC (1998). CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93, 805–814. [DOI] [PubMed] [Google Scholar]
- Saez L, and Young MW (1988). In situ localization of the per clock protein during development of Drosophila melanogaster. Mol. Cell. Biol. 8, 5378–5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P (1995). Transcription factors responsive to cAMP. Annu. Rev. Cell Dev. Biol. 11, 355–377. [DOI] [PubMed] [Google Scholar]
- Segal RA, and Greenberg ME (1996). Intracellular signaling pathways activated by neurotrophic factors. Annu. Rev. Neurosci. 19, 463–489. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Price J, and Young MW (1992). Ontogeny of a biological clock in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 89, 1423–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Price JL, Man B, and Young MW (1994). Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless [see comments]. Science 263, 1603–1606. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Rothenfluh-Hilfiker A, Hunter-Ensor M, Chen Y, Myers MP, and Young MW (1995). Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation [see comments]. Science 270, 808–810. [DOI] [PubMed] [Google Scholar]
- Sejnowski TJ (1995). Sleep and memory. Curr. Biol. 5, 832–834. [DOI] [PubMed] [Google Scholar]
- Self DW, and Nestler EJ (1995). Molecular mechanisms of drug reinforcement and addiction. Annu. Rev. Neurosci. 18, 463–495. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Kogan JH, Frankland PW, and Kida S (1998). CREB and memory. Annu. Rev. Neurosci. 21, 127–148. [DOI] [PubMed] [Google Scholar]
- Siwicki KK, Eastman C, Petersen G, Rosbash M, and Hall JC (1988). Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron 1, 141–150. [DOI] [PubMed] [Google Scholar]
- Stanewsky R, Jamison CF, Plautz JD, Kay SA, and Hall JC (1997). Multiple circadian-regulated elements contribute to cycling period gene expression in Drosophila. EMBO J. 16, 5006–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle JH, Foulkes NS, Molina CA, Simonneaux V, Pevet P, and Sassone-Corsi P (1993). Adrenergic signals direct rhythmic expression of transcriptional repressor CREM in the pineal gland [see comments]. Nature 365, 314–320. [DOI] [PubMed] [Google Scholar]
- Suri V, Qian Z, Hall JC, and Rosbash M (1998). Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron 21, 225–234. [DOI] [PubMed] [Google Scholar]
- Tan Y, Rouse J, Zhang A, Cariati S, Cohen P, and Comb MJ (1996). FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 15, 4629–4642. [PMC free article] [PubMed] [Google Scholar]
- Udvardy A, Maine E, and Schedl P (1985). The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol. 185, 341–358. [DOI] [PubMed] [Google Scholar]
- Usui T, Smolik SM, and Goodman RH (1993). Isolation of Drosophila CREB-B: a novel CRE-binding protein. DNA Cell Biol. 12, 589–595. [DOI] [PubMed] [Google Scholar]
- Van Gelder RN, Bae H, Palazzolo MJ, and Krasnow MA (1995). Extent and character of circadian gene expression in Drosophila melanogaster: identification of twenty oscillating mRNAs in the fly head. Curr. Biol. 5, 1424–1436. [DOI] [PubMed] [Google Scholar]
- Vazquez J, and Schedl P (1994). Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J. 13, 5984–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widnell KL, Russell DS, and Nestler EJ (1994). Regulation of expression of cAMP response element–binding protein in the locus coeruleus in vivo and in a locus coeruleus–like cell line in vitro. Proc. Natl. Acad. Sci. USA 91, 10947–10951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, and McNaughton BL (1994). Reactivation of hippocampal ensemble memories during sleep. Science 265, 676–679. [DOI] [PubMed] [Google Scholar]
- Yang Z, Emerson M, Su HS, and Sehgal A (1998). Response of the timeless protein to light correlates with behavioral entrainment and suggests a nonvisual pathway for circadian photoreception. Neuron 21, 215–223. [DOI] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, and Tully T (1994). Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell 79, 49–58. [DOI] [PubMed] [Google Scholar]
- Yin JC, Del Vecchio M, Zhou H, and Tully T (1995a). CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell 81, 107–115. [DOI] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Wilder EL, Klingensmith J, Dang D, Perrimon N, Zhou H, Tully T, and Quinn WG (1995b). A Drosophila CREB/CREM homolog encodes multiple isoforms, including a cyclic AMP–dependent protein kinase–responsive transcriptional activator and antagonist. Mol. Cell. Biol. 15, 5123–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Qian Z, Myers MP, and Rosbash M (1996). A light-entrainment mechanism for the Drosophila circadian clock. Nature 380, 129–135. [DOI] [PubMed] [Google Scholar]
- Zwiebel LJ, Hardin PE, Liu X, Hall JC, and Rosbash M (1991). A post-transcriptional mechanism contributes to circadian cycling of a per-β-galactosidase fusion protein. Proc. Natl. Acad. Sci. USA 88, 3882–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]