Abstract
Parkin, an E3 ubiquitin ligase, has been found to be responsible for autosomal recessive juvenile parkinsonism characterized primarily by selective loss of dopaminergic neurons with subsequent defects in movements. However, the molecular mechanisms underlying this neuron loss remain elusive. Here, we characterized Drosophila parkin loss-of-function mutants, which exhibit shrinkage of dopaminergic neurons with decreased tyrosine hydroxylase level and impaired locomotion. The behavioral defect of parkin mutant flies was partially restored by administering L-DOPA, and the dopamine level in the brains of parkin mutant flies was highly decreased. Intriguingly, we found that c-Jun N-terminal kinase (JNK) is strongly activated in the dopaminergic neurons of parkin mutants and that impaired dopaminergic neuron phenotypes are dependent on the activation of the JNK signaling pathway. In consistent with this, our epistatic analysis and mammalian cell studies showed that Parkin inhibits the JNK signaling pathway in an E3 activity-dependent manner. These results suggest that loss of Parkin function up-regulates the JNK signaling pathway, which may contribute to the vulnerability of dopaminergic neurons in Drosophila parkin mutants and perhaps autosomal recessive juvenile parkinsonism patients.
Keywords: Parkinson's disease, ubiquitination, tyrosine hydroxylase, degeneration, apoptosis
Parkinson's disease (PD) is the second most common neurodegenerative disease in North America, affecting >1 million people. The major symptoms of PD include rigidity, tremor, bradykinesia of the limbs, and postural instability. These symptoms result primarily from a deficiency of dopamine caused by selective degeneration of dopaminergic neurons in two regions of the brain, the substantia nigra pars compacta and striatum. Another pathological feature of this disease is the presence of inclusion bodies, called Lewy bodies, in those surviving neurons (1). The average onset of the disease is at the age of 60, but the pervasion of PD is not restricted to aging individuals; the onset of autosomal recessive juvenile parkinsonism (AR-JP) is usually between the age of 20 and 40 (2).
Even though PD is largely a sporadic disorder, mutations in the genes responsible for PD, either autosomal dominant or recessive, have been found in a number of affected families. To date, six genes, namely, α-synuclein (PARK1) (3), parkin (PARK2) (4), UCH-L1 (PARK5) (5), PINK1 (PARK6) (6), DJ-1 (PARK7) (7), and the most recently identified, LRRK2/dardarin (PARK8) (8, 9), have been isolated as pathological candidates for PD by family-based linkage analyses and positional cloning. Among them, mutations in parkin, PINK1, and DJ-1 were found to be associated with earlyonset autosomal recessive parkinsonism (4, 6, 7).
In particular, Parkin is an E3 ubiquitin ligase, encoded by parkin, the most common gene mutated in familial PD. Although its exact in vivo function is not clearly revealed, the structure of Parkin gives significant clues to its possible function in the ubiquitination pathway. Parkin is composed of an ubiquitin-like domain in its N terminus and two RING finger domains in its C terminus (10, 11). Like many other proteins with a RING finger domain, Parkin functions as an E3 ubiquitin ligase (11–13). This observation suggests that Parkin may play a role in controlling the level of other proteins or itself by regulated protein degradation. Indeed, recent studies have shown that Parkin ubiquitinates and degrades several proteins, including CDCrel-1 (13), parkin-associated endothelin receptor-like (Pael) receptor (14), α-synuclein (15), synphilin-1 (16), and cyclin E (17).
Furthermore, Parkin has been demonstrated to act as a protector of dopaminergic neurons against PD-related toxicities. Overexpressed Parkin protects dopaminergic neuroblastoma cells against dopamine-induced apoptosis (18) and inhibits dopaminergic neuron degeneration induced by Pael receptor (19). In a rat lentiviral model of PD, overexpression of Parkin protects dopaminergic neurons against the toxicity of mutated human A30P α-synuclein (20). Moreover, Drosophila parkin mutants show shrinkage of the dopmainergic neuron cell body and decreased tyrosine hydroxylase (TH) staining in the proximal dendrite (21). However, the molecular mechanism of how Parkin functions in protecting dopaminergic neurons has not been well understood.
Numerous in vitro and in vivo studies have implicated c-Jun N-terminal kinase (JNK), an established mediator of stress-induced apoptosis, in the neurodegenerative processes in PD pathogenesis (18, 22–26). For example, 1-methyl-4-phenylpyridinium ion (MPP+), a parkinsonian neurotoxin, activates JNK in human neuroblastoma cells (25), and overexpression of JNK interacting protein-1, a scaffold protein and inhibitor of JNK, or treatment of SP600125, a specific JNK inhibitor, protects dopaminergic neurons from cell death induced by MPP+ (22, 26).
In the present study, we characterized Drosophila parkin loss-of-function mutants to reveal the molecular mechanisms through which Parkin acts. The dopaminergic neuronal shrinkage and decreased level of TH found in the parkin mutants were demonstrated to arise from the activation of the JNK-dependent signaling cascade. Through epistatic analysis and mammalian cell culture studies, we confirmed that loss of Parkin function is tightly correlated with the deregulation of JNK activity.
Methods
Fly Stocks. The flies with a P-element in the parkin locus were obtained from a large-scale P-element-induced mutagenesis (GenExel, Taejon, Korea). The insertion sites of P-element in park1 and park2 are located at +988 and +433 of the parkin ORF, respectively. The loss-of-function mutants and revertants, parkrv34, were obtained from P-element excision experiments (27). In parkex68, one of the imprecise excision alleles of park1, 1,407 bp was deleted, including most of the parkin exons, as shown in Fig. 1E. We generated three transgenic lines: parkinWT, the full-length parkin; parkinΔN, N-terminal deleted parkin (only contains 108–482 aa); and parkinK71P, a point mutant similar to R42P in AR-JP patients. They were all Myc-tagged and subcloned into pUAST vector. The fly lines for hs-GAL4, ap-GAL4, and Ddc-GAL4 (28, 29) were obtained from the Bloomington Stock Center (Bloomington, IN). EP(2)0578 and EP(X)1516, which are DTRAF1 and DTRAF2 expression lines, respectively, were obtained from the Szeged Drosophila melanogaster P Insertion Mutant Stock Center in Hungary. The UAS-JNK, UAS-JNKDN, and UAS-hep lines were gifts from M. Mlodzik (Mount Sinai School of Medicine, New York) (30). The hep1 fly line was obtained from S. Noselli (Centre National de la Recherche Scientifique, Paris) (31). The UAS-puc line was obtained from M. Peifer (University of North Carolina, Chapel Hill) (32). The UAS-DASK1 and UAS-DASK1DN flies were provided by M. Miura (RIKEN Brain Science Institute, Tokyo) (33).
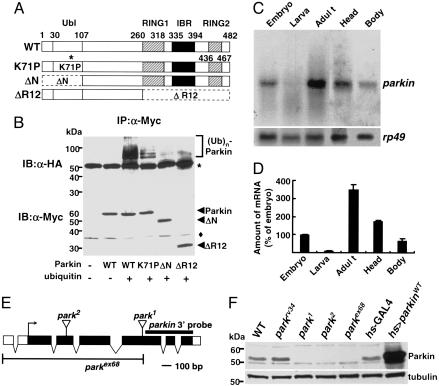
Fig. 1.
Drosophila parkin and its mutants. (A) Comparison between protein domains of Drosophila Parkin wild type and its mutants. Ubl, Ubiquitin-like domain; RING, RING finger domain; IBR, in-between-RING finger domain. (B) COS1 cells were transiently transfected with pcDNA3-Myc-parkinWT, parkinK71P, parkinΔN, and parkinΔR12 with (+) or without (-) pcDNA3-HA-ubiquitin as indicated. After 36 h of transfection, cells were lysed for measuring the E3 ligase activity of Myc-Parkin as described in Methods. Immunoblot analyses for immunoprecipitated proteins were completed by using anti-HA antibodies (Upper) from the immune complexes and anti-Myc antibodies (Lower) from the same cell lysates as described in Methods. The results shown are representative of three independent experiments. *, Antibody bands. Filled diamond, nonspecific bands. (C and D) Northern blot analysis of parkin in wild-type flies. Messenger RNA (2 μg) from the flies representing each developmental stage was subjected to Northern blot analysis. The rp49 was used as a loading control. The parkin mRNA band intensities was quantified and normalized to the amount of rp49 mRNA using IMAGEQUANT software (Molecular Dynamics). The values in D represent the mean of three independent experiments ± SD. (E) Genomic structures of Drosophila parkin and its mutants. Exons are drawn as boxes and coding regions are highlighted in black. The P-element insertion sites of park1 and park2 in the fifth and third exon, respectively, are indicated. The deleted region in parkex68 is displayed under the genomic structure. The region of parkin 3′ probe used in Northern blot analysis is indicated by a black line. (F) Western blot analysis showing the absence of Parkin in parkin mutants. Total body extracts (10 μg) from the male flies of WT (w1118), parkrv34 (parkrv34/parkrv34), park1 (park1/park1), park2 (park2/park2), parkex68 (parkex68/parkex68), hs-GAL4 (hs-GAL4/+), and hs>parkinWT (UAS-parkinWT/Y; hs-GAL4/+) were subjected to Western blot analysis using mouse polyclonal anti-Parkin antibody. For hs-GAL4 and hs>parkinWT flies, heat shock was applied at 37°C for 2 h. The anti-β-tubulin blot is shown as a loading control.
Histochemistry and Western and Northern Blot Analysis. Sections of paraplast-embedded adult fly heads (8 μm) were prepared and processed as described (29, 34) in a blind fashion. We used rabbit anti-TH (1:100, Pel-Freez Biologicals), sheep anti-TH (1:100, Pel-Freez Biologicals), and rabbit anti-phosphospecific JNK (1:100, Promega) antibodies as primary antibodies. Nuclei were stained with Hoechst dye 33258 (Sigma). The polyclonal antibody to Drosophila Parkin (1:200) was generated in mice by injecting GST-fused Parkin (amino acids 1–260) and further purified. Immunostaining of paraffin sections was performed by using horseradish peroxidase- and fluorophore-conjugated secondary antibodies. E7 mouse monoclonal anti-β-tubulin antibody (Development Studies Hybridoma Bank, University of Iowa, Iowa City), 9E10 mouse monoclonal anti-Myc antibody, 3F10 rat monoclonal anti-hemagglutinin (HA) antibody (Roche Diagnostics), and mouse anti-GST antibody (Upstate Biotechnology) were used as primary antibodies for immunoblot analysis. In situ hybridizations and Northern blot analysis were performed by using a parkin probe representing 3′ part of the coding region according to the standard procedures as described (35).
Cell Culture and Immunochemical Assays. COS1 cells were grown as described (36). The cells were transfected by Lipofectamine Plus method (Invitrogen). Immunoprecipitation and immunoblot analyses were carried out as reported (36).
In Vivo Ubiquitination Assay. COS1 cells transfected with Myc-tagged Parkin and/or HA-tagged ubiquitin were lysed and subjected to immunoprecipitation using anti-Myc antibody as described (36). The immune complexes were washed with lysis buffer twice, lysis buffer with 500 mM NaCl twice, and finally ST buffer (50 mM Tris·HCl pH 7.2/150 mM NaCl) twice. The assay samples were boiled in SDS sample buffer and subjected to immunoblot analyses using anti-HA antibody.
Protein Kinase Assay. COS1 cells were lysed and HA-JNK was immunoprecipitated as described (36). JNK activities were assayed in a reaction mixture consisting of 20 mM Hepes (pH 7.4), 10 mM MgCl2, 3 mM 2-mercaptoethanol, 1 μg of GST-c-Jun, 20 μMATP, and 5 μCi of [γ-32P]ATP (1 Ci = 37 GBq) at 30°C for 15 min. Phosphorylated proteins were visualized by autoradiography and quantified with a PhosphorImager (BAS1500, Fuji).
Climbing Assays and Feeding Drugs. Climbing assays were performed as described (29, 37, 38) with some modifications. Groups of 15 flies were transferred into climbing ability test vials and incubated for 1 h at room temperature for environmental acclimation. After tapping the flies down to the bottom, the numbers of the climbing flies in the limited time of 8 s were counted. Ten trials were performed for each time point. The climbing scores (percentage ratio of the number of climbed flies against the total number) were obtained for each test group, and the mean climbing score for five repeated tests was compared to that of wild type. All behavioral experiments were carried out in an isolated room at 25°C under red light (Kodak Safelight Filter 1A). To test the effects of DOPA on climbing ability, flies were fed with 1 mM L-DOPA or D-DOPA for 10 days after eclosion.
Enzyme Immunoassay (EIA) for Dopamine. Fifteen-day-old flies were collected and flash-frozen by liquid nitrogen. Then, the heads were dissected and subjected to EIAs for dopamine according to the manufacturer's instruction (LDN, Nordhorn, Germany).
Results
A sole Drosophila orthologue of mammalian parkin, CG10523, has recently been identified (21, 39). Its protein (482 aa) shows an overall similarity of 59% with its human homologue and contains all of the characteristic canonical motifs of human Parkin, including a ubiquitin-like domain, two RING finger domains, and an in-between RING fingers domain (Fig. 1A).
Because previous studies indicated that human Parkin has an E3 ubiquitin ligase activity (11–13), a function that requires an intact RING finger, we asked whether Drosophila Parkin is also functionally similar to its human orthologue. We measured the E3 ubiquitin ligase activity of Drosophila Parkin in COS1 cells as described in Methods. As expected, a polyubiquitin ladder with high molecular weights was observed in the presence of both Parkin and ubiquitin proteins (Fig. 1B). Interestingly, three mutant Parkin proteins, parkinK71P, a mutant that contains a point mutation similar to R42P mutation in AR-JP patients, parkinΔN, a ubiquitin-like domain deletion mutant, and parkinΔR12, a RING domain deletion mutant, showed a negligible ubiquitin ligase activity in vivo (Fig. 1B), emphasizing the evolutionarily conserved E3 ligase activity of Parkin in Drosophila.
Next, we examined the expression of parkin mRNA during development by Northern blot analysis and detected highly expressed ≈1.5-kb transcript of parkin in adult and embryo stages (Fig. 1 C and D) similar to the previous work (21, 39). Furthermore, we observed that the parkin transcript was more strongly expressed in the head compared to the body in adult flies (Fig. 1 C and D).
From a large-scale screening of the GenExel library (≈100,000 independent EP lines), we isolated two allelic mutants for parkin (named park1 and park2), each with an EP-element insertion in the coding sequence of the gene (Fig. 1E). In addition, parkex68 allele was generated through imprecise excision of the P-element in park1 flies and also the revertant of park1, parkrv34, was generated through precise transposon excision to use as a negative control. Indeed, RNA in situ hybridization (Fig. 2 F–I) and immunoblot analysis (Fig. 1F) clearly demonstrated that homozygous parkin mutant flies failed to produce parkin mRNA or protein, indicating that they are null alleles.
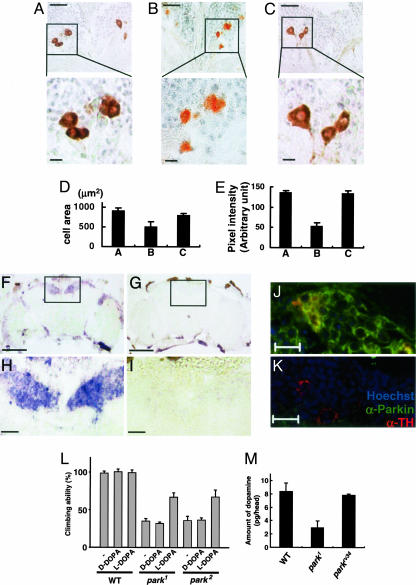
Fig. 2.
Parkin is essential for maintaining proper morphology or function of the DM dopaminergic neurons in Drosophila. (A–E) Frontal sections of the brains in the 15-day-old parkrv34/parkrv34 (A), park1/park1 (B), and UAS-parkinWT; Ddc-GAL4; park1/park1 (C) flies. (Scale bars, 20 μmin Upper and 5 μmin Lower.) Dorsal is up. The size (D) and color intensity (E) of TH-positive neurons were quantified by using PHOTOSHOP. Each bar represents mean ± SD from 10 different brains. (F–I) Expression of parkin mRNA in Drosophila brain. Frontal sections of adult brains from wild type (F and H) and park1 (G and I) flies were examined by RNA in situ hybridization as described in Methods. The surrounding cell bodies, the cortex region, of the wild-type brain were stained in blue. Intensively stained dorsomedial (DM) region is marked by squares (F and G, ×100). (Scale bar, 100 μm.) (H and I) Magnified views (×400) of the squared regions of F and G, respectively. (Scale bar, 20 μm.) (J and K) Frontal sections of parkrv34/parkrv34 (J) and park1/park1 (K) brains were coimmunostained with anti-Parkin antibody (green), rabbit anti-TH antibody (red), and Hoechst dye 33258 (blue). (Scale bar, 10 μm.) Dorsal is up. (L) Flies were fed with DOPA, and their climbing abilities were measured as described in Methods. Genotypes used were: WT (w1118), park1 (park1/park1), and park2 (park2/park2). Each bar represents mean ± SD of five independent experiments. (M) The amounts of dopamine in fly heads were measured by dopamine enzyme immunoassays as described in Methods. Values represent mean ± SD of four independent experiments. Genotypes used were: WT (w1118), park1 (park1/park1), and parkrv34 (parkrv34/parkrv34).
Our parkin mutant flies showed reduced longevity (data not shown), a drooped wing phenotype (data not shown), locomotor dysfunction (Fig. 2L), and muscle degeneration accompanied by apoptosis (data not shown), which are all consistent with the previous studies (21, 39). Furthermore, we examined the dopaminergic neurons of parkin mutant flies by performing histological analysis of the adult fly heads in a blind fashion. We particularly assessed the dorsomedial (DM) dopaminergic neurons because a previously published study reported that the dopaminergic neurons in the DM region are preferentially sensitive to α-synuclein toxicity than those in other regions of the brain (ref. 40 and references therein). Interestingly, we found a severe loss of TH immunostaining and shrunken morphology of the DM dopaminergic neurons of parkin mutant flies (Fig. 2 B, D, and E) compared to the age-matched control (Fig. 2 A, D, and E), which is in accord with the previous study (21). Surprisingly, we observed that this phenotype was completely restored in parkin mutants with Ddc-GAL4 driven-expression of parkinWT gene (Fig. 2 C–E), which indicates that Parkin plays an important role in maintaining proper morphology or function of the DM dopaminergic neurons in Drosophila.
Therefore, we investigated whether parkin is expressed in the DM dopaminergic neurons. parkin transcript was highly enriched in the DM region of the adult brain (compare Fig. 2 F and H with G and I). We also examined the expression of Parkin protein in the DM region with mouse polyclonal anti-Parkin antibody, whose specificity was confirmed by immunoblot analysis (Fig. 1F). Coimmunostaining with an anti-TH antibody, a dopaminergic neuron marker, clearly showed that Parkin is expressed in the DM dopaminergic neurons and prominently localized in the cytoplasm (compare Fig. 2 J with K). These results further support our hypothesis that Parkin is essential for maintaining proper morphology or function of the DM dopaminergic neurons in Drosophila.
As mentioned earlier, PD is characterized by motor disturbances, which result primarily from a deficiency of dopamine caused by selective degeneration of dopaminergic neurons in the brain (1). Therefore, we were interested to know whether the locomotion defective phenotype found in our parkin mutants was due to decreased dopamine level. To test this possibility, wild-type and parkin mutant flies were reared in each medium containing L-DOPA or D-DOPA, an inactive stereoisomer of L-DOPA, and then assayed their climbing ability. Interestingly, feeding L-DOPA to parkin mutant flies significantly restored the climbing ability (up to ≈70% of the unfed wild-type control), whereas D-DOPA had no effect (Fig. 2L). This result suggests that reduced dopamine level is at least partially responsible for the locomotive defects of parkin mutant flies.
To directly demonstrate that the dopamine level is reduced in parkin mutants, we measured the amount of dopamine in their heads through enzyme immunoassay for dopamine in a blind fashion as described in Methods. We found that the level of dopamine is significantly reduced in parkin mutants compared to the wild-type and parkin revertant control (Fig. 2M), further supporting our idea of strong interconnectivity between the dopamine level and the neuronal and locomotive phenotypes in parkin mutants.
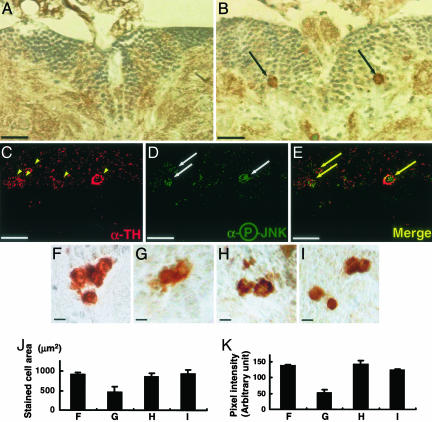
Many studies claimed that neuronal cell deaths in various neurodegenerative diseases are closely related to JNK activation (18, 22–24). Accordingly, we examined JNK activity in park1 brains by using anti-phosphospecific JNK antibodies. Immunohistochemical assays with serial brain sections of 15-day-old wild-type and park1 adult flies demonstrated that only park1 mutants, but not the control, contained DM neuronal cell bodies with highly phosphorylated JNK (compare Fig. 3 A with B). All of the phospho-JNK-positive cells were shown to be costained with anti-TH antibody in double-labeling immunohistochemical analyses (Fig. 3 C–E), indicating that the activation of JNK occurs specifically in the DM dopaminergic neurons of park1 mutants. Furthermore, we examined in a blind fashion whether down-regulation of the JNK pathway rescues the shrinkage features and decreased TH immunostaining in the DM dopaminergic neurons of park1 mutants. Ectopic expression of dominant negative form of JNK (JNKDN) induced by the Ddc-GAL4 driver or hemipterous, Drosophila JNKK, loss-of-function mutation (hep1)in park1 mutant brains restored the morphology of the DM dopaminergic neurons and their TH level (Fig. 3 F–K). These results demonstrate that the shrunken neurons and decreased TH level in parkin mutants result from up-regulation of the JNK signaling pathway.
Fig. 3.
JNK activation in the DM dopaminergic neurons of park1 mutants. (A and B) Frontal sections of the brain were immunostained with rabbit anti-phosphospecific JNK antibody in the brain DM regions of w1118 (A) and park1/park1 (B) and observed under the light microscope. The cells containing phosphorylated and activated JNK were visible only in the park1/park1 brain (black arrows). (Scale bar, 20 μm.) (C–E) The DM region of the 15-day-old park1/park1 brain was costained with sheep anti-TH antibody (C, yellow arrowheads) and rabbit anti-phosphospecific JNK antibody (D, white arrows). (E) Merged image of C and D. (Scale bar, 10 μm.) (F–K) The dopaminergic neurons in the DM regions of 15-day-old flies. (F) parkrv34/parkrv34. (G) park1/park1. (H) UAS-JNKDN/+; Ddc-GAL4/+; park1/park1. (I) hep1/+;park1/park1. TH-positive neuronal cells were marked with rabbit anti-TH antibody, and observed at ×400 under the light microscope. (Scale bar, 5 μm.) The size (J) and color intensity (K) of TH-positive neuronal cells are quantified by using photoshop. Each bar represents mean ± SD from 10 different brains. Dorsal is up in all of the brain sections.
Thorax closure, the joining of the parts of wing imaginal discs during metamorphosis, is tightly controlled by Drosophila JNK pathway. When the activity of the JNK pathway is down-regulated, the joining process is disrupted and a cleft is formed at the dorsal midline (41). As shown in Fig. 4C, inactivation of the JNK pathway by overexpression of JNKDN in the notum clearly showed a thoracic cleft phenotype. This phenotype has widely been used as a reliable marker for assessing JNK activity. To further investigate the relationship between Parkin and the JNK pathway, parkin was overexpressed in the notum of wing imaginal discs under the control of the ap-GAL4 driver and the thoracic cleft area was measured blinded to genotypes. Surprisingly, parkin expression induced a marked cleft in the thorax (Fig. 4E) in a parkin-dose-dependent manner (data not shown), implicating that Parkin inhibits the JNK signaling pathway. In contrast, overexpression of parkinΔN and parkinK71P, as expected, exerted no effect (Fig. 4 B and F) despite their detectable and relatively abundant expressions (Fig. 4I). Therefore, we further examined the genetic interactions between Parkin and the components of the JNK signaling pathway during thorax development. Interestingly, the parkin-induced cleft formation was further enhanced by coexpression of JNKDN (Fig. 4G). On the contrary, coexpression of JNK (Fig. 4H) completely suppressed the parkin-induced cleft formation, whereas expression of JNK alone showed no effect on the thorax closure (Fig. 4D). In addition, other signaling components of the JNK pathway such as hemipterous, puckered, DASK1, and DTRAF1 also consistently interacted with parkin in thorax development (Table 1). These data strongly suggest that Parkin is a negative regulator for the JNK signaling pathway.
Fig. 4.
Functional interactions between Parkin and JNK. Parkin inhibits the JNK pathway during thorax closure. (A–H) Scanning electron micrographs of the adult female thoraxes were shown at the same magnification (×80). (A) ap-GAL4/+. (B) UAS-parkinΔN/+; ap-GAL4/+. (C) UAS-JNKDN/+; ap-GAL4/+. (D) ap-GAL4/UAS-JNK. (E) UAS-parkinWT/+; ap-GAL4/+. (F) ap-GAL4/+; UAS-parkinK71P/+. (G) UAS-parkinWT/UAS-JNKDN; ap-GAL4/+. (H) UAS-parkinWT/+; ap-GAL4/UAS-JNK. The white dotted lines indicate thoracic cleft region. Anterior is up. (I) Western blot analysis of parkin expression detected by anti-Myc antibodies in flies of the following genotypes: hs-GAL4/+, UAS-parkinWT/+; hs-GAL4/+, hs-GAL4/+; UAS-parkinK71P/+, UAS-parkinΔN/+; hs-GAL4/+. Heat shock was applied at 37°C for 2 h. (J) Inhibition of JNK activity by human Parkin. Phosphorylated substrates were visualized by autoradiography (Upper Middle) and quantified with PhosphorImager analyses (Top). Immunoblot analyses were completed using the same cell lysates (Lower Middle and Bottom). The values in Upper Middle and Top represent the mean of three independent cell preparations ± SD. *, Nonspecific bands. (K) A schematic representation of the role of Parkin in Drosophila.
Table 1. Epistatic analyses between parkin and the JNK pathway.
| Genotype
|
Ratio of cleft formed area to total thorax area, %
|
||
|---|---|---|---|
| ap-GAL4 | UAS-parkinWT | Transgenes | |
| + | – | – | 0 |
| + | + | – | 8.5 ± 0.6 |
| + | UAS-parkinΔN | – | 0 |
| + | UAS-parkinK71P | – | 0 |
| + | – | UAS-JNKDN | 7.8 ± 0.5 |
| + | + | UAS-JNKDN | 18.6 ± 1.0 |
| + | – | UAS-puc | 11.1 ± 1.1 |
| + | + | UAS-puc | 14.7 ± 12 |
| + | – | hep1/+ | 0 |
| + | + | hep1/+ | 17.2 ± 1.3 |
| + | – | UAS-JNK | 0 |
| + | + | UAS-JNK | 0 |
| + | – | UAS-hep | 0 |
| + | + | UAS-hep | 0 |
| + | – | UAS-DASK1DN | 0 |
| + | + | UAS-DASK1DN | 19.3 ± 1.6 |
| + | – | UAS-DASK1 | 0 |
| + | + | UAS-DASK1 | 1.0 ± 1.2 |
| + | – | UAS-DTRAF1 | 0 |
| + | + | UAS-DTRAF1 | 0 |
| + | – | UAS-DTRAF2 | 0 |
| + | + | UAS-DTRAF2 | 7.9 ± 1.3 |
Scanning electron micrographs (SEM) of the adult female thoraxes were taken from various transgenic flies. + or - drawn under each genotype indicates the presence or absence, respectively, of that allele. The numbers corresponding to each genotype show the mean ± SD of cleft area to total dorsal area in percentage. The areas were measured by processing the SEM images using photoshop (Adobe Systems, San Jose, CA). n = 5.
To further clarify the regulation of the JNK pathway by Parkin, GST-tagged human Parkin (hParkin) and HA-tagged JNK were cotransfected into COS1 cells, and JNK activities were measured. Coexpression of the wild-type hParkin led to 50–70% reduction of the phosphotransferase activity of JNK (Fig. 4J, lane 3), whereas the R42P mutant, defective in E3-ligase activity, rather slightly increased the JNK activity (Fig. 4J, lane 4) compared to the control (Fig. 4J, lane 2). Collectively, these results strongly support that Parkin negatively modulates the JNK pathway in an E3 activity-dependent manner (Fig. 4K).
Discussion
To clarify our understanding of how mutations in parkin produce loss of dopaminergic neurons, we generated and characterized Drosophila parkin mutants. Our parkin mutants exhibited a drooped wing phenotype, short lifespan, impaired locomotion, and muscle degeneration associated with apoptosis, all of which were in accord with the phenotypes of previously generated parkin mutants (21, 39). In addition, we found shrinkage of the DM dopaminergic neurons with decreased TH immunostaining in our parkin mutant brains, which was also reported by Greene et al. (21). However, this is slightly controversial because Pesah et al. observed no morphological defects of the DM neurons in their parkin mutant allele under different experimental conditions (39). Meanwhile, from our rescuing experiment (Fig. 2C), we were able to demonstrate the importance of Parkin in maintaining proper morphology of dopaminergic neurons and the level of TH, which led us to conclude that the DM dopaminergic neurons of parkin mutants are morphologically and/or functionally degenerated. This conclusion was supported by a previous study using parkin RNA interference in Drosophila, which showed reduction of endogenous Parkin greatly enhance the Pael receptor toxicity in the DM clusters (19). Moreover, we observed the restoration effect of L-DOPA on the impaired locomotion of parkin mutants and their significantly reduced dopamine level in the brain, which indirectly supports our result of the reduced TH staining level. Our hypothesis of a role of dopamine in locomotion in Drosophila could be further supported by previous studies demonstrating that the flies with reduced synaptic dopamine caused by overexpression of tetanus toxin under TH-GAL4 driver showed climbing impairment (42), and that L-DOPA and SK & F 38393 (prototypical D1 receptor agonist) rescued the defective climbing ability induced by α-synuclein overexpression driven by the elav-GAL4 driver (43).
Furthermore, we found that JNK is highly activated in the dopaminergic neurons of parkin mutants and that the impairment of dopaminergic neurons depends on JNK activation, which indicates that the degenerative change in the morphology of the DM dopaminergic neurons could arise from JNK activation. Because sustained activation of JNK is frequently linked with apoptosis in the neurons, it is also possible that some of the DM dopaminergic neurons are in the process of apoptosis in parkin mutant brains.
One possible explanation of the JNK activation in parkin mutants could be an indirect effect of parkin mutation on JNK activation mediated by an accumulation of Parkin substrate(s). In other words, absence of Parkin may stimulate the aggregation of its substrates to cause cellular stress and JNK activation in dopaminergic neurons. In addition, our genetic results raise another possibility that Parkin may directly inhibit JNK activation. Thoracic cleft formation, a typical JNK loss-of-function phenotype induced by overexpression of Parkin in the Drosophila notum, was absolutely dependent on the ubiquitin ligase activity of the protein. According to this observation, it is possible that Parkin directly inactivates the JNK pathway via regulation of JNK signaling molecule(s) ubiquitination. This hypothesis is further supported by our cell biology results (Fig. 4J) and recent studies that showed the involvement of ubiquitination in regulating the JNK signaling cascade (44, 45). To fully understand how Parkin inhibits the JNK pathway, we must identify the exact targets of Parkin in the JNK pathway.
In summary, through systematic studies using the mutants, we demonstrated that loss of Parkin function activates the JNK pathway, which seems to promote degeneration of the dopaminergic neurons. We believe that our findings have helped to address the pathological mechanisms behind AR-JP and other types of hereditary PD at a molecular level and ultimately for developing treatment strategies for PD.
Acknowledgments
We thank Drs. B. A. Hay (California Institute of Technology, Pasadena), E. J. Choi (Korea University, Seoul), M. Mlodzik (Mount Sinai School of Medicine, New York), S. Noselli (Centre National de la Recherche Scientifique, Paris), M. Peifer (University of North Carolina, Chapel Hill), and M. Miura (RIKEN Brain Science Institute, Tokyo) for fly stocks and reagents, and Drs. H. J. Bellen and C. F. Boerkoel (Baylor College of Medicine, Houston) for the comments on the manuscript.
Author contributions: G.-H.C., S.K., J.P., J.C., and K.S.C. designed research; G.-H.C., S.K., J.P., E.L., M.K., S.B.L., J.M.K., and K.S.C. performed research; G.-H.C., S.K., J.P., and J.C. analyzed data; and G.-H.C., S.K., J.P., and J.C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PD, Parkinson's disease; AR-JP, autosomal recessive juvenile parkinsonism; TH, tyrosine hydroxylase; JNK, c-Jun N-terminal kinase; DM, dorsomedial.
References
- 1.Lang, A. E. & Lozano, A. M. (1998) N. Engl. J. Med. 339, 1044-1053. [DOI] [PubMed] [Google Scholar]
- 2.Saito, M., Maruyama, M., Ikeuchi, K., Kondo, H., Ishikawa, A., Yuasa, T. & Tsuji, S. (2000) Brain Dev. 22, S115-S117. [DOI] [PubMed] [Google Scholar]
- 3.Polymeropoulos, M. H., Lavedan, C., Leroy, E., Ide, S. E., Dehejia, A., Dutra, A., Pike, B., Root, H., Rubenstein, J., Boyer, R., et al. (1997) Science 276, 2045-2047. [DOI] [PubMed] [Google Scholar]
- 4.Kitada, T., Asakawa, S., Hattori, N., Matsumine, H., Yamamura, Y., Minoshima, S., Yokochi, M., Mizuno, Y. & Shimizu, N. (1998) Nature 392, 605-608. [DOI] [PubMed] [Google Scholar]
- 5.Leroy, E., Boyer, R., Auburger, G., Leube, B., Ulm, G., Mezey, E., Harta, G., Brownstein, M. J., Jonnalagada, S., Chernova, T., et al. (1998) Nature 395, 451-452. [DOI] [PubMed] [Google Scholar]
- 6.Valente, E. M., Abou-Sleiman, P. M., Caputo, V., Muqit, M. M., Harvey, K., Gispert, S., Ali, Z., Del Turco, D., Bentivoglio, A. R., Healy, D. G., et al. (2004) Science 304, 1158-1160. [DOI] [PubMed] [Google Scholar]
- 7.Bonifati, V., Rizzu, P., Van Baren, M. J., Schaap, O., Breedveld, G. J., Krieger, E., Dekker, M. C., Squitieri, F., Ibanez, P., Joosse, M., et al. (2002) Science 299, 256-259. [DOI] [PubMed] [Google Scholar]
- 8.Paisan-Ruiz, C., Jain, S., Evans, E. W., Gilks, W. P., Simon, J., van der Brug, M., de Munain, A. L., Aparicio, S., Gil, A. M., Khan, N., et al. (2004) Neuron 44, 595-600. [DOI] [PubMed] [Google Scholar]
- 9.Zimprich, A., Biskup, S., Leitner, P., Lichtner, P., Farrer, M., Lincoln, S., Kachergus, J., Hulihan, M., Uitti, R. J., Calne, D. B., et al. (2004) Neuron 44, 601-607. [DOI] [PubMed] [Google Scholar]
- 10.Morett, E. & Bork, P. (1999) Trends Biochem. Sci. 24, 229-231. [DOI] [PubMed] [Google Scholar]
- 11.Shimura, H., Hattori, N., Kubo, S., Mizuno, Y., Asakawa, S., Minoshima, S., Shimizu, N., Iwai, K., Chiba, T., Tanaka, K. & Suzuki, T. (2000) Nat. Genet. 25, 302-305. [DOI] [PubMed] [Google Scholar]
- 12.Imai, Y., Soda, M. & Takahashi, R. (2000) J. Biol. Chem. 275, 35661-35664. [DOI] [PubMed] [Google Scholar]
- 13.Zhang, Y., Gao, J., Chung, K. K., Huang, H., Dawson, V. L. & Dawson, T. M. (2000) Proc. Natl. Acad. Sci. USA 97, 13354-13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai, Y., Soda, M., Inoue, H., Hattori, N., Mizuno, Y. & Takahashi, R. (2001) Cell 105, 891-902. [DOI] [PubMed] [Google Scholar]
- 15.Shimura, H., Schlossmacher, M. G., Hattori, N., Frosch, M. P., Trockenbacher, A., Schneider, R., Mizuno, Y., Kosik, K. S. & Selkoe, D. J. (2001) Science 293, 263-269. [DOI] [PubMed] [Google Scholar]
- 16.Chung, K. K., Zhang, Y., Lim, K. L., Tanaka, Y., Huang, H., Gao, J., Ross, C. A., Dawson, V. L. & Dawson, T. M. (2001) Nat. Med. 7, 1144-1150. [DOI] [PubMed] [Google Scholar]
- 17.Staropoli, J. F., McDermott, C., Martinat, C., Schulman, B., Demireva, E. & Abeliovich, A. (2003) Neuron 37, 735-749. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, H., Ren, Y., Zhao, J. & Feng, J. (2004) Hum. Mol. Genet. 13, 1745-1754. [DOI] [PubMed] [Google Scholar]
- 19.Yang, Y., Nishimura, I., Imai, Y., Takahashi, R. & Lu, B. (2003) Neuron 37, 911-924. [DOI] [PubMed] [Google Scholar]
- 20.Lo Bianco, C., Schneider, B.L., Bauer, M., Sajadi, A., Brice, A., Iwatsubo, T. & Aebischer, P. (2004) Proc. Natl. Acad. Sci. USA 101, 17510-17515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene, J. C., Whitworth, A. J., Kuo, I., Andrews, L. A., Feany, M. B. & Pallanck, L. J. (2003) Proc. Natl. Acad. Sci. USA 100, 4078-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia, X. G., Harding, T., Weller, M., Bieneman, A., Uney, J. B. & Schulz, J. B. (2001) Proc. Natl. Acad. Sci. USA 98, 10433-10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng, J., Mao, X. O., Stevenson, F. F., Hsu, M. & Andersen, J. K. (2004) J. Biol. Chem. 279, 32626-32632. [DOI] [PubMed] [Google Scholar]
- 24.Hunot, S., Vila, M., Teismann, P., Davis, R. J., Hirsch, E. C., Przedborski, S., Rakic, P. & Flavell, R. A. (2004) Proc. Natl. Acad. Sci. USA 101, 665-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cassarino, D. S., Halvorsen, E. M., Swerdlow, R. H., Abramova, N. N., Parker, W. D., Jr., Sturgill, T. W. & Bennett, J. P., Jr. (2000) J. Neurochem. 74, 1384-1392. [DOI] [PubMed] [Google Scholar]
- 26.Wang, W., Shi, L., Xie, Y., Ma, C., Li, W., Su, X., Huang, S., Chen, R., Zhu, Z., Mao, Z., et al. (2004) Neurosci. Res. 48, 195-202. [DOI] [PubMed] [Google Scholar]
- 27.Robertson, H. M., Preston, C. R., Phillis, R. W., Johnson-Schlitz, D. M., Benz, W. K. & Engels, W. R. (1988) Genetics 118, 461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, H., Chaney, S., Roberts, I. J., Forte, M. & Hirsh, J. (2000) Curr. Biol. 10, 211-214. [DOI] [PubMed] [Google Scholar]
- 29.Feany, M. B. & Bender, W. W. (2000) Nature 404, 394-398. [DOI] [PubMed] [Google Scholar]
- 30.Weber, U., Paricio, N. & Mlodzik, M. (2000) Development (Cambridge, U.K.) 127, 3619-3629. [DOI] [PubMed] [Google Scholar]
- 31.Glise, B., Bourbon, H. & Noselli, S. (1995) Cell 83, 451-461. [DOI] [PubMed] [Google Scholar]
- 32.McEwen, D. G., Cox, R. T. & Peifer, M. (2000) Development (Cambridge, U.K.) 127, 3607-3617. [DOI] [PubMed] [Google Scholar]
- 33.Kuranaga, E., Kanuka, H., Igaki, T., Sawamoto, K., Ichijo, H., Okano, H. & Miura, M. (2002) Nat. Cell Biol. 4, 705-710. [DOI] [PubMed] [Google Scholar]
- 34.Auluck, P. K., Chan, H. Y., Trojanowski, J. Q., Lee, V. M. & Bonini, N. M. (2002) Science 295, 865-868. [DOI] [PubMed] [Google Scholar]
- 35.Cho, K. S., Lee, J. H., Kim, S., Kim, D., Koh, H., Lee, J., Kim, C., Kim, J. & Chung, J. (2001) Proc. Natl. Acad. Sci. USA 98, 6144-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, S., Jee, K., Kim, D., Koh, H. & Chung, J. (2001) J. Biol. Chem. 276, 12864-12870. [DOI] [PubMed] [Google Scholar]
- 37.Ganetzky, B. & Flanagan, J. R. (1978) Exp. Gerontol. 13, 189-196. [DOI] [PubMed] [Google Scholar]
- 38.Le Bourg, E. & Lints, F. A. (1992) Gerontology 38, 59-64. [DOI] [PubMed] [Google Scholar]
- 39.Pesah, Y., Pham, T., Burgess, H., Middlebrooks, B., Verstreken, P., Zhou, Y., Harding, M., Bellen, H. & Mardon, G. (2004) Development (Cambridge, U.K.) 131, 2183-2194. [DOI] [PubMed] [Google Scholar]
- 40.Chen, L. & Feany, M. B. (2005) Nat. Neurosci. 8, 657-663. [DOI] [PubMed] [Google Scholar]
- 41.Zeitlinger, J. & Bohmann, D. (1999) Development (Cambridge, U.K.) 126, 3947-3956. [DOI] [PubMed] [Google Scholar]
- 42.Friggi-Grelin, F., Iche, M. & Birman, S. (2003) Genesis 35, 175-184. [DOI] [PubMed] [Google Scholar]
- 43.Pendleton, R. G., Parvez, F., Sayed, M. & Hillman, R. (2002) J. Pharmacol. Exp. Ther. 300, 91-96. [DOI] [PubMed] [Google Scholar]
- 44.Li, X., Yang, Y. & Ashwell, J. D. (2002) Nature 416, 345-347. [DOI] [PubMed] [Google Scholar]
- 45.Liu, Y. & Min, W. (2002) Circ. Res. 90, 1259-1266. [DOI] [PubMed] [Google Scholar]