Abstract
Human noroviruses, the most common cause of nonbacterial gastroenteritis, are characterized by high infectivity rate, low infectious dose, and unusually high stability outside the host. However, human norovirus research is hindered by the lack of a cell culture system and a small animal model of infection. Norwalk virus (NV) is the prototype strain of human noroviruses. We report here replication of NV viral RNA and its packaging into virus particles in mammalian cells by intracellular expression of native forms of NV viral RNA devoid of extraneous nucleotide sequences derived from the expression vector by the use of replication-deficient vaccinia virus MVA encoding the bacteriophage T7 RNA polymerase (MVA/T7). Expressed genomic RNA was found to replicate; NV subgenomic RNA was transcribed from genomic RNA by use of NV nonstructural proteins expressed from genomic RNA and was subsequently translated into NV capsid protein VP1. Viral genomic RNA was packaged into virus particles generated in mammalian cells. The cesium chloride (CsCl) density gradient profile of virus particles containing genomic RNA was similar to that of NV purified from stool. These observations indicate that the NV cDNA constructed here is a biologically infectious clone, and that mammalian cells have the ability to replicate NV genomic RNA. This work establishes a mammalian cell-based system for analysis of human norovirus replication and, thus, makes it feasible to investigate antiviral agents in mammalian cells.
Keywords: human norovirus, norovirus RNA replication
Noncultivatable human norovirus belongs to the Norovirus genus of the family Caliciviridae and is classified as a Group B biodefense pathogen. Human noroviruses are responsible for almost all outbreaks (>95%) of nonbacterial gastroenteritis in the United States and Europe (1, 2). Incidents of the rapid spread of norovirus-related illness in the cruise ship industry have led to an increased recognition of the impact of norovirus infections on public health (3).
Norwalk virus (NV), the prototype strain of human norovirus, is a nonenveloped virus that contains a ≈7.7-kb positive-sense single-stranded RNA genome (4). The NV genome is polyadenylated at its 3′ end and encodes three primary ORFs (Fig. 1A) (4). ORF1 encodes a nonstructural polyprotein containing the following genes from the 5′ end to the 3′ end, respectively: p48, nucleotide triphosphatase (NTPase), p22, VPg, protease (Pro), and RNA polymerase (RNA Pol) (Fig. 1 A) (5). ORF2 and ORF3 encode structural proteins, the major capsid protein (VP1) and a minor structural protein (VP2), respectively (Fig. 1 A) (4). Expression of VP1 in insect cells infected with baculovirus recombinants results in the self-assembly of empty recombinant virus-like particles (VLPs) (6, 7). VP2 is also incorporated into VLPs when VP2 is coexpressed with VP1 (8), and its presence increases the yield and stability of VLPs (9).
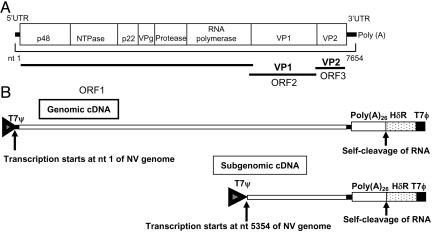
Fig. 1.
Plasmid constructs for the expression of native forms of Norwalk viral genomic and subgenomic RNA in mammalian cells. (A) Schematic of the complete NV genome, indicating the location of the nonstructural protein cleavage products within the ORF1 polyprotein, the viral structural proteins VP1 and VP2 encoded by ORF2 and ORF3, respectively, and the 5′ and 3′ untranslated regions (solid lines). (B) Diagram of the plasmids used for viral RNA expression (7). T7Ψ, HδR, and T7ϕ refer to the position of the T7 promoter, hepatitis δ virus antigenomic ribozyme, and T7 terminator sequence, respectively. Arrows indicate the transcription initiation site and the self-cleavage site. Numbers refer to the nucleotide positions of the NV genome (GenBank accession no. NC001959).
Since the initial cloning and sequencing of the NV genome (4, 10), many other norovirus sequences have been reported (11, 12). Despite the availability of cloned genomes, molecular studies of NV replication and development of antiviral drugs have been hampered by the lack of a suitable in vitro culture system and an animal model. The lack of a replication system for the viral genome in mammalian cells has also hindered the identification of a consensus nucleotide sequence representing a bona fide infectious molecular clone. To overcome these limitations, we have sought to establish a mammalian cell-based system for replication of human norovirus based on a system of reverse genetics. We report the replication of viral RNA and its packaging into virus particles in mammalian cells by intracellular expression of NV cDNA by using the attenuated recombinant modified vaccinia virus strain Ankara expressing the bacteriophage T7 RNA Pol (MVA/T7) (13, 14).
Materials and Methods
Cells, Viruses, and Antisera. HEK293T cells (kindly supplied by Richard E. Sutton, Baylor College of Medicine, Houston) and Syrian hamster kidney fibroblast BHK-21 cells (American Type Culture Collection, Rockville) were grown in DMEM and MEM, supplemented with 10% FBS, respectively. Growth of MVA/T7 virus (kindly provided by Bernard Moss, National Institutes of Health, Bethesda) and virus titration were performed in BHK-21 cells (14). Antisera to RNA-dependent RNA Pol and Pro were made to synthetic peptides (RLTQILKEYGLKPTRPDKTE) for RNA Pol and (DLGTIPGDCGAPYVHKRGND) for Pro as described in ref. 15.
Expression of NV Viral RNA by MVA/T7 System. 293T cells were infected with MVA/T7 at an moi of 10 TCID50 per cell, incubated for 1 h at 37°C, and then were transfected with plasmid DNA by using Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer.
Northern Blotting. Cytoplasmic RNA was isolated from cells similar to the method described by Makino et al. (16). Poly (A)+ RNA was selected from 9 μg of cytoplasmic RNA by using the PolyATtract mRNA isolation system (Promega) as recommended by the manufacturer, and then loaded onto a denaturing agarose gel for Northern blotting. After transfer onto ZetaProbe membranes (Bio-Rad Laboratories), RNA was probed with a (-)-sense [32P]-labeled RNA probe complementary to nt 6364–6771 of the NV genome. Prehybridization and hybridization were performed at 55°C according to the manufacturer's protocol. The membranes were washed four times for 5 min each at room temperature with 2× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/0.1% SDS, twice for 1 h each at 70°C with 2× SSC/0.1% SDS, and then twice for 1 h each at 70°C with 0.1× SSC/0.1% SDS. The membranes were exposed to film for signal detection. To prepare the RNA probe, a plasmid was constructed by inserting bases 6364–6771 of the NV genome into pSP73 (Promega). The RNA probe was prepared by in vitro transcription by using SP6 RNA Pol in the presence of 100 μCi (1 Ci = 37 GBq) of [α-32P]-UTP (10 mCi/ml).
Isolation of Virus or Virus Particles. At 48 h after infection, the culture supernatants were harvested from 12 flasks (75 cm2) and were passed through 0.1-μm pore-size sterile filters (MILLEXVV, Millipore) to eliminate any potentially contaminating MVA/T7 virus. The virus particles in the supernatants were isolated by pelleting through a 30% sucrose cushion, followed by isopycnic CsCl (0.43g/ml) gradient centrifugation and subsequent fractionation as described in ref. 17. Putative virus particles in individual fractions were pelleted (17) and suspended in 100 μl of water. The presence of virus particles in each fraction was examined by immunoblotting (8, 18) and negative staining electron microscopy (18). For immunoblotting, rabbit polyclonal antibodies against rNV VLPs (6) and VP2 (8) were used at a final concentration of 1:5,000 and 1:1,000, respectively.
Detection of Viral RNA Incorporated into Virus Particles by Quantitative RT-PCR. Isolated virus particles (25 μl) were mixed with 100 μl of buffer (50 mM Tris·HCl, pH 7.5/10 mM MnCl2) and incubated for 30 min at 37°C with 100 units/ml of DNase I (Roche Allied Science) and 20 μg/ml of RNase A (Sigma). An equal volume of 2× proteinase K buffer was added to this mixture and incubated for 1 h at 37°C with 200 μg/ml proteinase K. Viral RNA was extracted by using TRIzol LS reagent (Invitrogen) according to the manufacturer's protocol, followed by phenol/chloroform (pH 4.5) (Ambion), precipitated with ethanol, washed with 75% ethanol, dried, and then dissolved in 100 μl of water. Viral RNA (20 μl) was subjected to quantitative RT-PCR by using TaqMan One-Step RT-PCR Master Mix Reagents Kit (Applied Biosystems). The assays targeted genomic regions of both ORF1 (nt 4632–4715) and ORF2 (nt 5412–5522). The primers, probes, and conditions used for quantitative RT-PCR are described in Supporting Text, Fig. 5, and Tables 1 and 2, which are published as supporting information on the PNAS web site.
Results
Strategy for Expression of the Native Forms of NV Genomic and Subgenomic RNA. Two species of NV cDNA, genomic and subgenomic (Fig. 1B), were used because such viral RNAs have been detected in cells infected with animal caliciviruses (5). Construction of the subgenomic NV cDNA used for this experiment was based on the nucleotide sequences of subgenomic RNA found in animal caliciviruses (5). Genomic and subgenomic NV cDNAs having poly(A) tracts were positioned downstream of the T7 promoter and upstream of the hepatitis delta virus ribozyme (HδR) and the T7 RNA Pol termination signal (T7ϕ) to allow for generation of transcripts with the exact 5′ and 3′ ends of the NV genome (Fig. 1B). Because initiation of transcription of the plasmid constructs under the control of the T7 promoter is at a G nucleotide (19) and the initial nucleotide for NV RNA is a G, transcription of NV cDNAs occurs at the origin of the NV insertion. Autocatalytic cleavage of primary transcripts occurs between the final A of the poly(A) region and the first G of the HδR site, resulting in viral RNAs that terminate with a poly(A) tail.
Expression of Genomic RNA in Mammalian Cells. To determine whether the genomic cDNA construct of NV is a functional infectious clone, NV genomic RNA was expressed in 293T cells by use of the MVA/T7 system. Expression of the viral nonstructural proteins Pro and RNA-dependent RNA Pol was detected by radioimmunoprecipitation and immunofluorescence in cells expressing genomic RNA (Fig. 2A).
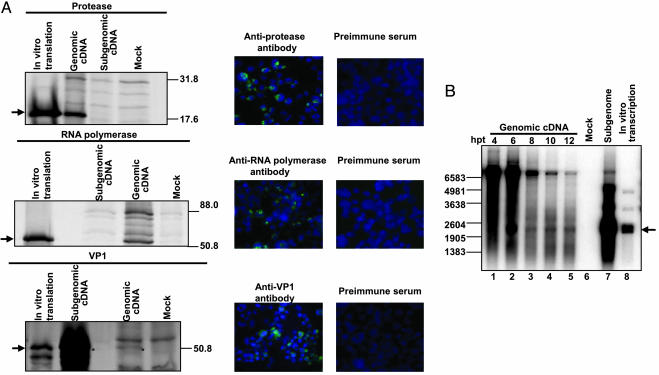
Fig. 2.
Expression and replication of genomic NV RNA in mammalian cells. 293T cells were infected with MVA/T7 and then transfected with plasmids encoding NV cDNAs. (A) Detection of nonstructural and structural NV proteins in cells expressing NV RNA by radioimmunoprecipitation (Left) and immunofluorescence (Right) with antisera to the proteins. Radiolabeled NV proteins synthesized by in vitro translation were processed in parallel. Molecular size markers (kilodaltons) are on the right. Arrows highlight the NV proteins. (B) Detection of subgenomic RNA synthesized in cells expressing genomic RNA. Poly(A)+ RNA, isolated at 4–12 h pt, and was subjected to Northern blotting. In vitro transcripts of subgenomic RNA were analyzed in parallel. Arrow indicates subgenomic RNA. RNA size markers (nucleotides) are on the left.
Because ORF1 of the NV genome encodes a nonstructural polyprotein (Fig. 1 A) that is cleaved into the corresponding nonstructural proteins by Pro (4, 20–25), detection of the correct sizes of Pro and RNA Pol, located at the extreme 3′ end of ORF1, by radioimmunoprecipitation indicates that the nonstructural polyprotein was translated from the genomic RNA expressed in 293T cells, and the individual nonstructural proteins were subsequently generated by cleavage with a biologically active Pro. Both Pro and RNA Pol also were detected by immunofluorescence in the cytoplasm of the expressing cells.
Expression of the viral capsid protein VP1 was also detected by radioimmunoprecipitation and immunofluorescence (Fig. 2 A). In animal caliciviruses such as feline calicivirus (FCV) and rabbit hemorrhagic disease virus, subgenomic RNA transcribed from genomic RNA serves as the template for translation of VP1 (5). In vitro translation of genomic RNA did not produce any VP1 (Fig. 6, which is published as supporting data on the PNAS web site), indicating that VP1 as detected in genomic RNA-expressing cells was not generated by an internal initiation-based translation mechanism. Thus, detection of VP1 in cells expressing genomic RNA suggested that it was translated from subgenomic RNA, which was transcribed from replicated genomic RNA by the expressed NV nonstructural proteins. Expression of poly(A)+ subgenomic RNA in the genomic RNA-expressing cells was examined at 4, 6, 8, 10, and 12 h after transfection (pt) by Northern blot analysis with a positive strand-specific RNA probe corresponding to the VP1 region of the NV genome (Fig. 2B). Genomic RNA (7.6 kb) was first detected at 4 h pt (lane 1), and the levels of genomic RNA declined over time (lanes 2 to 5). Run as controls, subgenomic cDNA-transfected cells (lane 7) and in vitro transcripts of subgenomic cDNA (lane 8) yielded two slightly different sized bands of 2.3 kb and 2.5 kb, in which the subgenomic RNA lacked and contained the HδR and T7ϕ, respectively. In cells transfected with genomic cDNA, expression of the expected 2.3 kb subgenomic RNA was first detected at 6 h pt (lane 2). Subgenomic RNA was not detected at 4 h pt despite expression of genomic RNA (lane 1) and was absent in mocktransfected cells (lane 6). Subgenomic RNA was not a degradation product or an intermediate of genomic RNA, because the expression level of subgenomic RNA dropped between 6 and 8 h pt (lanes 2 and 3), then remained relatively constant until 12 h pt (lanes 3–5) despite decreasing levels of genomic RNA observed over time (lanes 3 to 5). 5′ end mapping of the subgenomic RNA by primer extension indicated that the sequence of subgenomic RNA detected in cells expressing genomic RNA began at the conserved transcription initiation site of subgenomic calicivirus RNA (5) (Fig. 5). In addition, the subgenomic RNA expressed from genomic RNA was extracted from an agarose gel and then was subjected to 5′-RACE analysis (Fig. 7, which is published as supporting information on the PNAS web site). The 5′-end sequence of the subgenomic RNA began at the conserved transcription site of the calicivirus subgenomic RNA, which has similarity to the first four nucleotides of genomic RNA (GTAA), followed by the first in-frame AUG of ORF2. We also examined the expression of subgenomic RNA from genomic RNA, in which the RNA Pol was inactivated by deletion of the GDD amino acid sequence that is the conserved active site of RNA Pols. Expression of subgenomic RNA was not detected in cells expressing genomic RNA with an inactivated RNA Pol (Fig. 8, which is published as supporting information on the PNAS web site). These observations indicate that the detected subgenomic RNA was generated by transcription from genomic RNA and was not due to promiscuous T7 transcription (Fig. 9, which is published as supporting information on the PNAS web site).
Taken together, detection of capsid VP1 protein and subgenomic RNA in cells expressing genomic RNA indicates that replication of the genomic RNA expressed by the MVA/T7 system occurred, i.e., subgenomic RNA generated from genomic RNA by the expressed and functional nonstructural proteins, and was translated into VP1. Thus, a NV cDNA clone that is a biologically functional infectious clone has been constructed, and mammalian cells are able to replicate this clone.
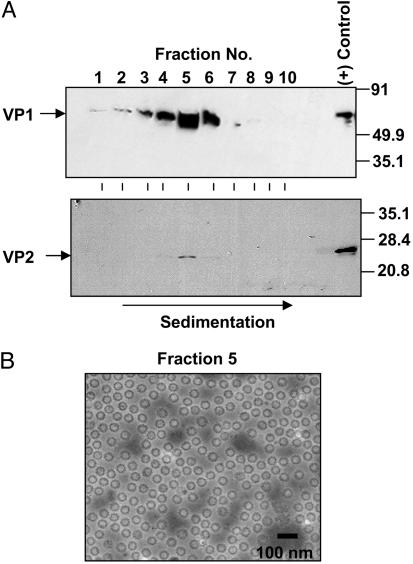
Packaging of Viral Genomic RNA into Virus Particles. Empty NV virus particles, which are morphologically and antigenically similar to wild-type virions, can be produced when the viral structural proteins VP1 and VP2 are expressed in insect Sf-9 cells by use of recombinant baculoviruses containing subgenomic cDNA (8). We expressed the native form of the viral subgenomic RNA in mammalian cells and examined whether virus particles were generated. Virus particles derived from subgenomic RNA-expressing cells were purified from culture supernatants by isopycnic CsCl gradient centrifugation, followed by fractionation, and the putative virus particles were pelleted from each fraction for analysis. Both VP1 and VP2 were detected in the same fraction by Western blotting (Fig. 3A, fractions 4–6); a peak of cosedimentation of VP1 and VP2 was detected in fraction 5. The presence of virus particles in fraction 5 was confirmed by negative stain electron microscopy (Fig. 3B). These results provide evidence that expression of a native form of NV subgenomic RNA in mammalian cells generates virus particles composed of both VP1 and VP2.
Fig. 3.
Generation of virus particles from cells expressing subgenomic RNA. Virus particles were isolated from the culture supernatants by isopycnic CsCl gradient centrifugation, and particles in individual gradient fractionations were sedimented. (A) The presence of virus particles in each fraction was examined by Western blotting with anti-rNV hyperimmune serum that reacts with VP1 (18) (Upper) and anti-VP2 antibody (24) (Lower). As a positive control, virus particles produced by the baculovirus expression system (24) were analyzed in parallel. Arrows indicate detected viral proteins. Size markers (kilodaltons) are on the right. (B) Electron micrograph of negatively stained purified virus particles from fraction 5. (Scale bar: 100 nm.)
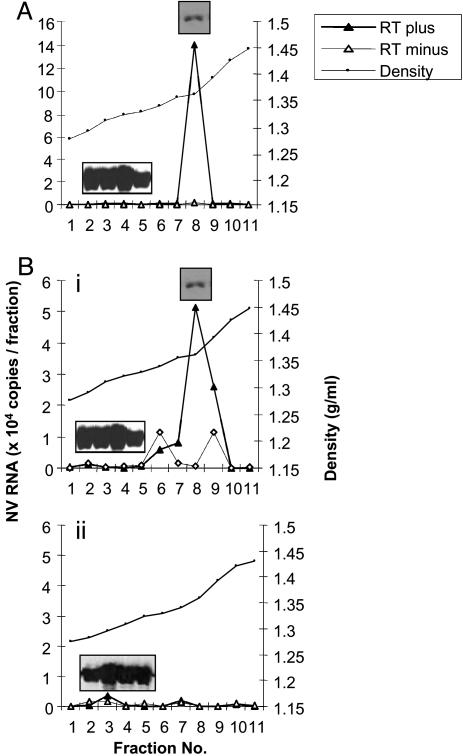
We next sought to determine whether viral genomic RNA was packaged into the virus particles produced by mammalian cells expressing viral RNA using the MVA/T7 system. For this purpose, virus particles, isolated from gradient fractions as described above, were treated with DNase I and RNase A to remove plasmid DNA or viral RNA that may have nonspecifically attached to the particle surface. Viral RNA was then extracted from virus particles and quantitated by real-time RT-PCR. To detect genomic RNA, primers specific to the RNA Pol region, present in genomic RNA but absent from subgenomic RNA, were used. When virus particles were fractionated from cells expressing genomic RNA alone, a weak positive signal indicating incorporation of viral genomic RNA into virus particles was detected in fraction 8 (data not shown). However, the presence of capsid protein VP1 could not be detected in fraction 8 because of the detection limits of Western blotting. The expression level of VP1 in genomic RNA expressing cells was much lower than that in subgenomic RNA-expressing cells (Fig. 2 A). Therefore, to augment the expression of VP1 and VP2 capsid proteins and increase the probability of packaging the viral genome, virus particles were isolated from cells coexpressing genomic and subgenomic RNA. A positive signal indicating incorporation of viral genomic RNA into virus particles was again detected in fraction 8 (Fig. 4A). This positive signal was detected in the presence of reverse transcriptase but not in its absence, indicating that this signal was due to the incorporation of viral genomic RNA but not NV cDNA. The capsid protein VP1 was also detected in fraction 8 by Western blotting with hyperimmune anti-NV serum (Fig. 4A). Virus particles were detected in fractions 2–5 by negative stain electron microscopy (data not shown); however, a positive signal indicating the packaging of genomic RNA was not detected in these fractions, indicating that these virus particles were empty. These results indicate that a small percentage of virus particles could package genomic RNA, and the density of these particles was shifted from fractions 2–5 (low density where empty particles band) to fraction 8 (high density). We also examined incorporation of viral RNA into virus particles, generated under two different expression conditions (subgenomic RNA alone or both subgenomic and genomic RNA), by use of primers specific to the capsid region that can detect both genomic and subgenomic RNA. Again, a positive signal was detected in fraction 8 in particles generated by coexpressing genomic and subgenomic RNA (Fig. 4Bi). However, virus particles generated by expression of subgenomic RNA alone did not show any positive RNA signal (Fig. 4Bii). Although high levels of capsid protein were detected in fractions 2–5 (Fig. 4Bii), the virus particles appeared empty by negative stain electron microscopy (data not shown). Thus, particles generated by expression of subgenomic RNA alone were empty virus particles that lacked the viral RNA. Virus particles containing the genomic RNA generated by coexpression of genomic and subgenomic RNA were isolated at a CsCl density of 1.32–1.36 g/ml, similar to the densities reported previously for NV purified from stool (10, 26–29). By contrast, the empty virus particles generated by expression of subgenomic had a CsCl density of 1.27–1.29 g/ml. Thus, the density of the fraction containing the genomic RNA was higher than that of empty virus particles and close to that of virus particles detected in human stool. These data strongly indicate that viral RNA is incorporated into virus particles generated by coexpressing genomic and subgenomic RNA.
Fig. 4.
Incorporation of viral RNA into virus particles. Virus particles were isolated from the culture supernatants of cells expressing subgenomic RNA or coexpressing both genomic and subgenomic RNA, followed by sedimentation as described in Fig. 3. The sedimented particles from individual fractions were treated with DNase I and RNase A, and nucleic acid was extracted and subjected to quantitative real-time RT-PCR, performed with or without reverse transcriptase. The copy number of viral RNA present in each fraction was determined by comparison with a standard curve, and the results are plotted against the density. Each fraction was also examined by Western blotting for the presence of VP1, and the results are shown in the inserts. (A) Detection of genomic RNA incorporated into virus particles generated by the coexpression of genomic and subgenomic RNA by using primers corresponding to the Pol region. (B) Detection of subgenomic RNA incorporated into virus particles generated by coexpression of genomic and subgenomic RNA (i) or subgenomic RNA alone (ii) by using primers corresponding to the capsid region.
Discussion
We developed a mammalian-based replication system for a noncultivatable human norovirus. This system is based on driving the expression of the NV viral RNA in the cell cytoplasm by T7 RNA Pol produced by the recombinant vaccinia virus strain MVA (13, 14). NV genomic RNA expressed in mammalian cells by use of this system can replicate: translation of the NV nonstructural proteins required for viral replication and transcription of subgenomic RNA from the expressed NV genomic RNA occurred. 5′-RACE analysis of subgenomic RNA extracted from an agarose gel indicated that the 5′ end sequence of subgenomic RNA was similar to the first four nucleotides of genomic RNA (GTAA) and then continued to the first in-frame AUG of ORF2. Thus, subgenomic RNA transcribed from the expressed genomic RNA began at the conserved predicted transcription initiation site for subgenomic RNAs in caliciviruses. In cells expressing genomic RNA that contain a mutated RNA Pol, expression of subgenomic RNA could not be detected. These observations indicate that the detected subgenomic RNA was generated by transcription from genomic RNA and was not due to the promiscuous T7 transcription or RNA degradation. Expression of VP1 was also detected in cells expressing genomic RNA, but not by in vitro translation of genomic RNA. Therefore, VP1 detected in cells expressing genomic RNA was due to the translation of subgenomic RNA and not due to an internal initiation-based translation mechanism, indicating that subgenomic RNA was biologically functional. We have not yet tried to detect synthesis of NV negative-strand RNA. Nevertheless, transcription of a biologically functional subgenomic RNA from the expressed genomic RNA suggests that the synthesis of NV negative-strand RNA from genomic RNA occurred by use of the expressed biologically functional NV nonstructural proteins.
NV genomic RNA was packaged into virus particles when genomic RNA was coexpressed with subgenomic RNA, whereas subgenomic RNA was not packaged when subgenomic RNA alone was expressed. This observation suggests that the genomic RNA packaging signal may reside in the ORF1 region and/or nonstructural proteins may be required for RNA packaging. Whether the produced virus particles containing the NV genome are infectious is not known because virus infectivity cannot be tested directly because of the lack of a system for cultivation of virus. However, the virus particles expressed in this system have the potential to be infectious, because the CsCl density of virus particles containing genomic RNA was similar to intact virus particles detected in human stool. In addition, empty virus particles generated in mammalian cells are antigenically and structurally similar to wild-type virus, and NV is a nonenveloped virus.
The RNA genomes of animal calicivirus, e.g., rabbit hemorrhagic disease virus and feline calicivirus, are covalently linked to the viral protein VPg at the 5′ end (30–33). In feline calicivirus, viral genome RNA without VPg is not infectious (34). Recent data indicate that VPg may function in the initiation of translation of NV RNA (35); however, the exact functions of VPg in NV replication still remain unclear. VPg proteins derived from positive-strand viruses in families other than Caliciviridae have multiple functions in viral RNA replication, translation, and encapsidation (36). The results in this article suggest that VPg is expressed from genomic RNA. Therefore, the expressed VPg may also have multiple functions and may play a role for successful NV replication, translation, and packaging of viral genome into virus particles. Further studies need to determine whether VPg is covalently linked to the NV genomic and subgenomic RNA and whether VPg is essential for NV replication.
Cultured cell lines susceptible to human norovirus infection are not currently available. This limitation may be due to the restriction of any step in the virus replication cycle, such as penetration of virus particles into the cell after binding to cellular receptors, uncoating of virus particles necessary for release of viral RNA into the cytoplasm, or later steps involving replication of the genome or virus assembly. The results in this article indicate that mammalian cells can replicate intracellularly expressed viral RNA and package viral RNA into virus particles. Thus, a lack of host factors to support intracellular expression of viral RNA is apparently not the cause of restriction for virus replication. For RNA viruses that lack an appropriate in vitro replication system, reverse genetics systems by using the MVA-T7 expression system are often used and have succeeded in generating infectious virus particles from cells lines not susceptible to these viruses, e.g., feline calicivirus (37). The origin of the 293T cells used for these experiments are human embryonic kidney cells, which likely is not the organ where NV replicates. However, these cells have a high transfection efficiency, and kidney cells frequently support the replication of enteric viruses. Recent reports indicate that down-regulation of IFN-mediated STAT1 enhances animal calicivirus replication (38, 39). Down-regulation of the IFN-mediated STAT1 signaling pathway in 293T cells may enhance the efficiency of NV genome replication. Testing NV genome replication in the human intestinal cell line, Caco-2, and other potentially susceptible cell lines may also lead to identification of other cells that support NV replication and that may replicate the viral genome more efficiently.
The mammalian-based system described here will allow characterization of the life cycle of human norovirus in mammalian cells and genetic manipulation of the NV genome to dissect specific viral and host functions required for efficient viral replication. In addition, this system should advance the molecular biological analysis of human calicivirus replication and facilitate the testing of antiviral agents in mammalian cells.
Supplementary Material
Acknowledgments
We thank B. Moss for providing the MVA/T7 virus; A. Ball for Vector 2.0 (University of Alabama, Birmingham); R. E. Sutton (Baylor College of Medicine) for HEK-293T cells; Richard E. Lloyd, Ronald T. Javier, C. Q. Zeng, A. Hutson, and T. D. Parker for critical comments; and William Jeong and Susana Guix for providing supporting data. This work was supported by Public Health Service Grants AI 38036 and AI 57788 (to M.K.E.), and M.A. was supported by National Institutes for Health Training Grant AI 07471.
Author contributions: M.A., R.L.A., and M.K.E. designed research; M.A., V.R., S.E.C., and F.H.N. performed research; M.A., R.L.A., and M.K.E. analyzed data; and M.A. and M.K.E. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MVA/T7, modified vaccinia virus strain Ankara encoding bacteriophage T7 RNA polymerase; NV, Norwalk virus; Pro, protease; pt, after transfection; RNA Pol, RNA polymerase.
References
- 1.Billgren, M., Christenson, B., Hedlund, K. O. & Vinje, J. (2002) J. Infect. 44, 26-32. [DOI] [PubMed] [Google Scholar]
- 2.Fankhauser, R. L., Noel, J. S., Monroe, S. S., Ando, T. & Glass, R. I. (1998) J. Infect. Dis. 178, 1571-1578. [DOI] [PubMed] [Google Scholar]
- 3.Hutson, A. M., Atmar, R. L. & Estes, M. K. (2004) Trends Microbiol. 12, 279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang, X., Wang, M., Wang, K. & Estes, M. K. (1993) Virology 195, 51-61. [DOI] [PubMed] [Google Scholar]
- 5.Green, K. Y., Chanock, R. M. & Kapikian, A. Z. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott Williams & Wilkins, Philadephia), pp. 841-874.
- 6.Jiang, X., Wang, M., Graham, D. Y. & Estes, M. K. (1992) J. Virol. 66, 6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White, L. J., Hardy, M. E. & Estes, M. K. (1997) J. Virol. 71, 8066-8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glass, P. J., White, L. J., Ball, J. M., Leparc-Goffart, I., Hardy, M. E. & Estes, M. K. (2000) J. Virol. 74, 6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertolotti-Ciarlet, A., Crawford, S. E., Hutson, A. M. & Estes, M. K. (2003) J. Virol. 77, 11603-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang, X., Graham, D. Y., Wang, K. N. & Estes, M. K. (1990) Science 250, 1580-1583. [DOI] [PubMed] [Google Scholar]
- 11.Lambden, P. R., Caul, E. O., Ashley, C. R. & Clarke, I. N. (1993) Science 259, 516-519. [DOI] [PubMed] [Google Scholar]
- 12.Karst, S. M., Wobus, C. E., Lay, M., Davidson, J. & Virgin, H. W., IV (2003) Science 299, 1575-1578. [DOI] [PubMed] [Google Scholar]
- 13.Sutter, G. & Moss, B. (1992) Proc. Natl. Acad. Sci. USA 89, 10847-10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyatt, L. S., Moss, B. & Rozenblatt, S. (1995) Virology 210, 202-205. [DOI] [PubMed] [Google Scholar]
- 15.Ball, J. M., Tian, P., Zeng, C. Q., Morris, A. P. & Estes, M. K. (1996) Science 272, 101-104. [DOI] [PubMed] [Google Scholar]
- 16.Makino, S., Taguchi, F., Hirano., N. & Fujiwara, K. (1984) Virology 139, 138-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutson, A. M., Atmar, R. L., Marcus, D. M. & Estes, M. K. (2003) J. Virol. 77, 405-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertolotti-Ciarlet, A., White, L. J., Chen, R., Prasad, B. V. & Estes, M. K. (2002) J. Virol. 76, 4044-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imburgio, D., Rong, M., Ma, K. & McAllister, W. T. (2000) Biochemistry 39, 10419-10430. [DOI] [PubMed] [Google Scholar]
- 20.Liu, B., Clarke, I. N. & Lambden, P. R. (1996) J. Virol. 70, 2605-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seah, E. L., Marshall, J. A. & Wright, P. J. (1999) J. Virol. 73, 10531-10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardy, M. E., Crone, T. J., Brower, J. E. & Ettayebi, K. (2002) Virus Res. 89, 29-39. [DOI] [PubMed] [Google Scholar]
- 23.Belliot, G., Sosnovtsev, S. V., Mitra, T., Hammer, C., Garfield, M. & Green, K. Y. (2003) J. Virol. 77, 10957-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seah, E. L., Marshall, J. A. & Wright, P. J. (2003) J. Virol. 77, 7150-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blakeney, S. J., Cahill, A. & Reilly, P. A. (2003) Virology 308, 216-224. [DOI] [PubMed] [Google Scholar]
- 26.Kapikian, A. Z., Gerin, J. L., Wyatt, R. G., Thornhill, T. S. & Chanock, R. M. (1973) Proc. Soc. Exp. Biol. Med. 142, 874-877. [DOI] [PubMed] [Google Scholar]
- 27.Thornhill, T. S., Wyatt, R. G., Kalica, A. R., Dolin, R., Chanock, R. M. & Kapikian, A. Z. (1977) J. Infect. Dis. 135, 20-27. [DOI] [PubMed] [Google Scholar]
- 28.Caul, E. O. & Appleton, H. (1982) J. Med. Virol. 9, 257-265. [DOI] [PubMed] [Google Scholar]
- 29.Madore, H. P., Treanor, J. J. & Dolin, R. (1986) J. Virol. 58, 487-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burroughs, J. N. & Brown, F. (1978) J. Gen. Virol. 41, 443-446. [DOI] [PubMed] [Google Scholar]
- 31.Schaffer, F. L., Ehresmann, D. W., Fretz, M. K. & Soergel, M. I. (1980) J. Gen. Virol. 47, 215-220. [DOI] [PubMed] [Google Scholar]
- 32.Meyers, G., Wirblich, C. & Thiel, H. J. (1991) Virology 184, 677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herbert, T. P., Brierley, I. & Brown, T. D. (1997) J. Gen. Virol. 78, 1033-1040. [DOI] [PubMed] [Google Scholar]
- 34.Sosnovtsev, S. & Green, K. Y. (1995) Virology 210, 383-390. [DOI] [PubMed] [Google Scholar]
- 35.Daughenbaugh, K. F., Fraser, C. S., Hershey, J. W. & Hardy, M. E. (2003) The EMBO J. 22, 2852-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadowy, E., Milner, M. & Haenni, A. L. (2001) Adv. Virus Res. 57, 185-262. [DOI] [PubMed] [Google Scholar]
- 37.Thumfart, J. O. & Meyers, G. (2002) J. Virol. 76, 6398-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wobus, C. E., Karst, S. M., Thackray, L. B., Chang, K. O., Sosnovtsev, S. V., Belliot, G., Krug, A., Mackenzie, J. M., Green, K. Y. & Virgin, H. W. I. (2004) PLoS Biology 2, e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang, K. O., Sosnovtsev, S. V., Belliot, G., Kim, Y., Saif, L. J. & Green, K. Y. (2004) PNAS 101, 8733-8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.