Summary
The vagus nerve is proposed to enable communication between the gut microbiome and the brain, but activity-based evidence is lacking. We find that mice reared germ-free exhibit decreased vagal tone relative to colonized controls, which is reversed via microbiota restoration. Perfusing antibiotics into the small intestines of conventional mice, but not germ-free mice, acutely decreases vagal activity which is restored upon re-perfusion with intestinal filtrates from conventional, but not germ-free, mice. Microbiome-dependent short-chain fatty acids, bile acids, and 3-indoxyl sulfate indirectly stimulate vagal activity in a receptor-dependent manner. Serial perfusion of each metabolite class activates both shared and distinct neuronal subsets with varied response kinetics. Metabolite-induced and receptor-dependent increases in vagal activity correspond with the activation of brainstem neurons. Results from this study reveal that the gut microbiome regulates select metabolites in the intestinal lumen that differentially activate vagal afferent neurons, thereby enabling the microbial modulation of chemosensory signals for gut-brain communication.
Subject areas: Neuroscience, Microbiome
Graphical abstract

Highlights
-
•
Microbiota colonization status modulates vagal afferent nerve activity
-
•
Gut microbes differentially regulate metabolites in the small intestine and cecum
-
•
Select microbial metabolites stimulate vagal afferent neurons with varied kinetics
-
•
Select microbial metabolites alter CNS neuronal activation via receptor-dependent signaling
Neuroscience; Microbiome
Introduction
The gut microbiota is emerging as a key modulator of brain function and behavior, as several recent studies reveal the effects of gut microbes on neurophysiology, complex animal behaviors, and endophenotypes of neurodevelopmental, neurological, and neurodegenerative diseases.1,2,3 Despite these findings supporting a “microbiome-gut-brain axis,” the mechanisms that underlie interactions between gut microbes and the brain remain poorly understood. While many studies highlight neuroimmune pathways for microbial influences on the brain,4,5 it is also believed that the gut microbiota may directly signal to the brain via gut-innervating vagal neurons.6 However, existing evidence for the vagal route largely derives from studies wherein subdiaphragmatic vagotomy abrogates behavioral alterations in response to microbiota perturbation in mice.2,7,8,9,10 This ablates signaling in both afferent and efferent directions, not only to the intestine but also to other peripheral organs. While the approach provides an important initial indication that the vagus nerve contributes to behavioral phenotypes that are modified by the microbiome, in vivo evidence of microbial signaling through vagal neurons is needed, and fundamental questions remain regarding the nature of microbial effects on vagal activity, the particular molecular constituents involved, and the diversity of neuronal responses elicited.
The gut microbiome is central to dietary metabolism and modulates hundreds of biochemicals in the intestine, as well as the blood and various distal extraintestinal tissues.11,12 Biochemical screens of supernatants from cultured human-derived gut microbes find that soluble microbial products (largely uncharacterized) have the capacity to directly bind to numerous G-protein-coupled receptors (GPCRs) that mediate neurotransmitter and neuropeptide signaling,13,14 some of which are reportedly expressed by vagal neurons.15,16,17,18,19 As such, microbial metabolites generated in the intestinal lumen have the potential to directly or indirectly activate vagal neurons via receptors on mucosal sensory afferents, or on enteroendocrine cells (EECs) that synapse onto the mucosal sensory afferents, as has been described for select lumenal nutrient stimuli20,21,22 and microbial antigens.23,24 In this study, we assess vagal afferent nerve activity in response to the presence, absence, depletion, and restoration of the gut microbiota. We further profile microbiome-dependent metabolites within the proximal small intestine and cecum, which are poised to signal to gut mucosal vagal afferents. We identify select microbial metabolites that induce vagal afferent neuronal activity with varied kinetics when administered to the lumen of the small intestine and that elevate vagal tone when orally supplemented to microbiota-deficient mice. Finally, we apply pharmacological approaches to probe the potential for the direct and/or indirect receptor-mediated signaling of microbial metabolites to vagal neurons. This study provides the fundamental functional and mechanistic evaluation of the gut microbiome as a regulator of lumenal metabolites that modulate vagal chemosensory signaling across the gut-brain axis.
Results
The gut microbiome promotes vagal afferent nerve activity
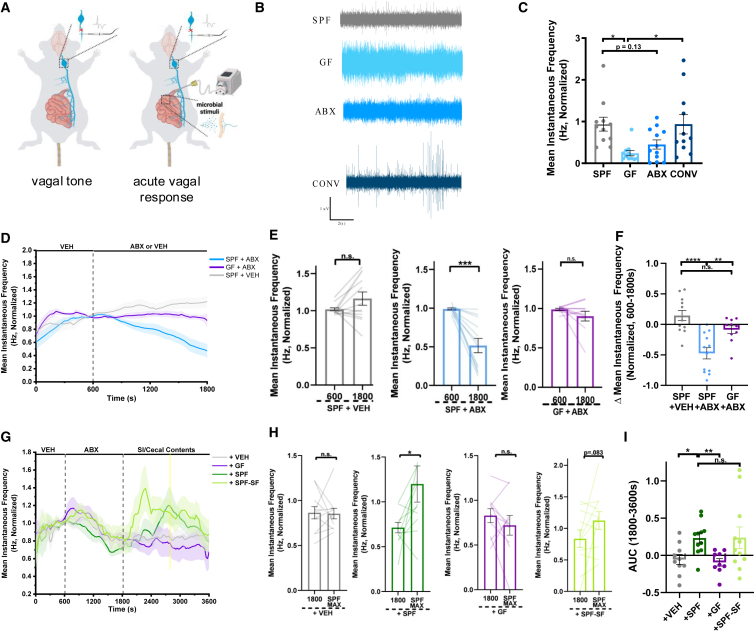
To determine the extent to which the microbiome contributes to gut-brain signaling via the vagus nerve, we began by applying whole nerve electrophysiology to measure bulk activity of vagal afferents in wildtype C57BL/6J mice reared in the presence vs. absence of microbial colonization (Figure 1A, left). vagal afferent nerve recordings were conducted over a 10-min period, allowing 5 min for signal stabilization,25 followed by 5 min of recording whereby the average spike frequency was quantified. Initial experiments indicated no differences in vagal tone between male and female mice (Figure S1), so all subsequent experiments were performed in males. Germ-free (GF) mice exhibited significantly decreased vagal tone as compared to conventionally colonized (specific pathogen-free, SPF) controls (Figures 1B and 1C). These reductions were reversed by colonizing GF mice with the SPF microbiota during adulthood (conventionalized, CONV), suggesting active interactions between the microbiome and vagus nerve that occur independently of developmental colonization. Treating adult SPF mice with oral broad-spectrum antibiotics (ABX; ampicillin, neomycin, vancomycin, and metronidazole) for 7 days to reduce bacterial load yielded modest, but not statistically significant, reductions in vagal tone. These data suggest a potential role for the microbiota in mediating various vagally mediated physiological processes associated with immune, metabolic, and neuronal function that may together influence homeostatic vagal function. We hypothesized that the inconsistent phenotype between the ABX and GF conditions may be due to the incomplete depletion of bacteria by ABX treatment, enrichment of native ABX-resistant bacteria over the one-week treatment period,26 off-target effects of ABX absorbed into the systemic circulation,27 metronidazole-induced neurotoxicity,28 the activity of non-bacterial members of the microbiota,29 and/or confounding effects of developmental GF rearing.30 To gain further insight into the acute effects of microbiota depletion on active vagal signaling in response to interoceptive signals, we recorded acute vagal afferent nerve responses during the introduction of a constant flow of the subset of the ABX cocktail that is non-absorbable (i.e., vancomycin and neomycin) directly into the lumen of the duodenum31 and through the first ∼10 cm of the small intestine, a site of dense vagal innervation16 (Figure 1A, right). In order to address the confounding effects of mechanical distention and the surgical procedure, the vehicle was perfused into the intestinal lumen for a period of 600s to allow for the stabilization of vagal recordings. All experiments were then normalized within the subject to the final 60s of the baseline perfusion period to capture the relative effect sizes of active vagal afferent nerve responses to lumenal stimuli. Subsequently perfusing nonabsorbable ABX into the intestinal lumen of SPF mice for 20 min decreased vagal afferent nerve activity, as compared to vehicle (VEH)-perfused controls (Figures 1D–1F). Intestinal perfusates from ABX-perfused mice exhibited significant decreases in the 16S rRNA gene sequence compared to VEH-perfused controls (Figure S2), consistent with reductions in mucosal bacteria. ABX-induced decreases in vagal afferent nerve activity were not seen in GF mice perfused with ABX (Figures 1D–1F). These results suggest that intestinal perfusion with non-absorbable ABX decreases vagal afferent nerve activity via the bactericidal actions of ABX on microbes in the small intestine.
Figure 1.
The gut microbiota and lumenal microbial metabolites promote vagal afferent nerve activity
(A) Diagram of in vivo whole nerve vagal electrophysiology for the quantification of vagal tone (left, for data in B–C) or acute vagal afferent nerve response to lumenal stimuli (right, for data in D-I) B) Representative electrophysiological recording traces over 10s in conventionally colonized (SPF), germ-free (GF), antibiotic-treated (ABX), and conventionalized GF (CONV) mice.
(C) Average vagal afferent tone of SPF, GF, ABX, and CONV mice over 300s of recording. (SPF, n = 11 mice; GF, n = 11 mice; ABX, n = 12 mice; CONV, n = 11 mice). Normalized to SPF average. One-Way ANOVA + Tukey.
(D) Vagal afferent nerve response in SPF or GF mice perfused intestinally with non-absorbable antibiotics (ABX, vancomycin/neomycin, 1 mg/mL) or vehicle (VEH) (SPF +VEH, n = 12 mice; SPF +ABX, n = 12 mice; GF + ABX, n = 10 mice). Normalized within-subject to the final 60s of the baseline vehicle recording period (t = 540s–600s).
(E) Vagal afferent nerve firing rates before and after intestinal perfusion of SPF or GF mice with ABX or VEH. (SPF +VEH, n = 12 mice; SPF +ABX, n = 12 mice; GF + ABX, n = 10 mice). Paired t-test.
(F) Change in vagal afferent nerve activity after intestinal perfusion with ABX or VEH (t = 1800) relative to stable baseline (t = 600s) (SPF +VEH, n = 12 mice; SPF +ABX, n = 12 mice; GF + ABX, n = 10 mice). One-way ANOVA + Tukey.
(G) Vagal afferent nerve response in SPF mice intestinally perfused with ABX, followed by re-perfusion with pooled small intestinal (SI) and cecal contents from SPF mice (+SPF) or GF mice (+GF), sterile filtered SI/cecal contents from SPF mice (+SPF-SF), or VEH (+VEH, n = 10 mice; +SPF, n = 11 mice; +GF, n = 9 mice; +SPF-SF, n = 11 mice). Normalized within-subject to the final 60s of the baseline vehicle recording period (t = 540s–600s).
(H) Vagal afferent nerve firing rate after intestinal ABX perfusion (t = 1800s) and after re-perfusion with VEH, SI/cecal contents from SPF mice, SI/cecal contents from GF mice, or sterile filtered SI/cecal contents from SPF mice, at the time of maximum mean firing rate for perfusion of SPF SI/Cecal contents (SPF MAX, t = 2760s) (+VEH, n = 10 mice; +SPF, n = 11 mice; +GF, n = 9 mice; +SPF-SF, n = 11 mice). Paired t-test.
(I) Vagal afferent nerve activity as measured by area under the curve (AUC) (from 1800 to 3600s) in response to intestinal perfusion with VEH (n = 10 mice), SPF SI/cecal contents (n = 11 mice), GF SI/cecal contents (n = 9 mice), and SPF-SF SI/cecal contents (n = 11 mice). Brown-Forsythe and Welch ANOVA + Games-Howell. All data displayed as mean ± SEM, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
The gut microbiota influences many aspects of host biology, in large part by bacterial metabolism of dietary substrates, synthesis of secondary metabolites, and modification of host-derived molecules in the intestine.4,32,33,34,35 To acutely evaluate the effects of lumenal microbial molecules on vagal afferent nerve activity, we administered a solution of small intestinal (SI) and cecal contents collected from donor SPF or GF mice into the SI lumen following ABX perfusion in SPF mice. SI perfusion with non-absorbable ABX reduced vagal afferent nerve activity, as reported above, whereas re-perfusion of SI/cecal contents from SPF mice acutely increased activity toward levels seen at pre-ABX baselines (“+ SPF” in Figures 1G–1I). No such effect was seen with re-perfusion of SI/cecal contents from GF mice (“+ GF” in Figures 1G–1I) or vehicle (“+VEH,” in Figures 1G–1I), suggesting that the observed vagal response is due the presence of SI and cecal microbes and/or microbial molecules. To investigate the contribution of microbial small molecules, in particular, we sterile-filtered the equivalent solution of SPF SI/cecal contents with a 0.22um polyethersulfone membrane filter in order to exclude bacteria, fungi, and other larger microorganisms, as well as larger macronutrients such as lipids and proteins and administered it to the SI lumen of ABX-perfused SPF mice. Sterile-filtered SPF SI/cecal contents increased vagal afferent nerve activity to levels similar to SPF on average, albeit with shorter latency and more variability (“+ SPF-SF” in Figures 1G–1I). These differences could be due to differential kinetics of small molecule signaling resulting from a lack of macromolecules present in SPF-SF samples20 and/or variability in the fidelity of small molecules upon filtration or the distribution of small-molecule activated receptors along the length of the proximal and medial small intestine.36 Taken together, these data provide evidence that active signaling from the gut microbiota modulates vagal afferent activity in vivo and that these effects are mediated, at least in part, by microbial small molecules within the lumen of the small intestine and cecum.
Microbiome-dependent bile acids, short-chain fatty acids, and 3-indoxyl sulfate indirectly stimulate vagal afferent nerve activity in a receptor-dependent manner
The gut microbiota regulates numerous metabolites within the host.11,12 However, most characterizations to date have profiled metabolites in fecal or serum samples, excluding signaling molecules localized to the small intestine and cecum that are poised to interact with villus-innervating vagal neurons.37 To identify candidate microbial metabolites in the small intestine and cecum that may modify vagal afferent activity, we performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) based untargeted metabolomic profiling of lumenal contents from the duodenum (proximal SI) and cecum of SPF, GF, ABX, and CONV mice. 931 metabolites were identified from mouse proximal SI and cecal contents (Tables S1 and S2). Principal component analysis revealed distinct clustering of GF and ABX samples away from SPF and CONV samples along PC1 (Figure 2A), indicating that the acute ABX depletion of the gut microbiota yields SI and cecal metabolomic profiles that are similar to those seen with GF rearing and that the adult inoculation of GF mice with a conventional microbiota induces SI and cecal metabolomic profiles that are similar to those seen with conventional colonization (SPF). This is consistent with our finding that the acute depletion and re-introduction of the microbiota or microbial metabolites alters vagal afferent nerve activity (Figure 1). However, there were also notable differences within the cecal datasets in particular, with the discrimination of ABX from GF profiles and, to a lesser degree, CONV from SPF profiles, along PC2 (Figure 2A, bottom). These differences highlight potential developmental influences of microbial colonization on host physiology38 and/or incomplete depletion and/or restoration of microbial communities within the lower GI tract relative to the proximal small intestine.39 Based on the ability of both GF status and acute perfusion of nonabsorbable ABX to decrease vagal afferent nerve activity and of CONV to elevate activity toward levels seen in SPF controls (Figures 1A–1F), we then filtered the datasets to identify metabolites that were commonly differentially regulated by both microbiota-deficient conditions relative to both colonized conditions. In SI contents, there were 79 shared metabolites that were significantly modulated by both GF and ABX conditions relative to both SPF and CONV conditions, and in cecal contents, there were 521 (Figure 2B, Tables S1 and S2). Based on the observation that SI/cecal contents and filtrates from SPF mice stimulate vagal afferent nerve activity compared to filtrates from GF controls (Figures 1G–1I), we focused in particular on the subset of 49 SI and 335 cecal metabolites that were significantly decreased by microbiota deficiency relative to colonized conditions (Figure 2B; Tables S1 and S2). These included microbial metabolites that were extremely low or undetectable in microbiota-deficient conditions (which we refer to as “microbiome-dependent”), as well as metabolites that were partially downregulated (but still detectable) in microbiota-deficient conditions (which we refer to as “microbially modulated”).
Figure 2.
The gut microbiome regulates metabolites in the lumen of the small intestine and cecum
(A) PCA analysis of metabolomic data from small intestinal (SI, top) or cecal (bottom) lumenal contents from SPF, GF, ABX, and CONV mice. (n = 6 mice for all groups).
(B) Venn diagram of differentially modulated metabolites in SI (top) and cecal (bottom) lumenal contents. Red numbers indicate downregulated metabolites and blue numbers indicate upregulated metabolites. (n = 6 mice for all groups).
(C) Random forest (RF) analysis of metabolomic data from SI lumenal contents reveals the top 30 metabolites that distinguish GF/ABX from SPF/CONV samples with 100% predictive accuracy. Red asterisks indicate metabolites included in experiments in Figure 3. Hash symbols indicate metabolites included in experiments in Figure S3 (n = 6 mice for all groups).
(D and E) Lumenal levels of microbially modulated bile acids and the short-chain fatty acid butyrate from SI (orange tones) and/or cecum (blue tones) of SPF, GF, ABX, and CONV mice. (n = 6 mice for all groups). Welch’s t-test.
(F) Lumenal levels of microbially modulated metabolites with unknown signaling to vagal neurons from SI (orange tones) or cecum (blue tones) of SPF, GF, ABX, and CONV mice. (n = 6 mice for all groups). Welch’s t-test. All data displayed as mean ± SEM, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p <0 .0001.
To identify the subset of microbial metabolites that have the potential to signal directly to vagal neurons, we cross-referenced existing bulk and single-cell RNA sequencing datasets15,16,18,19,40 for reported expression of known or putative receptors for the candidate SI and cecal metabolites. This identified select species of microbiome-dependent bile acids (BAs), a subset of which were identified as key drivers for classifying microbiota status via random-forest analysis in small-intestinal samples (Figures 2C and 2D). Importantly, we observed reductions in both primary BAs synthesized by the liver and microbially derived secondary BAs. Absence (GF) or depletion (ABX) of the microbiota results in an ablation of or a reduction in microbial synthesis of microbially derived secondary BAs in the small intestinal lumen. Moreover, bile acids themselves exert effects on gut microbial composition resulting in altered feedback mechanisms regulating primary bile acid synthesis. Thus, our results reflect alterations in both microbial synthesis of secondary BAs and compositional effects on host BA synthesis. Additional classes of metabolites that were uncovered included the microbiome-dependent short chain fatty acid (SCFA) butyrate (Figure 2E) and microbially modulated tryptophan derivatives (TRPs, Figure S3A), fatty acid ethanolamides (FAEs, Figure S3B), monohydroxy fatty acids (MFAs, Figure S3C), as well as succinate and glutamate (Figures S3D–S3E). To initially assess the ability of these metabolites to actively modify vagal activity from the SI lumen, we recorded vagal afferent nerve activity while perfusing physiologically relevant concentrations of metabolite pools into the SI. There were no statistically significant changes in vagal afferent nerve activity as measured by AUC over the entire 30-min stimulus window with SI perfusion of the detected TRPs, FAEs, MFAs, or succinate (Figures S4A–S4D). Consistent with existing literature demonstrating vagal responses to gastric delivery of glutamate,41 SI perfusion of glutamate robustly increased vagal afferent nerve activity (Figure S4E), possibly due to the increased recruitment of vagal afferent neurons and/or repeated stimulation of metabotropic glutamate receptors. We further observed that SI perfusion of select microbiome-dependent BAs elicited more rapid, transient vagal afferent nerve activity that returns to levels similar to baseline (Figures 3A–3C) relative to other metabolites tested, which parallels existing literature on the systemic administration of select primary and secondary BAs.33 In contrast, SI perfusion of microbiome-dependent SCFAs (acetate, propionate, and butyrate) led to slower onset and gradual increases in vagal afferent nerve activity (Figures 3D–3F). This latency could be due to metabolite-specific differences in the rate of intestinal absorption,42 differential spatial localization of metabolite absorption and functional activity along the length of the gastrointestinal tract,43,44 and/or indirect signaling of the metabolites to non-neuronal mediators21,45,46 or enteric nervous cells.47,48
Figure 3.
Select lumenal microbial metabolites increase vagal afferent nerve activity with varied response kinetics
(A) Vagal afferent nerve firing rate in SPF mice after intestinal perfusion with vehicle (VEH: PBS, n = 10 mice) or pooled bile acids (BAs: cholate, 1240nM; glycocholate, 3.5nM; chenodeoxycholate, 42nM; alpha-muricholate, 142nM; beta-muricholate, 1080nM; deoxycholate, 390nM; taurodeoxycholate, 260nM; ursodeoxycholate, 74nM; taurohyodeoxycholate, 18.8nM; 7-ketodeoxycholate, 100nM; lithocholate, 390nM; taurolithocholate, 0.33nM; n = 9 mice), with or without pre- and co-perfusion with TGR5 antagonist (SBI-115, 200uM, n = 8 mice). Yellow shading indicates perfusion of pure antagonist prior to the co-perfusion of antagonist with metabolites.
(B) Normalized vagal afferent nerve firing rate before and during treatment with BAs (left, n = 9 mice), or BAs and SBI-115 (right, n = 8 mice). BA max, t = 1114s. Wilcoxon matched-pairs signed rank test.
(C) Vagal afferent nerve firing rate as measured by area under the curve (AUC) (from 600 to 2400s) in response to intestinal perfusion with VEH (PBS, n = 10 mice) or pooled BAs (n = 9 mice), with or without pre- and co-perfusion with TGR5 antagonist (SBI-115, 200uM, n = 8 mice). Brown-Forsythe and Welch ANOVA + Games-Howell.
(D) Vagal afferent nerve firing rate in SPF mice after intestinal perfusion with VEH (PBS, n = 10 mice) or pooled short-chain fatty acids (SCFAs: acetate, 80uM; butyrate, 22uM; propionate, 10uM, pooled, n = 10 mice), with or without pre- and co-perfusion with FFAR2 antagonist (GLPG0794, 10uM, n = 8 mice). Yellow shading indicates perfusion of pure antagonist prior to the co-perfusion of antagonist with metabolites.
(E) Normalized vagal afferent nerve firing rate before and during treatment with SCFAs (left, n = 10 mice), or SCFAs and GLPG0794 (right, n = 8 mice). SCFA max, t = 2400s. Paired t-test.
(F) Vagal afferent nerve firing rate as measured by AUC (from 600 to 2400s) in response to intestinal perfusion with VEH (PBS, n = 10 mice) or pooled SCFAs (10uM, n = 10 mice), with or without pre- and co-perfusion with FFAR2 antagonist (GLPG0794, 10uM, n = 8 mice). One-way ANOVA + Tukey.
(G) Vagal afferent nerve firing rate in SPF mice after intestinal perfusion with VEH (1uM KCl, n = 7 mice) or 3-indoxyl sulfate (3-IS, 1uM, n = 10 mice), with or without pre- and co-perfusion with TRPA1 antagonist (A967079, 10uM, n = 7 mice). Yellow shading indicates perfusion of pure antagonist prior to co-perfusion of antagonist with metabolites.
(H) Normalized vagal afferent nerve firing rate before and during treatment with 3IS (left, n = 10 mice), or 3IS and A967079 (right, n = 7 mice). 3IS max, t = 2400s. Paired t-test.
(I) Vagal afferent activity as measured by AUC (from 600 to 2400s) in response to intestinal perfusion with VEH (1uM KCl, n = 7 mice) or 3-indoxyl sulfate (3-IS, 1uM, n = 10 mice), with or without pre- and co-perfusion with TRPA1 antagonist (A967079, 10uM, n = 7 mice). One-way ANOVA + Tukey. All time-series data were normalized within-subject to the final 60s of the 600s vehicle recording period (first 300s of recording not shown). All data displayed as mean ± SEM, ∗p < 0.05.
Microbiome-dependent BAs and SCFAs in the intestinal lumen have the potential to bind to cognate receptors expressed by various cell types in the gastrointestinal tract (e.g., vagal, enteroendocrine, epithelial, immune).49,50 To determine the relative contributions of different cognate GPCRs to vagal responses induced by lumenal microbial metabolites, we perfused select receptor antagonists immediately before and during the administration of their corresponding metabolites into the SI lumen of SPF mice. BAs signal through the membrane-bound Takeda G protein-coupled receptor 5 (TGR5), which is expressed by intestinal epithelial cells, enteric neurons, and various intestinal innate immune cells in mice.34,49,51 Intestinal pre- and co-perfusion of the TGR5 antagonist m-tolyl 5-chloro-2-[ethylsulonyl] pyrimidine-4-carboxylate (SBI-115)52 prevented the initial rapid, transient increases in vagal afferent nerve activity induced by microbiome-dependent BAs (Figures 3A–3C). We do not observe a difference in total area under the curve (AUC) across the entire stimulus window, as the administration of SBI-115 leads to a delayed rise in vagal activity that was not observed with perfusion of microbiome-dependent BAs alone. This partial ablation suggests that TGR5 antagonism may elicit compensatory vagal responses to microbiome-dependent BAs through farsenoid X receptor (FXR) or other signaling mechanisms independent of local TGR5. SCFAs signal to free fatty acid receptor 2 (FFAR2), which is expressed by intestinal epithelial cells43 and free fatty acid receptor 3 (FFAR3), which is expressed by gut-innervating vagal neurons.53 Intestinal pre- and co-perfusion of the FFAR2 antagonist 4-[[(R)-1-(benzo[b]thiophene-3-carbonyl)-2-methyl-azetidine-2-carbonyl]-(3-chloro-benzyl)-amino]-butyric acid 99 (GLPG0974)54 prevented the increase in vagal afferent nerve activity induced by SCFAs (Figures 3D–3F). Mouse vagal afferent neurons do not express appreciable levels of FFAR215 or TGR5 (Figure S5), suggesting that the antagonists prevent metabolite-induced vagal activity by acting through a cellular mediator. To test the potential for the direct action of microbial metabolites on vagal neurons, we measured calcium responses in acutely dissociated vagal nodose neurons during bath perfusion of microbial BAs or SCFAs. There was little to no activation in response to BAs and SCFAs, with only 2% and 1% of imaged neurons showing a calcium response, respectively (Figures S6A–S6C). These results suggest that microbiome-dependent BAs and SCFAs activate vagal afferent neurons via indirect activation of intestinal epithelial cells or other cellular mediators. Small intestinal EECs release neurotransmitters and neuropeptide hormones in response to nutrient- and microbial-derived cues such as glucose, BAs, and SCFAs (Figure S7A).21,22,45,46,55,56,57 Moreover, EECs are enriched for FFAR2 and TGR5 compared to other epithelial cell subtypes (Figures S7B–S7C).36 We, therefore, tested the potential for the indirect action of microbial metabolites on vagal neurons via EECs. Calcium imaging of intestinal secretin tumor (STC-1) EECs revealed that BAs and SCFAs significantly increase EEC activity compared to PBS alone, with 32% and 26% of sampled EECs eliciting a calcium response, respectively (Figures S7D–S7E). Taken together, these results demonstrate that microbiome-dependent BAs and SCFAs likely elevate vagal afferent activity via the activation of intestinal EECs.
In addition to testing microbial metabolites with reported receptor expression by vagal neurons, we also evaluated the effects of select microbiome-dependent metabolites that have as yet unknown signaling mechanisms on vagal activity. We focused in particular on metabolites that i) are reproducibly dependent upon the microbiome across various studies and biological contexts58 and ii) have been reportedly linked to brain function and/or behavior.1,59,60,61,62 Of these, 3-indoxyl sulfate (3IS), hippurate, and trimethylamine-N-oxide (TMAO) are microbiome-dependent metabolites in SI lumen, imidazole propionate is a microbiome-dependent metabolite in the cecum, and phenethylamine is microbially modulated in the cecum (Figure 2F). These metabolites are also reduced in the serum of microbiome-deficient mice,11 suggesting that they are typically absorbed from the intestine and poised to interact with mucosal vagal afferents. Perfusing physiologically relevant concentrations of hippurate, TMAO, imidazole propionate, and phenethylamine individually through the small intestine had no measurable effect on vagal afferent nerve activity (Figures S4F–S4I). In contrast, SI perfusion with 3IS elicited rapid and sustained increases in vagal afferent nerve activity relative to vehicle controls (Figures 3G–3I). Microbial indole (the metabolic precursor to 3IS) and its derivative indole-3-carboxaldehyde are reported to activate vagal afferent neurons in zebrafish via the indirect stimulation of colonic EECs in a transient receptor potential ankyrin1 (TRPA1)-dependent manner.45 To evaluate this possible signaling mechanism for 3IS in the small intestine, we pre- and co-perfused the TRPA1 antagonist (1E,3E)-1-(4-Fluorophenyl)-2-methyl-1-penten-3-one oxime (A967079)) with 3IS into the SI lumen, which completely prevented 3IS-induced vagal afferent nerve activity (Figures 3G–3I). TRPA1 is expressed by vagal neurons, (Figure S5) however, applying 3IS directly to dissociated vagal neurons yielded calcium responses in only 8% of imaged neurons (Figures S6A–S6C). Conversely, TRPA1 is enriched in intestinal EECs (Figures S7B–S7C)36 and application of 3IS to STC-1 cells results in the activation of 32% of sampled EECs (Figures S7D–S7E), suggesting an indirect mechanism of metabolite-induced elevation in vagal nerve activity via EEC activation. Overall, these results reveal that microbial BAs, SCFAs, and 3IS promote vagal afferent nerve activity through indirect receptor-dependent signaling from the SI lumen.
Lumenal bile acids, short-chain fatty acids, and 3-indoxyl sulfate excite both distinct and shared the subsets of vagal afferent neurons with varied temporal responses
Different lumenal stimuli can activate distinct populations of vagal neurons with differing response kinetics. In particular, recent work has identified populations of vagal afferent neurons that respond exclusively to fats versus sugars.22 Microbiome-dependent BAs, SCFAs, and 3IS are related to the dietary metabolism of fats, complex carbohydrates, and proteins, respectively,32,45,63 raising the question of whether they promote vagal nerve activity via shared vs. distinct vagal afferent neurons. To gain insight, we imaged the calcium activity of vagal afferent neurons in response to acute SI perfusion of microbiome-dependent metabolites in mice expressing GCaMP6s in Phox2b+ sensory neurons (Figures 4A and 4B). Microbiome-dependent BAs, SCFAs, and 3IS elicited calcium responses in 57%, 48%, and 58% of detected vagal afferent neurons responsive to electrical stimuli at the end of the imaging session, respectively, on average across independent animals (Figure 4C). The latency to maximum calcium response in metabolite-responsive units varied within each subclass of microbial metabolite, where SCFAs and 3IS similarly elicited primarily delayed calcium responses, while BAs elicited a bimodal distribution of acute and delayed calcium responses (Figure 4D). Upon perfusing pairs of metabolites for 2 min in counterbalanced sequence separated by at least 10 min of PBS perfusion, of the metabolite-responsive neurons alone, BAs and SCFAs elicited calcium responses in largely distinct vagal afferent neurons, with 43% responsive to BAs only, 38% responsive to SCFAs, and 19% dually responsive to both BAs and SCFAs (“BAs <> SCFAs” in Figure 4E). In contrast, sequential perfusion of 3IS and SCFAs yielded many shared neuronal responses, where 40% of responsive vagal afferent neurons were dually activated by 3IS and SCFAs, 27% to SCFAs only, and 33% to 3IS only (“3IS <> SCFAs” in Figure 4E). Similarly, with BAs and 3IS, we observed 43% dual responders, 35% responsive to 3IS only, and 22% responsive to BAs only (“BAs <> 3IS” in Figure 4E). Together, these data reveal distinct and shared neuronal populations for sensing different microbiome- and macronutrient-dependent metabolites, with greater distinct neuronal responses to microbial BAs and SCFAs, than either with 3IS.
Figure 4.
Lumenal BAs, SCFAs, and 3IS activate distinct subsets of vagal neurons with heterogeneous kinetics and restore vagal afferent nerve tone
(A) Diagram of the experimental setup for in vivo calcium imaging and associated micrograph of the imaging window.
(B) Representative heatmaps for cells responding to only pooled bile acids (BAs: cholate, 1240nM; glycocholate, 3.5nM; chenodeoxycholate, 42nM; alpha-muricholate, 142nM; beta-muricholate, 1080nM; deoxycholate, 390nM; taurodeoxycholate, 260nM; ursodeoxycholate, 74nM; taurohyodeoxycholate, 18.8nM; 7-ketodeoxycholate, 100nM; lithocholate, 390nM; taurolithocholate, 0.33nM; n = 4 mice, n = 131 units, right), only pooled short-chain fatty acids (SCFAs: acetate, 80uM; butyrate, 22uM; propionate, 10uM, n = 4 mice, n = 82 cells, middle), or only 3-indoxyl sulfate (3IS, 1uM, n = 4 mice, n = 132 cells, left). Recording duration for all experiments was 12 min. Only metabolite-responsive neurons are displayed. Each ROI was normalized to the average of the 120s imaging period prior to stimulus onset.
(C) Percentage of metabolite-responsive neurons out of total excitable neurons in response to lumenal perfusion of BAs (n = 5 mice), SCFAs (n = 5 mice), or 3IS (n = 4 mice).
(D) Latency to maximum change in fluorescence for metabolite-responding neurons with lumenal perfusion of BAs (n = 4 mice), SCFAs (n = 4 mice), or 3IS (n = 4 mice). One-way ANOVA + Tukey.
(E) Percentage of single- or dual-responding neurons following serial perfusion of BAs (n = 5 mice), SCFAs (n = 5 mice), and 3IS (n = 4 mice). Metabolites were sequentially perfused for 2 min with at least 10 min of PBS perfusion between metabolite trials. Electrical stimuli were applied at the end of each individual metabolite trial. Order of metabolites for perfusion was counterbalanced between experiments.
(F) Vagal afferent nerve activity of GF mice following three-day treatment via oral gavage with VEH (12uM KCl, 1.34mM NaCl, 0.012% DMSO in PBS, n = 7 mice) or pooled microbial metabolites (MMs: cholate, 14.88uM; glycocholate, 42nM; chenodeoxycholate, 504nM; alpha-muricholate, 1.7uM; beta-muricholate, 12.96uM; deoxycholate, 4.68uM; taurodeoxycholate, 3.12uM; ursodeoxycholate, 888nM; taurohyodeoxycholate, 226nM; 7-ketodeoxycholate, 1.2uM; lithocholate, 4.68uM; taurolithocholate, 3.96nM, acetate, 0.96mM; butyrate, 264uM; propionate, 120uM, 3IS, 12uM, n = 9 mice). All data displayed as mean ± SEM, ∗p < 0.05, ∗∗∗p < 0.001.
Supplementation of microbially modulated metabolites into germ-free animals increases homeostatic vagal afferent nerve activity
Diet-induced decreases in gut microbial diversity64,65,66 are associated with alterations in vagal sensitivity to peripheral signals.64,67,68,69 Our initial experiments demonstrated a role for the microbiota in modulating baseline vagal afferent nerve activity in gnotobiotic mice, whereby microbiome-deficient mice exhibited decreased homeostatic vagal nerve activity compared to colonized controls (Figure 1C). Therefore, we sought to uncover whether the oral administration of microbial metabolites could phenocopy the increased vagal activity seen in response to persistent microbial colonization. To address this, GF mice were supplemented via twice-daily oral gavage with a cocktail of microbial metabolites (MMs: BAs, SCFAs, and 3IS, pooled) or vehicle solution for three days, followed by whole-nerve vagal electrophysiological recordings. Indeed, supplementation with MMs significantly increased vagal afferent nerve tone compared to vehicle-treated controls (Figure 4F). Thus, microbiome-dependent dietary metabolites may be an important link between diet-induced alterations in the gut microbiota and associated vagally regulated host physiological and behavioral outcomes.
Receptor-mediated signaling of bile acids, short-chain fatty acids, and 3-indoxyl sulfate from the small intestinal lumen activates neurons in the nucleus of the solitary tract
Changes in the gut microbiota have been associated with the altered activation of neurons of the nucleus of the solitary tract (NTS), which receives direct visceral afferents from nodose neurons.70,71 Consistent with the ability of microbial metabolites in the SI lumen to stimulate vagal afferent neuronal activity (Figures 3 and 4), we find that acute lumenal perfusion of microbiome-dependent BAs, SCFAs, and 3IS each increase neuronal expression of the activation marker cFos in the NTS (Figures 5A and 5B), to levels similar to those seen with intestinal perfusion of sucrose.21,72 As with the vagal afferent nerve and neuronal responses, the microbial metabolite-driven increases in NTS neuronal activation were prevented by the pre- and co-administration of antagonists for TGR5, FFAR2, and TRPA1 with their respective metabolite ligands. There were no significant differences in the dorsal motor nucleus of the vagus (DMV) (Figures S8A–S8C;73) suggesting that the luminal application of microbial metabolites primarily stimulates ascending vagal pathways. Circulating microbial metabolites regulate satiety via vagal receptor-dependent pathways and alterations in agouti-related protein/neuropeptide Y (AgRP/NPY) and proopiomelanocortin (POMC) neurons of the hypothalamus.33,53,74,75 We, therefore, sought to determine whether the lumenal application of BAs, SCFAs, and 3IS modulates neuronal activation in the arcuate nucleus of the hypothalamus (ARH) via their cognate receptors. We observed a modest decrease in ARH cFos levels with lumenal perfusion of 3IS, and to a lesser degree, BAs, and that these effects were attenuated with pre- and co-perfusion of the selective antagonists for TRPA1 and TGR5, respectively (Figures S8D–S8E). Conversely, lumenal SCFAs had no appreciable effect on ARH cFos levels (Figures S8D–S8E), suggesting a need for blood-brain barrier (BBB) translocation or engagement of vagal FFAR3 via circulating propionate in order to exert their anorexigenic effects previously described.53,75 Together, these data indicate that microbial BAs, SCFAs, and 3IS in the SI lumen may alter brain activity via receptor-mediated modulation of vagal afferent signaling.
Figure 5.
Microbial metabolites alter cFos levels in NTS neurons in a receptor-dependent manner
(A) Representative image of full ROIs in mNTS for cFos quantification (dotted line) and inset images in B (solid line). Scale bar = 100uM.
(B and C) Significant increase in the % of cFos+ NTS neurons following lumenal perfusion of BAs (cholate, 1240nM; glycocholate, 3.5nM; chenodeoxycholate, 42nM; alpha-muricholate, 142nM; beta-muricholate, 1080nM; deoxycholate, 390nM; taurodeoxycholate, 260nM; ursodeoxycholate, 74nM; taurohyodeoxycholate, 18.8nM; 7-ketodeoxycholate, 100nM; lithocholate, 390nM; taurolithocholate, 0.33nM, pooled, n = 3 mice), SCFAs (acetate 80uM, butyrate 22uM, propionate 10uM, pooled, n = 3 mice), and 3-IS (1uM, n = 4 mice) compared to vehicle (VEH, n = 3 mice) or corresponding receptor antagonists alongside each respective metabolite class (SBI-115, 200uM, n = 3 mice, GLPG0794, 10uM, n = 3 mice, A967079, 10uM, n = 3 mice). Scale bar = 50uM. One-way ANOVA + Tukey. All data displayed as mean ± SEM, ∗p < 0.05, ∗∗p < 0.01.
Discussion
Results from this study demonstrate that microbial colonization status, as well as acute manipulation of the gut microbiota and microbial metabolites, modulate vagal activity. In particular, we find that the gut microbiota regulates numerous small molecules in the small intestine and cecum. Moreover, administering select microbiome-dependent BAs, SCFAs, and 3IS at physiological concentrations and rates of peristalsis into the lumen of the small intestine stimulates vagal afferent neuronal activity. The vagal responses are elicited within relatively short timescales (<∼9 min) and are abrogated by the pre- and co-administration of select receptor antagonists, suggesting active signaling between the gut microbiome and vagal afferents via excitatory metabolites.
The functional evidence provided in this study aligns with prior reports indicating that subdiaphragmatic vagotomy abrogates the effects of microbial interventions on behaviors such as anxiety,7 depression,70 cognition,76 feeding,46 and social behaviors.2,53 Additionally, a few prior studies have reported that the gut microbiota and various microbial products regulate the transcriptome and excitability of vagal neurons.71 For instance, nodose neurons from mice reared GF exhibited altered gene expression profiles when compared to those from SPF mice,77 suggesting that the microbiome modulates the cellular state of vagal afferent neurons. In addition, bacterial supernatants from a cultured microbial community increased the intrinsic excitability of nodose neurons in vitro, through a mechanism that implicated a role for bacterial cysteine proteases.78 Another study reported that microbial single-stranded RNAs elevated vagal activity via Piezo1-mediated sensing by enterochromaffin cells.23 Moreover, the tryptophan metabolite indole has been reported to induce serotonin release onto colonic vagal afferents via TRPA1-mediated EEC activation.45 Together, these findings suggest that there exist multiple signaling factors and pathways by which the host-associated microbiota can impact vagal activity.
Aligning with the complexity of microbial influences on vagal activity, we observed that mice reared as GF exhibited significantly reduced vagal afferent nerve activity relative to mice reared with a conventional SPF microbiota, and while this effect was abrogated by colonizing GF mice with an SPF microbiota during adulthood, it was not fully recapitulated by the depletion of the gut bacteria by oral ABX (Figure 1C). Many factors could contribute to this discrepancy. First, GF mice lack microbiota across all exposed body sites, whereas oral ABX treatment only partially ablates bacterial members of the oral and gastrointestinal microbiomes.39 As such, it is possible that the reductions in vagal tone seen in GF mice could be mediated by changes in both intestinal and non-intestinal vagal afferents and/or the presence of residual microbes or microbial products in the intestine of SPF mice treated twice daily with ABX by oral gavage. Moreover, the reported alterations in nodose gene expression in GF mice relative to SPF mice77 raise the question of whether there are early influences of microbiota status on vagal neuronal development, which are not captured by oral ABX treatment during adulthood, such as alterations in immune function that can subsequently influence vagal activity. Additionally, we performed experiments in non-fasted mice due to the effects of fasting on vagal activation and microbiota composition.79,80,81,82,83 It is therefore possible that variations in feeding may contribute to variability across conditions. Further studies are needed to uncover the relative contributions of different vagal neuronal subtypes as well as additional cellular mediators and body states that may regulate microbiome-induced alterations in vagal tone, as well as to what degree microbes endogenous to different parts of the gastrointestinal tract contribute to these alterations.
Despite the modest effects of oral ABX treatment in mice on reducing vagal tone (Figure 1C), we found that restricted perfusion of non-absorbable ABX through the lumen of the small intestine acutely reduced vagal afferent nerve activity. We additionally observed that vagal afferent nerve activity was restored by the re-introduction of SPF SI and cecal contents into the small intestine and that these increases in activity were driven, at least in part, by the small molecule fraction. This was not seen with lumenal perfusion of SI and cecal contents from GF mice, indicating a role for small molecules that are modulated by the gut microbiome. Notably, we did not observe an overt vagal nutrient response with perfusion of intestinal contents from GF mice, which could reflect the rapid digestion, absorption, and/or relatively low homeostatic concentration of microbiome-independent nutrients, such as glucose and sucrose, in the SI,84 as compared to the those exogenously delivered in other studies.20,21,22 Overall, these findings reveal that soluble microbiome-dependent metabolites from the lumen of the small intestine can acutely stimulate vagal afferent activity.
By untargeted metabolomic profiling of SI and cecal contents from conventionally colonized (SPF, CONV) and microbiota deficient (ABX, GF) mice (Figure 2), followed by the in vivo screening of select microbial metabolites with or without pharmacological antagonists (Figure 3), we identified particular subclasses of microbiome-dependent molecules that activate vagal afferent neurons in a receptor-dependent manner upon administration to the small intestine. The metabolites—specific microbial BAs, SCFAs, and 3IS—promoted vagal afferent nerve activity with differing response kinetics, which could be due to differences in their physiological concentrations, rate of absorption, spatial localization of cognate receptors, and potential indirect activation via various non-vagal cellular mediators, among other factors. We did additionally observe a modest, transient increase in vagal afferent nerve activity at the time of the average maximal change in frequency with the lumenal perfusion of TRPs (Figure S4A), suggesting the possibility that other microbial metabolites may modulate vagal neuronal activity with smaller overall effect sizes. Indeed, neuronal activation via GPCR signaling is dependent upon the concentration of the ligand, whereby differences can induce switching from G-protein coupled to G-protein independent signaling85 resulting in alterations in the downstream signal transduction pathways engaged during neuronal activation. Additionally, we observed 30 small intestinal and 186 cecal metabolites that were increased in GF and ABX mice as compared to SPF and CONV samples (Figure 2B). It is indeed possible that these metabolites may play a role in microbiome-driven changes in vagal afferent nerve activity that we observe in conditions of microbial colonization. Therefore, further experiments screening the inhibitory effects of select microbial metabolites are warranted.
Microbiome-dependent BAs and SCFAs are reported to be absorbed by the intestinal epithelium, which offers the potential to activate gut-innervating vagal afferents through direct receptor binding.15,16,18,19 However, our observed lack of vagal neuronal activation during the in vitro stimulation of primarily dissociated nodose neurons in response to BAs and SCFAs (Figures S6A–S6C), and the absence of their cognate receptors on mouse vagal neurons (Figure S5),15,40 suggests these particular subclasses of microbial metabolites likely act through indirect interactions with diverse EECs, subsets of which can synapse directly onto vagal neurons, or other cellular mediators.86 Additionally, 3IS has not been shown to be readily re-absorbed following secretion into the intestinal lumen and was rarely observed to activate vagal neurons in vitro (Figures S6A–S6C), suggesting this metabolite also acts through the indirect pathway in order to activate vagal afferents. It is also possible that select intestinal metabolites may access systemic circulation and act at extra-intestinal sites to modulate vagal activity, presumably with a time delay. In particular, lumenal microbiome-dependent BAs more acutely increased vagal afferent nerve activity, primarily via indirect activation of TGR5, which is expressed on various non-vagal cellular mediators along the gastrointestinal tract (Figures 3A–3C and S5A). Moreover, the direct application of BAs onto TGR5-expressing EECs induced calcium activity (Figures S7B–S7E). Duodenal and jejunal EECs are enriched for CCK,87 suggesting that this may align with prior studies demonstrating that BAs work synergistically with epithelial-derived CCK following absorption from the intestinal lumen,33 and that CCK signaling is dynamic and rapidly desensitizes.88 In contrast, perfusion of SCFAs into the SI lumen increased vagal afferent nerve activity following a latency period in an FFAR2-dependent manner (Figures 3D–3F). SCFAs also increased activity in EECs expressing FFAR2 (Figures S7B–S7E), suggesting the indirect activation of FFAR2-expressing epithelial cells and subsequent GLP-1 release.89 Finally, we found that microbiome-dependent 3IS in the small intestine elicited sustained vagal afferent nerve activity in a TRPA1-dependent manner (Figures 3G–3I). Further, 3IS activated EECs, but not vagal neurons alone (Figures S6 and S7). This may align with a previous study wherein indole stimulated TRPA1+ colonic EECs to release serotonin and activate colon-innervating neurons.45 Our observations, considered together with existing vagal single cell transcriptomic data, suggest the indirect activation of vagal afferent neurons via EECs and/or other cellular mediators (via FFAR2, TGR5, or TRPA1) by lumenal microbial metabolites. Future studies interrogating additional cell-type specific roles of metabolite receptors expressed on multiple cell types, as well as dissecting which individual metabolites within each pool acting as key drivers, will aid in uncovering the precise differential effects of indirect versus direct signaling on vagal responses to microbial stimuli.
While whole nerve vagal electrophysiological recordings provide insight into the effects of lumenal stimuli on population responses of vagal afferent neurons, the low resolution of this method overlooks more nuanced changes in vagal afferent neuronal activity at the single-unit level. Recent studies demonstrate that lumenal nutrient cues, such as fats and carbohydrates, engage parallel vagal afferent neurons via labeled lines.20,21,22 However, further characterization of how different subclasses of diet- and microbiome-dependent small molecules engage gut-brain circuits involved in nutrient sensing is needed. We assessed the effects of acute lumenal perfusion of select microbial BAs, SCFAs, and 3IS (involved in dietary fat, carbohydrate, and protein metabolism, respectively) on vagal afferent neuronal calcium activity in vivo. We found that all three classes of microbial metabolites resulted in increased calcium activity in nodose neurons with varied kinetics (Figure 4B)-- BAs elicited a bimodal distribution of immediate vs. delayed responses, whereas SCFAs and 3IS mostly elicited delayed responses (Figure 4D), which aligns with the slow, gradual onset of vagal afferent nerve activity in response to SCFA and sustained onset of afferent nerve activity with 3IS perfusion. This increased latency to peak calcium response suggests that our observed activation is likely due to the indirect activation of vagal afferents via enteroendocrine-enteric circuitry, absorption of metabolized factors, or possibly neuroimmune crosstalk. Therefore, future studies are warranted in order to dissect the particular mechanisms mediating rapid-versus latent-responding vagal neurons in response to lumenal microbially modulated metabolites. When assessing single-unit responses to sequential perfusion of two metabolite classes, microbiome-dependent BAs and SCFAs elicited calcium responses via largely non-overlapping subpopulations of vagal afferent neurons, whereas 3IS and BAs or SCFAs acted primarily via shared subpopulations (Figure 4E). These findings are supported by previous work demonstrating a shared role for both TGR5-and TRPA1-mediated alterations in digestion and satiety via epithelial CCK signaling,33,90,91,92,93 as well as TRPA1-and FFAR2-mediated alterations in host metabolism and feeding behaviors that have been reported to act via epithelial secretion of GLP-1.89,94,95,96 In contrast, previous work demonstrates that BA- and SCFA-mediated alterations in feeding behaviors act via distinct receptor-dependent signaling pathways.53,74 More broadly, diet-induced decreases in microbial diversity are associated with alterations in vagal innervation and responses to interoceptive signals.64,67,68,69 Our initial experiments demonstrated that GF mice exhibit decreased baseline vagal activity compared to colonized controls (Figures 1B and 1C). Following our identification of macronutrient-derived microbial metabolites that increase vagal activity, we sought to determine whether the oral supplementation of these metabolites can phenocopy increases in baseline vagal activity observed in response to microbial colonization. We found that indeed, supplementation with MMs increased vagal afferent nerve tone compared to vehicle controls (Figure 4F), providing additional insight into the role of microbiome-dependent dietary metabolites as potential mediators of vagal physiology.
Despite evidence for vagal chemosensory pathways mediating communication from the intestinal lumen to the brain,22,71,97 effects of specific microbial metabolites and their associated receptors on CNS neuronal activity remain unclear. We, therefore, addressed the effects of lumenal perfusion of BAs, SCFAs, and 3IS alongside their respective receptor antagonists for TGR5, FFAR2, and TRPA1 on medial NTS neuronal activation by the immunofluorescence detection of the immediate-early gene cFos. We found that all metabolite classes significantly increased NTS neuronal activation, which could be prevented by pre-and co-perfusion of antagonists (Figure 5). Sensory information processed in the NTS is directed to several other brain regions, including the DMV and ARH.73 We found that none of the tested metabolite classes modified DMV neuronal cFos levels compared to VEH perfusion (Figures S8A–S8C), suggesting that BAs, SCFAs, and 3IS in the SI lumen do not play a predominant role in acute vagovagal circuit modulation. In contrast, lumenal BAs and 3IS elicited a modest reduction in neuronal activation in the ARH via TGR5 and TRPA1, respectively, while SCFAs elicited no overt effect (Figures S8D–S8F). This aligns with reports that microbial metabolites elicit anorexigenic effects via hypothalamic regulation, many of which are vagally mediated.33,53,74,75 Notably, BAs and 3IS exhibit the highest percentage of dually responsive vagal neurons (Figure 4E), suggesting that these metabolites may engage shared circuitry in order to modulate CNS physiology. Circulating deoxycholic acid promotes satiety synergistically with peripheral CCK via vagal TGR5 and modulation of ARH and POMC neuronal activation.53 BAs also directly modulate AgRP/NPY neuronal gene expression, suggesting circuit-specific effects of BAs on ARH neuronal activity responsible for driving satiety.74 Therefore, further investigation of the ARH neuronal subtype-specific effects of vagal afferent signaling and behavioral outcomes in response to lumenal BAs is warranted. Previous work has suggested a role for the metabolic precursor to 3IS, indole, in regulating host feeding behaviors via indole-mediated GLP-1 release from enteroendocrine L-cells.98 Here, we demonstrate that lumenal 3IS functions as a potential modulator of host hypothalamic circuitry via vagal afferent signaling and TRPA1. However, little is known regarding how this signaling pathway may alter host behaviors. Therefore, future studies aimed at uncovering the precise role of the TRPA1-mediated vagal chemosensation of lumenal 3IS on host behavioral outcomes is necessary. Together, these data suggest that lumenal metabolites modulate CNS neuronal activity via receptor-dependent vagal signaling and may have consequential effects on CNS responses that contribute to behavior. However, more work is needed to determine additional downstream targets and differential effects of these gut-to brain signaling pathways on CNS physiology and associated behaviors.
Findings from this study highlight lumenal microbial metabolites derived from various sources of dietary macronutrients-- fats (BAs), complex carbohydrates (SCFAs), and proteins (3IS)—that differentially activate vagal afferent neurons via receptor-mediated signaling to convey information to the brain. Following the ingestion of dietary fats, BAs are released from the liver into the SI lumen where they undergo chemical transformations, such as deconjugation and dehydroxylation, which are carried out by gut microbes.99 Enzymes capable of catalyzing such reactions are found across all major bacterial phyla,100 suggesting a broad role for the microbiota in regulating the lumenal BA pool. Similarly, SCFAs are derived from the microbial fermentation of dietary fibers that are otherwise non-digestible by the host, with differential production by bacterial members of the phyla Bacteroidetes (Bacteriodata; acetate, propionate) and Firmicutes (Bacillota; butyrate).101,102 Despite being predominantly produced and concentrated in the colon, SCFAs have been shown to be present in the ileum and are able to translocate to more distal body sites via enterohepatic circulation.103 Tryptophan derivatives, such as indole, are produced by pathobionts, such as Escherichia coli, Enterococcus faecalis, and Edwardsiella tarda,104 before being hydroxylated and sulfated in the liver and secreted as 3IS into the small intestine. As levels of dietary metabolites and microbiome composition can fluctuate depending on meal time,105,106 further examination into circadian effects of diet- and microbiome-dependent metabolites on vagal afferent neuronal activity is warranted. Moreover, the scope of this study is limited to understanding the role of small-intestinal lumenal metabolites on vagal afferent neuronal activation. Previous studies demonstrate that vagal afferent neurons innervate the colon at lower densities107 where microbial density is highest. Microbially derived metabolites have been demonstrated to activate colonic vagal afferent and enteric neurons via EEC-mediated serotonin release.45 Moreover, small-intestinal microbial metabolites such as BAs are able to reach the colon following enterohepatic circulation in order to undergo further bacterial transformations108 where they can then act as signaling molecules. As such, future studies examining the role of colonic lumenal microbial metabolite-neuronal interactions is warranted.
Understanding the temporal variation in the bioavailability of neuromodulatory microbial metabolites and in vagal activity could reveal important insights into the functional role of the vagal interoception of different types of microbial metabolites. As proof of principle, vagal TGR5 signaling mediated the anorexigenic effects of circulating BAs,33 whereas vagal FFAR3 signaling mediated the effects of circulating SCFAs on satiety.53 However, further studies on whether the vagal circuits engaged by lumenal microbial metabolites modulate analogous or additional CNS-associated behaviors remains to be determined. Circuit tracing studies have uncovered polysynaptic connections from gut-innervating vagal afferent neurons to higher order brain regions such as substantia nigra and hippocampus,109 suggesting the potential for microbial metabolites to impact complex behavioral responses beyond those involved in feeding. BAs, SCFAs, and 3IS have been associated with alterations in behavioral endophenotypes of anxiety and depression,59,110,111 cognitive impairment,112 and motor deficits,3,111 each which has been linked to vagal signaling and alterations in the gut microbiota.3,9,10 Overall, more research is needed to determine brain and behavioral responses to the vagal chemosensation of lumenal microbial metabolites, and to further evaluate the potential to leverage the microbiome to modify neuronal signaling across the gut-brain axis.
Limitations of the study
In summary, we demonstrate that the microbiome regulates select metabolites in the SI lumen that differentially activate chemosensory vagal neurons in a receptor-dependent manner, thereby enabling communication to downstream CNS neuronal targets. While our study provides initial evidence for the indirect signaling of microbial metabolites through enteroendocrine cells, additional experiments involving cell-type specific TGR5, FFAR2, and TRPA1 knockout mouse models will more precisely uncover the cellular mediators underpinning the receptor-dependent vagal chemosensation of lumenal SCFAs, BAs, and 3IS. In addition, cell-type specific histological or functional examination of the CNS neuronal subtypes that are differentially modulated by these chemosensory circuits can lend more direct insight into the behavioral and physiological outcomes associated with microbiota-gut-brain signaling via the vagus nerve. Understanding the precise signaling pathways and cellular mediators involved in microbiome-gut-brain communication may help inform new avenues for treating neurological symptoms that modulated by the vagus nerve.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Elaine Hsiao (ehsiao@g.ucla.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
All source data are included as supplementary tables in this article.
-
•
All original code has been deposited at Zenodo: https://doi.org/10.5281/zenodo.14497742 and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact upon request.
Acknowledgments
We thank members of the Hsiao laboratory for their guidance and review of the article; members of the UCLA Goodman Luskin Microbiome Center Gnotobiotics Core Facility for technical support; Dr. Stephen Liberles for critical training on intestinal perfusion and vagal nerve electrophysiology, Dr. Daniel Aharoni for helpful advice regarding calcium imaging data analysis; Dr. Scott Kanoski and Dr. Diego Bohorquez for helpful feedback on the project; and Dr. Baljit Khakh for allowing usage of his osmometer. This work was supported by funds from an NIH Ruth L. Kirschstein National Research Service Awards (F31 NS118966 and T32 GM007185), UCLA Hyde Pre-doctoral Fellowship, and UCLA Dissertation Year Fellowship to K.G.J., UCLA Hyde Pre-doctoral Fellowship to S.A.K., American Health Assistance Foundation to C.S., and NINDS grant (R01 NS115537) to E.Y.H. E.Y.H. is a New York Stem Cell Foundation – Robertson Investigator. This research was supported in part by the New York Stem Cell Foundation. This project has been made possible in part by grant number 2018-191860 from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation.
Author contributions
K.G.J., S.A.K., C.S., K.B.Y., T.O., D.M., and E.L. performed the experiments and analyzed the data. H.E.V., J.P., L.Y., S.C.M., and F.E.S. provided key technical guidance and resources. K.G.J. and E.Y.H. designed the study. K.G.J. and E.Y.H. wrote the article. All authors discussed the results and commented on the article.
Declaration of interests
The authors declare no competing interests. Findings reported in the article are the subject of UCLA provisional patent application PCT/US24/43222.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Guinea pig anti-NeuN | Sigma | RRID: AB_11205592 |
| Rabbit anti-cFos | Abcam | RRID: AB_2905616 |
| Prolong Gold Antifade Mountant with DAPI | ThermoFisher | Cat# P36931 |
| Chemicals and peptides and recombinant proteins | ||
| Ampicillin | Sigma | Cat# A9518 |
| Metronidazole | Sigma | Cat# M1547 |
| Neomycin | Sigma | Cat# N1876 |
| Vancomycin | Chem-Impex International | Cat# 00315 |
| 7-ketodeoxycholic Acid | Sigma | Cat# SMB00806 |
| Alpha-muricholic Acid | Cayman Chemical | Cat# 20291 |
| Beta-muricholc Acid | Sigma | Cat# ML2372 |
| Sodium Cholate Hydrate | Sigma | Cat# C9282 |
| Sodium Chenodeoxycholate | Sigma | Cat# C8261 |
| Deoxycholate | Sigma | Cat# D6750 |
| Sodium Glycocholate Hydrate | Sigma | Cat# G7132 |
| Lithocholic Acid | Sigma | Cat# L6250 |
| Sodium Taurodeoxycholate Hydrate | Sigma | Cat# T0557 |
| Sodium Taurohyodeoxycholate hydrate | Sigma | Cat# T0682 |
| Sodium Taurolithocholate | Sigma | Cat# T7515 |
| Ursodeoxycholic Acid | Sigma | Cat# U5127 |
| SBI-115 | MedChem Exppress | Cat# HY-111534 |
| Sodium Acetate | Sigma | Cat# S2889 |
| Sodium Butyrate | Sigma | Cat# B5887 |
| Sodium Propionate | Sigma | Cat# P1880 |
| GLPG0974 | MedChem Exppress | Cat# HY-12940 |
| Indoxyl Sulfate Potassium Salt | Sigma | Cat# I3875 |
| A967079 | MedChem Exppress | Cat# HY-108463 |
| L-Kynurenine | Sigma | Cat# K8625 |
| Kynurenic Acid | Sigma | Cat# K3375 |
| Xanthurenic Acid | Sigma | Cat# D120804 |
| Dihydrocaffeic Acid | Sigma | Cat# 102601 |
| Tryptamine | Sigma | Cat# 193747 |
| Oleylethanolamide | Sigma | Cat# O0383 |
| Palmitoylethanolamide | Sigma | Cat# P0359 |
| Linoleylethanolamide | Sigma | Cat# L1164 |
| 9-s-HODE | Sigma | Cat# SML0503 |
| 13-s-HODE | Sigma | Cat# H9146 |
| Succinic Acid | Sigma | Cat# S3674 |
| L-Glutamic Acid | Sigma | Cat# G1251 |
| Hippurate | Sigma | Cat# H9380 |
| Imidazole Propionic acid | Sigma | Cat# 77951 |
| Phenethylamine | Sigma | Cat# 241008 |
| Trimethylamine-N-Oxide | Sigma | Cat# 317594 |
| D-glucose | Sigma | Cat# G5767 |
| DMEM + GlutaMAX | Gibco | Cat# 10569010 |
| 100x Penicillin/Streptomycin | Gibco | Cat# 15140122 |
| FBS Certified One Shot | Gibco | Cat# A3160401 |
| PBS without Ca/Mg | Gibco | Cat# 10010023 |
| 0.5M EDTA | Invitrogen | Cat# AM9260G |
| 0.25% Trypsin-EDTA | Gibco | Cat# 25200056 |
| Poly-D-lysine | Gibco | Cat# A3890401 |
| Matrigel Matrix | Corning | Cat# 354234 |
| L-WRN conditioned medium | ATCC | Cat# CRL-3276 |
| Advanced DMEM/F12 | Gibco | Cat# 12634010 |
| EGF | Preprotech | Cat# 315-09 |
| Y-27632 | R&D Systems | Cat# 1254 |
| EP4 inhibitor | R&D Systems | Cat# 2514 |
| HBSS | ThermoFisher | Cat#14170112 |
| Collagenase 1A | Sigma-Aldrich | Cat# C96891 |
| Dispase II | Sigma-Aldrich | Cat# D4693 |
| Fluo-8 AM | AAT Bioquest | Cat# 21080 |
| Fluo-4 AM | ThermoFisher | Cat# F14217 |
| Critical commercial assays | ||
| RNeasy Micro Kit | QIAGEN | Cat# 74004 |
| RNeasy Plus Mini Kit | QIAGEN | Cat# 74134 |
| Dneasy PowerSoil Kit | QIAGEN | Cat# 47014 |
| Deposited data | ||
| Metabolomics Data | This study | N/A |
| Spike Sorting Code | This study, Zenodo | https://doi.org/10.5281/zenodo.14497742 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Jackson Laboratories | RRID:IMSR_JAX:000664 |
| Mouse: Phox2b-cre | Jackson Laboratories | RRID:IMSR_JAX:016223 |
| Mouse: Ai96 | Jackson Laboratories | RRID:IMSR_JAX:028866 |
| Experimental models: Cell lines | ||
| STC-1 | ATCC | Cat# CRL-3254, RRID:CVCL_J405 |
| Oligonucleotides | ||
| Primers for tissue quantitative real-time PCR, See Table S3 | ||
| Software and algorithms | ||
| LabVIEW | National Instruments | RRID:SCR_014325 |
| MATLAB | MathWorks | RRID:SCR_001622 |
| OriginPro | Origin Lab | RRID:SCR_014212 |
| PRISM 6 | GraphPad | RRID:SCR_002798 |
| Minian | Dong et al.113 | RRID:SCR_022601 |
| FIJI | Schindelin et al.114 | RRID:SCR_002285 |
| Other | ||
| QuantStudio 5 Real-Time PCR System | Applied Biosystems | N/A |
| Zeiss LSM780 | Zeiss | N/A |
| Leica Dmi8 | Leica | N'A |
Experimental model and study participant details
Mice
All experimental procedures were carried out in accordance with US NIH guidelines for the care and use of laboratory animals and approved by the UCLA Institutional Animal Care and Use Committees. Mice used for data collection were males, at least 6 weeks of age. C57BL/6J mice were purchased from Jackson laboratories (stock no. 000664), reared as SPF or rederived as GF and bred in flexible film isolators at the UCLA Center for Health Sciences Barrier Facility. Ai96 (JAX stock no. 028866), Phox2b-cre (JAX stock no.016223), were obtained from Jackson laboratories and bred at the UCLA Biomedical Sciences Research Building barrier facility. Mice were housed on a 12-h light-dark schedule in a temperature-controlled (22°C–25°C) and humidity-controlled environment with ad libitum access to water and standard chow (Lab Diet 5010).
Method details
Antibiotic treatment and conventionalization
Adult SPF mice were gavaged twice daily for 1 week with a cocktail of vancomycin (50 mg/kg), neomycin (100 mg/kg) and metronidazole (100 mg/kg) every 12 h daily for 7 days. Ampicillin (1 mg/mL) was provided ad libitum in drinking water. For conventionalization, fecal samples were collected from adult SPF C57BL/6J mice and homogenized in 1mL pre-reduced PBS per pellet. 100uL of the homogenate was administered by oral gavage to recipient GF mice.
Preparation of antibiotics, small-intestinal, and cecal contents
Vancomycin and neomycin were diluted in water to a final concentration of 1 mg/mL for all antibiotic perfusion experiments, then sterile filtered. Vehicle for all antibiotic perfusion experiments was water. For SPF SI and cecal content preparations adult SPF male mice were euthanized, and SI and cecal lumenal contents were snap-frozen in liquid nitrogen. Equal weights of frozen SI and cecal content were then combined and diluted to a concentration of 0.1 g/mL wet weight in sterile-filtered PBS. Samples were then centrifuged at 500g for 5 min to pellet out any large dietary components, and supernatants were used for lumenal perfusion. GF SI/cecal contents were collected in the same way from donor GF adult male mice. Sterile-filtered SI/cecal contents were prepared by vacuum-filtering SPF SI/cecal content supernatants through a 0.2μm filter. Sterile-filtered PBS was used as vehicle in all SI/cecal perfusion experiments.
In vivo vagal electrophysiology
Baseline vagal tone recordings: Adult SPF male and female (Figure S1), Adult SPF, GF, ABX, or CONV male (Figure 1C), or adult GF male (Figure 4F) mice were anesthetized with isoflurane (5%) and maintained at 1.8% throughout the experiment. The left cervical vagus nerve was exposed, transected inferior to the nodose ganglion and placed across two platinum iridium wires (insulated except for a short segment in contact with the nerve) for recording of baseline vagal tone. Recordings were conducted for 10 min. Vagal recordings with lumenal perfusion: the cervical vagus nerve was prepared as above in adult male SPF mice. Additionally, a 20-gauge gavage needle attached to a peristaltic pump (Cole Parmer) with separate tubing for each infusion solution was inserted into the duodenal lumen and secured with sutures. An outflow port was generated by transecting the small intestine ∼10cm distal to the inflow site. During recording, vehicle was first perfused through the lumen at a constant rate for 10 min to establish a baseline following surgery at a flow rate of 250 μl/minute.20 Following the baseline period, stimuli were introduced into the small-intestinal lumen and perfused at the same rate for the remainder of the experiment. Data Acquisition: a differential amplifier was used (A-M Systems LLC). The gain was set to 1000x and a bandpass filter was applied (300Hz-5kHz). The signal was digitized at 20kHz using a data acquisition board (National Instruments) under the control of LabView software Data Analysis: Spikes were detected using an SO-CFAR threshold (window duration of 1501/8000, guard duration 10/8000) to generate an adaptive threshold 4SD above RMS noise.115 Firing rates were calculated by generating 10s (baseline vagal tone) or 30s (perfusion experiments) bins and then applying a Savitszky-Golay filter (OriginPro) of 10 points. Baseline vagal tone was defined as the average of the final 300s of recording. For experiments assessing sex differences all raw values were normalized to the SPF male cohort average for the rig on which the recordings took place. For comparison between SPF, GF, ABX, and CONV mice, all raw values were normalized to the SPF cohort average. For metabolite supplementation experiments, all raw values were normalized to the VEH cohort average. Perfusion experiments: baseline values were defined as the average frequency of the final 60s of recording in the initial baseline period (F0). Frequency of the recording was then normalized to the baseline value within-subject (F/Fo). Area under the curve was calculated for each stimulus window and defined as the integral of frequencies over the stimulus window.
16s rRNA gene qPCR for intestinal perfusate samples
To quantify bacterial loads in perfusate outflow samples, 500uL of perfusate outflow contents were collected over the course of 2 min directly from the small-intestinal outflow opening at t = 0–120s, t = 600-720s, t = 1200-1320s, and t = 1800-1920s. DNA was then isolated using QIAGEN DNeasy Kit according to the manufacturers protocol and the concentration of each pellet was quantified via spectrophotometry (BioTek Synergy H1 Multimode Reader). qPCR was performed using PowerUp SYBR Green Master Mix, according to the manufacturer’s protocol (Applied Biosystems, Carlsbad, USA) with universal primer sets targeting the bacterial 16S rRNA gene.
Metabolomics
Samples were collected from adult SPF, GF, ABX, or CONV mice. Lumenal contents were collected from the first 3cm of small intestine and the entirety of the cecum, then snap frozen in liquid nitrogen and stored at −80°C. Samples were prepared using the automated MicroLab STAR system (Hamilton Company) and analyzed on gas chromatography (GC)-mass spectrometry (MS), liquid chromatography (LC)-MS and LC-MS/MS platforms by Metabolon, inc. Protein fractions were removed by serial extractions with organic aqueous solvents, concentrated using a TurboVap system (Zymark) and vacuum dried. For LC/MS and LC-MS/MS, samples were reconstituted in acidic or basic LC-compatible solvents containing >11 injection standards and run on a Waters ACQUITY UPLC and Thermo-Finnigan LTQ mass spectrometer, with a linear ion-trap front-end and a Fourier transform ion cyclotron resonance mass spectrometer back-end. For GC/MS, samples were derivatized under dried nitrogen using bistrimethyl-silyl-trifluoroacetamide and analyzed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer using electron impact ionization. Chemical entities were identified by comparison to metabolomic library entries of purified standards. Following log transformation and imputation with minimum observed values for each compound, data were analyzed using one-way ANOVA to test for group effects. P and q-values were calculated based on two-way ANOVA contrasts. Principal components analysis was used to visualize variance distributions. Supervised Random Forest analysis was conducted to identify metabolomics prediction accuracies.
Preparation of metabolite pools, single metabolites, and receptor antagonists
All working metabolite solutions were made up in PBS and up to 0.05% DMSO, brought to a pH of 7.3, and sterile filtered. The concentrations for each metabolite pool, individual metabolites, and receptor antagonists were determined from serum metabolomics data in the lab (data not shown) and existing literature quantifying candidate metabolites in murine fecal, serum, and lumenal content samples or based on dose-response curves assessing metabolite-induced effects on our candidate receptors and are as follows: Tryptophan metabolites (kynurenine 100uM,116 kynurenic acid 16.1uM; 117 xanthurenate 1uM; 118 dihydrocaffeate 30nm, tryptamine 0.03uM, pooled), FAEs (oleylethanolamide (OEA), palmitoylethanolamide (PEA), linoleylethanolamide (LEA) 10uM, pooled),119 HODEs (9-s- and 13-s- HODE,1uM, pooled), succinate (2mM), glutamate (50mM), bile acids (BAs, cholate 1240nM, glycocholate 3.5nM, chenodeoxycholate 42nM, alpha-muricholate 142nM, beta-muricholate 1080nM, deoxycholate 390nM, taurodeoxycholate 260nM, ursodeoxycholate 74nM, taurohyodeoxycholate 18.8nM, 7-ketodeoxycholate 100nM, lithocholate 390nM, taurolithocholate 0.33nM, pooled), m-tolyl 5-chloro-2-[ethylsulonyl] pyrimidine-4-carboxylate (SBI-115, 200uM),52 short-chain fatty acids (SCFAs, acetate 80uM, butyrate 22uM, propionate 10uM, pooled), 4-[[(R)-1-(benzo[b]thiophene-3-carbonyl)-2-methyl-azetidine-2-carbonyl]-(3-chloro-benzyl)-amino]-butyric acid 99 (GLPG0974, 10uM),120 hippurate (2uM),121 Trimethylamine-N-oxide (TMAO, 3uM),122 imidazole propionate (200nM),123 phenethylamine (PEA, 100uM),13 3-indoxyl sulfate (3IS, 1uM),124 or (1E,3E)-1-(4-Fluorophenyl)-2-methyl-1-penten-3-one oxime (A967079, 10uM).46 Vehicle for tryptophan metabolites, FAEs, MFAs, succinate, glutamate, SCFAs, TMAO, hippurate, imidazole propionate, and phenethylamine was sterile-filtered PBS. Vehicle for BAs was 0.05% DMSO in PBS, Vehicle for 3-IS was 1uM KCl in PBS.
In vitro calcium imaging
Adult male SPF mice were euthanized and both left and right vagal ganglia were quickly excised and immersed in ice-cold HBSS, then mechanically desheathed with a razor blade. Ganglia were then digested for 1 h at 37C and 5% CO2 in HBSS with 1 mg/ml collagenase and dispase II. Following enzymatic digestion, the neurons were pelleted via centrifugation at 500g for 2 min and the solution was replaced with HDMEM. Neurons were then manually triturated, and placed onto poly-L-lysine coated coverslips. Coverslips were then incubated for 2 h at 37C and 5% CO2. The HDMEM was then carefully pipetted off of the coverslips and replaced with a solution of the calcium indicator Fluo-4 and probenecid to prevent extrusion (Life Technologies). Neurons were then incubated for 30 min at 37C and 5% CO2, followed by an additional 15-min incubation at room temperature. Following loading with fluo-4, coverslips were loaded into a perfusion system and imaged with a Leica Dmi8 Fluorescence microscope. Neurons were initially bath perfused over the course of 10 min with live-cell imaging solution with 2% glucose to allow for acclamation. Cells were then imaged for 2 min to establish a baseline signal before a 2 min bath perfusion of High K+ (60mM), tryptophan metabolites (kynurenine 100uM, kynurenic acid 16.1uM, xanthurenate 1uM, dihydrocaffeate 30nm, tryptamine 0.03uM, pooled), endocannabinoids (oleylethanolamide (OEA), palmitoylethanolamide (PEA), linoleylethanolamide (LEA) 10uM, pooled), HODEs (9-s- and 13-s- HODE,1uM, pooled), succinate (2mM), glutamate (50mM), bile acids (cholate 1240nM, glycocholate 3.5nM, chenodeoxycholate 42nM, alpha-muricholate 142nM, beta-muricholate 1080nM, deoxycholate 390nM, taurodeoxycholate 260nM, ursodeoxycholate 74nM, taurohyodeoxycholate 18.8nM, 7-ketodeoxycholate 100nM, lithocholate 390nM, taurolithocholate 0.33nM, pooled), short-chain fatty acids (acetate 80uM, butyrate 22uM, propionate 10uM, pooled), or 3-indoxyl sulfate (1uM), followed by 30 min of bath perfusion of LCIS with 2% glucose to allow for response recovery. Data Analysis: individual neuronal ROI’s were drawn and fluorescence signal was quantified using LASX software (Leica). The baseline period for the calcium signal was calculated as the average of the 60 s before stimulus onset (Fo). Responses were reported in units of baseline fluorescence (ΔF/F). Cells were considered responsive to stimuli if the maximal ΔF/F signal following stimulus onset was >50% of the baseline value.
STC-1 and enteroid cell culture, qPCR, and calcium imaging
STC-1 cells (ATCC #CRL-3254) were thawed and passaged until >70% confluence for maintenance in DMEM with GlutaMAX (Gibco #10569010), 1% Penicillin-Streptomycin (Gibco #15140122), and 10% FBS at 37C and 5% CO2. qRT-PCR experiments: STC-1 cells were collected 1 day after passaging, lysed, and processed with RNeasy Plus Mini kit (Q\IAGEN #74134) per the manufacturers protocol for RNA isolation. Enteroids were grown as previously described (Miyoshi 2017). Briefly, stem cell-enriched mouse duodenal spheroids were maintained in 50% L-WRN conditioned medium, dissociated, and grown in differentiation medium for 48h prior to RNA isolation, followed by processing with RNeasy Plus Mini kit. qRT-PCR was carried out with iTaq Universal SYBR Green Supermix (Bio-Rad) and primers targeting enterocyte (Ace2) and enteroendocrine cell (Chga) markers, and microbial metabolite receptors (Gpbar1, Ffar2, Trpa1). Transcript levels were normalized to β2 microglobulin and relative expression values were calculated using the ΔΔCt method. Calcium Imaging: STC-1 cells were re-suspended in culture media and incubated overnight at 37C and 5% CO2 in poly-D-lysine coated wells. On the day of imaging, culture media was replaced with Fluo-8 in PBS with Ca2+ and Mg2+ and incubated at 37C for 1 h prior to imaging. During imaging, Fluo-8 solution was replaced with PBS and imaged for 60 s to establish a baseline signal, followed by addition of 2x microbial metabolites (bile acids: cholate 2480nM, glycocholate 14nM, chenodeoxycholate 168nM, alpha-muricholate 568nM, beta-muricholate 2480nM, deoxycholate 780nM, taurodeoxycholate 520nM, ursodeoxycholate 148nM, taurohyodeoxycholate 37.6nM, 7-ketodeoxycholate 200nM, lithocholate 780nM, taurolithocholate 0.66nM, pooled, short-chain fatty acids: acetate 160uM, butyrate 44uM, propionate 20uM, pooled, or 3-indoxyl sulfate: 2uM) for a final 1x working concentration, High K+ (60mM), or D-glucose (20mM). Cells were continually imaged throughout the experiment for 180 s at 1Hz on a Leica Dmi8 fluorescence microscope. Data Analysis: 100 randomly sampled ROIs were drawn and fluorescence signal was quantified using LASX software (Leica). The baseline period for the calcium signal was calculated as the average of the 60s prior to stimulus onset (Fo). Responses were reported in units of baseline fluorescence (ΔF/F). Graphs are displayed beginning at experimental t = 20s for visualization purposes. Cells were considered responsive to stimuli if the maximal ΔF/F signal following stimulus onset was >50% of the baseline value.
qPCR of vagal nodose neurons
Vagal nodose ganglia were bilaterally harvested, pooled (n = 2 per subject), and snap-frozen in RNAlater (ThermoFisher #AM7020). RNA was then isolated using the RNeasy Micro Kit (QUIAGEN #74004) per the manufacturers protocol. qRT-PCR was carried out using PowerUp SYBR Green Master Mix (ThermoFisher #A25742) and primers targeting microbial metabolite receptors (Gpbar, Ffar2, Trpa1). Transcripts were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression and relative expression values were calculated using the ΔCt method.
In vivo calcium imaging
Vagal afferent neuron imaging was performed as previously described.20 In brief, SPF mice were anesthetized with a cocktail of ketamine and xylazine (100 mg/kg and 10 mg/kg, intraperitoneal), tracheotomized, and maintained on 1.5% isoflurane for the remainder of the surgery and imaging session via ventilator (Kent Scientific). Body temperature was maintained at 37°C using an electrical heating pad and rectal temperature probe (Kent Scientific). The vagus nerve was transected superior to the nodose ganglion and vagal ganglia were then embedded between two 5mm-diameter glass coverslips (neuVitro) with silicone adhesive (KWIK-SIL, World Precision Instruments). Imaging was conducted on a Zeiss LSM 780 confocal microscope at a frame rate of 1Hz. Analysis of imaging data: Imaging data were analyzed using custom Python scripts based on the Minian pipeline.113 Imaging data was registered to collect for motion artifacts and denoised using a median filter. A constrained non-negative matrix factorization (CNMF) algorithm was used to identify single cells and extract calcium activity. CNMF output regions of interest were then manually inspected to remove non-neuronal signals. Metabolite stimuli were perfused into the small intestinal lumen for 2 min and separated by at least 10 min of PBS perfusion. Order was randomized to account for order-specific effects on neuronal activity. For single-unit analysis in response to sequential metabolite application, neuronal regions of interest in metabolite-responsive neurons were cross-registered across stimulus runs and compared to determine single- or dual-responsivity to metabolite classes. The baseline period for the calcium signal was calculated as the average of the last 120 s before stimulus onset (Fo). Responses were reported in units of baseline fluorescence. Cells were considered responsive to stimuli if the maximal ΔF/F signal following stimulus onset was i) greater than 2 standard deviations above the baseline fluorescence, and ii) the mean ΔF/F signal over a 20s window around peak response was >50% of the baseline value. Only units that displayed a ΔF/F signal >4 standard deviations over the baseline fluorescence in response to an electrical stimulus were included in analysis. For visualization, metabolite-responding units were hierarchically clustered over the entire time-course of each experiment.
Immunohistochemistry for cFos
Lumenal perfusion of stimuli: Mice were anesthetized with 5% isoflurane and maintained at 2% for the entirety of the surgery. To begin intralumenal perfusion, the small intestine was transected at the pyloric sphincter, the junction between the stomach and duodenum, to create an inflow port. Subsequently, a gavage needle attached to a peristaltic pump was inserted into the duodenal lumen. An outflow port was made by transecting the small intestine 3 cm distal to the inflow port. All lumenal contents were flushed from the small intestine with PBS. Animals were then continually perfused with PBS at 250uL/min flow rate for a 10-min baseline period, followed by perfusion of stimuli for 30 min at a constant flow rate to account for any mechanical distension. Following stimulus perfusion, mice remained under anesthesia for 1 h to allow for cFos induction before tissue harvesting. Following the 1-h rest period, animals were sacrificed, and tissues were harvested and fixed via intracardial perfusion of ice-cold PBS followed by 4% paraformaldehyde (PFA). Brains were then post-fixed in 4% PFA at 4°C for 3 h followed by an overnight incubation in 30% sucrose at 4°C for cryoprotection. Ganglia were positioned bulb-side down in optimal cutting temperature compound (OCT compound) and frozen for cryostat sectioning. Tissues were then sectioned at 30μm and mounted on a microscope slide for immunohistochemical processing (IHC). Slides were thawed for 10 min at room temperature in a humidified chamber to prevent the tissue from drying and then permeabilized in 0.5% Triton/0.05% tween 20 in PBS (PBS-TT). Blocking solution consisting of 5% normal goat serum (NGS) in PBS-TT was applied to the tissue and allowed to incubate at room temperature for 2 h to prevent non-specific antibody binding and reduce background staining. The tissue sections were then incubated overnight at 4°C with primary antibodies (rabbit anti-cFos 1:500, and guinea pig anti-NeuN, 1:500) in blocking solution (5% NGS + PBS-TT). The following day, slides were washed three times, 5 min per wash, with PBS-TT before incubating at room temperature for 2 h with secondary antibodies (goat anti-rabbit 488 1:1000, goat anti-guinea pig 568 1:1000, and DAPI 1:1000) in blocking solution (5% NGS + PBS-TT). Confocal Imaging and Quantification Analysis: Images were obtained using a 20× air objective (NA 0.8) on an upright Zeiss LSM 780 confocal microscope. Z-stacks were acquired for three technical replicates of NTS brain tissue or two technical replicates of DMV and ARH tissue and maximum-intensity projections were generated for subsequent analysis in ImageJ. NTS, DMV, and ARH sections were selected and ROI’s were drawn based on of the Allen Mouse Brain Atlas. First, NeuN positive (NeuN+) cells that were each confirmed to colocalize with a DAPI nucleus were counted using the multi-point tool to obtain the total number of neurons. Subsequently, cFos+ cells were counted using the multi-point tool by confirming colocalization of a cFos immunofluorescence signal with NeuN and DAPI. For each image, the total number of cFos+ neurons were divided by the total number of NeuN+ cells to obtain the percentage of cFos+ neurons. Finally, the percentage of cFos+ neurons for all technical replicates of NTS slices per animal were averaged to obtain a biological n = 1 and find the overall percentage of cFos+ neurons.
Metabolite supplementation
Adult male GF C57BL/6J mice were treated twice-daily via oral gavage with 200uL of either vehicle (VEH: 12uM KCl, 1.34mM NaCl, 0.012% DMSO in PBS), or a cocktail of microbial metabolites (MMs: cholate, 14.88uM; glycocholate, 42nM; chenodeoxycholate, 504nM; alpha-muricholate, 1.7uM; beta-muricholate, 12.96uM; deoxycholate, 4.68uM; taurodeoxycholate, 3.12uM; ursodeoxycholate, 888nM; taurohyodeoxycholate, 226nM; 7-ketodeoxycholate, 1.2uM; lithocholate, 4.68uM; taurolithocholate, 3.96nM, acetate, 0.96mM; butyrate, 264uM; propionate, 120uM, 3IS, 12uM). Concentrations of metabolites were determined in order to account for average total fluid intake over a 24 h period.125 On the day of each experiment, mice were additionally given a single 200uL oral gavage of VEH or MMs 1 h prior to in vivo vagal whole-nerve electrophysiological recordings.
Quantification and statistical analysis
Statistical analysis was performed using Prism software version 8.2.1 (GraphPad). Data were assessed for normal distribution and plotted in the figures as mean ± SEM. For each figure, n = the number of independent biological replicates. Outliers were identified with ROUT using a threshold of q = 2% for all electrophysiological recordings. Differences between two treatment groups were assessed using two-tailed, unpaired Student’s t test with Welch’s correction. Differences among >2 groups with only one variable were assessed using one-way ANOVA. If groups were determined to have significantly different variances, groups were assessed with Brown-Forsythe and Welch ANOVA + Games-Howell testing or Wilcoxon matched-pairs signed rank test. Significant differences emerging from the above tests are indicated in the figures by ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Notable non-significant (and non-near significant) differences are indicated in the figures by “n.s.”. Individual statistical analyses performed can be found in the figure legends.
Published: December 27, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111699.
Contributor Information
Kelly G. Jameson, Email: kgjameson@g.ucla.edu.
Elaine Y. Hsiao, Email: ehsiao@g.ucla.edu.
Supplemental information
References
- 1.Vuong H.E., Pronovost G.N., Williams D.W., Coley E.J.L., Siegler E.L., Qiu A., Kazantsev M., Wilson C.J., Rendon T., Hsiao E.Y. The maternal microbiome modulates fetal neurodevelopment in mice. Nature. 2020;586:281–286. doi: 10.1038/s41586-020-2745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sgritta M., Dooling S.W., Buffington S.A., Momin E.N., Francis M.B., Britton R.A., Costa-Mattioli M. Mechanisms Underlying Microbial-Mediated Changes in Social Behavior in Mouse Models of Autism Spectrum Disorder. Neuron. 2019;101:246–259.e6. doi: 10.1016/j.neuron.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sampson T.R., Debelius J.W., Thron T., Janssen S., Shastri G.G., Ilhan Z.E., Challis C., Schretter C.E., Rocha S., Gradinaru V., et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson's Disease. Cell. 2016;167:1469–1480.e12. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erny D., Hrabě de Angelis A.L., Jaitin D., Wieghofer P., Staszewski O., David E., Keren-Shaul H., Mahlakoiv T., Jakobshagen K., Buch T., et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015;18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh V., Roth S., Llovera G., Sadler R., Garzetti D., Stecher B., Dichgans M., Liesz A. Microbiota Dysbiosis Controls the Neuroinflammatory Response after Stroke. J. Neurosci. 2016;36:7428–7440. doi: 10.1523/JNEUROSCI.1114-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fulling C., Dinan T.G., Cryan J.F. Gut Microbe to Brain Signaling: What Happens in Vagus. Neuron. 2019;101:998–1002. doi: 10.1016/j.neuron.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Bravo J.A., Forsythe P., Chew M.V., Escaravage E., Savignac H.M., Dinan T.G., Bienenstock J., Cryan J.F. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. USA. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bharwani A., West C., Champagne-Jorgensen K., McVey Neufeld K.A., Ruberto J., Kunze W.A., Bienenstock J., Forsythe P. The vagus nerve is necessary for the rapid and widespread neuronal activation in the brain following oral administration of psychoactive bacteria. Neuropharmacology. 2020;170 doi: 10.1016/j.neuropharm.2020.108067. [DOI] [PubMed] [Google Scholar]
- 9.Bercik P., Park A.J., Sinclair D., Khoshdel A., Lu J., Huang X., Deng Y., Blennerhassett P.A., Fahnestock M., Moine D., et al. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neuro. Gastroenterol. Motil. 2011;23:1132–1139. doi: 10.1111/j.1365-2982.2011.01796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee K.E., Kim J.K., Han S.K., Lee D.Y., Lee H.J., Yim S.V., Kim D.H. The extracellular vesicle of gut microbial Paenalcaligenes hominis is a risk factor for vagus nerve-mediated cognitive impairment. Microbiome. 2020;8:107. doi: 10.1186/s40168-020-00881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wikoff W.R., Anfora A.T., Liu J., Schultz P.G., Lesley S.A., Peters E.C., Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quinn R.A., Melnik A.V., Vrbanac A., Fu T., Patras K.A., Christy M.P., Bodai Z., Belda-Ferre P., Tripathi A., Chung L.K., et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature. 2020;579:123–129. doi: 10.1038/s41586-020-2047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H., Nwe P.K., Yang Y., Rosen C.E., Bielecka A.A., Kuchroo M., Cline G.W., Kruse A.C., Ring A.M., Crawford J.M., Palm N.W. A Forward Chemical Genetic Screen Reveals Gut Microbiota Metabolites That Modulate Host Physiology. Cell. 2019;177:1217–1231.e18. doi: 10.1016/j.cell.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen L.J., Esterhazy D., Kim S.H., Lemetre C., Aguilar R.R., Gordon E.A., Pickard A.J., Cross J.R., Emiliano A.B., Han S.M., et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature. 2017;549:48–53. doi: 10.1038/nature23874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kupari J., Haring M., Agirre E., Castelo-Branco G., Ernfors P. An Atlas of Vagal Sensory Neurons and Their Molecular Specialization. Cell Rep. 2019;27:2508–2523e2504. doi: 10.1016/j.celrep.2019.04.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai L., Mesgarzadeh S., Ramesh K.S., Huey E.L., Liu Y., Gray L.A., Aitken T.J., Chen Y., Beutler L.R., Ahn J.S., et al. Genetic Identification of Vagal Sensory Neurons That Control Feeding. Cell. 2019;179:1129–1143.e23. doi: 10.1016/j.cell.2019.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Q., Yu C.D., Wang R., Xu Q.J., Dai Pra R., Zhang L., Chang R.B. A multidimensional coding architecture of the vagal interoceptive system. Nature. 2022;603:878–884. doi: 10.1038/s41586-022-04515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prescott S.L., Umans B.D., Williams E.K., Brust R.D., Liberles S.D. An Airway Protection Program Revealed by Sweeping Genetic Control of Vagal Afferents. Cell. 2020;181:574–589.e14. doi: 10.1016/j.cell.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao J., Campbell J.N., Tsai L.T., Wu C., Liberles S.D., Lowell B.B. Highly selective brain-to-gut communication via genetically defined vagus neurons. Neuron. 2021;109:2106–2115.e4. doi: 10.1016/j.neuron.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams E.K., Chang R.B., Strochlic D.E., Umans B.D., Lowell B.B., Liberles S.D. Sensory Neurons that Detect Stretch and Nutrients in the Digestive System. Cell. 2016;166:209–221. doi: 10.1016/j.cell.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchanan K.L., Rupprecht L.E., Kaelberer M.M., Sahasrabudhe A., Klein M.E., Villalobos J.A., Liu W.W., Yang A., Gelman J., Park S., et al. The preference for sugar over sweetener depends on a gut sensor cell. Nat. Neurosci. 2022;25:191–200. doi: 10.1038/s41593-021-00982-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M., Tan H.E., Lu Z., Tsang K.S., Chung A.J., Zuker C.S. Gut-Brain Circuits for Fat Preference. Nature. 2022;610:722–730. doi: 10.1038/s41586-022-05266-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugisawa E., Takayama Y., Takemura N., Kondo T., Hatakeyama S., Kumagai Y., Sunagawa M., Tominaga M., Maruyama K. RNA Sensing by Gut Piezo1 Is Essential for Systemic Serotonin Synthesis. Cell. 2020;182:609–624.e21. doi: 10.1016/j.cell.2020.06.022. [DOI] [PubMed] [Google Scholar]
- 24.Uhlig F., Grundy L., Garcia-Caraballo S., Brierley S.M., Foster S.J., Grundy D. Identification of a Quorum Sensing-Dependent Communication Pathway Mediating Bacteria-Gut-Brain Cross Talk. iScience. 2020;23 doi: 10.1016/j.isci.2020.101695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiman A.A., Ratze D.C., Welle E.J., Patel P.R., Richie J.M., Bottorff E.C., Seymour J.P., Chestek C.A., Bruns T.M. Multi-channel intraneural vagus nerve recordings with a novel high-density carbon fiber microelectrode array. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-72512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blair J.M.A., Webber M.A., Baylay A.J., Ogbolu D.O., Piddock L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 27.Hauser W.E., Jr., Remington J.S. Effect of antibiotics on the immune response. Am. J. Med. 1982;72:711–716. doi: 10.1016/0002-9343(82)90534-4. [DOI] [PubMed] [Google Scholar]
- 28.Chaturvedi S., Malik M.Y., Rashid M., Singh S., Tiwari V., Gupta P., Shukla S., Singh S., Wahajuddin M. Mechanistic exploration of quercetin against metronidazole induced neurotoxicity in rats: Possible role of nitric oxide isoforms and inflammatory cytokines. Neurotoxicology. 2020;79:1–10. doi: 10.1016/j.neuro.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Liang G., Bushman F.D. The human virome: assembly, composition and host interactions. Nat. Rev. Microbiol. 2021;19:514–527. doi: 10.1038/s41579-021-00536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kennedy E.A., King K.Y., Baldridge M.T. Mouse Microbiota Models: Comparing Germ-Free Mice and Antibiotics Treatment as Tools for Modifying Gut Bacteria. Front. Physiol. 2018;9:1534. doi: 10.3389/fphys.2018.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiner A., Turjeman S., Koren O. Gut microbes and host behavior: The forgotten members of the gut-microbiome. Neuropharmacology. 2023;227 doi: 10.1016/j.neuropharm.2023.109453. [DOI] [PubMed] [Google Scholar]
- 32.Silva Y.P., Bernardi A., Frozza R.L. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020;11:25. doi: 10.3389/fendo.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X., Li J.Y., Lee A., Lu Y.X., Zhou S.Y., Owyang C. Satiety induced by bile acids is mediated via vagal afferent pathways. JCI Insight. 2020;5 doi: 10.1172/jci.insight.132400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas C., Gioiello A., Noriega L., Strehle A., Oury J., Rizzo G., Macchiarulo A., Yamamoto H., Mataki C., Pruzanski M., et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metabol. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahlstrom A., Sayin S.I., Marschall H.U., Backhed F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metabol. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zarei I., Koistinen V.M., Kokla M., Klåvus A., Babu A.F., Lehtonen M., Auriola S., Hanhineva K. Tissue-wide metabolomics reveals wide impact of gut microbiota on mice metabolite composition. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-19327-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox L.M., Yamanishi S., Sohn J., Alekseyenko A.V., Leung J.M., Cho I., Kim S.G., Li H., Gao Z., Mahana D., et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tirelle P., Breton J., Riou G., Déchelotte P., Coëffier M., Ribet D. Comparison of different modes of antibiotic delivery on gut microbiota depletion efficiency and body composition in mouse. BMC Microbiol. 2020;20:340. doi: 10.1186/s12866-020-02018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egerod K.L., Petersen N., Timshel P.N., Rekling J.C., Wang Y., Liu Q., Schwartz T.W., Gautron L. Profiling of G protein-coupled receptors in vagal afferents reveals novel gut-to-brain sensing mechanisms. Mol. Metabol. 2018;12:62–75. doi: 10.1016/j.molmet.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uneyama H., Niijima A., San Gabriel A., Torii K. Luminal amino acid sensing in the rat gastric mucosa. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;291:G1163–G1170. doi: 10.1152/ajpgi.00587.2005. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt M.G., Jr., Soergel K.H., Wood C.M., Steff J.J. Absorption of short-chain fatty acids from the human ileum. Am. J. Dig. Dis. 1977;22:340–347. doi: 10.1007/BF01072192. [DOI] [PubMed] [Google Scholar]
- 43.Karaki S.I., Tazoe H., Hayashi H., Kashiwabara H., Tooyama K., Suzuki Y., Kuwahara A. Expression of the short-chain fatty acid receptor, GPR43, in the human colon. J. Mol. Histol. 2008;39:135–142. doi: 10.1007/s10735-007-9145-y. [DOI] [PubMed] [Google Scholar]
- 44.Tazoe H., Otomo Y., Karaki S.I., Kato I., Fukami Y., Terasaki M., Kuwahara A. Expression of short-chain fatty acid receptor GPR41 in the human colon. Biomed. Res. 2009;30:149–156. doi: 10.2220/biomedres.30.149. [DOI] [PubMed] [Google Scholar]
- 45.Ye L., Bae M., Cassilly C.D., Jabba S.V., Thorpe D.W., Martin A.M., Lu H.Y., Wang J., Thompson J.D., Lickwar C.R., et al. Enteroendocrine cells sense bacterial tryptophan catabolites to activate enteric and vagal neuronal pathways. Cell Host Microbe. 2021;29:179–196.e9. doi: 10.1016/j.chom.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bellono N.W., Bayrer J.R., Leitch D.B., Castro J., Zhang C., O'Donnell T.A., Brierley S.M., Ingraham H.A., Julius D. Enterochromaffin Cells Are Gut Chemosensors that Couple to Sensory Neural Pathways. Cell. 2017;170:185–198.e16. doi: 10.1016/j.cell.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poole D.P., Godfrey C., Cattaruzza F., Cottrell G.S., Kirkland J.G., Pelayo J.C., Bunnett N.W., Corvera C.U. Expression and function of the bile acid receptor GpBAR1 (TGR5) in the murine enteric nervous system. Neuro Gastroenterol. Motil. 2010;22:814–825, e227-8. doi: 10.1111/j.1365-2982.2010.01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nohr M.K., Pedersen M.H., Gille A., Egerod K.L., Engelstoft M.S., Husted A.S., Sichlau R.M., Grunddal K.V., Poulsen S.S., Han S., et al. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology. 2013;154:3552–3564. doi: 10.1210/en.2013-1142. [DOI] [PubMed] [Google Scholar]
- 49.Fiorucci S., Biagioli M., Zampella A., Distrutti E. Bile Acids Activated Receptors Regulate Innate Immunity. Front. Immunol. 2018;9:1853. doi: 10.3389/fimmu.2018.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim C.H. Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell. Mol. Immunol. 2021;18:1161–1171. doi: 10.1038/s41423-020-00625-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sayin S.I., Wahlström A., Felin J., Jäntti S., Marschall H.U., Bamberg K., Angelin B., Hyötyläinen T., Orešič M., Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metabol. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 52.Masyuk T.V., Masyuk A.I., Lorenzo Pisarello M., Howard B.N., Huang B.Q., Lee P.Y., Fung X., Sergienko E., Ardecky R.J., Chung T.D.Y., et al. TGR5 contributes to hepatic cystogenesis in rodents with polycystic liver diseases through cyclic adenosine monophosphate/Galphas signaling. Hepatology. 2017;66:1197–1218. doi: 10.1002/hep.29284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cook T.M., Gavini C.K., Jesse J., Aubert G., Gornick E., Bonomo R., Gautron L., Layden B.T., Mansuy-Aubert V. Vagal neuron expression of the microbiota-derived metabolite receptor, free fatty acid receptor (FFAR3), is necessary for normal feeding behavior. Mol. Metabol. 2021;54 doi: 10.1016/j.molmet.2021.101350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pizzonero M., Dupont S., Babel M., Beaumont S., Bienvenu N., Blanqué R., Cherel L., Christophe T., Crescenzi B., De Lemos E., et al. Discovery and optimization of an azetidine chemical series as a free fatty acid receptor 2 (FFA2) antagonist: from hit to clinic. J. Med. Chem. 2014;57:10044–10057. doi: 10.1021/jm5012885. [DOI] [PubMed] [Google Scholar]
- 55.Kaelberer M.M., Buchanan K.L., Klein M.E., Barth B.B., Montoya M.M., Shen X., Bohórquez D.V. A gut-brain neural circuit for nutrient sensory transduction. Science. 2018;361 doi: 10.1126/science.aat5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bala V., Rajagopal S., Kumar D.P., Nalli A.D., Mahavadi S., Sanyal A.J., Grider J.R., Murthy K.S. Release of GLP-1 and PYY in response to the activation of G protein-coupled bile acid receptor TGR5 is mediated by Epac/PLC-epsilon pathway and modulated by endogenous H2S. Front. Physiol. 2014;5:420. doi: 10.3389/fphys.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Masse K.E., Lu V.B. Short-chain fatty acids, secondary bile acids and indoles: gut microbial metabolites with effects on enteroendocrine cell function and their potential as therapies for metabolic disease. Front. Endocrinol. 2023;14 doi: 10.3389/fendo.2023.1169624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Popkov V.A., Zharikova A.A., Demchenko E.A., Andrianova N.V., Zorov D.B., Plotnikov E.Y. Gut Microbiota as a Source of Uremic Toxins. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23010483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brydges C.R., Fiehn O., Mayberg H.S., Schreiber H., Dehkordi S.M., Bhattacharyya S., Cha J., Choi K.S., Craighead W.E., Krishnan R.R., et al. Indoxyl sulfate, a gut microbiome-derived uremic toxin, is associated with psychic anxiety and its functional magnetic resonance imaging-based neurologic signature. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-99845-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun C.Y., Li J.R., Wang Y.Y., Lin S.Y., Ou Y.C., Lin C.J., Wang J.D., Liao S.L., Chen C.J. Indoxyl sulfate caused behavioral abnormality and neurodegeneration in mice with unilateral nephrectomy. Aging (Albany NY) 2021;13:6681–6701. doi: 10.18632/aging.202523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le J., Peng R., Li Y. Trimethylamine-N-Oxide and Precursors as Novel Potential Biomarkers for Anxiety Disorder. Lab. Med. 2022;53:177–182. doi: 10.1093/labmed/lmab063. [DOI] [PubMed] [Google Scholar]
- 62.Chen J.J., Bai S.J., Li W.W., Zhou C.J., Zheng P., Fang L., Wang H.Y., Liu Y.Y., Xie P. Urinary biomarker panel for diagnosing patients with depression and anxiety disorders. Transl. Psychiatry. 2018;8:192. doi: 10.1038/s41398-018-0245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao K., Mu C.L., Farzi A., Zhu W.Y. Tryptophan Metabolism: A Link Between the Gut Microbiota and Brain. Adv. Nutr. 2020;11:709–723. doi: 10.1093/advances/nmz127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sen T., Cawthon C.R., Ihde B.T., Hajnal A., DiLorenzo P.M., de La Serre C.B., Czaja K. Diet-driven microbiota dysbiosis is associated with vagal remodeling and obesity. Physiol. Behav. 2017;173:305–317. doi: 10.1016/j.physbeh.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de La Serre C.B., Ellis C.L., Lee J., Hartman A.L., Rutledge J.C., Raybould H.E. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;299:G440–G448. doi: 10.1152/ajpgi.00098.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Do M.H., Lee E., Oh M.J., Kim Y., Park H.Y. High-Glucose or -Fructose Diet Cause Changes of the Gut Microbiota and Metabolic Disorders in Mice without Body Weight Change. Nutrients. 2018;10 doi: 10.3390/nu10060761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Al Helaili A., Park S.J., Beyak M.J. Chronic high fat diet impairs glucagon like peptide-1 sensitivity in vagal afferents. Biochem. Biophys. Res. Commun. 2020;533:110–117. doi: 10.1016/j.bbrc.2020.08.045. [DOI] [PubMed] [Google Scholar]
- 68.de Lartigue G., Barbier de la Serre C., Espero E., Lee J., Raybould H.E. Diet-induced obesity leads to the development of leptin resistance in vagal afferent neurons. Am. J. Physiol. Endocrinol. Metab. 2011;301:E187–E195. doi: 10.1152/ajpendo.00056.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Lartigue G., Barbier de la Serre C., Espero E., Lee J., Raybould H.E. Leptin resistance in vagal afferent neurons inhibits cholecystokinin signaling and satiation in diet induced obese rats. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siopi E., Galerne M., Rivagorda M., Saha S., Moigneu C., Moriceau S., Bigot M., Oury F., Lledo P.M. Gut microbiota changes require vagus nerve integrity to promote depressive-like behaviors in mice. Mol. Psychiatr. 2023;28:3002–3012. doi: 10.1038/s41380-023-02071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muller P.A., Schneeberger M., Matheis F., Wang P., Kerner Z., Ilanges A., Pellegrino K., Del Mármol J., Castro T.B.R., Furuichi M., et al. Microbiota modulate sympathetic neurons via a gut-brain circuit. Nature. 2020;583:441–446. doi: 10.1038/s41586-020-2474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.D'Agostino G., Lyons D.J., Cristiano C., Burke L.K., Madara J.C., Campbell J.N., Garcia A.P., Land B.B., Lowell B.B., Dileone R.J., Heisler L.K. Appetite controlled by a cholecystokinin nucleus of the solitary tract to hypothalamus neurocircuit. Elife. 2016;5 doi: 10.7554/eLife.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davis S.F., Derbenev A.V., Williams K.W., Glatzer N.R., Smith B.N. Excitatory and inhibitory local circuit input to the rat dorsal motor nucleus of the vagus originating from the nucleus tractus solitarius. Brain Res. 2004;1017:208–217. doi: 10.1016/j.brainres.2004.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perino A., Velázquez-Villegas L.A., Bresciani N., Sun Y., Huang Q., Fénelon V.S., Castellanos-Jankiewicz A., Zizzari P., Bruschetta G., Jin S., et al. Central anorexigenic actions of bile acids are mediated by TGR5. Nat. Metab. 2021;3:595–603. doi: 10.1038/s42255-021-00398-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frost G., Sleeth M.L., Sahuri-Arisoylu M., Lizarbe B., Cerdan S., Brody L., Anastasovska J., Ghourab S., Hankir M., Zhang S., et al. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat. Commun. 2014;5:3611. doi: 10.1038/ncomms4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vazquez E., Barranco A., Ramirez M., Gruart A., Delgado-Garcia J.M., Jimenez M.L., Buck R., Rueda R. Dietary 2'-Fucosyllactose Enhances Operant Conditioning and Long-Term Potentiation via Gut-Brain Communication through the Vagus Nerve in Rodents. PLoS One. 2016;11 doi: 10.1371/journal.pone.0166070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yoshioka Y., Tachibana Y., Uesaka T., Hioki H., Sato Y., Fukumoto T., Enomoto H. Uts2b is a microbiota-regulated gene expressed in vagal afferent neurons connected to enteroendocrine cells producing cholecystokinin. Biochem. Biophys. Res. Commun. 2022;608:66–72. doi: 10.1016/j.bbrc.2022.03.117. [DOI] [PubMed] [Google Scholar]
- 78.Pradhananga S., Tashtush A.A., Allen-Vercoe E., Petrof E.O., Lomax A.E. Protease-dependent excitation of nodose ganglion neurons by commensal gut bacteria. J. Physiol. 2020;598:2137–2151. doi: 10.1113/JP279075. [DOI] [PubMed] [Google Scholar]
- 79.Kentish S., Li H., Philp L.K., O'Donnell T.A., Isaacs N.J., Young R.L., Wittert G.A., Blackshaw L.A., Page A.J. Diet-induced adaptation of vagal afferent function. J. Physiol. 2012;590:209–221. doi: 10.1113/jphysiol.2011.222158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen J., Cheng M., Wang L., Zhang L., Xu D., Cao P., Wang F., Herzog H., Song S., Zhan C. A Vagal-NTS Neural Pathway that Stimulates Feeding. Curr. Biol. 2020;30:3986–3998.e5. doi: 10.1016/j.cub.2020.07.084. [DOI] [PubMed] [Google Scholar]
- 81.Hu X., Xia K., Dai M., Han X., Yuan P., Liu J., Liu S., Jia F., Chen J., Jiang F., et al. Intermittent fasting modulates the intestinal microbiota and improves obesity and host energy metabolism. NPJ Biofilms Microbiomes. 2023;9:19. doi: 10.1038/s41522-023-00386-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li L., Su Y., Li F., Wang Y., Ma Z., Li Z., Su J. The effects of daily fasting hours on shaping gut microbiota in mice. BMC Microbiol. 2020;20:65. doi: 10.1186/s12866-020-01754-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maifeld A., Bartolomaeus H., Löber U., Avery E.G., Steckhan N., Markó L., Wilck N., Hamad I., Šušnjar U., Mähler A., et al. Fasting alters the gut microbiome reducing blood pressure and body weight in metabolic syndrome patients. Nat. Commun. 2021;12:1970. doi: 10.1038/s41467-021-22097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferraris R.P., Yasharpour S., Lloyd K.C., Mirzayan R., Diamond J.M. Luminal glucose concentrations in the gut under normal conditions. Am. J. Physiol. 1990;259:G822–G837. doi: 10.1152/ajpgi.1990.259.5.G822. [DOI] [PubMed] [Google Scholar]
- 85.Sun Y., Huang J., Xiang Y., Bastepe M., Jüppner H., Kobilka B.K., Zhang J.J., Huang X.Y. Dosage-dependent switch from G protein-coupled to G protein-independent signaling by a GPCR. EMBO J. 2007;26:53–64. doi: 10.1038/sj.emboj.7601502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beumer J., Puschhof J., Bauzá-Martinez J., Martínez-Silgado A., Elmentaite R., James K.R., Ross A., Hendriks D., Artegiani B., Busslinger G.A., et al. High-Resolution mRNA and Secretome Atlas of Human Enteroendocrine Cells. Cell. 2020;182:1062–1064. doi: 10.1016/j.cell.2020.08.005. [DOI] [PubMed] [Google Scholar]
- 87.Xie C., Jones K.L., Rayner C.K., Wu T. Enteroendocrine Hormone Secretion and Metabolic Control: Importance of the Region of the Gut Stimulation. Pharmaceutics. 2020;12 doi: 10.3390/pharmaceutics12090790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kowalski C.W., Lindberg J.E.M., Fowler D.K., Simasko S.M., Peters J.H. Contributing mechanisms underlying desensitization of cholecystokinin-induced activation of primary nodose ganglia neurons. Am. J. Physiol. Cell Physiol. 2020;318:C787–C796. doi: 10.1152/ajpcell.00192.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tolhurst G., Heffron H., Lam Y.S., Parker H.E., Habib A.M., Diakogiannaki E., Cameron J., Grosse J., Reimann F., Gribble F.M. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61:364–371. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cho H.J., Callaghan B., Bron R., Bravo D.M., Furness J.B. Identification of enteroendocrine cells that express TRPA1 channels in the mouse intestine. Cell Tissue Res. 2014;356:77–82. doi: 10.1007/s00441-013-1780-x. [DOI] [PubMed] [Google Scholar]
- 91.Fothergill L.J., Callaghan B., Rivera L.R., Lieu T., Poole D.P., Cho H.J., Bravo D.M., Furness J.B. Effects of Food Components That Activate TRPA1 Receptors on Mucosal Ion Transport in the Mouse Intestine. Nutrients. 2016;8 doi: 10.3390/nu8100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nakajima S., Hira T., Yahagi A., Nishiyama C., Yamashita T., Imagi J., Hara H. Unsaturated aldehydes induce CCK secretion via TRPA1 in STC-1 cells. Mol. Nutr. Food Res. 2014;58:1042–1051. doi: 10.1002/mnfr.201300412. [DOI] [PubMed] [Google Scholar]
- 93.Purhonen A.K., Louhivuori L.M., Kiehne K., Kerman K.E.O., Herzig K.H. TRPA1 channel activation induces cholecystokinin release via extracellular calcium. FEBS Lett. 2008;582:229–232. doi: 10.1016/j.febslet.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 94.Christiansen C.B., Gabe M.B.N., Svendsen B., Dragsted L.O., Rosenkilde M.M., Holst J.J. The impact of short-chain fatty acids on GLP-1 and PYY secretion from the isolated perfused rat colon. Am. J. Physiol. Gastrointest. Liver Physiol. 2018;315:G53–G65. doi: 10.1152/ajpgi.00346.2017. [DOI] [PubMed] [Google Scholar]
- 95.Emery E.C., Diakogiannaki E., Gentry C., Psichas A., Habib A.M., Bevan S., Fischer M.J.M., Reimann F., Gribble F.M. Stimulation of GLP-1 secretion downstream of the ligand-gated ion channel TRPA1. Diabetes. 2015;64:1202–1210. doi: 10.2337/db14-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chepurny O.G., Holz G.G., Roe M.W., Leech C.A. GPR119 Agonist AS1269574 Activates TRPA1 Cation Channels to Stimulate GLP-1 Secretion. Mol. Endocrinol. 2016;30:614–629. doi: 10.1210/me.2015-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fernandes A.B., Alves da Silva J., Almeida J., Cui G., Gerfen C.R., Costa R.M., Oliveira-Maia A.J. Postingestive Modulation of Food Seeking Depends on Vagus-Mediated Dopamine Neuron Activity. Neuron. 2020;106:778–788.e6. doi: 10.1016/j.neuron.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chimerel C., Emery E., Summers D.K., Keyser U., Gribble F.M., Reimann F. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep. 2014;9:1202–1208. doi: 10.1016/j.celrep.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chadaideh K.S., Carmody R.N. Host-microbial interactions in the metabolism of different dietary fats. Cell Metabol. 2021;33:857–872. doi: 10.1016/j.cmet.2021.04.011. [DOI] [PubMed] [Google Scholar]
- 100.Jones B.V., Begley M., Hill C., Gahan C.G.M., Marchesi J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Louis P., Flint H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 102.Louis P., Young P., Holtrop G., Flint H.J. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 2010;12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- 103.Cummings J.H., Pomare E.W., Branch W.J., Naylor C.P., Macfarlane G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee J.H., Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 105.Zeb F., Wu X., Chen L., Fatima S., Ijaz Ul H., Chen A., Xu C., Jianglei R., Feng Q., Li M. Time-restricted feeding is associated with changes in human gut microbiota related to nutrient intake. Nutrition. 2020;78 doi: 10.1016/j.nut.2020.110797. [DOI] [PubMed] [Google Scholar]
- 106.Folz J., Culver R.N., Morales J.M., Grembi J., Triadafilopoulos G., Relman D.A., Huang K.C., Shalon D., Fiehn O. Human metabolome variation along the upper intestinal tract. Nat. Metab. 2023;5:777–788. doi: 10.1038/s42255-023-00777-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Berthoud H.R., Neuhuber W.L. Functional and chemical anatomy of the afferent vagal system. Auton. Neurosci. 2000;85:1–17. doi: 10.1016/S1566-0702(00)00215-0. [DOI] [PubMed] [Google Scholar]
- 108.Schaap F.G., Trauner M., Jansen P.L.M. Bile acid receptors as targets for drug development. Nat. Rev. Gastroenterol. Hepatol. 2014;11:55–67. doi: 10.1038/nrgastro.2013.151. [DOI] [PubMed] [Google Scholar]
- 109.Han W., Tellez L.A., Perkins M.H., Perez I.O., Qu T., Ferreira J., Ferreira T.L., Quinn D., Liu Z.W., Gao X.B., et al. A Neural Circuit for Gut-Induced Reward. Cell. 2018;175:887–888. doi: 10.1016/j.cell.2018.10.018. [DOI] [PubMed] [Google Scholar]
- 110.van de Wouw M., Boehme M., Lyte J.M., Wiley N., Strain C., O'Sullivan O., Clarke G., Stanton C., Dinan T.G., Cryan J.F. Short-chain fatty acids: microbial metabolites that alleviate stress-induced brain-gut axis alterations. J. Physiol. 2018;596:4923–4944. doi: 10.1113/JP276431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lirong W., Mingliang Z., Mengci L., Qihao G., Zhenxing R., Xiaojiao Z., Tianlu C. The clinical and mechanistic roles of bile acids in depression, Alzheimer's disease, and stroke. Proteomics. 2022;22 doi: 10.1002/pmic.202100324. [DOI] [PubMed] [Google Scholar]
- 112.Val-Laillet D., Guérin S., Coquery N., Nogret I., Formal M., Romé V., Le Normand L., Meurice P., Randuineau G., Guilloteau P., et al. Oral sodium butyrate impacts brain metabolism and hippocampal neurogenesis, with limited effects on gut anatomy and function in pigs. Faseb. J. 2018;32:2160–2171. doi: 10.1096/fj.201700547RR. [DOI] [PubMed] [Google Scholar]
- 113.Dong Z., Mau W., Feng Y., Pennington Z.T., Chen L., Zaki Y., Rajan K., Shuman T., Aharoni D., Cai D.J. Minian, an open-source miniscope analysis pipeline. Elife. 2022;11 doi: 10.7554/eLife.70661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zanos T.P., Silverman H.A., Levy T., Tsaava T., Battinelli E., Lorraine P.W., Ashe J.M., Chavan S.S., Tracey K.J., Bouton C.E. Identification of cytokine-specific sensory neural signals by decoding murine vagus nerve activity. Proc. Natl. Acad. Sci. USA. 2018;115:E4843–E4852. doi: 10.1073/pnas.1719083115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Refaey M.E., McGee-Lawrence M.E., Fulzele S., Kennedy E.J., Bollag W.B., Elsalanty M., Zhong Q., Ding K.H., Bendzunas N.G., Shi X.M., et al. Kynurenine, a Tryptophan Metabolite That Accumulates With Age, Induces Bone Loss. J. Bone Miner. Res. 2017;32:2182–2193. doi: 10.1002/jbmr.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang J., Simonavicius N., Wu X., Swaminath G., Reagan J., Tian H., Ling L. Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J. Biol. Chem. 2006;281:22021–22028. doi: 10.1074/jbc.M603503200. [DOI] [PubMed] [Google Scholar]
- 118.Fazio F., Lionetto L., Curto M., Iacovelli L., Cavallari M., Zappulla C., Ulivieri M., Napoletano F., Capi M., Corigliano V., et al. Xanthurenic Acid Activates mGlu2/3 Metabotropic Glutamate Receptors and is a Potential Trait Marker for Schizophrenia. Sci. Rep. 2015;5 doi: 10.1038/srep17799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fu J., Gaetani S., Oveisi F., Lo Verme J., Serrano A., Rodríguez De Fonseca F., Rosengarth A., Luecke H., Di Giacomo B., Tarzia G., Piomelli D. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature. 2003;425:90–93. doi: 10.1038/nature01921. [DOI] [PubMed] [Google Scholar]
- 120.Bolognini D., Barki N., Butcher A.J., Hudson B.D., Sergeev E., Molloy C., Moss C.E., Bradley S.J., Le Gouill C., Bouvier M., et al. Chemogenetics defines receptor-mediated functions of short chain free fatty acids. Nat. Chem. Biol. 2019;15:489–498. doi: 10.1038/s41589-019-0270-1. [DOI] [PubMed] [Google Scholar]
- 121.Huang M., Wei R., Wang Y., Su T., Li P., Chen X. The uremic toxin hippurate promotes endothelial dysfunction via the activation of Drp1-mediated mitochondrial fission. Redox Biol. 2018;16:303–313. doi: 10.1016/j.redox.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chan M.M., Yang X., Wang H., Saaoud F., Sun Y., Fong D. The Microbial Metabolite Trimethylamine N-Oxide Links Vascular Dysfunctions and the Autoimmune Disease Rheumatoid Arthritis. Nutrients. 2019;11 doi: 10.3390/nu11081821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Koh A., Molinaro A., Stahlman M., Khan M.T., Schmidt C., Manneras-Holm L., Wu H., Carreras A., Jeong H., Olofsson L.E., et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell. 2018;175:947–961e917. doi: 10.1016/j.cell.2018.09.055. [DOI] [PubMed] [Google Scholar]
- 124.Schroeder J.C., Dinatale B.C., Murray I.A., Flaveny C.A., Liu Q., Laurenzana E.M., Lin J.M., Strom S.C., Omiecinski C.J., Amin S., Perdew G.H. The uremic toxin 3-indoxyl sulfate is a potent endogenous agonist for the human aryl hydrocarbon receptor. Biochemistry. 2010;49:393–400. doi: 10.1021/bi901786x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bachmanov A.A., Reed D.R., Beauchamp G.K., Tordoff M.G. Food intake, water intake, and drinking spout side preference of 28 mouse strains. Behav. Genet. 2002;32:435–443. doi: 10.1023/a:1020884312053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All source data are included as supplementary tables in this article.
-
•
All original code has been deposited at Zenodo: https://doi.org/10.5281/zenodo.14497742 and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact upon request.