Abstract
The marine microbiome arouses an increasing interest, aimed at better understanding coral reef biodiversity, coral resilience, and identifying bioindicators of ecosystem health. The present study is a microbiome mining of three environmentally contrasted sites along the Hermitage fringing reef of La Réunion Island (Western Indian Ocean). This mining aims to identify bioindicators of reef health to assist managers in preserving the fringing reefs of La Réunion. The watersheds of the fringing reefs are small, steeply sloped, and are impacted by human activities with significant land use changes and hydrological modifications along the coast and up to mid-altitudes. Sediment, seawater, and coral rubble were sampled in austral summer and winter at each site. For each compartment, bacterial, fungal, microalgal, and protist communities were characterized by high throughput DNA sequencing methodology. Results show that the reef microbiome composition varied greatly with seasons and reef compartments, but variations were different among targeted markers. No significant variation among sites was observed. Relevant bioindicators were highlighted per taxonomic groups such as the Firmicutes:Bacteroidota ratio (8.4%:7.0%), the genera Vibrio (25.2%) and Photobacterium (12.5%) dominating bacteria; the Ascomycota:Basidiomycota ratio (63.1%:36.1%), the genera Aspergillus (40.9%) and Cladosporium (16.2%) dominating fungi; the genus Ostreobium (81.5%) in Chlorophyta taxon for microalgae; and the groups of Dinoflagellata (63.3%) and Diatomea (22.6%) within the protista comprising two dominant genera: Symbiodinium (41.7%) and Pelagodinium (27.8%). This study highlights that the identified bioindicators, mainly in seawater and sediment reef compartments, could be targeted by reef conservation stakeholders to better monitor La Réunion Island’s reef state of health and to improve management plans.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00248-025-02495-3.
Keywords: Microbiome, Bioindicators, Fringing coral reef, La Réunion Island
Introduction
Coral reefs are one of the most diverse ecosystems on the planet, providing a vital habitat for a multitude of marine species [1]. However, these ecosystems are facing increasing threats, including climate change, and impacts of local human activities. Those disturbances are compromising their health, their stability, and in turn, their abilities to provide ecosystem services [2]. At the heart of their functioning lies an often overlooked but crucial component: the microbiome. The microbiome, a complex set of microorganisms (i.e., viruses, bacteria including cyanobacteria, fungi, algae, and protists) interacting with the environment and host organisms, plays a crucial role in coral health and reef resilience to global change and anthropogenic pressures [3]. Thus, the coral microbiome composition, function, and diversity have received a growing interest to better understand mechanisms by which corals respond to environmental stresses [4] and to develop more effective conservation strategies based on microbial bioindicators [5]. By monitoring changes in the microbiome diversity, community composition, structure, and functions of the microbiome in a given environment or organism, it is possible to identify early indicators of stress or decline as shown in a few previously studied coral reefs recently [3, 6, 7]. Most of the attention has been given to two reef compartments: the coral holobiont, which comprises the animal tissue, its microbial endosymbionts [8–10], and the surrounding water column (e.g., [11, 12]). The coral skeleton comprises microbes within its pores and cracks (chasmo- and crypto-microbial organisms; see [13]) and those dissolving and living inside the aragonite (eu-endoliths or bioeroding microflora; see [14–16]) have received in contrast, much less attention. But this coral compartment has raised more interest a decade ago as some microorganisms such as bioeroding microflora (see review by [16–19]) may play an important role in coral survival and resilience in a changing and warming environment by recycling nutrients [9, 20], providing photoassimilates [21] or by reducing skeletal reflectance during bleaching events [22].
In general, most studies on reef microbiome focused on one taxon of microbes (mainly bacteria) in a specific compartment (coral tissues, sponges, or water column for instance), and/or rarely how the diversity of a specific taxon diversity varied among sites or environmental conditions [23]. Very recently, studies investigated reef microbiomes, mainly bacteria, at different spatial scales: from multiple organisms at one specific site, to multiple compartments at several reef sites among ocean basins [24]. This allowed revealing the major contribution of the reef bacterial microbiome to the Earth’s prokaryotic diversity [25] and its variability among ocean basins and to a lesser degree, among sites (due to environmental variations). But rare studies [7, 10, 26] have investigated simultaneously the variability of the diversity of several microbial taxa in diverse compartments using multi-markers at different sites and over time. Indeed, to our knowledge, only Marcelino and Verbruggen [10] studied the endolithic algae in skeletons of several coral genera in different habitats and sites while [7, 26] investigated the bacterial diversity among reef compartments, sites, and over time. Glasl et al. [7] showed that the bacterial microbiome of the water compartment appears as the most relevant compartment to monitor reef environmental changes. However, those authors did not explore the potential of the other microbial taxa (fungi, algae, and protists) as they only sequenced the bacterial 16S rRNA genes. Although Glasl et al. [7] were the first to our knowledge to provide a data baseline of the variability of bacterial communities in diverse compartments (water, sediments, live corals, sponges, macroalgae) at three different reef sites on the Great Barrier Reef over 16 months, to our knowledge, no study has been carried out to understand the variability of the whole microbial community, including at the time bacteria, fungi, algae, and protists, of different reef compartments at different sites and during two seasons. Indeed, other taxa such as fungi, algae, and protists play crucial roles in reef ecosystem functioning and therefore may have significant potential as bioindicators of reef health [23, 27, 28]. For instance, as many marine fungi could have a terrestrial origin and are organotrophs (thus depending on organic matter [29, 30]), they may be good indicators of terrigenous inputs. Microalgae, on the other hand, are especially influenced by light, temperature, and nutrients and thus can be influenced by freshwater and terrigenous runoffs or upwellings [16, 31, 32]. Additionally, some bioeroding microalgae (e.g., Ostreobium sp., [33]) were shown to respond positively to changes in seawater pH and nutrients [33, 34] suggesting a potential utility as bioindicators of reef environment changes. Protists also hold unexplored potential in this context, as they play critical roles in ecosystem functioning [23, 35] such as primary production, organic matter remineralization (nutrient cycling), and coral thermal-stress adaptation [36–38].
It is crucial to consider the temporal dimension when studying microbial communities, as natural successions occur over time due to biotic interactions, such as competition and predation, alongside abiotic factors like temperature, pH, and nutrient availability [26, 39, 40]. These dynamic changes make it challenging to disentangle the influences of biotic vs. abiotic drivers on microbial composition. To identify reliable bioindicators of environmental changes, it is essential to be able to differentiate the role of natural community shifts from the influence of environmental changes [39]. In tropical coastal environments such as coral reefs, seasonality is well marked and plays a pivotal role in shaping microbial communities [26] as variations in temperature, rainfall, and nutrient influx between summer and winter influence significantly microbial dynamics and ecosystem processes [39, 40].
Here, by DNA high-throughput sequencing using four different primers (16S, 18S, ITS, and tufA, this latter being specific to endolithic phototrophic chlorophytes in corals; see [41]), we examined the microbiome composition (pro- and eukaryotic microorganisms) of seawater, sediments, and coral rubble compartments at three environmentally contrasted sites on the coral fringing reef system located in the west coast of La Réunion Island, at both seasons (austral summer and austral winter). Our main goal was to identify potential bioindicators within each microbial studied taxon to improve the management strategies of local authorities and to better protect the ecosystem.
Material and Methods
Site Description and Sampling Methods
Located at 55° east, 21° south, La Réunion Island is part of the Mascarenes archipelago in the southwestern Indian Ocean, approximately 700 km east of Madagascar and 180 km southwest of Mauritius (Fig. 1). This young and active volcanic tropical island, estimated to be around 2 million years old, covers an area of 2500 km2 and reaches an altitude of 3800 m, representing only about 3% of the volcanic cone [42]. The watersheds, which are small and steeply sloped, are heavily impacted by human activities with significant land use changes with the expansion of agricultural areas and urbanization and hydrological modifications up to mid-altitudes. The coral reefs, which are primarily fringing reefs, and the highlands of the island are respectively protected and managed by the La Réunion Marine Nature Reserve.
Fig. 1.
Location map of La Réunion Island in the southwest Indian Ocean close to Madagascar and Mauritius Island, and map of La Réunion Island including the two major cities and the study site location (red rectangle). Location of the three sampling sites (Toboggan, Copacabana, and Livingstone), near l’Hermitage-les-Bains city, on La Réunion Island. The right side shows the three sampling conditions (seawater, coral rubble, and sediment). Landsat satellite image, worldwide map from creative commons, pictures from PLS
La Réunion Island is therefore considered a biodiversity hotspot [43]. Like most young volcanic tropical islands, recent fringing reefs have emerged (12 km2 surface, 25 km long, and around 10,000 years old). The largest reef part (9 km long, 500 m wide on average, and 1 m depth on average) on La Réunion Island is located on the western coast of the island, at La Saline (Fig. 1; [44]). Groundwater, flowing into this fringing reef from volcanic aquifers [45], introduces nitrate discharges from human activities [46] and leads to eutrophication of some parts of the fringing reefs with significant carbon, nitrogen, phosphorus, and oxygen flows [44]. Groundwater inputs also influence the distribution of fluorescent dissolved organic matter (FDOM) and contribute to the release of pollutants such as hydrocarbons in the reef ecosystem [47, 48]. Eutrophication can affect the reef carbonate budget as it increases rates of bioerosion [15, 49] and can stimulate to a certain extent coral calcification under certain [50]. This local disturbance also influences the distribution of coral and algal communities [51]. Due to its geomorphology, especially the presence of several channels, but also its shallow depth and hydrodynamism, Lagoutte et al. [52] showed that part of the water exiting the fringing system through channels is immediately re-entrained onto the reef flat. Seawater parameters on the fringing reef of La Saline are therefore strongly influenced by both benthic community metabolic activity and physical parameters (groundwater discharge, reef geomorphology, hydrodynamism, depth, and water residence time). These stressors, notably freshwater discharges (rivers and groundwater), have been extensively documented in prior studies [47]. Submarine groundwater discharge introduces significant levels of nutrients and pollutants in the southern part of the back-reef area, making it representative of degraded conditions. Eutrophication of this part of the reef results in a higher phytoplankton biomass, massive presence of fleshy algal formations, and low coral coverage. In contrast, the Toboggan site is located in an area with lower anthropogenic influence; this site serves as a comparative example of relatively pristine conditions [47].
Samples of seawater, sediment, and coral rubble (i.e., loose coral rubble pieces on the reef floor) were thus collected at three sites on the La Saline fringing reef: Toboggan (21°04′49″ S; 55°13′16″ E), Copacabana (21°05′38″ S; 55°13′58″ E), and Livingstone (21°05′52″ S; 55°14′18″ E) (Fig. 1). Collection of samples occurred in July 2021 (austral winter) and January 2022 (austral summer). Samples were processed according to the methods provided by [35, 53]. Briefly, all samples were collected at one m depth. Dead rubble and sediments (around 100 g) were collected in sterile 250-ml polypropylene straight red cap containers (Corning/Dutscher, Bernolsheim, France) while seawater was sampled in sterile 5-l flasks (the sample was taken at a depth of 1 m, bearing in mind that the water column at these points is around 1.5 to 2 m deep at high tide). All samples were collected in triplicates. Coral rubble and sediments were conserved at − 20 °C while seawater samples were filtered immediately after collection, onto sterile 0.22 µm nitrocellulose membranes (Millipore/Merck, Burlington, MA, USA) and then stocked at − 20 °C in sterile 15-ml Falcon tube (Falcon/Dutscher, Bernolsheim, France).
Molecular Methods
DNA Extraction
Sediment samples were thawed at room temperature and then mixed thoroughly to ensure homogeneity. Coral debris samples were ground into powder using a Mixer Mill MM 400 (Retsch Gmbh, Haan, Germany). For sediment and ground coral rubble debris samples, DNA extractions were performed using 2 g of sediment or coral debris powder with the Qiagen DNAeasy PowerSoil ProKit (Qiagen, Hilden, Germany). For seawater samples filtered on membranes, the DNA extractions were performed with the Qiagen DNAeasy Power Water ProKit (Qiagen, Hilden, Germany). DNA samples were controlled for quality and quantification using a NanoDrop (Thermo Fischer scientific, Illkirch-Graffenstaden, France). Negative extraction controls were performed alongside sample extractions for seawater, sediment, and coral rubble samples, adding nothing in place of the sample in the first extraction step.
Libraries Generation and Sequencing
DNA samples were sent for new generation sequencing (NGS) to the Microsynth Sequencing Platform (Microynsth, Vaux en Velin, France). Microbiome diversity was assessed as follows: the V3V4 region of the bacterial 16S RNA gene was used to characterize the bacterial community using the primers 341F 5′-CCTACGGGNGGCWGCAG-3′ and 805R 5′-GACTACHVGGGTATCTAATCC-3′ [54]. The ITS2 region of the 18S nuclear ribosomal RNA gene was used to characterize the fungal community using the primers 18S-Fwd-ITS7 5′-GTGARTCATCGAATCTTTG-3′ [55] and 18S-Rev-ITS4 5’-TCCTCCGCTTATTGATATGC-3′ [56]. The V3/V4 of the 18S nuclear ribosomal RNA gene was used to characterize the Eukaryota community including alga and protist using the primers 515F 5′-GTGCCAGCMGCCGCGG-3′ [57] and Ek-NSR951 5 ′-TTGGYRAATGCTTTCGC-3′ [58]. The plastid elongation factor tufA (chloroplast) for the microalgae community was amplified using the primers env_tufAF 5′-GGGTDGAHAADATTTWYNMNYTRATGR-3′ and env_tufAR 5 ′-TNACATCHGTWGTWCKNACATARAAYTG-3 [41].
PCR amplification n°1 was run by 30 cycles for 16S V3–V4, by 35 cycles for 18S V4, by 35 cycles for ITS2, and by 35 cycles for tufA. Quality control was carried out with quantification by PicoGreen and qualification on QIAxcel. The PCR mix was 5 µl or 25 ng of gDNA, “5X HOT BIOAmp® BlendMaster Mix—12.5 mM MgCl2” from Microsynth—“10X GC-rich Enhancer” from Microsynth—BSA 20 mg/ml; with a final volume of 25 µl, indexing PCR n°2 amplification 10 cycles of PCR n°1 and quality control. Next, an equimolar standardization of the sequencing libraries and the creation of the equimolar library pool were carried out. Next, deposit the sequencing library pool with 5% phiX on a V3 Flow Cell. This was sequenced on a MiSeq flow cell in paired-end 2 × 300 bp (301 × 8 × 8 × 301). A total of 4,473,087; 4,112,153; 4,035,361; and 7,615,342 paired reads were obtained respectively for ITS2 (18S), V4 (16S), V1/V3 (18S), and tufA independent sequencing runs. Sequences are available under the NCBI BioProject PRJNA985136, BioSample SUB13558791, from SRR24958245 to SRR24958195 for the 16S, from SRR25108721 to SRR25108698 for the ITS, from SRR25080909 to SRR25080857 for the tufA, and from SRR24961358 to SRR24961336 for the 18S.
Bioinformatics
Working Environment
The pipeline was run on the Nouméa Institut de Recherche pour le Développement’s cluster, running under CentOS Linux release 8.3.2011, and then downstream analysis proceeded on macOS Mojave 10.14.6 (× 86_64-apple-darwin17.0 (64-bit)). A tailored bioinformatic workflow developed for this project can be found in Supplementary Fig. S1. All scripts created and used for this pipeline can be found at https://github.com/PLStenger/BioIndic_La_Reunion_Island_Lagoon.
Pre-processing
First, raw Illumina sequences from the V4 (16S), ITS2 (18S), tufA (chloroplast), and V1/V3 (18S) datasets quality were assessed with FastQC V. 0.11.9 [59], and multi reports were generated using MultiQC V. 1.10.1 [60]. Reads were cleaned and adaptors were removed with Trimmomatic [61] (V. 0.39—illuminaclip 2:30:10; leading 30, trailing 30, and minlen 150). FastQC V. 0.11.9 [59] and MultiQC V. 1.10.1 [60] were reused to check the data after this cleaning step. The number of raw sequences is available as Supplementary Table S1.
Qiime2 Framework
Microbiome analysis was performed using the QIIME 2 framework V. 2021.4.0 [62]. Dereplicated and trimmed sequences were imported into the framework as paired-end (Phred33V2) sequences and denoised using the DADA2 plugin, based on the DADA2 V. 1.8 R library [63], which removed singletons, chimeras, and sequencing errors and processed the sequences into a table of exact amplicon sequence variants (ASVs) [64]. Negative control library sequences, as putative contaminant sequences, were removed from each sample sequence [65]. ASVs that were present in only a single sample were filtered based on the idea that these may not represent real biological diversity but rather PCR or sequencing errors [66]. ASV abundance of raw data is available in Supplementary Table S1. After this contingency step, all samples were rarefied according to the alpha-rarefaction QIIME2 tool with a maximum depth of 16,708, 18,908, 64,625, and 13,250, respectively, for V4 (16S), ITS2 (18S), tufA (chloroplast), and V1/V3 (18S) datasets, with the Shannon entropy (a measure of richness and diversity that accounts for both the abundance and evenness of taxa) [67] and Faith PD (a measure of biodiversity that incorporates phylogenetic differences between species using the sum of the lengths of branches) [68]. A depth value of 137, 4202, 7,181, and 2,945, respectively, for V4 (16S), ITS2 (18S), tufA (chloroplast), and V1/V3 (18S) datasets (Supplementary Table S1) were obtained for these rarefactions Supplementary Fig. S2. A multiple sequence alignment was produced using MAFFT V. 7.310 [69], and a rooted phylogenetic tree relating the ASV sequences to one another was constructed using FastTree V. 2.1.10 [70]. Naive Bayes feature classifiers were trained using the q2-feature-classifier tool to assign taxonomy to the sequences [71]. For the fungal classifier training (only for ITS2 (18S)), the new fungal UNITE ITS reference set [72, 73] was used. QIIME pre-formatted database with dynamic homology clustering was used. As recommended by the QIIME 2 development team, the fungal classifier was trained on the full reference sequences. For bacterial classifier training (only for V4 (16S)), the SILVA-138-SSURef-Full-Seqs 1QIIME pre-formatted database (SILVA-138-SSURef-Full-Seqs.qza as DataSeq.qza and Silva-200 v138-full-length-seq-taxonomy.qza as RefTaxo.qza; see https://github.com/mikerobeson/make_SILVA_db) was used. For hook the V4 part of the 16S region, we used the forward 341F sequence ('CCTACGGGNGGCWGCAG') and the reverse 805R sequence (“GACTACHVGGGTATCTAATCC”) in the feature-classifier extract-reads tool. For the tufA marker, we used the rescript get-ncbi-data QIIME2 plugin to obtain the largest possible number of comparable sequences, as recent as possible with the query “(tufA[ALL] OR TufA[ALL] OR TUFA[ALL] OR tufa[ALL] NOT bacteria[ORGN])).” For eukaryota classifier training (only for V3/V4 (18S)), the same pre-formatted database as for V4 (16S) was used. For hook the V3/V4 part of the 18S region, we used the forward 515F sequence (“GTGCCAGCMGCCGCGG”) and the reverse 951R sequence (“TTGGYRAATGCTTTCGC”) in the feature-classifier extract-reads tool. The choice of phylum kept for this study was based on [74–76].
Statistical Analysis
First, we assessed and compared the classical triptych “microbial diversity, community composition, and structure” (see sections below for details) among the three studied sites (Toboggan, Copacabana, and Livingstone), reef compartments (coral rubble, seawater, and sediment), and seasons (austral summer and winter) for each marker (bacteria studied with the marker16S-V3/V4, fungi with the marker18S-ITS2, microalgae with the marker tufA and protists with the marker 18S-V4). As the methodology presented above did not yield significant results for site comparisons (results not shown here—but see Supplementary Table S1 and Fig. S7–S10), we developed the methodology below only for comparisons between compartments and seasons. In total, six conditions compartment + season were thus compared: S-CO (summer-coral), W-CO (winter-coral), S-SD (summer-sediment), W-SD (winter-sediment), S-SW (summer-seawater), and W-SW (winter-seawater). We developed this methodology below to highlight variations and then potential bioindicators in microbial diversity and community composition differences among compartments and between seasons and the potential interactions between those two factors.
In the second step, we use Bayesian statistics (with a priori) (see sections below for details) with a focus on the compartments and then on the season to focus not only on the most abundant groups but also on those that are most significantly linked to a given condition (SW, CO or SD in compartments, then austral summer or winter in seasons). These will enable us to add reliable bioindicators strongly linked to these conditions. All statistical analyses were performed using the R software environment V. 4.1.0 [77].
Diversity of Microorganisms in Reef Compartment Through the Seasons
The alpha diversity of each sample (the basic unit of study corresponding to a spatio (site COP, LIV, or TOB)-temporal point (season S or W) of a particular compartment (SW, CO or SD): S-CO, W-CO, S-SD, W-SD, S-SW, and W-SW) was characterized by using the observed number of ASVs [78] and the Chao1 index (expected richness taking into account the occurrence of rare species [79]). Diversity was assessed by considering both species richness and the distribution of ASVs among species. The evenness of species abundance was measured using Pielou evenness [80]. Shannon entropy [67] was used as a measure of diversity accounting for both richness and evenness. Additionally, phylogenetic diversity was quantified using Faith’s PD [68], which incorporates the evolutionary relationships among species. Then the effects of reef compartments and seasons on the above diversity metrics were tested using Kruskal–Wallis tests. Diversity index and the significance of their comparison (i.e., effects of reef compartments + seasons − sites results not shown) in post hoc analyses were obtained using the agricolae R package V. 1.3–5 [81]—see Table 1 and Supplementary Fig. S3–6. Boxplots were realized with the ggplot2 R package V. 3.3.5 [82].
Table 1.
Bacteria diversity index. Lowercase letters indicate significant differences among conditions based on pairwise comparisons following a Kruskal-Wallis test (P value < 0.05). S, austral summer; W, austral winter; CO, coral rubble; SW, seawater; SD, sediments
| Condition | Observed ASV | Chao1 | Simpson | Shannon | Faith PD | Pielou evenness |
|---|---|---|---|---|---|---|
| S-CO | 6.13 ± 2.10 (c) | 6.13 ± 2.10 (c) | 0.72 ± 0.14 (c) | 2.18 ± 0.62 (c) | 1.06 ± 0.29 (ab) | 0.87 ± 0.06 (a) |
| S-SW | 12.33 ± 3.84 (c) | 12.56 ± 4.30 (c) | 0.87 ± 0.06 (c) | 3.23 ± 0.53 (c) | 0.85 ± 0.32 (a) | 0.91 ± 0.06 (a) |
| S-SD | 3.67 ± 1.51 (b) | 3.67 ± 1.51 (b) | 0.53 ± 0.20 (b) | 1.40 ± 0.63 (b) | 0.85 ± 0.33 (ab) | 0.78 ± 0.21 (a) |
| W-CO | 8.13 ± 10.56 (a) | 8.56 ± 11.79 (a) | 0.72 ± 0.14 (a) | 2.24 ± 1.14 (a) | 0.79 ± 0.23 (ab) | 0.92 ± 0.04 (a) |
| W-SW | 16.33 ± 6.16 (c) | 17.49 ± 7.44 (c) | 0.87 ± 0.04 (c) | 3.39 ± 0.43 (c) | 1.05 ± 0.28 (ab) | 0.86 ± 0.04 (a) |
| W-SD | 29.44 ± 3.84 (ab) | 32.40 ± 6.94 (ab) | 0.94 ± 0.02 (b) | 4.38 ± 0.26 (b) | 0.71 ± 0.13 (a) | 0.90 ± 0.03 (a) |
Community Composition for Deciphering the Reef Compartments and Seasonal Effect on Communities
The phyla microbial community composition of microorganisms (each sample in Supplementary Fig. S7 to S10 and each compartment + season (S-CO, W-CO, S-SD, W-SD, S-SW, and W-SW) in Figs. 2A, 4A, 6A, and 8A—sites not shown, Supplementary Tables 2, 3, 6, 7, 10, and 12) was visualized using bar plots realized with the ggplot2 R package V. 3.3.5 [82]. Each time a focus on genera was done on the most abundant phyla in each kingdom/marker (Figs. 2B, 4B, 6B, and 8B). To test the significance of differences in the relative abundance of microorganisms among conditions (i.e., reef compartments + seasons), the Wilcoxon test was used.
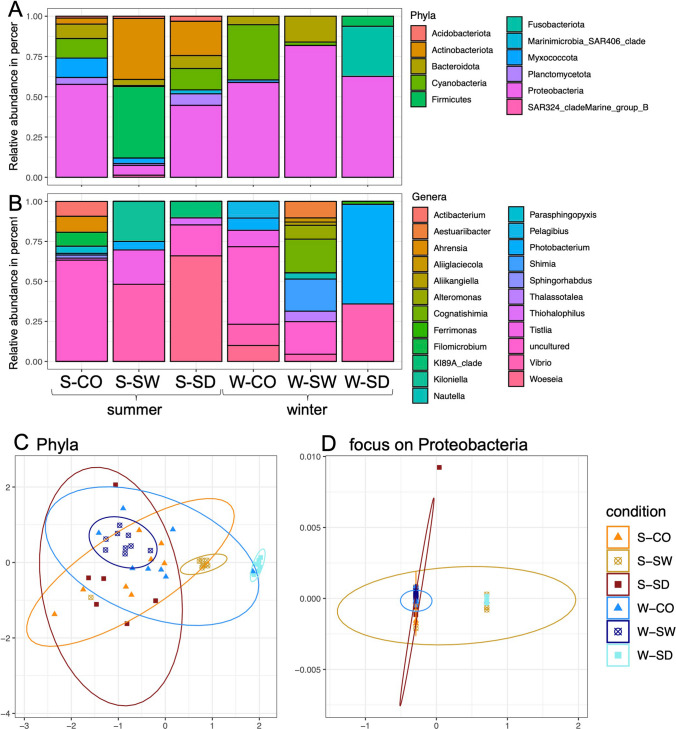
Fig. 2.
Bar plot for bacterial communities. A Bacterial phyla relative abundance; B focus on the more abundant phylum Proteobacteria genera relative abundance; C, D NMDS for bacterial phyla communities (D) and focus on the Proteobacteria phylum, the most abundant one (E). S-CO, summer-coral; S-SW, summer-seawater; S-SD, summer-sediment; W-CO, winter-coral; W-SW, winter-seawater; W-SD, winter-sediment
Fig. 4.
Bar plot for fungal communities. A Fungal phyla relative abundance; B focus on the more abundant phylum Ascomycota genera relative abundance; C, D NMDS for bacterial phyla communities (C) and focus on the Ascomycota phylum, the most abundant one (D). S-CO, summer-coral; S-SW, summer-seawater; S-SD, summer-sediment; W-CO, winter-coral; W-SW, winter-seawater; W-SD, winter-sediment
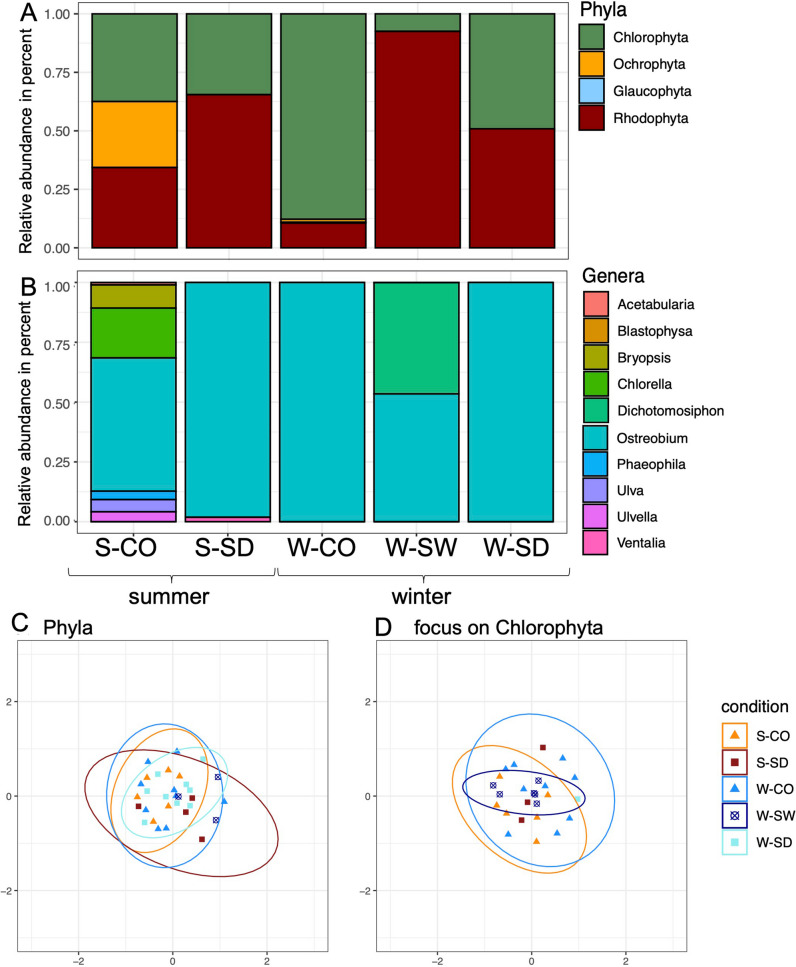
Fig. 6.
Bar plot for microalgae communities. A Microalgae phyla relative abundance; B focus on the more abundant phylum Chlorophyta genera relative abundance; C, D NMDS for microalgae phyla communities (C) and focus on the Chlorophyta phylum, the most abundant one (D). S-CO, summer-coral; S-SW, summer-seawater; S-SD, summer-sediment; W-CO, winter-coral; W-SW, winter-seawater; W-SD, winter-sediment
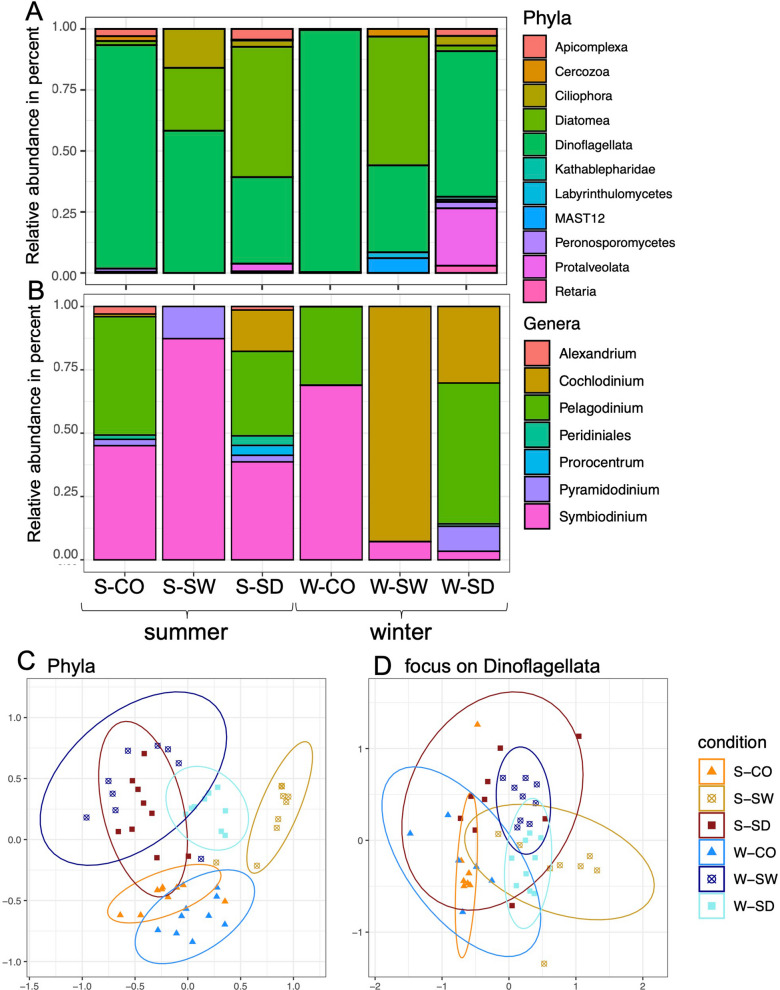
Fig. 8.
Bar plot for protista communities. A Protista phyla relative abundance; B focus on the more abundant phylum Dinoflagellata genera relative abundance; C, D: NMDS for protista phyla communities (C) and focus on the Dinoflagellata phylum, the most abundant one (D). S-CO, summer-coral; S-SW, summer-seawater; S-SD, summer-sediment; W-CO, winter-coral; W-SW, winter-seawater; W-SD, winter-sediment
Community Structure
To compare community structure among conditions (S-CO, W-CO, S-SD, W-SD, S-SW, and W-SW), Bray–Curtis dissimilarities [83] were calculated with the q2-diversity tool [62]. Bray–Curtis dissimilarities were further visualized using non-metric multidimensional scaling (NMDS) using the vegan R package V. 2.5–7 (function “metaMDS,” [84]) and the ggplot2 R package V. 3.3.5 [82] for phyla (Figs. 2C, 4C, 6C, and 8C) and with a focus on the genera of the most relative abundant phylum (Figs. 2D, 4D, 6D and 8D). Differences in microbial community composition were tested using PERMANOVA, with 9999 permutations using the vegan R package V. 2.5–7 [84], and the post hoc analyses were performed using the pairwiseAdonis R package V. 0.4 [85] (Supplementary Tables 4, 8, 11 and 13).
Community Structuring by Condition, Compartment, and Season: Insights for Bioindicator Identification
The previous methods (“diversity, composition, and structure”) have enabled us to find variations and possible bioindicator groups within the variables reef compartments by seasons; we then used a sharper statistical technique which finds, with a priori (Bayesian statistics) ASVs or groups of ASVs (according to taxonomic assignation, this can be species, a genus grouping several species, etc.) which were significantly linked to a particular condition (here, reef compartments or seasons and not both). This will enable us to focus not only on the most abundant groups (classic tryptic analyses) but above all on those species or groups that are most significantly linked to a particular condition. This strategy makes it possible to find reliable bioindicators in relation to these conditions. To identify these groups of ASVs that responded most to variation in environmental conditions between the compartments and seasons, we used the same techniques as in [26, 86–88]. To catch the largest differences in ASVs abundances (“targeted analysis”), we followed the method of Glasl et al. [26] with the Anaconda R package version 0.1.5 [89, 90].
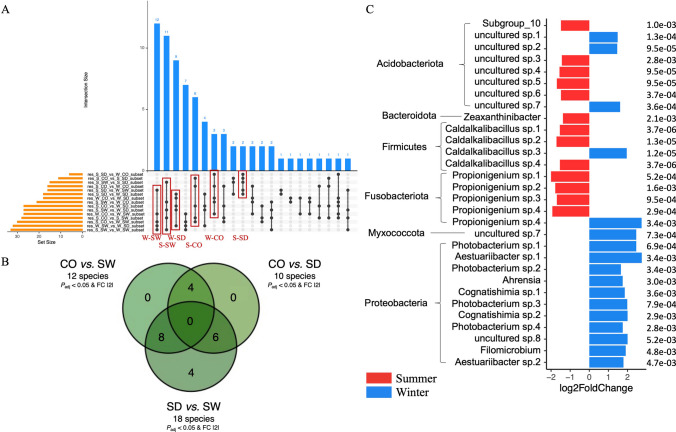
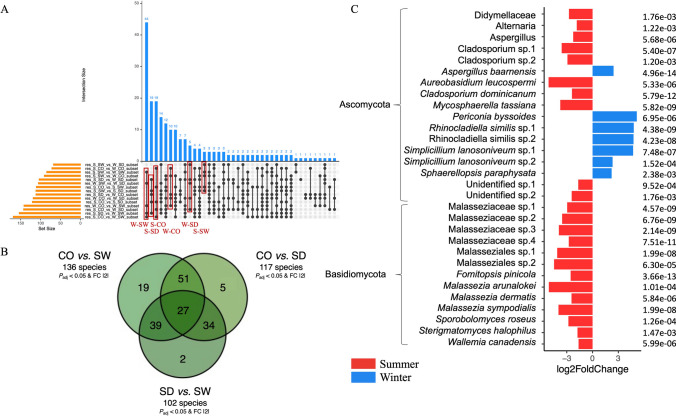
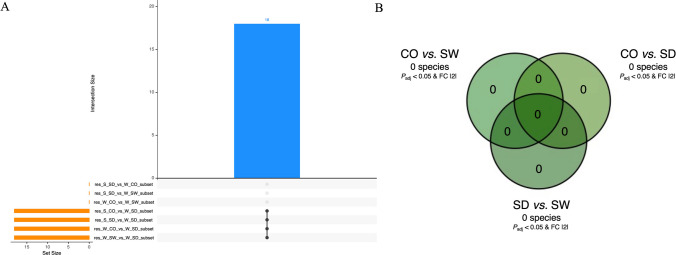
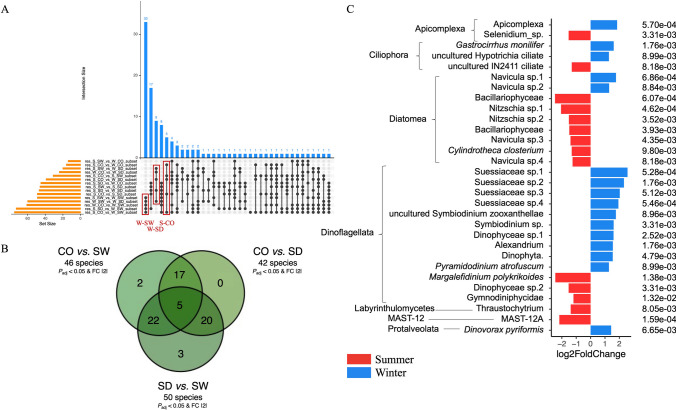
However, before looking for these ASVs specific to compartments and seasons, we used this same method (the one from Glasl et al. [26] with the Anaconda R package) to look for ASVs specific to the compartment + season conditions already studied in the “diversity, composition and structure” triptych (e.g., S-CO, S-SW, S-SD, W-CO, W-SW, and W-SD). In this way, we aimed to find ASVs specific (if any) to (one or more of) these conditions. These ASVs, which would therefore have been deemed too specific to be classified as compartment- or season-specific bioindicators (because they could be putatively linked only to a too-specific condition like S-CO for example, and not only S or CO), would therefore be excluded from subsequent analyses (aimed at identifying only compartment- or season-specific ASVs) if such ASVs were also found (this analysis mainly served as a backup for the next two). In brief, across these three Bayesian analyses (e.g., condition-specific ASVs as a safeguard, compartment-specific ASVs, and season-specific ASVs), we conducted targeted differential enrichment analyses of taxonomic ranks using ASVs [26, 86–88]. Differential analysis was performed by estimating the variance-mean dependence in ASV counts using a negative binomial model to identify significantly and differentially represented ASVs among conditions [26, 86–88]. In the first analysis (condition-specific ASVs—the safeguard), given the six conditions (e.g., S-CO, S-SW, S-SD, W-CO, W-SW, and W-SD), this resulted in 15 comparisons. The findings were presented as an upset plot. Specific ASVs were derived by comparing the results of these comparisons. More precisely, the shared denominator corresponding to the condition of interest needed to be present in five comparisons (because it is the maximum number of times each condition appears—as an example, see the first red rectangle in the left in Fig. 3A; this is for 12 ASVs condition-specific (the blue column), where black dots appear for five comparisons involving W-SW, so these 12 ASVs are linked to the W-SW condition—because W-SW is the shared denominator in these five comparisons), and identifying these ASVs required a “comparison of the comparisons” (same method as in [91]) (indeed, to find these condition-specific ASVs, we need to compare these 15 comparisons to determine which conditions have which set of ASVs). An ASVs was considered significantly over- or under-represented in a condition based on the criteria of adjusted P value < 0.05 and Log2FoldChange >|2| (Figs. 3A, 5A, 7A, and 9A). All of these significative ASVs are listed in Supplementary Tables 5 (bacteria), 9 (fungi), and 13 (protista) (none significant ASV was found for microalgae). For the second analysis (compartment-specific ASVs), given the three conditions (e.g., CO, SW, and SD), this resulted in three comparisons (CO vs. SW, SD vs. SW, and CO vs. SD), represented in a Venn diagram (same method as in [91]) (Figs. 3B, 5B, 7B, and 9B). For the third analysis (season-specific ASVs), given the two conditions (e.g., austral summer and austral winter), this involved a single comparison (S vs. W), with the results represented in a bar plot (same method as in [26]) (Figs. 3C, 5C, and 9C).
Fig. 3.
Identification of condition-, compartment-, and season-specific ASVs using targeted Bayesian analysis. A Upset plot of the condition-specific bacterial ASVs identified in the six experimental conditions (S-CO, S-SW, S-SD, W-CO, W-SW, and W-SD) (in red) collected from the comparison of comparisons (the common denominator corresponding to the condition sought must be present in five comparisons, and to retrieve these ASVs a comparison of these comparisons must be made) based on the criteria of adjusted P value < 0.05 and Log2FoldChange >|2|. B Compartment-specific ASVs were determined across coral rubble, sediment, and seawater compartments, highlighting bacterial taxa with differential representation (adjusted P value < 0.05 and Log2FoldChange >|2|). C Season-specific ASVs were determined across the austral summer vs. winter revealed comparison highlighting bacterial taxa with differential representation (adjusted P value < 0.05 and Log2FoldChange >|2|). No overlaps with condition-specific ASVs (A) were detected in analyses (B and C). S-CO, summer-coral; S-SW, summer-seawater; S-SD, summer-sediment; W-CO, winter-coral; W-SW, winter-seawater; W-SD, winter-sediment
Fig. 5.
Identification of condition-, compartment-, and season-specific ASVs using targeted Bayesian analysis. A Upset plot of the condition-specific fungal ASVs identified in the six experimental conditions (S-CO, S-SW, S-SD, W-CO, W-SW, and W-SD) (in red) collected from the comparison of comparisons (the common denominator corresponding to the condition sought must be present in five comparisons, and to retrieve these ASVs a comparison of these comparisons must be made) based on the criteria of adjusted P value < 0.05 and Log2FoldChange >|2|. B Compartment-specific ASVs were determined across coral rubble, sediment, and seawater compartments, highlighting fungal taxa with differential representation (adjusted P value < 0.05 and Log2FoldChange >|2|). C Season-specific ASVs were determined across the austral summer vs. winter revealed comparison highlighting fungal taxa with differential representation (adjusted P value < 0.05 and Log2FoldChange >|2|). No overlaps with condition-specific ASVs (A) were detected in analyses (B and C). S-CO, summer-coral; S-SW, summer-seawater; S-SD, summer-sediment; W-CO, winter-coral; W-SW, winter-seawater; W-SD, winter-sediment
Fig. 7.
Identification of condition-, compartment-, and season-specific ASVs using targeted Bayesian analysis. A Upset plot of the condition-specific microalgae ASVs identified in the six experimental conditions (S-CO, S-SW, S-SD, W-CO, W-SW, and W-SD) based on the criteria of adjusted P value < 0.05 and Log2FoldChange >|2|. B Compartment-specific ASVs were determined across coral rubble, sediment, and seawater compartments, highlighting no taxa with differential representation (adjusted P value < 0.05 and Log2FoldChange >|2|). S-CO, summer-coral; S-SW, summer-seawater; S-SD, summer-sediment; W-CO, winter-coral; W-SW, winter-seawater; W-SD, winter-sediment
Fig. 9.
Identification of condition-, compartment-, and season-specific ASVs using targeted Bayesian analysis. A Upset plot of the condition-specific Protista ASVs identified in the six experimental conditions (S-CO, S-SW, S-SD, W-CO, W-SW, and W-SD) (in red) collected from the comparison of comparisons (the common denominator corresponding to the condition sought must be present in five comparisons, and to retrieve these ASVs a comparison of these comparisons must be made) based on the criteria of adjusted P value < 0.05 and Log2FoldChange >|2|. B Compartment-specific ASVs were determined across coral rubble, sediment, and seawater compartments, highlighting protista taxa with differential representation (adjusted P value < 0.05 and Log2FoldChange >|2|). C Season-specific ASVs were determined across the austral summer vs. winter revealed comparison highlighting protista taxa with differential representation (adjusted P value < 0.05 and Log2FoldChange >|2|). No overlaps with condition-specific ASVs (A) were detected in analyses (B) and (C). S-CO, summer-coral; S-SW, summer-seawater; S-SD, summer-sediment; W-CO, winter-coral; W-SW, winter-seawater; W-SD, winter-sediment
Results
For bacteria, 1,143,990 total sequences were grouped into 4502 ASVs, tallied at an overall mean of 4859 ± 2190 ASVs/sample. Bacterial ASVs richness did not differ significantly among sites (Supplementary Table S1a, P > 0.05), but differed among compartments and between seasons (Supplementary Table S1b and c, P > 0.05).
For fungi, 1,222,568 total sequences were grouped into 1603 ASVs, tallied at an overall mean of 15,721 ± 3418 ASVs/sample. Fungal ASVs richness did not differ significantly among sites and between seasons (Supplementary Table S1a and c, P > 0.05), but differed among compartments (Supplementary Table S1b, P > 0.05).
For protista, 1,090,485 total sequences were grouped into 2784 ASVs, tallied at an overall mean of 10,713 ± 2742 ASVs/samples. Protista ASVs richness did not differ significantly among sites, compartments, nor between seasons (Supplementary Table S1a, b and c, P > 0.05).
For microalgae, 2,435,431 total sequences were grouped into 7312 ASVs, tallied at an overall mean of 13,542 ± 16,035 ASVs/samples. Microalgae ASVs richness did not differ significantly among sites (Supplementary Table S1a, P > 0.05), but differed among compartments and between seasons (Supplementary Table S1b and c, P > 0.05).
For ease of reading, the acronyms used are replaced here for each studied condition: S-CO, summer-coral; S-SW, summer-seawater; S-SD, summer-sediment; W-CO, winter-coral; W-SW, winter-seawater; W-SD, winter-sediment.
Bacteria
Diversity of Microorganisms in Reef Compartment Through the Seasons
In terms of observed amplicon sequence variants (ASVs) (Table 1, Supplementary Fig. S3), the sediments showed the highest ASVs bacterial diversity during austral winter (W-SD) (P value < 0.001). For Chao1 and Shannon diversity, sediments and seawater during austral winter conditions (W-SD and W-SW) detained the highest number of ASVs (P value < 0.001). The Simpson diversity was lower in S-SD compared to the other conditions (P value < 0.001). The Faith PD was higher in coral rubble during austral summer (S-CO) and in seawater in austral winter (W-SW) (P value < 0.05).
In summary, the various diversity indices reveal several significant differences in microbial diversity among reef compartments and seasonal conditions, with sediments and seawater in austral winter displaying higher diversity levels according to several metrics, while coral rubble in austral summer and seawater in austral winter show distinct patterns of phylogenetic diversity and evenness.
Community Composition for Deciphering the Reef Compartments and Seasonal Effects
Proteobacteria was the most abundant phylum (mean relative abundance 51.9%, N = 6) (P value < 0.05, Wilcoxon test) (Fig. 2A, Supplementary Table S2), except in austral summer seawater (S-SW) with a relative abundance of 6.08%. Cyanobacteria was the second most abundant phylum (mean relative abundance 10.4%, N = 6). Cyanobacteria were abundant in coral rubble in austral winter (W-CO), representing 34.3% of the total ASVs abundance (P value < 0.05, Wilcoxon test). In contrast, cyanobacteria were present in small proportions in austral summer in coral rubble (S-CO, 12.1%) and sediments (S-SD, 13.3%). Firmicutes represented the third most abundant phylum (mean relative abundance 8.4% N = 6) and is present in seawater in austral summer (S-SW) at 44.4% and less in sediment in austral winter (W-SD) at 6.3%.
The phylum Fusobacteria was present in austral winter in sediments (W-SD, 31.1%). Actinobacteria were only observed in austral summer (21.0%, N = 3), particularly in seawater (S-SW, 37.9%). Myxococcota were observed in austral summer, abundant in coral rubble (S-CO, 12.0%).
Focusing on the most abundant phylum, i.e., Proteobacteria, (Fig. 2B, Supplementary Table S3) revealed that genera Vibrio (25.2% N = 6), followed by Woeseia (12.6% N = 6) and then Photobacterium (12.5% N = 6) are the most abundant. The relative abundance of Proteobacteria was variable among reef compartments and seasons, with no significant structuration. In the austral summer in seawater (S-SW), Vibrio is the most abundant genus of proteobacteria with 48.2%. In austral summer in sediment (S-SD), it is Woeseia with 65.9% followed by the Vibrio genus with 13.2%. In the same season in seawater (W-SW), it is Cognatishimia with 21.0%, and in sediment (W-SD), it is Photobacterium with 62.2%.
Bacterial Structure of Reef Compartment and Seasonal Data
The NMDS (stress = 0.14) for bacterial phyla communities (Fig. 2D) showed a distinction between seawater and sediment for the microbial communities between the two seasons, whereas for coral rubble, the seasonal separation distinguished few samples in austral summer. The PERMANOVA (nPerm = 9999) confirmed significant effects of the conditions (seasons and compartments) on the structuration of communities (R2 0.39811; P value 0.001, Supplementary Table S4a), and the pairwise Adonis tests were significant in all comparisons (see Supplementary Table S4b). For the Proteobacteria phylum (a focus on the most represented phylum) (Fig. 2E) (stress = 0.0001), the PERMANOVA (nPerm = 9999) confirmed significant differences between the conditions (R2 0.30479; P value 0.001, Supplementary Table S4c), and the pairwise Adonis test showed that some comparisons were significant (see Supplementary Table S4d). So, these analyses show significant seasonal structuring of bacterial communities in seawater and sediment, with less distinct separation in coral rubble.
Bacterial Community Structuring by Condition, Compartment, and Season: Insights for Bioindicator Identification
Using a targeted Bayesian analysis (P value adjusted < 0.05 & Log2FoldChange >|2|), we identified condition-specific ASVs (S-CO, S-SW, S-SD, W-CO, W-SW, and W-SD) that could be too specific to serve as bioindicators (Fig. 3A). If these ASVs will be found in the two next analyses (e.g., Bayesian analysis for compartment-specific ASVs and Bayesian analysis for season-specific ASVs), they will be deleted from these two next analyses (the ASV keys are authoritative, but not the taxonomic assignment, which means that species may nevertheless be found here and not subsequently removed if they do not correspond exactly to the same ASV). Specific ASVs were found for each of the six conditions: W-SW, 12; S-SW, 11; W-SD, 9; S-CO, 6; W-CO, 3; S-SD, 2. ASVs keys, Log2FoldChanges, and taxonomic assignments can be found at Supplementary Table S5.
Using a targeted Bayesian analysis, we identified compartment-specific ASVs (coral rubble vs. seawater; coral rubble vs. sediment; sediment vs. seawater) that could serve as bioindicators (P value adjusted < 0.05 and Log2FoldChange >|2|). Four ASVs were significantly over-represented in coral rubble compared to other compartments (Fig. 3B). These included Hyphomonas sp. (Alphaproteobacteria), Pelagibius sp. (Alphaproteobacteria), a bacterium from the Sandaracinaceae family (Myxococcota), and a bacterium from the Rhizobiaceae family (Alphaproteobacteria). Six ASVs were significantly under-represented in sediment compared to other compartments. These included five Propionigenium sp. (Fusobacteriota) and a bacterium from the Actinomarinales order (Actinobacteriota). Eight ASVs were significantly under-represented in seawater compared to other compartments. These included four ASVs assigned to Caldalkalibacillus thermarum (Firmicutes) and four ASVs assigned to bacteria from the Micrococcaceae family (Actinobacteriota). As no ASVs from the first Bayesian analysis (Fig. 3A) were found here, no ASVs were removed from this analysis.
Through a targeted Bayesian analysis, we focused on season-specific (austral winter vs. summer) species that could serve as strong bioindicators (P value adjusted < 0.05 and Log2FoldChange >|2|). Forty-six ASVs had been significantly (P value adjusted < 0.05) found differentially over-represented between austral summer and winter (13 ASVs were over-represented in austral summer and 17 ASVs in austral winter), but for better visualization, only the 30 most significant ASVs are shown in Fig. 3C. More specifically, Caldalkalibacillus sp. (Firmicutes), Propionigenium sp. (Fusobacteriota), Subgroup_10 sp. (Acidobacteriota), and Zeaxanthinibacter sp. (Bacteroidota) were over-represented in austral summer, while in austral winter, Aestuariibacter sp., Ahrensia sp., Cognatishimia sp., Filomicrobium sp., Photobacterium sp. (all Proteobacteria), Acidobacteriota, Caldalkalibacillus sp. (Firmicutes), and Propionigenium sp. (Fusobacteriota) were predominant. As no ASVs from the first Bayesian analysis (Fig. 3A) were found here, no ASVs were removed from this analysis.
Fungi
Diversity of Microorganisms in Reef Compartment Through the Seasons
In terms of observed ASVs (Table 2, Supplementary Fig. S6), coral rubble showed higher ASVs fungal diversity in austral summer (S-CO) and lesser in seawater in austral summer (S-SW) (P value < 0.001). Chao1 and Shannon’s index demonstrated that corals and sediment in austral summer (S-CO and S-SD), as well as seawater in austral winter (W-SW), detained rarer ASVs than others (P value < 0.001). The Faith PD showed higher diversity in seawater in austral winter (W-SW) (P value < 0.001).
Table 2.
Fungi diversity index. Lowercase letters indicate significant differences among conditions based on pairwise comparisons following a Kruskal-Wallis test (P value < 0.05). S, austral summer; W, austral winter; CO, coral rubble; SW, seawater; SD, sediment
| Condition | Observed ASV | Chao1 | Simpson | Shannon | Faith PD | Pielou evenness |
|---|---|---|---|---|---|---|
| S-CO | 52.67 ± 7.81 (a) | 52.67 ± 7.81 (a) | 0.94 ± 0.02 (a) | 4.78 ± 0.39 (a) | 8.10 ± 0.98 (b) | 0.84 ± 0.04 (a) |
| S-SW | 11.00 ± 5.81 (a) | 11.00 ± 5.81 (a) | 0.83 ± 0.05 (a) | 2.88 ± 0.57 (a) | 2.98 ± 1.30 (b) | 0.86 ± 0.05 (a) |
| S-SD | 47.11 ± 12.95 (c) | 47.11 ± 12.95 (c) | 0.93 ± 0.03 (b) | 4.50 ± 0.65 (b) | 9.44 ± 1.90 (c) | 0.81 ± 0.06 (a) |
| W-CO | 36.89 ± 16.01 (bc) | 36.94 ± 16.02 (bc) | 0.86 ± 0.11 (ab) | 3.78 ± 1.17 (ab) | 7.41 ± 2.60 (b) | 0.73 ± 0.14 (a) |
| W-SW | 46.89 ± 12.40 (ab) | 46.89 ± 12.40 (ab) | 0.85 ± 0.11 (ab) | 3.84 ± 1.00 (ab) | 13.36 ± 2.46 (b) | 0.70 ± 0.17 (a) |
| W-SD | 27.67 ± 12.68 (ab) | 27.67 ± 12.68 (ab) | 0.91 ± 0.05 (ab) | 3.98 ± 0.69 (ab) | 7.94 ± 2.20 (a) | 0.85 ± 0.05 (a) |
In summary, the analysis of various diversity indexes revealed significant variations in microbial diversity among the different conditions (i.e., compartments at different seasons), with notable differences in diversity levels between coral rubble, seawater, and sediment in both austral summer and winter seasons.
Community Composition for Deciphering the Reef Compartments and Seasonal Effects
Ascomycota (63.1% N = 6) and Basidiomycota (36.1% N = 6) were the two most abundant fungal phyla (P value < 0.05, Wilcoxon test), and both showed a change in their relative abundances through conditions (Fig. 4A, Supplementary Table S6). A third phylum appeared in the sediment in austral summer (S-SD), in seawater in austral winter (W-SW), and in sediment in austral winter (W-SD) conditions: Chytridiomycota (0.8%, N = 6). In coral rubble (S-CO and W-CO), Ascomycota was dominant whatever the season (P value < 0.05, Wilcoxon test). For seawater and sediment, the Ascomycota/Basidiomycota (A/B) abundance ratio depends on the season (Fig. 4A). This A/B abundance ratio was very similar for both seawater and sediment in austral winter (S-SW and S-SD). The A/B richness ratio for coral rubble and seawater was 50% for both austral summer and winter.
Focusing on the most abundant phylum, i.e., Ascomycota, (Fig. 4B, Supplementary Table S7) revealed that the most abundant genera are Aspergillus (40.9% N = 6), followed by Cladosporium (16.2%, N = 6) and then Rhinocladiella (6.6% N = 6) (P value < 0.05, Wilcoxon test). The relative abundance of Ascomycota was variable among reef compartments and seasons, with no significant differences. In coral rubble, Rhinocladiella is the second most relatively abundant represented genera in austral summer with 31.7% (S-CO) of relative abundance, and the Leptospora genus is the second most relatively abundant represented genus in austral winter with 23.3% (W-CO) of relative abundance. In sediment, the Cladosporium genus is the most abundant in austral winter with 47.0% (W-SD) of relative abundance and the second most relatively abundant represented genus in austral summer with 18.7% of relative abundance (S-SD). Aspergillus genus is the second most relatively abundant represented genus in sediment in austral winter with 16.5% (W-SD) of relative abundance. In seawater, Penicillium is the most relatively abundant represented genus in austral summer with 19.6% (S-SW) of relative abundance and the Periconia genus is the second most relatively abundant represented genus in austral winter with 17.67% (W-SW) of relative abundance.
Fungal Structure of Reef Compartment and Seasonal Data
The NMDS (stress = 0.23) (Fig. 4D) highlighted that the seawater fungal community was significantly different from that of sediments, during both seasons. On the contrary, the coral rubble fungal community did not vary with seasonality. For phyla, the PERMANOVA (nPerm = 9999) confirmed significant effects of the conditions (seasons and compartments) on the structuration of communities (R2 0.35514; P value 0.001, Supplementary Table S8a), and the pairwise Adonis test showed most comparisons are significant (see Supplementary Table S8b). For the Ascomycota phylum (a focus on the most represented phylum) (Fig. 4E) (stress = 0.25), the PERMANOVA (nPerm = 9999) confirms significant differences between the conditions (seasons and compartments) (R2 0.28262; P value 0.001, Supplementary Table S8c), and the pairwise Adonis test shows that some comparisons are significant (see Supplementary Table S8d). In summary, seawater fungal communities significantly differed from sediment communities across both seasons, while coral rubble showed no seasonal variation (Fig. 4).
Fungal Community Structuring by Condition, Compartment, and Season: Insights for Bioindicator Identification
Using a targeted Bayesian analysis (P value adjusted < 0.05 and Log2FoldChange >|2|), we identified condition-specific ASVs (S-CO, S-SW, S-SD, W-CO, W-SW, and W-SD) that could be too specific to serve as bioindicators (Fig. 5A). If these ASVs will be found in the two next analyses (e.g., Bayesian analysis for compartment-specific ASVs and Bayesian analysis for season-specific ASVs), they will be deleted from these two next analyses (the ASV keys are authoritative, but not the taxonomic assignment, which means that species may nevertheless be found here and not subsequently removed if they do not correspond exactly to the same ASV). Specific ASVs were found for each of the six conditions: W-SW, 44; S-SD, 19; S-CO, 19; W-CO, 10; W-SD, 5; S-SW, 3. ASVs keys, Log2FoldChanges, and taxonomic assignments can be found at Supplementary Table S9.
Using a targeted Bayesian analysis, we identified compartment-specific ASVs (coral rubble vs. seawater; coral rubble vs. sediment; sediment vs. seawater) that could serve as bioindicators (P value adjusted < 0.05 and Log2FoldChange >|2|). Twenty-seven ASVs were significantly over- or under-represented across all three compartments (Fig. 5B), most of which were unidentified fungi, although four belonged to the family Malasseziaceae (Basidiomycota). Coral rubble contained 51 unique ASVs, including Cladosporium dominicanum and Aspergillus unguis (both Ascomycota), which were under-represented compared to other compartments. Sediment-specific ASVs (n = 34) included Mycosphaerella tassiana (Ascomycota), Fomitopsis pinicola (Basidiomycota), and Cladosporium sp. (Ascomycota), all over-represented in sediment. Seawater contained 39 unique ASVs, such as Periconia byssoides, Aspergillus baarnensis, Aureobasidium leucospermi (all Ascomycota), and several Malasseziaceae species, including Malassezia sympodialis and Malassezia arunalokei (Basidiomycota), which were over-represented in seawater. ASVs under-represented in seawater (present in coral rubble and sediment but absent in seawater) were all unidentified fungi. As no ASVs from the first Bayesian analysis (Fig. 5A) were found here, so no ASVs were removed from this analysis.
Through a targeted Bayesian analysis, we focused on season-specific (austral winter vs. summer) species that could serve as strong bioindicators (P value adjusted < 0.05 and Log2FoldChange >|2|). A total of 135 ASVs had been significantly found differentially over-represented between austral summer and winter, but for better visualization, only the 30 most significant ASVs are shown in Fig. 5C. More specifically, in austral summer, Alternaria sp., Aspergillus sp., Cladosporium sp., Aureobasidium leucospermi, Cladosporium dominicanum, Mycosphaerella tassiana (all in Ascomycota), Malassezia arunalokei, M. dermatis and M. sympodialis, Fomitopsis pinicola, Sporobolomyces roseus, Sterigmatomyces halophilus, and Wallemia canadensis (all Basidiomycota) were particularly over-represented (P value < 0.05). In austral winter, Aspergillus baarnensis, Periconia byssoides, Rhinocladiella similis, Simplicillium lanosoniveum, and Sphaerellopsis paraphysata (all in Ascomycota) were particularly over-represented (P value < 0.05). As no ASVs from the first Bayesian analysis (Fig. 5A) were found here, so no ASVs were removed from this analysis.
Microalgae
For clarity, we note that an insufficient number of samples met the rarefaction threshold for the S-SW condition, which explains its exclusion from the analysis here (see the “Qiime2 framework” section).
Diversity of Microorganisms in Reef Compartment Through the Seasons
The Faith PD diversity index showed significant changes between conditions (P value < 0.05) (Table 3, Supplementary Fig. S5). Post hoc tests showed that sediment in austral winter (W-SD) was significantly different from seawater in austral winter (W-SW), and corals in the same season (W-CO) (P value < 0.05). The microalgal diversity in seawater in austral winter (W-SW) was also significantly different from corals in austral summer (S-CO) (P value < 0.05). Other comparisons were not significant. Other diversity indexes did not show significant results.
Table 3.
Microalgae diversity index. Lowercase letters indicate significant differences among conditions based on pairwise comparisons following a Kruskal-Wallis test (P value < 0.05). S, austral summer; W, austral winter; CO, coral rubble; SW, seawater; SD, sediments
| Condition | Observed ASV | Chao1 | Simpson | Shannon | Faith PD | Pielou evenness |
|---|---|---|---|---|---|---|
| S-CO | 6.75 ± 2.87 (a) | 0.74 ± 0.34 (a) | 0.63 ± 0.24 (a) | 0.61 ± 0.28 (a) | 0.50 ± 0.18 (a) | 6.75 ± 2.87 (a) |
| S-SD | 8.17 ± 2.93 (a) | 0.92 ± 0.37 (a) | 0.82 ± 0.14 (a) | 0.75 ± 0.12 (a) | 0.60 ± 0.20 (a) | 8.17 ± 2.93 (a) |
| W-CO | 8.44 ± 3.05 (a) | 0.95 ± 0.38 (a) | 0.69 ± 0.10 (a) | 0.70 ± 0.10 (a) | 0.47 ± 0.14 (a) | 8.44 ± 3.05 (a) |
| W-SW | 9.50 ± 2.12 (a) | 1.08 ± 0.27 (a) | 0.65 ± 0.07 (a) | 0.73 ± 0.06 (a) | 0.39 ± 0.01 (a) | 9.50 ± 2.12 (a) |
| W-SD | 14.22 ± 19.06 (a) | 1.32 ± 1.45 (a) | 0.82 ± 0.18 (a) | 0.74 ± 0.20 (a) | 0.67 ± 0.20 (a) | 10.78 ± 10.32 (a) |
Community Composition for Deciphering the Reef Compartments and Seasonal Effects
Rhodophyta was the most abundant phylum (50.7% N = 5), followed by Chlorophyta (43.3% N = 5) and then Ochrophyta (5.9% N = 5) (P value < 0.05, Wilcoxon test) (Fig. 6A, Supplementary Table S10). In coral rubble, Chlorophyta was the most abundant phylum in austral summer (37.5%) (S-CO), and in austral winter (87.8%) (W-CO). Ochrophyta was only found in coral rubble (28.2% in austral summer and 1.2% in austral winter) (S/W-CO). In sediment, Rhodophyta was the most abundant phylum in austral summer (65.4%) (S-SD), and in austral winter (50.9%) (W-SD). In seawater (data only for austral winter), Rhodophyta was the most abundant phylum with 92.5% (W-SW).
Figure 6B shows a focus on genera of the second abundant phylum (Chlorophyta) because of the biomonitoring and ecological relevance of this phylum (see the “Discussion” section). Information on genera of the most abundant phylum (Rhodophyta) can be found in Supplementary Table S8. For instance, Rhodophyta presented here four genera (Compsothamnion, Neogoniolithon, Wrangelia, and “uncultured”), and Chlorophyta presented 10 genera here (Fig. 6B, Supplementary Table S10).
In Chlorophyta (Fig. 6B, Supplementary Table S8), Ostreobium was the most abundant genus (81.5% N = 5), followed by Dichotomosiphon genus (9.3% N = 5) and then Chlorella genus (4.2% N = 5). During austral winter, the Ostreobium genus represented 100% of the relative abundance in coral rubble and sediment (W-CO/SD), and this genus represented 53.4% of the relative abundance in seawater (W-SW). In austral summer, the Ostreobium genus represented 55.7% of the relative abundance in coral rubble (S-CO) and 98.1% in sediment (S-SD).
Using a targeted Bayesian analysis, we focused on season-specific and then compartment-specific species as potential bioindicators (austral winter vs. summer and coral rubble vs. seawater vs. sediment), but no ASVs showed significant over- or under-representation between the seasons nor compartments (Fig. 6).
Microalgae Structure of Reef Compartments and Seasonal Data
No structure appeared in the NMDS (Fig. 6C) (stress = 0.30), and the PERMANOVA (nPerm = 9999) did not show significant differences between the conditions for phyla (R2 0.16321; P value 1, Supplementary Table S11a), as well as for the Chlorophyta phylum (focused on the most abundant phylum) (Fig. 4D) (stress = 0.29) (R2 0.16602; P value 0.614, Supplementary Table S11b).
Microalgae Community Structuring by Condition, Compartment, and Season: Insights for Bioindicator Identification
Using a targeted Bayesian analysis (P value adjusted < 0.05 and Log2FoldChange >|2|), we did not identify condition-specific ASVs that could be too specific to serve as bioindicators (Fig. 7A), neither in compartment-specific ASVs (Fig. 7B) or season-specific ASVs.
Protista
Diversity of Microorganisms in Reef Compartment Through the Seasons
In terms of observed ASVs (Table 4, Supplementary Fig. S6), coral rubble showed higher diversity in austral summer (S-CO) (P value < 0.05). The Chao1 index demonstrated that coral rubble and sediment during austral summer (S-CO and S-SD), and sediment during austral winter (W-SD) detained rarer ASVs than others (P value < 0.05). The Shannon diversity index also shows that coral rubble and sediment during austral summer (S-CO and S-SD) are more diverse and that seawater during austral winter (S-SW) has less overall diversity. The Faith PD index demonstrated higher diversity in sediment during austral winter (W-SD).
Table 4.
Protista diversity index. Lowercase letters indicate significant differences among conditions based on pairwise comparisons following a Kruskal-Wallis test (P value < 0.05). S, austral summer; W, austral winter; CO, coral rubble; SW, seawater; SD, sediments
| Condition | Observed ASV | Chao1 | Simpson | Shannon | Faith PD | Pielou evenness |
|---|---|---|---|---|---|---|
| S-CO | 52.78 ± 20.06 (a) | 52.78 ± 20.06 (a) | 0.96 ± 0.04 (a) | 5.11 ± 0.79 (a) | 6.36 ± 1.94 (ab) | 0.91 ± 0.04 (a) |
| S-SW | 19.89 ± 16.24 (a) | 19.89 ± 16.24 (a) | 0.87 ± 0.06 (a) | 3.46 ± 0.89 (a) | 3.03 ± 2.26 (ab) | 0.84 ± 0.09 (a) |
| S-SD | 49.33 ± 13.17 (b) | 49.33 ± 13.17 (b) | 0.95 ± 0.02 (b) | 4.98 ± 0.35 (b) | 7.14 ± 1.57 (c) | 0.89 ± 0.05 (a) |
| W-CO | 43.89 ± 12.42 (a) | 43.94 ± 12.52 (a) | 0.93 ± 0.04 (ab) | 4.55 ± 0.73 (ab) | 5.78 ± 1.01 (a) | 0.84 ± 0.08 (a) |
| W-SW | 37.11 ± 10.74 (ab) | 37.11 ± 10.74 (ab) | 0.93 ± 0.04 (ab) | 4.41 ± 0.76 (ab) | 5.67 ± 1.53 (bc) | 0.86 ± 0.07 (a) |
| W-SD | 51.44 ± 11.35 (ab) | 51.56 ± 11.30 (ab) | 0.94 ± 0.02 (ab) | 4.81 ± 0.50 (ab) | 8.75 ± 1.71 (bc) | 0.85 ± 0.05 (a) |
In summary, the analysis of protista diversity among various conditions revealed that coral rubble in austral summer (S-CO) exhibits the highest diversity, while seawater in austral summer (S-SW) had the lowest diversity, as indicated by multiple diversity indexes, with sediment generally showing higher diversity than seawater, and sediment in austral winter displaying a distinct pattern of higher phylogenetic diversity.
Community Composition for Deciphering the Reef Compartments and Seasonal Effects
Dinoflagellata was the most abundant protista phylum (63.3% N = 6), followed by Diatomea (22.6% N = 6) and then Protalveolata (4.5% N = 6) (P value < 0.05) (Fig. 8 A, Supplementary Table S12).
Dinoflagellata was the most relatively abundant protista phylum (Fig. 8A, Supplementary Table S12) with 91.6%, followed by Apicomplexa (3.0%) and then Cercozoa (2.0%) in austral summer in coral rubble (S-CO). In seawater (S-SW), the Dinoflagellata was the most relatively abundant protista phylum with 58.3%, followed by Diatomea (25.8%) and then Ciliophora (16.0%). In sediment (S-SD), the Diatomea was the most relatively abundant protista phylum with 53.4%, followed by Dinoflagellata (35.5%) and then Apicomplexa (4.4%).
During austral winter in coral rubble (W-CO), Dinoflagellata was the most relatively abundant protista phylum with 99.2%, followed by Apicomplexa (0.4%) and then Protalveolata (0.2%). In seawater (W-SW), the Diatomea was the most relatively abundant protista phylum with 52.8%, followed by Dinoflagellata (35.7%) and then MAST12 (6.1%). In sediment (W-SD), the Dinoflagellata was the most relatively abundant protista phylum with 59.7%, followed by Peronosporomycetes (23.5%), and then Ciliophora (3.9%).
Focusing on the most abundant phylum, i.e., Dinoflagetella (Fig. 8B, Supplementary Table S10), Symbiodinium was the most abundant genus (41.7% N = 6) followed by Pelagodinium (27.8% N = 6) and then Cochlodinium (23.4% N = 6). During austral summer, in coral rubble (S-CO), Pelagodinium was the most relatively abundant Dinoflagellata genus with 46.8%, followed by Symbiodinium (45.1%) and then Alexandrium (3.0%). In seawater (S-SW), Symbiodinium was the most relatively abundant Dinoflagellata genus with 87.3%, followed by Pyramidodinium (12.7%). In sediment (S-SD), Symbiodinium was the most relatively abundant Dinoflagellata genus with 38.7%, followed by Pelagodinium (33.4%) and then Cochlodinium (16.3%). During austral winter in coral rubble (W-CO), Symbiodinium was the most relatively abundant Dinoflagellata genus with 68.9%, followed by Pelagodinium (30.9%) and then Pyramidodinium (0.2%). In seawater (W-SW), Cochlodinium was the most relatively abundant Dinoflagellata genus with 92.8%, followed by Symbiodinium (7.2%) and then Pelagodinium (0.1%). In sediment (W-SD), Pelagodinium was the most relatively abundant Dinoflagellata genus with 55.6%, followed by Cochlodinium (30.2%) and then Pyramidodinium (9.9%).
Protista Structure of Reef Compartment and Seasonal Data
The NMDS (stress = 0.23) for protista phyla communities (Fig. 8D) showed a separation between all conditions. For phyla, the PERMANOVA (nPerm = 9999) confirmed significant differences between the conditions (R2 0.43375; P value 0.001, Supplementary Table S13a) and the pairwise Adonis test shows most comparisons were significant (Supplementary Table S13b). For the Dinoflagellata phylum (focus on the most abundant phylum) (Fig. 8E) (stress = 0.17), the PERMANOVA (nPerm = 9999) confirmed significant differences between the conditions (R2 0.35421; P value 0.001, Supplementary Table S13c) and the pairwise Adonis test showed most comparisons were significant (Supplementary Table S13d). So, analyses revealed significant differences in protista communities across all conditions, as well as for the dominant Dinoflagellata phylum (Fig. 8).
Protista Community Structuring by Condition, Compartment, and Season: Insights for Bioindicator Identification
Using a targeted Bayesian analysis (P value adjusted < 0.05 and Log2FoldChange >|2|), we identified condition-specific ASVs (W-SW, W-SD, and S-CO) that could be too specific to serve as bioindicators (Fig. 9A). If these ASVs will be found in the two next analyses (e.g., Bayesian analysis for compartment-specific ASVs and Bayesian analysis for season-specific ASVs), they will be deleted from these two next analyses (the ASV keys are authoritative, but not the taxonomic assignment, which means that species may nevertheless be found here and not subsequently removed if they do not correspond exactly to the same ASV). Specific ASVs were found for each of the six conditions: W-SW, 33; W-SD, 9; and S-CO, 5. ASVs keys, Log2FoldChanges, and taxonomic assignments can be found in Supplementary Table S14.
Using a targeted Bayesian analysis, we identified compartment-specific ASVs (coral rubble vs. seawater; coral rubble vs. sediment; sediment vs. seawater) that could serve as bioindicators (P value adjusted < 0.05 and Log2FoldChange >|2|). Five ASVs were significantly over- or under-represented across all three compartments (Fig. 9B). These included three from the Suessiaceae family (Dinoflagellata), one from the Dinophyceae class (Dinoflagellata), and one from the Bacillariophyceae family (Diatomea). Seventeen ASVs were significantly over-represented in corals, including Margalefidinium polykrikoides (Dinoflagellata), Bacillariophyceae (Diatomea), and Navicula sp. (Diatomea). Twenty ASVs were significantly over-represented in sediment, including Psammodictyon pustulatum (Diatomea), Dinovorax pyriformis (Protalveolata), and Pyramidodinium atrofuscum (Dinoflagellata). Twenty-two ASVs were significantly over-represented in seawater, including Pelagodinium sp. (Dinoflagellata), Symbiodinium uncultured zooxanthellae (Dinoflagellata), and MAST-12A (Stramenopile). As no ASVs from the first Bayesian analysis (Fig. 9A) were found here, no ASVs were removed from this analysis.
Through a targeted Bayesian analysis, we focused on season-specific (austral winter vs. summer) species that could serve as strong bioindicators (P value adjusted < 0.05 and Log2FoldChange >|2|). Fifty-eight ASVs had been significantly (P value adjusted < 0.05) found differentially over- or under-represented between austral summer and winter, but for better visualization, only the 30 most significant ASVs are shown in Fig. 9C. More specifically, during austral summer, 14 ASVs were significantly over-represented (P value adjusted < 0.05), including Navicula sp., Bacillariophyceae, Nitzschia sp., Cylindrotheca closterium (all Diatomea), Margalefidinium polykrikoides, Gymnodiniphycidae, Dinophyceae (all Dinoflagellata), Selenidium sp. (Apicomplexa), uncultured IN2411 ciliate (Ciliophora), Thraustochytrium sp. (Labyrinthulomycetes), and MAST-12A genera (MAST-12 phylum). During austral winter, 16 ASVs were significantly over-represented (P value adjusted < 0.05), more specifically, Suessiaceae, Symbiodium sp., Alexandrium sp., Dinophyta sp., Pyramidodinium atrofuscum (all Dinoflagellata), Gastrocirrhus moniliferis, Hypotrichia ciliate (both Ciliophora), Navicula sp., (Diatomea), and Dinovorax pyriformis (Protalveolata). As no ASVs from the first Bayesian analysis (Fig. 5A) were found here, no ASVs were removed from this analysis.
Discussion
Introduction to Bioindicators in Coral Reefs
Microbial bioindicators, including DNA-based markers, have become indispensable for diagnosing the ecological state of coral reef ecosystems and their responses to climate and local stressors. These tools leverage the sensitivity of microbial communities to environmental changes, providing early detection of ecosystem modifications such as nutrient enrichment, sedimentation, and temperature anomalies. Recent studies [5–7, 26] highlighted the growing potential of eDNA bioindicators in revealing these changes, underscoring their relevance for monitoring and managing reef health.
Given the diversity and complexity of tropical aquatic ecosystems, it is increasingly evident that bioindication approaches must be tailored to specific environmental contexts [5, 92, 93]. The initial step toward effective microbial bioindication involves defining microbial baselines—establishing the diversity, composition, and structure of natural microbial communities across the most relevant reef compartments under a range of environmental conditions. This foundational data is critical for distinguishing between natural variability and stressor-induced shifts. A subsequent step is characterizing temporal variability, as seasonal changes have been shown to significantly influence coral reef microbiomes [26]. Both steps are essential for identifying reliable bioindicators that reflect the ecological state and resilience of reef systems.
In contrast to the findings of Laroche et al. [94], where a number of the putative indicators identified with eDNA in their study exhibited strong site specificity, we were able to eliminate this problem with Bayesian analyses: statistical analyses, including both classical and Bayesian approaches, showed no significant differences in community structure across sites, whether by community type or by compartment. Moreover, the focus on finding compartment- or season-specific indicator ASVs with this type of statistical analysis and their comparison with the ASVs found in the guard safe analysis (the six initial conditions, allowing specific ASV patterns to be found) showing no overlap between the different ASVs perfectly demonstrates the power and relevance of this type of analysis here. Nevertheless, we agree with the conclusions of Laroche et al. [94] that some taxa (taxonomic assignment, not ASVs) were recurrent under certain conditions, and others reflected very specific responses to given conditions (compartments or season), underlining the importance of establishing spatial and temporal baselines to improve the robustness of bioindicator-based monitoring strategies.
In this study, we applied a holistic and intensive approach to characterize the microbiome of the La Saline fringing reef, integrating three ecosystem compartments (seawater, sediment, and coral rubble), two seasons (austral summer and winter), and four genetic markers targeting bacteria, fungi, protists, and microalgae. This multi-faceted approach allowed us for the first time for each kingdom targeted to (i) Document microbial diversity, composition, and structure for each taxonomic kingdom in the La Réunion reef ecosystem; (ii) assess variability across compartments and seasons; and (iii) identify taxa, classes, and genera with potential as bioindicators to enhance reef health monitoring and management strategies. By leveraging eDNA-based bioindicators, this study aims to enhance coral reef monitoring with a framework tailored to ecosystem-specific and temporal dynamics, emphasizing their crucial role in advancing reef management and conservation efforts.
Bacteria
The marine bacterial microbiome in La Réunion Island’s fringing reef exhibited a varying diversity among the studied compartments and seasons. Consistent with studies by [26] and [76], the highest bacterial diversity was observed in sediments during the austral winter. Phylogenetically distant bacterial species in coral rubble during austral summer and seawater during austral winter suggest that substrate type influences microbiome composition [7, 95].
The distribution and relative abundance of certain microbial groups are strongly influenced by the characteristics of specific reef compartments. Endolithic Cyanobacteria, for instance, are predominantly more abundant in hard substrates such as coral rubble compared to sediment or seawater. In seawater, they typically occur as sparse spores or filaments, resulting in a lower genetic signature due to probable dilution effects [14, 96, 97]. Conversely, in sediments, the abundance of endolithic Cyanobacteria is significantly reduced, as most of these species are photophilic, requiring high light availability [14, 17]. Sediments, being more mobile than rubble, present a less stable substrate for their colonization, except species like Plectonema terebrans, which is sciaphilic and adapted to lower light conditions [14, 96]. In seawater, only planktonic Cyanobacteria are naturally expected, while benthic Cyanobacteria dominate in sediment environments. Interestingly, some species may exhibit ubiquity, being present across all compartments, although their relative abundances and ecological roles may vary significantly. Such observations underscore the compartment-specific dynamics that shape the presence and functional distribution of microbial groups within reef ecosystems.
The prevalence of Firmicutes underscores their ecological importance in coral ecosystems. Indeed, [98] and [99] showed the important role of those taxa in reef nutrient cycling, nitrogen fixation, and overall reef resilience, supporting coral health and ecosystem stability. Similarly, as observed in previous research [24, 100, 101], Proteobacteria dominated our studied bacterial communities, highlighting probable specialization within ecological niches [102].
The relative abundance of Proteobacteria shows that it is the most abundant phylum (except in summer seawater) and variable among reef compartments and seasons, with no significant structuration. Cyanobacteria, the second abundant phylum, were mostly abundant in coral rubble during winter, and Firmicutes, the third abundant phylum, in seawater in austral summer. These findings suggest that microbial community composition in coral reefs is highly dynamic [103] and influenced by both seasonal changes and specific microhabitats [26], reflecting the adaptive responses of different bacterial phyla to environmental fluctuations and their potential functional roles in maintaining reef ecosystem stability [102, 104]. Indeed, as an example, Zubia et al. [104] demonstrated a significant shift in the diversity and composition of benthic epilithic bacterial communities in degraded reefs compared to pristine reefs in Moorea, French Polynesia.
By grouping all compartments, we observed significant seasonal changes in bacterial microbiome composition, with distinct taxa dominating respectively in austral summer and winter. During the austral winter, Proteobacteria such as Aestuariibacter, Ahrensia, Cognatishimia, Filomicrobium, and Photobacterium were more abundant, indicating their ecological importance in colder months. Indeed, Aestuariibacter likely plays a role in organic matter degradation (nutrient cycling) [105, 106] and Ahrensia is involved in sulfur oxidation (sulfur cycling) [107]. These processes, potentially more active in winter, suggest a crucial role for Aestuariibacter and Ahrensia in maintaining ecosystem function during this season, particularly by contributing to nutrient and sulfur cycling under conditions that may favor organic matter accumulation and reduced sulfur compound availability. Cognatishimia’s ability to thrive across a wide temperature and salinity range suggests a role in maintaining coral homeostasis under varying environmental conditions [108], Filomicrobium participates in denitrification (contributing to nitrogen removal and balancing nitrogen levels in the ecosystem) [109], and Photobacterium are known for nitrate reduction (supports nutrient cycling) [110]. While Filomicrobium and Photobacterium play key roles in nitrogen cycling through denitrification and nitrate reduction, their hypothesized higher abundance in austral winter aligns with increased nutrient inputs and organic matter from seasonal rainfall. This seasonal nutrient influx likely intensifies microbial recycling processes, underscoring the importance of these taxa in sustaining ecosystem nutrient balance during periods of heightened biogeochemical activity. In contrast, Caldalkalibacillus (Firmicutes), Propionigenium (Fusobacteriota), Acidobacteriota Subgroup_10, and Zeaxanthinibacter (Bacteroidota) are over-represented during the austral summer, indicating their adaptation to warmer conditions (or maybe it is also linked to the fact that the others are eaten by predators that are there in the summer) [111, 112]. These findings underscore the impact of seasonality on the marine microbiome’s composition and function, warranting further research into the ecological roles of seasonally abundant taxa.
In austral summer seawater, Vibrio, a gram-negative bacteria genus [113], is the most abundant among Proteobacteria. Although most Vibrio species are harmless, some can cause diseases in humans, animals, and plants [114], making biomonitoring relevant for this genus. Vibrio are well-established indicators of marine fecal contamination, reflecting water quality issues linked to anthropogenic pressures [115]. In coral reefs, Vibrio serves as a marker of temperature increases and stress events, such as coral bleaching, which are closely tied to global warming [6, 116]. Their prevalence often escalates in response to rising sea surface temperatures and deteriorating reef health. Vibrio collectively represents pressures from pollution, global warming, and ecological disruptions [117]. Some species, such as Vibrio coralliilyticus and Vibrio shilonii, are well-documented coral pathogens. For instance, Vibrio coralliilyticus is a temperature-sensitive opportunistic pathogen that infects multiple coral species and poses a global threat, particularly when temperatures exceed 27 °C [118, 119]. Cyanobacteria play a vital role in contemporary coral reef ecosystems, dominating communities and microbial mats [98]. Cyanobacteria play also important roles in reef primary production [120, 121], nutrient cycling (especially within coral tissues, in sediments and dead corals), nitrogen fixation, and overall coral health and ecosystem stability [9]. Our data indicate a seasonal shift in their abundance among reef compartments, with a higher abundance in coral rubble, particularly in austral winter, then shifting toward a higher relative abundance in austral summer sediments [26]. In austral summer, Actinobacteria, notably abundant in seawater, contrasts with the seasonal pattern observed in the Pearl River Estuary (China) which becomes dominant only in austral winter [122]. Also in austral summer, considering Firmicute’s potential as a bioindicator in polluted areas and its association with coral disease [123, 124] maintaining this phylum as a bioindicator is relevant. Additionally, Glasl et al. [26] have detected a shift in the Firmicutes:Bacteroidota ratio (F:B) over summer, underscoring how seasonal changes in dominant microbial taxa reshape the functional repertoire of host-associated and seawater microbiomes, thereby illustrating the impact of environmental perturbations on microbially mediated processes within coral reef ecosystems. Indeed, an increase in Firmicutes relative to Bacteroidetes may indicate eutrophication or anthropogenic pollution [125, 126], as Firmicutes are often associated with environments impacted by organic enrichment [127, 128]. In our results, we observe a higher F:B ratio in sediments and seawater during the austral summer (same findings as in Glasl et al. [26] for the summer period). This suggests that the sediment microbial community may reflect the increased organic input during warmer periods, possibly due to enhanced microbial metabolism and runoff. Targeted analysis revealed the significative presence of four Photobacterium in austral winter, which are known to be an emerging pathogen often associated with fish, sometimes leading to the death of many fish on the shore [129, 130]. As was recently the case on the beaches of La Réunion Island, bio-monitoring, therefore, seems appropriate. Two significant occurrences of Cognatishimia were noted, and according to [131], the abundance of this species is associated with the good health of some marine life, potentially serving as a positive environmental indicator.
Fungi
As in previous studies [132, 133], Ascomycota and Basidiomycota were the dominant phyla, with Ascomycota consistently prevailing in corals year-round. Specific genera, such as Aspergillus, Cladosporium, and Rhinocladiella, exhibit varying degrees of abundance in different conditions, as in [134]. The seasonal presence of Chytridiomycota in sediment and seawater during summer hints at their ecological roles in nutrient cycling and organic matter decomposition, as in summer, there are more terrigenous inputs due to precipitations (wet season) in the lagoon, enriched in nutrients and organic matter [135, 136]. In contrast to distinct seawater and sediment community structures among seasons, coral rubble communities were more stable, echoing prior studies on environmental influences on microbial fungal dynamics [29, 30, 134]. Indeed, coral rubble condition was stable year-round, probably because fungi present in this type of hard carbonate substrates are bioeroding (cryptic) fungi [96, 137, 138]. Bioeroding communities in dead reef carbonates are known to be relatively mature (stable) after 6 to 12 months of colonization [14, 139]. Moreover bioeroding fungi rely on the organic matter provided by the matrix of the coral itself and from other bioeroding microbes, especially the bioeroding microalga Ostreobium sp. [137, 140], which frees themselves from the presence of organic matter in the water column.
Fungi represent an overlooked kingdom in marine studies [141], and the seasonal variations of their distribution and abundance need to be more studied [30, 137, 142]. Our results suggest distinct seasonal dynamics in different fungal taxa. For example, the over-representation of Aureobasidium leucospermi and Cladosporium dominicanum during austral summer is contrasted by a significant proliferation of well-known species like Aspergillus baarnensis or Periconia byssoides during austral winter. Though these species are still poorly studied, their change in abundance between seasons suggests that important ecological factors may significantly modulate their abundance. Targeted analysis further revealed interesting patterns, including the presence of multiple closely related but distinct species of Malassezia sympodialis in austral winter and the abundance of Simplicillium lanosoniveum strains during the same season, extending to previous studies [143, 144]. In austral summer, seawater exhibits the fewest ASVs compared to coral and sediments, while in austral winter, it surpasses both. This statistically significant seasonal effect is also noted in fungal alpha diversity in seawater and sediment samples in Norway [142]. Our results indicate that the dominant species of marine fungi (mainly Ascomycota and Basidiomycota) are not unique to corals and are frequently found in terrestrial environments [145]. Marine communities, however, include a few specific genera such as Rhodotorula and Chytridiomycota [141, 145, 146], although no ASVs belonging to Rhodotorula were detected in our study, Chytridiomycota were found in sediment and seawater. Moreover, in terms of interactions, coral fungi do not differ fundamentally from the symbiotic, ubiquitous, or pathogenic interactions observed for terrestrial fungi [146]. However, variations in their diversity can be used as environmental indicators [147], for example, during bleaching episodes. A recent study showed that, during a bleaching episode, pathogens such as those in the Apiotrichum genus thrive, while other genera with probiotic roles, such as Fusicolla, regress [147]. This reinforces the importance of monitoring fungal community dynamics in coral reefs.
The Ascomycota:Basidiomycota (A:B) ratio shifts between austral summer and winter in seawater and sediments, as noted previously by [142] between March and September, albeit to a lesser extent. This shift may be attributed to a marine dinoflagellate bloom, disrupting the environment. It is established that inferred changes in community composition reveal potential interactions among taxa [148]. The A:B ratio is already used for terrestrial bio-monitoring with eDNA technics [66], but using this in the marine environment will probably need repeated sampling through time to validate this bioindicator [149]. Indeed, the A:B ratio, while primarily well studied in terrestrial environments [66, 150], could gain recognition in marine systems as a potential bioindicator. Similarly, to land, Ascomycota dominance often reflects high nutrient availability and adaptability to variable conditions, as well as in degraded environments [66, 151], while Basidiomycota dominance is linked to organic matter degradation and healthy environment [149, 152]. For example, Rosenberg and Ben-Haim (2005) [153] noted shifts in Ascomycota species in response to stress events, such as coral bleaching, particularly genera like Aspergillus, increased disproportionately. In our study, the A:B ratio varied across reef compartments and seasons, with Ascomycota dominating in coral rubble and sediments, especially during the austral summer. So, we observed that Ascomycota’s adaptive capacity allows it to thrive under fluctuating environmental conditions, highlighting its utility as a potential bioindicator in reef ecosystems. Within Ascomycota, Aspergillus dominates in relative abundance, except in austral winter sediment where Cladosporium prevails. Given the pathogenicity of some Aspergillus to gorgonians [154] and the strong pathogenic effect of some Cladosporium on shoreline plants [155], their biomonitoring may be relevant, notably in the context of marine pollution [156]. The third phylum, Chytridiomycota, is abundant in austral summer sediments and present in austral winter sediments and seawater. As a species-rich phylum of basal fungi, Chytridiomycota plays vital roles in terrestrial and aquatic ecosystems, but its diversity and richness are largely unknown [157]. The species matching our data is Dinomyces arenysensis, the first chytrid identified to infect marine dinoflagellates [158]. Since the importance of coral–dinoflagellate symbiosis is no longer in question [159], specific monitoring of this species over time could be relevant. Targeted analyses revealed the prominence of the Malassezia taxon. Multiple strains/subspecies/varieties of M. sympodialis are present in austral winter, with one being highly significant in austral summer. The role of this yeast fungus commonly found on human skin, is controversial as it occurs on both healthy and diseased skin [160], suggesting potential applications for health monitoring. The genus Rhinocladiella is notably abundant in austral winter, with R. similis identified significantly by at least two strains/subspecies/varieties during this period. Considering their increasing reports in healthcare settings [161], bio-monitoring of this species could be relevant in future studies.
Microalgae
The highest microalgal diversity was observed in sediment compartments during the austral summer, while coral rubble exhibited lower diversity compared to seawater in the same season (as indicated by Faith’s PD results). These findings suggest the presence of evolutionarily distant species across compartments, reflecting distinct ecological niches and adaptation strategies [162–164]. Analysis of the microalgae composition highlighted the dominance of three major phyla: Rhodophyta, Chlorophyta, and Ochrophyta, with the two first being the most abundant. Rhodophyta is the most abundant phylum in sediment and seawater, while Chlorophyta dominates in coral rubble. Ostreobiaceae was the most abundant family within Chlorophyta. Most of the ASVs were assigned to Ostreobium sp., which is a genus of bioeroding algae that are commonly found in reef environments [41, 165]. The genus-level analysis focused on Chlorophyta revealed Ostreobium sp. as the most abundant genus, followed by Dichotomosiphon and Chlorella. Ostreobium sp. exhibits high relative abundance in various conditions, including coral rubble, sediment, and seawater. Notably, during austral winter, Ostreobium sp. accounted for 100% of the relative abundance in coral rubble and sediment, and 53.4% in seawater, while in austral summer, it represented 55.7% in coral rubble and 98.1% in sediment. These findings highlight for the first time the presence of these bioeroding microalgae in different reef compartments both in winter and summer. Only Grange [166] showed previously the dynamics of dead coral colonization by Ostreobium sp. over several months during both seasons. This author showed that the colonization of dead coral blocks by Ostreobium sp. was delayed in winter compared to summer probably due to lower light conditions, as well as lower temperature and nutrient concentrations. The fact that tufA sequences of Ostreobium sp. were also present in great abundance within microalgae communities in both coral rubble and sediments suggests that those compartments are “reservoir” and that this genus is well present year-round in coral reefs [166, 167]. Moreover, its presence in seawater in winter (> 50%) confirms Massé et al.’s [168] results.
Microalgae show more diverse abundant algae classes in corals in austral summer. While the Rhodophyta are not necessarily well-assigned taxonomically in this study case, the Chlorophyta are well documented. Ostreobiaceae is the most abundant family in Chlorophyta. Most of the ASVs are assigned to Ostreobium sp., which is a genus of green algae that is commonly found in marine and estuarine environments [169]. Effective management strategies are necessary to balance the ecological benefits of Ostreobium sp. with its potential negative impacts and to ensure the health and sustainability of marine ecosystems [9].
Those microboring algae are known to play important roles in reef functioning including benthic primary production [170], carbonate dissolution [33], and nutrient recycling [9]. Furthermore, Ostreobium sp. can contribute to biomineralization, producing calcium carbonate that aids in forming and stabilizing marine structures, including coral reefs, and assisting in coral bleaching recovery [22]. Ostreobium sp. can sometimes become overabundant and form dense mats or coatings that can smother other organisms and reduce biodiversity [171]. Additionally, it can contribute to the build-up of organic matter, which can lead to the development of hypoxic or anoxic conditions in many skeletal zones of corals [19]. They are greatly influenced by environmental factors such as ocean acidification [33], ocean temperature [166], and eutrophication [34]. They can also bloom within a few days inside skeletons of living corals that are under thermal stress due to ocean warming or marine heat waves (e.g., [21]). Thus, monitoring their relative abundance in coral reefs, in seawater, sediments, and coral rubble or dead corals may be of interest to detect possible environmental changes that may greatly impact coral survival such as marine heat waves. But before being able to reach this goal it is important to better understand the natural dynamics of microboring communities dominated by Ostreobium sp., in various places, and at different time scales. Overall, Ostreobium sp. is an interesting and important group of green algae, and ongoing research is helping us to better understand its ecological and evolutionary significance, as well as its potential impacts on marine ecosystems [22, 169, 171].
Protista
The observed variations in diversity metrics, such as the Chao1 index and Simpson evenness index, further emphasized the importance of considering both rare and abundant protist species in understanding the overall diversity and ecological dynamics of microbiomes [172]. The abundance of protists in marine environments is linked to their crucial ecological roles such as primary producers, symbiotic relationships, oxygen production, or again indicator species [173, 174]. The dominance of Dinoflagellata and Diatomea, within the protists community is not surprising as Dinoflagellata form a major group of endosymbionts in living corals [175] and Diatomea are very commonly found within sediments [176]. Surprisingly a relatively unknown phyla, Protalveolata, was also found in great abundance in the present protists community [99, 177]. Analysis of key genera in the Dinoflagetella phylum, including Symbiodinium, Pelagodinium, and Cochlodinium, highlighted their prevalence in the tropical lagoon’s marine microbiome. Symbiodinium, in particular, stands out, underscoring its crucial role in coral physiology and other organisms and in general in ecosystem dynamics [178].
During austral summer, several ASVs belonging to the Diatomea phylum, such as Navicula sp. and Bacillariophyceae, were significantly over-represented. Additionally, ASVs from the Dinoflagellata phylum, including Margalefidinium polykrikoides and Gymnodiniphycidae, showed higher abundance during this season, as well as other phyla, such as Apicomplexa, Ciliophora, Labyrinthulomycetes, and MAST-12. In contrast, austral winter was characterized by an abundance of Dinoflagellata ASVs, particularly from the Suessiaceae family and Symbiodium species. In austral winter, ASVs from Apicomplexa, Ciliophora, Diatomea, and Protalveolata phyla are more abundant, aligning with previous studies on seasonal variations in protista marine microbiomes. This underscores the importance of understanding the ecological dynamics [179] and highlights the necessity for further investigations into the functional roles and ecological implications of these taxa in the marine environment.
Protista is increasingly used in eDNA studies as a bioindicator [180–182]. One of the major interests of the marker we used is its ability to capture sequences relating to the Kingdom of Chromista phytoplankton. The usefulness of Dinoflagellata, Diatomea, Kathablepharidae, Protalveolata, and Peronosporomycetes as a bioindicator is no longer a question [31, 183, 184]. A deep look into the abundant Dinoflagellata showed that some pathogenic genera were found in the La Réunion Island lagoon. Alexandrium is a genus of marine dinoflagellates that includes several species of harmful algae, also known as “red tide” algae. While most species of Alexandrium are harmless, some can produce potent neurotoxins that can cause serious health problems in humans and marine animals [185] and can have devastating impacts on ecosystems [186]. Pelagodinium, a genus found abundant in sediment here, exhibits resilience through the formation of dormant cysts [187], enabling survival in fluctuating environments and potentially contributing to harmful algal blooms [188]. Cochlodinium, a marine dinoflagellate genus that can generate harmful algal blooms, posing environmental and human health risks [189], is abundant in austral winter seawater in this study. Biomonitoring may be relevant for future assessments. Marine stramenopiles (MAST12 phyla) are becoming more and more studied as bioindicators [190, 191]. As heterotrophic protists, they play a pivotal role in microbial food webs, particularly in the recycling of organic matter and the regulation of bacterial populations. Their distribution and abundance are often tightly linked to specific environmental conditions, such as nutrient availability, temperature, and hydrodynamic processes, making them effective proxies for assessing ecosystem health and detecting shifts in ecological balance [190, 191]. They are highly abundant in La Réunion lagoon’s seawater in austral winter, and 600 times less so in sediment at the same time. In contrast, they are not detected in corals or elsewhere in austral summer. So, their seasonal and compartment-specific patterns further underscore their potential as indicators of biogeochemical processes and habitat quality.
Conclusions
The present study is one of the first to mine the microbiome of bacteria, fungi, microalga, and protists simultaneously in a fringing reef ecosystem. The lagoon ecosystem of La Réunion Island revealed variable microbial diversities, compositions, and community structuration among all three compartments (coral rubble, seawater, and sediment), during both austral summer and winter, highlighting the potential for a multi-taxa bioindication of the ecological state of this ecosystem. The fact that no significant variation was observed between the studied sites suggests the reef microbiome is influenced by more contrasted environmental changes than those observed at La Saline fringing reefs. Indeed, the absence of this variation between stations separated by a few kilometers is somewhat unexpected, as spatial heterogeneity in marine microbiomes is typically observed over such distances due to localized environmental factors, such as variations in hydrodynamics, nutrient gradients, and habitat structure. This suggests that microbial communities within the reef may be well integrated across the study area, possibly facilitated by water movement, which could act as a dispersal mechanism. However, it is also possible that the spatial resolution of sampling, or temporal variability in environmental conditions, may have obscured finer-scale differences in community structure. Future studies with higher-resolution sampling or incorporating temporal dynamics could provide further insights into the drivers of microbial distribution at this scale (Table 5).
Table 5.
List of potential bio-indicators
| Kingdom | Potential bio-indicators | Role | Sources |
|---|---|---|---|
| Bacteria | Vibrio | Potentially pathogenic for environment and human’s health | [6, 115–117, 192] |
| Photobacterium | Potentially pathogenic | [129, 130] | |
| Cyanobacteria | Consider seasonal variations when using them as bio-indicators | [98] | |
| Actinobacteria | Consider seasonal variations when using them as bio-indicators | [122] | |
| Firmicutes | Consider seasonal variations when using them as bio-indicators | [123, 124] | |
| Firmicutes:Bacteroidota | Increase in stress | [26, 125, 126, 128] | |
| Cognatishimia | Positive indicator of the environment | [131] | |
| Fungi | Ascomycota:Basidiomycota | Increase in stress | [66, 150–153] |
| Aspergillus | Potentially pathogenic to gorgonians, bio-indicator in polluted area | [154] | |
| Cladosporium | Potentially pathogenic to shore plants | [155] | |
| Dinomyces arenysensis | Potentially pathogenic to marine dinoflagellates | [158] | |
| Malassezia sympodialis | Potentially pathogenic for human’s health | [143, 144] | |
| Simplicillium lanosoniveum | Potentially pathogenic for human’s health | [143, 144] | |
| Rhinocladiella | Potentially pathogenic for human’s health | [161] | |
| Microalgae | Ostreobium | Depending of abundance, need more studies | [22, 169, 171] |
| Protista | Symbiodinium | Seasonal variations underscore the need for further investigations | [178] |
| Pelagodinium | Seasonal variations underscore the need for further investigations | [188] | |
| Cochlodinium | Seasonal variations underscore the need for further investigations | [189] | |
| Dinoflagellata | Bio-indicators for water quality, harmful algal blooms, and overall ecosystem health | [185, 186] | |
| Marine stramenopiles | Bio-indicators for water quality, harmful algal blooms, and overall ecosystem health | [190, 191] |
Austral winter sediments show elevated diversity and unique phylum-level patterns, shaped by seasonal shifts and reef compartments, emphasizing the significance of substrate-specific bacterial taxa in the marine environment. Over-represented species in corals play crucial roles in coral health, symbiosis, sediment degradation, and organic matter recycling. Conversely, under-represented species in sediments have important ecological roles in that habitat. Seasonal fluctuations add complexity to the marine bacterial microbiome, with distinct taxa dominating during austral summer and winter, indicating their ecological relevance in specific seasons. Notably, potentially pathogenic genera like Vibrio and Photobacterium are present and underscore the importance of implementing biomonitoring strategies to ensure environmental safety [193]. Furthermore, the seasonality of certain bacterial phyla, including Cyanobacteria, Actinobacteria, and Firmicutes, highlights the need to consider seasonal variations when using them as bioindicators. The presence of Cognatishimia also suggests its potential as a positive indicator of the environment.
Marine fungal communities, primarily Ascomycota and Basidiomycota, display probable dynamic responses to environmental factors, necessitating further research on their ecological roles and dynamics. As confirmed by [145], Ascomycota and Basidiomycota are ubiquitous in a wide range of ecosystems, including marine and terrestrial environments, where they play essential roles in nutrient cycling and decomposition. In addition, [29] and [146] have documented the presence of these large fungal groups, particularly in coral reefs, highlighting their importance in marine ecosystem dynamics, including in tropical areas where reefs are present. Our study highlights the potential of fungi, particularly Aspergillus and Cladosporium, as bioindicators of water quality and environmental health in coral reef ecosystems. These taxa exhibited significant variability across compartments and seasons, reflecting their sensitivity to changes in nutrient and organic matter inputs. Such patterns suggest that fungal bioindicators could be instrumental in detecting pollution, particularly from wastewater discharges, which may indicate deficiencies in sanitation infrastructure. These findings emphasize the relevance of fungal monitoring for local managers and authorities, providing a valuable tool to inform decisions related to urban planning and wastewater management to safeguard reef ecosystems. Seasonal variations underscore the importance of monitoring different strains and varieties for a comprehensive understanding of ecological roles and potential pathogenicity. These findings contribute to our knowledge of fungal diversity in marine environments, emphasizing the necessity of studying seasonal shifts and community composition. The study highlights indicators for assessing fungal dynamics in marine environments. The Ascomycota:Basidiomycota (A:B) ratio exhibits seasonal shifts in seawater and sediments, potentially serving as a bioindicator pending validation. Monitoring pathogenic genera such as Aspergillus and Cladosporium is relevant for evaluating ecosystem health, while attention to Chytridiomycota, especially Dinomyces arenysensis, may offer insights into coral-dinoflagellate symbiosis. The detection of strains/subspecies/varieties such as M. sympodialis, Simplicillium lanosoniveum, and Rhinocladiella highlights emerging species with implications for human health and biological control.
The microalgae in La Réunion reef ecosystems are underscored by the dominance of Rhodophyta and Chlorophyta phyla, with Ostreobium as a prevalent genus. However, the absence of discernible patterns in microbial community structure suggests a relatively homogeneous composition among the investigated conditions, aligning with the observed high spatial and temporal variability in marine ecosystems.
In brief, the study reveals the diverse abundance of algae classes in corals during austral summer, although the limitation of missing data for one condition restricts interpretation. Effective management strategies are vital to balance the ecological benefits and potential negative impacts of Ostreobium, emphasizing the need for further research to explore seasonal and compartmental variations. Ostreobium, as a diverse and ecologically significant group of green algae, plays a crucial role in primary production, biomineralization, and ecosystem functioning in marine environments. However, its overabundance and potential impact on biodiversity and organic matter accumulation necessitate further investigation and monitoring for the health and resilience of marine ecosystems.
The ecological significance of protista in the marine ecosystem is evident through the dominance of Dinoflagellata and Diatomea phyla, featuring genera like Symbiodinium, Pelagodinium, and Cochlodinium. The consistent presence of certain ASVs among reef compartments hints at their potential ecological importance. Seasonal variations between austral summer and winter underscore the need for further investigations into the functional roles and ecological implications of key protista taxa. Dinoflagellata and marine stramenopiles emerge as promising bioindicators for water quality, harmful algal blooms, and overall ecosystem health, emphasizing the importance of ongoing research and biomonitoring efforts for a deeper understanding of protista in marine environments.
Monitoring the identified bioindicators over time provides a crucial perspective for understanding the ecological dynamics of marine ecosystems. Correlating these indicators with lagoon physico-chemical parameters, including pH, salinity, temperature, and nutrients, offers a holistic view of the interplay between microbial communities and environmental conditions. Analyzing these correlations helps unveil patterns, shedding light on the impact of seasonal variations and human activities on the lagoon ecosystem’s health and resilience. Integrating anthropogenic parameters, such as human occupation, enhances the analysis, providing insights into the potential influence of human activities on microbial diversity. This combined approach is vital for developing effective management and conservation strategies that strike a balance between the ecological integrity of the marine environment and human needs.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
PJ, AT, FG, CP, and GP conceived the study; PJ, AT, and FG performed fieldwork. PJ performed the laboratory work. PLS completed sample processing, analysis, and interpretation of results. PLS, PJ, AT, and FG wrote the manuscript while CP and GP contributed to the review of the manuscript prior to submission.
Funding
The project BIOINDIC (part 1, 2021) funded by by the Fédération des Laboratoires de l’Observations des Milieux naturels et des changements globaux (OMNCG), Université de La Réunion, France, supported the study. This research was also supported by funds from Observatoire des Sciences de l’Univers à La Réunion (OSU-R) and funds from Institut de la Recherche pour le Developpement (IRD; UMRs ENTROPIE and LOCEAN).
Data Availability
Raw sequences generated and analyzed during the current study are available under the NCBI BioProject PRJNA985136, BioSample SUB13558791, from SRR24958245 to SRR24958195 for the 16S, from SRR25108721 to SRR25108698 for the ITS, from SRR25080909 to SRR25080857 for the tufA, and from SRR24961358 to SRR24961336 for the 18S.
Declarations
Competing Interests
The authors declare no competing interests.
References
- 1.Hoegh-Guldberg O, Poloczanska ES, Skirving W, Dove S (2017) Coral reef ecosystems under climate change and ocean acidification. Front Mar Sci 4. 10.3389/fmars.2017.00158
- 2.Sweet M, Burian A, Bulling M (2021) Corals as canaries in the coalmine: towards the incorporation of marine ecosystems into the ‘One Health’ concept. J Invertebr Pathol 186:107538. 10.1016/j.jip.2021.107538 [DOI] [PubMed] [Google Scholar]
- 3.Voolstra CR, Raina JB, Dörr M et al (2024) The coral microbiome in sickness, in health and in a changing world. Nat Rev Microbiol 22:460–475. 10.1038/s41579-024-01015-3 [DOI] [PubMed] [Google Scholar]
- 4.Mohamed AR, Ochsenkühn MA, Kazlak AM et al (2023) The coral microbiome: towards an understanding of the molecular mechanisms of coral-microbiota interactions. FEMS Microbiol Rev 47:1–26. 10.1093/femsre/fuad005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roitman S, Joseph Pollock F, Medina M (2018) Coral microbiomes as bioindicators of reef health. 39–57. 10.1007/13836_2018_29
- 6.Glasl B, Webster NS, Bourne DG (2017) Microbial indicators as a diagnostic tool for assessing water quality and climate stress in coral reef ecosystems. Mar Biol 164:1–18. 10.1007/s00227-017-3097-x27980349 [Google Scholar]
- 7.Glasl B, Bourne DG, Frade PR et al (2019) Microbial indicators of environmental perturbations in coral reef ecosystems. Microbiome 7:1–13. 10.1186/s40168-019-0705-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reigel AM, Paz-García DA, Hellberg ME (2021) Microbiome of a reef-building coral displays signs of acclimation to a stressful shallow hydrothermal vent habitat. Front Mar Sci 8. 10.3389/fmars.2021.652633
- 9.Massé A, Tribollet A, Meziane T et al (2020) Functional diversity of microboring Ostreobium algae isolated from corals. Environ Microbiol 22:4825–4846. 10.1111/1462-2920.15256 [DOI] [PubMed] [Google Scholar]
- 10.Marcelino VR, Verbruggen H (2016) Multi-marker metabarcoding of coral skeletons reveals a rich microbiome and diverse evolutionary origins of endolithic algae. Sci Rep 6:1–9. 10.1038/srep31508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epstein HE, Smith HA, Cantin NE et al (2019) Temporal variation in the microbiome of Acropora coral species does not reflect seasonality. Front Microbiol 10:1–14. 10.3389/fmicb.2019.01775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lima LFO, Alker AT, Papudeshi B et al (2023) Coral and seawater metagenomes reveal key microbial functions to coral health and ecosystem functioning shaped at reef scale. Microb Ecol 86:392–407. 10.1007/s00248-022-02094-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golubic S, Friedmann EI, Schneider J (1981) The lithobiontic ecological niche, with special reference to microorganisms. J Sediment Res 51:475–478. 10.1306/212F7CB6-2B24-11D7-8648000102C1865D [Google Scholar]
- 14.Tribollet A (2008) The boring microflora in modern coral reefs: a review of its roles. Current developments in bioerosion. Berlin-Heiderlberg, Springer-Verlag, Wisshak M, pp 67–94 [Google Scholar]
- 15.Tribollet A, Golubic S (2011) Reef bioerosion: agents and processes. In: Springer (ed) Coral reefs: an ecosystem in transition. Dordrecht, pp 435–449
- 16.Pernice M, Raina JB, Rädecker N et al (2020) Down to the bone: the role of overlooked endolithic microbiomes in reef coral health. ISME J 14:325–334. 10.1038/s41396-019-0548-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tribollet A, Veinott G, Golubic S, Dart R (2008) Infestation of the North American freshwater mussel Elliptiocomplanata (Head Lake, Canada) by the euendolithic cyanobacterium Plectonematerebrans Bornet et Flahault. Arch Hydrobiol Suppl Algol Stud 128:65–77. 10.1127/1864-1318/2008/0128-0065 [Google Scholar]
- 18.Verbruggen H, Tribollet A (2011) Boring algae. Curr Biol 21:R876–R877. 10.1016/j.cub.2011.09.014 [DOI] [PubMed] [Google Scholar]
- 19.Ricci F, Rossetto Marcelino V, Blackall LL et al (2019) Beneath the surface: community assembly and functions of the coral skeleton microbiome. Microbiome 7:1–10. 10.1186/s40168-019-0762-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrer LM, Szmant AM (1988) Nutrient regeneration by the endolithic community in coral skeletons. In: Proceedings of the 6th international Coral Reef Symposium. Townsville, Australia: AIMS., p (Vol. 2) 1–4
- 21.Fine M, Loya Y (2002) Endolithic algae: an alternative source of photoassimilates during coral bleaching. Proc R Soc B Biol Sci 269:1205–1210. 10.1098/rspb.2002.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galindo-Martínez CT, Weber M, Avila-Magaña V et al (2022) The role of the endolithic alga Ostreobium spp. during coral bleaching recovery. Sci Rep 12:1–12. 10.1038/s41598-022-07017-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourne DG, Morrow KM, Webster NS (2016) Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol 70:317–340. 10.1146/annurev-micro-102215-095440 [DOI] [PubMed] [Google Scholar]
- 24.Cleary DFR, Polónia ARM, Becking LE et al (2018) Compositional analysis of bacterial communities in seawater, sediment, and sponges in the Misool coral reef system, Indonesia. Mar Biodivers 48:1889–1901. 10.1007/s12526-017-0697-0 [Google Scholar]
- 25.Galand PE, Ruscheweyh HJ, Salazar G et al (2023) Diversity of the Pacific Ocean coral reef microbiome. Nat Commun 14:1–13. 10.1038/s41467-023-38500-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glasl B, Robbins S, Frade PR et al (2020) Comparative genome-centric analysis reveals seasonal variation in the function of coral reef microbiomes. ISME J 14:1435–1450. 10.1038/s41396-020-0622-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menaa F, Wijesinghe PAUI, Thiripuranathar G et al (2020) Ecological and industrial implications of dynamic seaweed-associated microbiota interactions. Mar Drugs 18:1–25. 10.3390/md18120641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Domart-Coulon IJ, Sinclair CS, Hill RT et al (2004) A basidiomycete isolated from the skeleton of Pocilloporadamicornis (Scleractinia) selectively stimulates short-term survival of coral skeletogenic cells. Mar Biol 144:583–592. 10.1007/s00227-003-1227-0 [Google Scholar]
- 29.Raghukumar C, Ravindran J (2012) Fungi and their role in corals and coral reef ecosystems. Biology of marine fungi. Springer, Berlin Heidelberg, pp 89–113 [DOI] [PubMed]
- 30.Richards TA, Jones MDM, Leonard G, Bass D (2012) Marine fungi: their ecology and molecular diversity. Ann Rev Mar Sci 4:495–522. 10.1146/annurev-marine-120710-100802 [DOI] [PubMed] [Google Scholar]
- 31.Amengual-Morro C, Moyà Niell G, Martínez-Taberner A (2012) Phytoplankton as bioindicator for waste stabilization ponds. J Environ Manage 95:S71–S76. 10.1016/j.jenvman.2011.07.008 [DOI] [PubMed] [Google Scholar]
- 32.Wu G, Huang A, Wen Y et al (2022) Euendolithic Cyanobacteria and Proteobacteria together contribute to trigger bioerosion in aquatic environments. Front Microbiol 13:1–12. 10.3389/fmicb.2022.938359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tribollet A, Chauvin A, Cuet P (2019) Carbonate dissolution by reef microbial borers: a biogeological process producing alkalinity under different pCO 2 conditions. Facies 65. 10.1007/s10347-018-0548-x
- 34.Carreiro-Silva M, McClanahan TR, Kiene WE (2009) Effects of inorganic nutrients and organic matter on microbial euendolithic community composition and microbioerosion rates. Mar Ecol Prog Ser 392:1–15. 10.3354/meps08251 [Google Scholar]
- 35.Brandt MI, Pradillon F, Trouche B et al (2021) Evaluating sediment and water sampling methods for the estimation of deep-sea biodiversity using environmental DNA. Sci Rep 11:1–14. 10.1038/s41598-021-86396-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laroche O, Kersten O, Smith CR, Goetze E (2020) From sea surface to seafloor: a benthic allochthonous eDNA survey for the abyssal ocean. Front Mar Sci 7:1–16. 10.3389/fmars.2020.0068232802822 [Google Scholar]
- 37.Thakur NL, Müller WE (2004) Biotechnological potential of marine microbes. Curr Sci 1506–1512. 10.1007/978-3-642-53971-8_26
- 38.Bonacolta AM, Weiler BA, Porta-Fitó T et al (2023) Beyond the Symbiodiniaceae: diversity and role of microeukaryotic coral symbionts. Coral Reefs 42:567–577. 10.1007/s00338-023-02352-0 [Google Scholar]
- 39.Aranda Lastra M, van Oppen MJH (2022) Synthesis: coral reef conservation and restoration in the Omics Age. Springer International Publishing, Cham [Google Scholar]
- 40.Grange JS, Rybarczyk H, Tribollet A (2015) The three steps of the carbonate biogenic dissolution process by microborers in coral reefs (New Caledonia). Environ Sci Pollut Res 22:13625–13637. 10.1007/s11356-014-4069-z [DOI] [PubMed] [Google Scholar]
- 41.Sauvage T, Schmidt WE, Suda S, Fredericq S (2016) A metabarcoding framework for facilitated survey of endolithic phototrophs with tufA. BMC Ecol 16:1–21. 10.1186/s12898-016-0068-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villeneuve N, Bachèlery P, Kemp J (2014) La réunion island: a typical example of a basaltic shield volcano with rapid evolution. World geomorphological landscapes, pp 261–270. 10.1007/978-94-007-7022-5_25.hal-02109094
- 43.Myers N, Mittermeier RA, Mittermeier CG et al (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858. 10.1038/35002501 [DOI] [PubMed] [Google Scholar]
- 44.L’Helguen S, Chauvaud L, Cuet P et al (2014) A novel approach using the 15N tracer technique and benthic chambers to determine ammonium fluxes at the sediment-water interface and its application in a back-reef zone on Reunion Island (Indian Ocean). J Exp Mar Bio Ecol 452:143–151. 10.1016/j.jembe.2013.12.001 [Google Scholar]
- 45.Join J, Coudray J, Longworth K (1997) Using principal components analysis and Na/Cl ratios to trace groundwater circulation in a volcanic island: the example of Reunion. J Hydrol 190:1–18. 10.1016/S0022-1694(96)03070-3 [Google Scholar]
- 46.Naim O, Tourrand C, Bigot L et al (2013) Fringing Reefs of Reunion island and eutrophication effects - Part 2: long-term monitoring of primary producers. Atoll Res Bull 597:1–36 [Google Scholar]
- 47.Tedetti M, Bigot L, Turquet J et al (2020) Influence of freshwater discharges on biogeochemistry and benthic communities of a coral reef ecosystem (La Réunion Island, Indian Ocean). Front Mar Sci 7:1–18. 10.3389/fmars.2020.59616532802822 [Google Scholar]
- 48.Guigue C, Bigot L, Turquet J et al (2015) Hydrocarbons in a coral reef ecosystem subjected to anthropogenic pressures (La Reúnion Island, Indian Ocean). Environ Chem 12:350–365. 10.1071/EN14194 [DOI] [PubMed] [Google Scholar]
- 49.Chazottes V, Le Campion-Alsumard T, Peyrot-Clausade M, Cuet P (2002) The effects of eutrophication-related alterations to coral reef communities on agents and rates of bioerosion (Reunion Island, Indian Ocean). Coral Reefs 21:375–390. 10.1007/s00338-002-0259-0 [Google Scholar]
- 50.Chauvin A, Denis V, Cuet P (2011) Is the response of coral calcification to seawater acidification related to nutrient loading? Coral Reefs 30:911–923. 10.1007/s00338-011-0786-7 [Google Scholar]
- 51.Zubia M, Depetris M, Flores O et al (2018) Macroalgae as a tool for assessing the ecological status of coral reefs under the Water Framework Directive: a case study on the reef flats of La Réunion (Indian Ocean). Mar Pollut Bull 137:339–351. 10.1016/j.marpolbul.2018.10.029 [DOI] [PubMed] [Google Scholar]
- 52.Lagoutte E, Tribollet A, Bureau S et al (2024) Biogeochemical evidence of flow re-entrainment on the main fringing reef of La Reunion Island. Mar Chem 259. 10.1016/j.marchem.2024.104352
- 53.Bruce K, Blackman R, Bourlat SJ, et al (2021) A practical guide to DNA-based methods for biodiversity assessment, Advanced B. 10.3897/ab.e68634
- 54.Klindworth A, Pruesse E, Schweer T et al (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:1–11. 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ihrmark K, Bödeker ITM, Cruz-Martinez K et al (2012) New primers to amplify the fungal ITS2 region - evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol 82:666–677. 10.1111/j.1574-6941.2012.01437.x [DOI] [PubMed] [Google Scholar]
- 56.White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc a Guid to methods Appl 18:315–322 [Google Scholar]
- 57.Maddison DR, Baker MD, Ober KA (1999) Phylogeny of carabid beetles as inferred from 18S ribosomal DNA (Coleoptera: Carabidae). Syst Entomol 24:103–138. 10.1046/j.1365-3113.1999.00088.x [Google Scholar]
- 58.Mangot JF, Domaizon I, Taib N et al (2013) Short-term dynamics of diversity patterns: evidence of continual reassembly within lacustrine small eukaryotes. Environ Microbiol 15:1745–1758. 10.1111/1462-2920.12065 [DOI] [PubMed] [Google Scholar]
- 59.Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinforma. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 60.Ewels P, Magnusson M, Lundin S, Käller M (2016) MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolger AM, Lohse M, Usadel B (2014) Genome analysis Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolyen E, Rideout JR, Dillon MR et al (2018) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Callahan BJ, McMurdie PJ, Holmes SP (2017) Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. 10.1038/ismej.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galan M, Razzauti M, Bard E et al (2016) 16S rRNA amplicon sequencing for epidemiological surveys of bacteria in wildlife. mSystems 1. 10.1128/msystems.00032-16 [DOI] [PMC free article] [PubMed]
- 66.Fernandez Nuñez N, Maggia L, Stenger P-L et al (2021) Potential of high-throughput eDNA sequencing of soil fungi and bacteria for monitoring ecological restoration in ultramafic substrates: the case study of the New Caledonian biodiversity hotspot. Ecol Eng 173. 10.1016/j.ecoleng.2021.106416
- 67.Shannon C, Weaver W (1949) The mathematical theory of communication. University of illinois Press, Champaign [Google Scholar]
- 68.Faith DP (1992) Conservation evaluation and phylogenetic diversity. Biol Conserv 61:1–10. 10.1016/0006-3207(92)91201-3 [Google Scholar]
- 69.Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Price MN, Dehal PS, Arkin AP (2009) Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bokulich NA, Kaehler BD, Rideout JR et al (2018) Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:1–17. 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abarenkov K, Zirk A, Piirmann T et al (2021) Full UNITE+INSD dataset for eukaryotes. Version 10.05.2021. UNITE Community
- 73.Porter TM (2021) terrimporter/UNITE_ITSClassifier: UNITE v2.0. Zenodo
- 74.Burki F, Roger AJ, Brown MW, Simpson AGB (2020) The new tree of eukaryotes. Trends Ecol Evol 35:43–55. 10.1016/j.tree.2019.08.008 [DOI] [PubMed] [Google Scholar]
- 75.Kolodziej K, Stoeck T (2007) Cellular identification of a novel uncultured marine stramenopile (MAST-12 clade) small-subunit rRNA gene sequence from a Norwegian estuary by use of fluorescence in situ hybridization-scanning electron microscopy. Appl Environ Microbiol 73:2718–2726. 10.1128/AEM.02158-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Winkel M, Trivedi CB, Mourot R et al (2022) Seasonality of glacial snow and ice microbial communities. Front Microbiol 13. 10.3389/fmicb.2022.876848 [DOI] [PMC free article] [PubMed]
- 77.R Development Core Team R (2011) R: a language and environment for statistical computing. R Found Stat Comput 1:409 [Google Scholar]
- 78.DeSantis TZ, Hugenholtz P, Larsen N et al (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Stat 11:265–270 [Google Scholar]
- 80.Pielou EC (1966) The measurement of diversity in different types of biological collections. J Theor Biol 13:131–144. 10.1016/0022-5193(66)90013-0 [Google Scholar]
- 81.de Mendiburu F (2021) agricolae: statistical procedures for agricultural research (No Title)
- 82.Wickham H, Chang W, Henry L, Pedersen T, Takahashi K, Wilke C, Woo K (2019) R Package ‘ggplot2’ v. 3.1.1. Cran R
- 83.Sørensen TA (1948) A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biol Skar 5:1–34 [Google Scholar]
- 84.Oksanen AJ, Blanchet FG, Friendly M et al (2020) Vegan: community ecology package, pp 1–298. http://vegan.r-forge.r-project.org/
- 85.Martinez Arbizu P (2020) pairwiseAdonis: Pairwise multilevel comparison using Adonis. R package version 0.4
- 86.Mbareche H, Dumont-leblond N, Bilodeau GJ, Duchaine C (2020) An overview of bioinformatics tools for DNA meta-barcoding analysis of microbial communities of bioaerosols : digest for microbiologists. Life 10:1–20. 10.3390/life10090185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruiz-Ramos D V, Meyer RS, Toews D et al (2022) Environmental DNA (eDNA) detects temporal and habitat effects on community composition and endangered species in ephemeral ecosystems : a case study in vernal pools. Environ DNA 1–17. 10.1002/edn3.360
- 88.Senn S, Bhattacharyya S, Presley G et al (2022) The functional biogeography of eDNA metacommunities in the post-fire landscape of the Angeles National Forest. Microorganisms 10. 10.3390/microorganisms10061218 [DOI] [PMC free article] [PubMed]
- 89.Stenger P-L (2022) The R Package “Anaconda”: targeted differential and global enrichment analysis of taxonomic rank by shared ASVS 1–28. Cran R. 10.13140/RG.2.2.11117.67048
- 90. Stenger P-L, Léopold A, Dinh K, Mournet P, Robert N, Drouin J et al (2025) Advancing biomonitoring of eDNA studies with the Anaconda R package: integrating soil and One Health perspectives in the face of evolving traditional agriculture practices. PLoS ONE 20(1):e0311986. 10.1371/journal.pone.0311986 [DOI] [PMC free article] [PubMed]
- 91.Stenger P-L, Ky C-L, Reisser C et al (2021) Molecular pathways and pigments underlying the colors of the pearl oyster Pinctada margaritifera var. cumingii (Linnaeus 1758). Genes (Basel) 12. 10.3390/genes12030421 [DOI] [PMC free article] [PubMed]
- 92.Bridge TCL, Done TJ, Friedman A et al (2011) Variability in mesophotic coral reef communities along the great barrier reef, Australia. Mar Ecol Prog Ser 428:63–75. 10.3354/meps09046 [Google Scholar]
- 93.Rivest EB, Comeau S, Cornwall CE (2017) The role of natural variability in shaping the response of coral reef organisms to climate change. Curr Clim Chang Rep 271–281. 10.1007/s40641-017-0082-x
- 94.Laroche O, Wood SA, Tremblay LA et al (2018) A cross-taxa study using environmental DNA/RNA metabarcoding to measure biological impacts of offshore oil and gas drilling and production operations. Mar Pollut Bull 127:97–107. 10.1016/j.marpolbul.2017.11.042 [DOI] [PubMed] [Google Scholar]
- 95.Oberbeckmann S, Osborn AM, Duhaime MB (2016) Microbes on a bottle: Substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PLoS ONE 11:1–24. 10.1371/journal.pone.0159289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Le Campion-Alsumard T, Golubic S, Hutchings P (1995) Microbial endoliths in skeletons of live and dead corals: Porites lobata (Moorea, French Polynesia). Mar Ecol Prog Ser:149–157. https://www.jstor.org/stable/44634827
- 97.Schneider J, Le C-A (1999) Construction and destruction of carbonates by marine and freshwater cyanobacteria. Eur J Phycol 34:417–426. 10.1080/09670269910001736472 [Google Scholar]
- 98.Charpy L, Casareto BE, Langlade MJ, Suzuki Y (2012) Cyanobacteria in coral reef ecosystems: a review. J Mar Biol 2012:1–9. 10.1155/2012/259571 [Google Scholar]
- 99.Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG (2004) Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science (80-) 305:997–1000. 10.1126/science.1099128 [DOI] [PubMed] [Google Scholar]
- 100.Delmont TO, Quince C, Shaiber A et al (2018) Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat Microbiol 3:804–813. 10.1038/s41564-018-0176-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.González JM, Moran MA (1997) Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol 63:4237–4242. 10.1128/aem.63.11.4237-4242.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dussud C, Meistertzheim AL, Conan P et al (2018) Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters. Environ Pollut 236:807–816. 10.1016/j.envpol.2017.12.027 [DOI] [PubMed] [Google Scholar]
- 103.Tout J, Jeffries TC, Webster NS et al (2014) Variability in microbial community composition and function between different niches within a coral reef. Microb Ecol 67:540–552. 10.1007/s00248-013-0362-5 [DOI] [PubMed] [Google Scholar]
- 104.Zubia M, Vieira C, Palinska KA et al (2019) Benthic cyanobacteria on coral reefs of Moorea Island (French Polynesia): diversity response to habitat quality. Hydrobiologia 843:61–78. 10.1007/s10750-019-04029-8 [Google Scholar]
- 105.Erni-Cassola G, Wright RJ, Gibson MI, Christie-Oleza JA (2020) Early colonization of weathered polyethylene by distinct bacteria in marine coastal seawater. Microb Ecol 79:517–526. 10.1007/s00248-019-01424-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Whitaker, (2008) Exoenzyme dynamics of surface ocean microbial communities in response to oil exposure. Texas A&M University, College Station [Google Scholar]
- 107.Li Y, Tang K, Zhang L et al (2018) Coupled carbon, sulfur, and nitrogen cycles mediated by microorganisms in the water column of a shallow-water hydrothermal ecosystem. Front Microbiol 9:1–13. 10.3389/fmicb.2018.02718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu X, Liu S, Yu Z et al (2024) Cognatishimia coralii sp. nov., a marine bacterium isolated from seawater surrounding corals. Int J Syst Evol Microbiol 74:006467. 10.1099/ijsem.0.006467 [DOI] [PubMed] [Google Scholar]
- 109.Chen Y, Dong X, Sun Z et al (2024) Potential coupling of microbial methane, nitrogen, and sulphur cycling in the Okinawa Trough cold seep sediments. Microbiol Spectr 12:1–15. 10.1128/spectrum.03490-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nicholas DJD, Redmond WJ, Wright MA (1964) Effects of cultural conditions on nitrate reductase in Photobacteriumsepia. Microbiology 35:401–410. 10.1099/00221287-35-3-401 [DOI] [PubMed] [Google Scholar]
- 111.Abirami B, Radhakrishnan M, Kumaran S, Wilson A (2021) Impacts of global warming on marine microbial communities. Sci Total Environ 791:147905. 10.1016/j.scitotenv.2021.147905 [DOI] [PubMed] [Google Scholar]
- 112.Auladell A, Barberán A, Logares R et al (2022) Seasonal niche differentiation among closely related marine bacteria. ISME J 16:178–189. 10.1038/s41396-021-01053-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brumfield KD, Usmani M, Chen KM et al (2021) Environmental parameters associated with incidence and transmission of pathogenic Vibrio spp. Environ Microbiol 23:7314–7340. 10.1111/1462-2920.15716 [DOI] [PubMed] [Google Scholar]
- 114.Baker-Austin C, Oliver JD, Alam M et al (2018) Vibrio spp. infections. Nat Rev Dis Prim 4:1–19 [DOI] [PubMed] [Google Scholar]
- 115.Holcomb DA, Stewart JR (2020) Microbial indicators of fecal pollution: recent progress and challenges in assessing water quality. Curr Environ Heal Reports 7:311–324. 10.1007/s40572-020-00278-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang W, Tang K, Wang X (2024) High temperatures increase the virulence of Vibrio bacteria towards their coral host and competing bacteria via type VI secretion systems. PLoS Biol 22:4–7. 10.1371/journal.pbio.3002788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sampaio A, Silva V, Poeta P, Aonofriesei F (2022) Vibrio spp.: life strategies, ecology, and risks in a changing environment. Diversity 14:1–26. 10.3390/d14020097 [Google Scholar]
- 118.Ben-Haim Y, Zicherman-Keren M, Rosenberg E (2003) Temperature-regulated bleaching and lysis of the coral Pocilloporadamicomis by the novel pathogen Vibriocoralliilyticus. Appl Environ Microbiol 69:4236–4242. 10.1128/AEM.69.7.4236-4242.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Munn CB (2015) The role of vibrios in diseases of corals. Microbiol Spectr 3. 10.1128/microbiolspec.ve-0006-2014 [DOI] [PubMed]
- 120.Adey WH (1998) Coral reefs: algal structured and mediated ecosystems in shallow, turbulent, alkaline waters. J Phycol 34:393–406 [Google Scholar]
- 121.Tribollet A, Langdon C, Golubic S, Atkinson M (2006) Endolithic microflora are major primary producers in dead carbonate substrates of Hawaiian coral reefs. J Phycol 42:292–303. 10.1111/j.1529-8817.2006.00198.x [Google Scholar]
- 122.Li Y, Huang D, Sun W et al (2022) Characterizing sediment bacterial community and identifying the biological indicators in a seawater-freshwater transition zone during the wet and dry seasons. Environ Sci Pollut Res 29:41219–41230. 10.1007/s11356-021-18053-6 [DOI] [PubMed] [Google Scholar]
- 123.Frias-Lopez J, Klaus JS, Bonheyo GT, Fouke BW (2004) Bacterial community associated with black band disease in corals. Appl Environ Microbiol 70:5955–5962. 10.1128/AEM.70.10.5955-5962.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Torres GG, Figueroa-Galvis I, Muñoz-García A et al (2019) Potential bacterial bioindicators of urban pollution in mangroves. Environ Pollut 255. 10.1016/j.envpol.2019.113293 [DOI] [PubMed]
- 125.Dai W, Zhang J, Tu Q et al (2017) Bacterioplankton assembly and interspecies interaction indicating increasing coastal eutrophication. Chemosphere 177:317–325. 10.1016/j.chemosphere.2017.03.034 [DOI] [PubMed] [Google Scholar]
- 126.Zhang H, Yang L, Li Y et al (2022) Pollution gradients shape the co-occurrence networks and interactions of sedimentary bacterial communities in Taihu Lake, a shallow eutrophic lake. J Environ Manage 305:114380. 10.1016/j.jenvman.2021.114380 [DOI] [PubMed] [Google Scholar]
- 127.Li H, Peng J, Weber KA, Zhu Y (2011) Phylogenetic diversity of Fe(III)-reducing microorganisms in rice paddy soil: enrichment cultures with different short-chain fatty acids as electron donors. J Soils Sediments 11:1234–1242. 10.1007/s11368-011-0371-2 [Google Scholar]
- 128.Kondo R, Mori Y, Sakami T (2012) Comparison of sulphate-reducing bacterial communities in Japanese fish farm sediments with different levels of organic enrichment. Microbes Environ 27:193–199. 10.1264/jsme2.ME11278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Romalde JL (2002) Photobacteriumdamselae subsp.piscicida: an integrated view of a bacterial fish pathogen. Int Microbiol 5:3–9. 10.1007/s10123-002-0051-6 [DOI] [PubMed] [Google Scholar]
- 130.Terceti MS, Ogut H, Osorio R (2016) Photobacteriumdamselae subsp.damselae, an emerging fish pathogen in the Black Sea: evidence of a multiclonal origin. Appl Environ Microbiol 82:3736–3745. 10.1128/AEM.00781-16.Editor [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Reyes G, Betancourt I, Andrade B et al (2022) Microbiome of Penaeusvannamei larvae and potential biomarkers associated with high and low survival in shrimp hatchery tanks affected by acute hepatopancreatic necrosis disease. Front Microbiol 13:1–14. 10.3389/fmicb.2022.838640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.van Oppen MJH, Blackall LL (2019) Coral microbiome dynamics, functions and design in a changing world. Nat Rev Microbiol 17:557–567. 10.1038/s41579-019-0223-4 [DOI] [PubMed] [Google Scholar]
- 133.Schultz J, Modolon F, Rosado AS et al (2022) Methods and strategies to uncover coral-associated microbial dark matter. mSystems 7. 10.1128/msystems.00367-22 [DOI] [PMC free article] [PubMed]
- 134.Priest T, Fuchs B, Amann R, Reich M (2021) Diversity and biomass dynamics of unicellular marine fungi during a spring phytoplankton bloom. Environ Microbiol 23:448–463. 10.1111/1462-2920.15331 [DOI] [PubMed] [Google Scholar]
- 135.Manohar CS, Raghukumar C (2013) Fungal diversity from various marine habitats deduced through culture-independent studies. FEMS Microbiol Lett 341:69–78. 10.1111/1574-6968.12087 [DOI] [PubMed] [Google Scholar]
- 136.Pang KL, Hassett BT, Shaumi A et al (2021) Pathogenic fungi of marine animals: a taxonomic perspective. Fungal Biol Rev 38:92–106. 10.1016/j.fbr.2021.03.008 [Google Scholar]
- 137.Golubic S, Radtke G, Le Campion-Alsumard T (2005) Endolithic fungi in marine ecosystems. Trends Microbiol 13:229–235. 10.1016/j.tim.2005.03.007 [DOI] [PubMed] [Google Scholar]
- 138.Priess K, Le Campion-Alsumard T, Thomassin BA et al (2000) Fungi in corals: black bands and density-banding of Poriteslutea and P.lobata skeleton. Mar Biol 136:19–27. 10.1007/s002270050003 [Google Scholar]
- 139.Tribollet A, Atkinson MJ, Langdon C (2006) Effects of elevated pCO2 on epilithic and endolithic metabolism of reef carbonates. Glob Chang Biol 12:2200–2208. 10.1111/j.1365-2486.2006.01249.x [Google Scholar]
- 140.Tribollet A, Payri C (2001) Bioerosion of the coralline alga Hydrolithon onkodes by microborers in the coral reefs of Moorea, French Polynesia. Oceanol Acta 24:329–342. 10.1016/S0399-1784(01)01150-1 [Google Scholar]
- 141.Breyer E, Baltar F (2023) The largely neglected ecological role of oceanic pelagic fungi. Trends Ecol Evol 38:870–888. 10.1016/j.tree.2023.05.002 [DOI] [PubMed] [Google Scholar]
- 142.Husanovic E (2021) Assessing the effect of salmon aquaculture on fungal diversity in seawater and sediments through eDNA metabarcoding. Master's thesis. UiT The Arctic University of Norway
- 143.Aleem AA (1980) Distribution and ecology of marine fungi in Sierra Leone (tropical West Africa). Bot Mar 23:679–688. 10.1515/botm-1980-1104 [Google Scholar]
- 144.Vrijmoed LLP, Hodgkiss IJ, Thrower LB (1982) Seasonal patterns of primary colonization by lignicolous marine fungi in Hong Kong. Hydrobiologia 89:253–262. 10.1007/BF00005712 [Google Scholar]
- 145.Debeljak P, Baltar F (2023) Fungal diversity and community composition across ecosystems. J Fungi 9. 10.3390/jof9050510 [DOI] [PMC free article] [PubMed]
- 146.Roik A, Reverter M, Pogoreutz C (2022) A roadmap to understanding diversity and function of coral reef-associated fungi. FEMS Microbiol Rev 46:1–26. 10.1093/femsre/fuac028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen X, Liao X, Chang S et al (2024) Comprehensive insights into the differences of fungal communities at taxonomic and functional levels in stony coral Acropora intermedia under a natural bleaching event. Mar Environ Res 196:106419. 10.1016/j.marenvres.2024.106419 [DOI] [PubMed] [Google Scholar]
- 148.Djurhuus A, Closek CJ, Kelly RP et al (2020) Environmental DNA reveals seasonal shifts and potential interactions in a marine community. Nat Commun 11:1–9. 10.1038/s41467-019-14105-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Warnasuriya SD, Udayanga D, Manamgoda DS, Biles C (2023) Fungi as environmental bioindicators. Sci Total Environ 892:164583. 10.1016/j.scitotenv.2023.164583 [DOI] [PubMed] [Google Scholar]
- 150.Gourmelon V, Maggia L, Powell JR et al (2016) Environmental and geographical factors structure soil microbial diversity in new caledonian ultramafic substrates: a metagenomic approach. PLoS ONE 11:1–25. 10.1371/journal.pone.0167405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sun Q, Liu Y, Yuan H, Lian B (2017) The effect of environmental contamination on the community structure and fructification of ectomycorrhizal fungi. Microbiologyopen 6:1–8. 10.1002/mbo3.396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ludley KE, Robinson CH (2008) “Decomposer” Basidiomycota in Arctic and Antarctic ecosystems. Soil Biol Biochem 40:11–29. 10.1016/j.soilbio.2007.07.023 [Google Scholar]
- 153.Rosenberg E, Ben-Haim Y (2005) Microbial diseases of corals. Ocean Heal Pathog Mar Environ 4:415–430. 10.1007/0-387-23709-7_18 [Google Scholar]
- 154.Ein-Gil N, Ilan M, Carmeli S et al (2009) Presence of Aspergillussydowii, a pathogen of gorgonian sea fans in the marine sponge Spongiaobscura. ISME J 3:752–755. 10.1038/ismej.2009.18 [DOI] [PubMed] [Google Scholar]
- 155.Liu Y, Li Y, Lin Q, Zhang Y (2017) Assessment of the pathogenicity of marine Cladosporium spp. towards mangroves. For Pathol 47:1–5. 10.1111/efp.12322 [Google Scholar]
- 156.Arvanitidou M, Kanellou K, Katsouyannopoulos V, Tsakris A (2002) Occurrence and densities of fungi from northern Greek coastal bathing waters and their relation with faecal pollution indicators. Water Res 36:5127–5131. 10.1016/S0043-1354(02)00235-X [DOI] [PubMed] [Google Scholar]
- 157.Blaalid R, Khomich M (2021) Current knowledge of Chytridiomycota diversity in Northern Europe and future research needs. Fungal Biol Rev 36:42–51. 10.1016/j.fbr.2021.03.001 [Google Scholar]
- 158.Lepelletier F, Karpov SA, Alacid E et al (2014) Dinomycesarenysensis gen. et sp. nov. (Rhizophydiales, Dinomycetaceae fam. nov.), a chytrid infecting marine dinoflagellates. Protist 165:230–244. 10.1016/j.protis.2014.02.004 [DOI] [PubMed] [Google Scholar]
- 159.Stat M, Morris E, Gates RD (2008) Functional diversity in coral-dinoflagellate symbiosis. Proc Natl Acad Sci U S A 105:9256–9261. 10.1073/pnas.0801328105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Theelen B, Cafarchia C, Gaitanis G et al (2018) Malassezia ecology, pathophysiology, and treatment. Med Mycol 56:S10–S25. 10.1093/mmy/myx134 [DOI] [PubMed] [Google Scholar]
- 161.Abdolrasouli A, Gibani MM, de Groot T et al (2021) A pseudo-outbreak of Rhinocladiellasimilis in a bronchoscopy unit of a tertiary care teaching hospital in London, United Kingdom. Mycoses 64:394–404. 10.1111/myc.13227 [DOI] [PubMed] [Google Scholar]
- 162.First MR, Hollibaugh JT (2010) Environmental factors shaping microbial community structure in salt marsh sediments. Mar Ecol Prog Ser 399:15–26. 10.3354/meps08385 [Google Scholar]
- 163.Plante CJ, Fleer V, Jones ML (2016) Neutral processes and species sorting in benthic microalgal community assembly: effects of tidal resuspension. J Phycol 52:827–839. 10.1111/jpy.12445 [DOI] [PubMed] [Google Scholar]
- 164.Rojo C, Mesquita-Joanes F, Monrós JS et al (2016) Hydrology affects environmental and spatial structuring of microalgal metacommunities in tropical pacific coast wetlands. PLoS ONE 11:1–15. 10.1371/journal.pone.0149505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Odum HT, Odum EP (1955) Trophic structure and productivity of a windward coral reef community on Eniwetok Atoll. Ecol Monogr 25:291–320 [Google Scholar]
- 166.Grange J (2015) The temporal dynamic of microboring successions and their associated biogenic dissolution in coral reef ecosystem. Sorbonne University, Paris [Google Scholar]
- 167.Chazottes V, Le C-A, Peyrot-Clausade M (1995) Bioerosion rates on coral reefs: interactions between macroborers, microborers and grazers (Moorea, French Polynesia). “Palaeogeography. Palaeoclimatol Palaeoecol 113:189–198. 10.1016/0031-0182(95)00043-L [Google Scholar]
- 168.Massé A, Domart-Coulon I, Golubic S et al (2018) Early skeletal colonization of the coral holobiont by the microboring Ulvophyceae Ostreobium sp. Sci Rep 8:1–11. 10.1038/s41598-018-20196-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tandon K, Pasella MM, Iha C et al (2022) Every refuge has its price: Ostreobium as a model for understanding how algae can live in rock and stay in business. Semin Cell Dev Biol 134:27–136. 10.1016/j.semcdb.2022.03.010 [DOI] [PubMed] [Google Scholar]
- 170.Enochs IC, Manzello DP, Tribollet A et al (2016) Elevated colonization of microborers at a volcanically acidified coral reef. PLoS ONE 11:1–16. 10.1371/journal.pone.0159818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Iha C, Dougan KE, Varela JA et al (2021) Genomic adaptations to an endolithic lifestyle in the coral-associated alga Ostreobium. Curr Biol 31:1393-1402.e5. 10.1016/j.cub.2021.01.018 [DOI] [PubMed] [Google Scholar]
- 172.Jost L (2006) Entropy and diversity. Oikos 113:363–375. 10.1111/j.2006.0030-1299.14714.x [Google Scholar]
- 173.Burki F, Sandin MM, Jamy M (2021) Diversity and ecology of protists revealed by metabarcoding. Curr Biol 31:R1267–R1280. 10.1016/j.cub.2021.07.066 [DOI] [PubMed] [Google Scholar]
- 174.Caron DA, Alexander H, Allen AE et al (2017) Probing the evolution, ecology and physiology of marine protists using transcriptomics. Nat Rev Microbiol 15:6–20. 10.1038/nrmicro.2016.160 [DOI] [PubMed] [Google Scholar]
- 175.Hoegh-Guldberg O (1999) Climate change, coral bleaching and the future of the world’s coral reefs. Mar Freshw Res 50:839–866. 10.1071/MF99078 [Google Scholar]
- 176.Ryves DB, Jewson DH, Sturm M et al (2003) Quantitative and qualitative relationships between planktonic diatom communities and diatom assemblages in sedimenting material and surface sediments in Lake Baikal, Siberia. Limnol Oceanogr 48:1643–1661. 10.4319/lo.2003.48.4.1643 [Google Scholar]
- 177.Maier SR, Kutti T, Bannister RJ et al (2020) Recycling pathways in cold-water coral reefs: use of dissolved organic matter and bacteria by key suspension feeding taxa. Sci Rep 10:1–13. 10.1038/s41598-020-66463-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Carlos AA, Baillie BK, Maruyama T (2000) Diversity of dinoflagellate symbionts (zooxanthellae) in a host individual. Mar Ecol Prog Ser 195:93–100 [Google Scholar]
- 179.Rizos I, Debeljak P, Finet T et al (2023) Beyond the limits of the unassigned protist microbiome: inferring large-scale spatio-temporal patterns of Syndiniales marine parasites. ISME Commun 3:1–11. 10.1038/s43705-022-00203-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Castro-Cubillos ML, Taylor JD, Mastretta-Yanes A et al (2022) Monitoring of benthic eukaryotic communities in two tropical coastal lagoons through eDNA metabarcoding: a spatial and temporal approximation. Sci Rep 12:1–14. 10.1038/s41598-022-13653-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lanzén A, Dahlgren TG, Bagi A, Hestetun JT (2021) Benthic eDNA metabarcoding provides accurate assessments of impact from oil extraction, and ecological insights. Ecol Indic 130. 10.1016/j.ecolind.2021.108064
- 182.Lim NKM, Tay YC, Srivathsan A et al (2016) Next-generation freshwater bioassessment: eDNA metabarcoding with a conserved metazoan primer reveals species-rich and reservoir-specific communities. R Soc Open Sci 3. 10.1098/rsos.160635 [DOI] [PMC free article] [PubMed]
- 183.Jakhar P (2013) Role of phytoplankton and zooplankton as health indicators of aquatic ecosystem : a review. Int J Innov Res Stud 2:490–500 [Google Scholar]
- 184.Poulíčková A, Duchoslav M, Dokulil M (2004) Littoral diatom assemblages as bioindicators of lake trophic status: a case study from perialpine lakes in Austria. Eur J Phycol 39:143–152. 10.1080/0967026042000201876 [Google Scholar]
- 185.Wang X, Li Z, Su J et al (2010) Lysis of a red-tide causing alga, Alexandriumtamarense, caused by bacteria from its phycosphere. Biol Control 52:123–130. 10.1016/j.biocontrol.2009.10.004 [Google Scholar]
- 186.Anderson DM, Alpermann TJ, Cembella AD et al (2012) The globally distributed genus Alexandrium: multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 14:10–35. 10.1016/j.hal.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shang L, Hu Z, Deng Y et al (2019) Metagenomic sequencing identifies highly diverse assemblages of dinoflagellate cysts in sediments from ships’ ballast tanks. Microorganisms 7:1–28. 10.3390/microorganisms7080250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Decelle J, Siano R, Probert I et al (2012) Multiple microalgal partners in symbiosis with the acantharian Acanthochiasma sp. (Radiolaria). Symbiosis 58:233–244. 10.1007/s13199-012-0195-x [Google Scholar]
- 189.Harun SNR, Mohammad-Noor N, Ahmad Z et al (2015) First report of Cochlodiniumpolykrikoides (Dinophyceae), a harmful algal bloom (HAB) species in the coastal waters of Peninsular Malaysia. Malaysian J Sci 34:87–92. 10.22452/mjs.vol34no1.9 [Google Scholar]
- 190.Canals O, Massana R, Riera JL et al (2018) Microeukaryote community in a partial nitritation reactor prior to anammox and an insight into the potential of ciliates as performance bioindicators. N Biotechnol 43:3–12. 10.1016/j.nbt.2017.05.003 [DOI] [PubMed] [Google Scholar]
- 191.Lee SR, Song EH, Lee T (2018) Eukaryotic plankton species diversity in the western channel of the Korea Strait using 18S rDNA sequences and its implications for water masses. Ocean Sci J 53:119–132. 10.1007/s12601-018-0005-3 [Google Scholar]
- 192.Ben-haim Y, Zicherman-keren M, Rosenberg E (2003) Temperature-regulated bleaching and tissue lysis of Pocilloporadamicornis by the novel pathogen Vibriocoralliilyticus. Appl Environ Microbiol 69:4236–4242. 10.1128/AEM.69.7.4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Moi IM, Roslan NN, Leow ATC et al (2017) The biology and the importance of Photobacterium species. Appl Microbiol Biotechnol 101:4371–4385. 10.1007/s00253-017-8300-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences generated and analyzed during the current study are available under the NCBI BioProject PRJNA985136, BioSample SUB13558791, from SRR24958245 to SRR24958195 for the 16S, from SRR25108721 to SRR25108698 for the ITS, from SRR25080909 to SRR25080857 for the tufA, and from SRR24961358 to SRR24961336 for the 18S.