Abstract
The cell cycle and the circadian clock are endogenous pacemakers, which coexist in most eukaryotic cells and share a number of conceptual features. In the zebrafish, light directly regulates the timing of both clocks, although the signaling and transcriptional pathways that convey photic information to essential nuclear regulators have yet to be deciphered. We have previously established the Z3 cell line, which recapitulates the features of zebrafish circadian clock and represents an ideal system to study light-dependent signaling and gene regulation. We conducted a search for light-responsive transcription factors and found that AP-1 DNA binding is highly induced. Light induces the expression of zWee1, a cell cycle gene essential for G2/M transition, and zCry1a, a clock gene of the feedback regulatory loop. We have found consensus AP-1 sites in the regulatory regions of both zWee1 and zCry1a genes, and we show that light inducibility of both genes is abrogated by inhibition of AP-1 function. Light also elicits chromatin remodeling by stimulating hyperacetylation at Lys-14 of histone H3 at both zWee1 and zCry1a promoters, as assessed by chromatin immunoprecipitation assays by using anti-Fos antibody. These findings provide strong evidence that circadian and cell cycle clocks share unique light-responsive pathways in zebrafish.
Circadian rhythms are based on intracellular time-tracking systems that enable organisms to anticipate environmental changes and thereby adapt their behavior and physiology to the appropriate time of day (1). The principal cue that influences circadian rhythmicity is light (2). The molecular pathways that light uses to influence the clock mechanism are thereby of central importance. A specific set of transcription factors constitutes the molecular architecture of the circadian clock. These are organized in regulatory positive-negative feedback loops, which function in a cell-autonomous manner (3, 4).
The zebrafish constitutes an attractive vertebrate model for circadian studies (5, 6). Gene duplication during evolution has generated four Per, seven Cry, and three Bmal genes (7-10) in the zebrafish, whose regulation and function display some unique features (5). We have previously established the zebrafish embryonic cell line, Z3, which recapitulates the light response characteristics of the vertebrate clock (11). Oscillations of clock gene expression can be entrained to new light-dark cycles in Z3 cells, which also show self-sustained rhythmicity (11). We have established that light induces clock gene expression via CRY (cryptochrome) photoreceptors and activation of the mitogen-activated protein kinase-signaling cascade (12). To date, however, the molecular mechanism of light-responsive transcription of clock genes remains obscure.
In mammals, a pulse of light during the subjective night elicits potent induction of c-fos, fos-B, and jun-B gene expression in the suprachiasmatic nucleus (13, 14). c-Fos is an oncoprotein that associates with Jun proteins to form the AP-1 (activator protein 1) complex. AP-1 regulates transcription of numerous genes by binding promoter sequence whose consensus is TGA(C/G)TCA (15). c-Fos induction has been implicated in the light-induced phase-shift of the circadian clock (16), through yet unidentified mechanisms.
Interestingly, AP-1 modulates a wide range of cellular processes, including cell proliferation, apoptosis, and differentiation (17, 18). In mammals, AP-1 transiently inhibits G2-M progression, also through stimulation of Wee1 via an AP-1 element in its promoter (19, 20). The WEE-l kinase controls the timing of G2-M transition by inhibiting Cdc2/cyclin B through direct phosphorylation, leading to inhibition of mitotic cell division (21). Interestingly, Wee1 displays robust circadian oscillation in mammalian liver (22). Thus, at least in some circumstances, the circadian clock system appears to influence the timing of cell cycle entry by modulating Wee1 expression. Increasing evidence points to functional links between cell cycle and circadian rhythms (23, 24). These links are conceptually strengthened by the similarity in the molecular architecture of both systems. Both rely on interconnected autoregulatory loops, constituted by sequential phases of transcription-translation and protein modifications and degradation. In zebrafish, light directly regulates circadian rhythmicity in most tissues (8, 11, 25, 26), and a link with the cell cycle has also been proposed (27). Here we show that in zebrafish light induces the activity of AP-1, which in turn controls some specific circadian and cell cycle genes. Thus, a unique light-responsive transcriptional pathway is shared by the two cellular systems of cyclic regulation.
Materials and Methods
Cell Culture and Fish. Z3 cells were grown as described (11, 28). Zebrafish were maintained at 26-29°C, fed once daily, and kept in constant darkness. Animals were killed by rapid emersion in chilled water.
Cloning and Characterization of Zebrafish Wee1 and Cry1a Promoters. Zebrafish Wee1 and Cry1a gene promoters were isolated from zebrafish genomic DNA by PCR. 5′-RACE was used to identify transcription initiation sites. Total RNA was isolated from Z3 cells exposed to a 2-h light pulse. Adaptor-specific primers for primary and secondary PCR were used together with Wee1-specific and Cry1a-specific primers. 5′-RACE products were gel-extracted and subcloned into pCR2.1-TOPO (Invitrogen) for sequencing.
Nuclear Extracts. Nuclear extracts were prepared as described (20), with some modifications. Z3 cells were harvested and resuspended in hypotonic buffer containing 25 mM Hepes (pH 7.5), 70 mM KCl, 1.5 mM MgCl2, 0.5 mM sodium orthovanadate, 0.5 mM NaF, 0.5 mM phenylmethylsulfonyl fluoride, and 1 mM DTT in the presence of protease inhibitors, and incubated on ice for 10 min. Nuclei were separated and suspended in hypertonic buffer (20 mM Hepes, pH 7.9/25% glycerol/0.4 M NaCl/1 mM EDTA/1 mM EGTA/1 mM DTT/0.5 mM PMSF) in the presence of protease inhibitors.
RNA Analyses. Miniaturized RNase protection assay was performed as described (11). Zebrafish Per2, Cry1a, and Wee1 riboprobes were prepared by using an in vitro transcription kit (Promega). Per2 and Cry1a probes were as described in refs. 11 and 12; the Wee1 probe spans nucleotides 1,981-2,369. Primer sequences used for PCR analysis are available on request.
Western Blot Analysis. Z3 cells were homogenized in lysis buffer (150 mM NaCl/5 mM EDTA/0.5% Nonidet P-40/50 mM Tris·HCl, pH 7.5) containing protease inhibitors and then clarified by centrifugation. After electrophoresis and transfer, membranes were blocked with 7% nonfat milk and incubated with rabbit anti-c-Fos or anti-c-Jun Abs (Santa Cruz Biotechnology) overnight at 4°C.
Chromatin Immunoprecipitation (ChIP) Analysis. ChIP assays were as described (29) with some modifications. The same amount of cross-linked chromatin-protein complex were used for incubation with 5 μg of antihuman pan-acetylated histone H3 (Upstate Biotechnology), with 5 μl of antihuman dimethyl-K4 histone H3 (Upstate Biotechnology) or with anti 5 μg of anti c-Fos Ab (Santa Cruz Biotechnology). Quantitative real-time PCRs were performed by using a LightCycler and continuous SYBR Green I monitoring according to the manufacturer's (Roche) recommendations. Relative proportions of immunoprecipitated promoter fragments were determined based on the threshold cycle value for each PCR (30).
Results
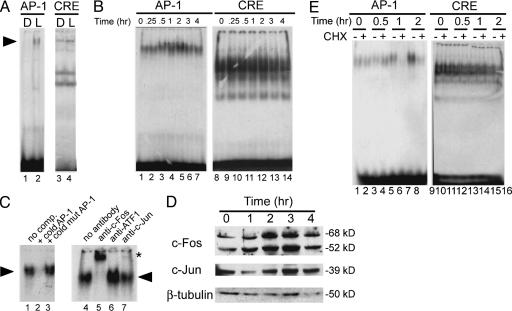
AP-1-Binding Activity Is Induced by Light in Zebrafish Eye. Circadian gene transcription is responsive to light in peripheral tissues of the zebrafish (25, 26). Within a program aimed at identifying light-inducible transcription factors in zebrafish (our unpublished results), we designed a set of experiments based on EMSA to search for light-responsive DNA sequences. A battery of putative binding sites for transcription factors was used. Here we show results obtained with four sequences corresponding to AP-1, cyclic AMP-responsive element (CRE), E-box, and CAAT/enhancer binding protein-binding sites. Because oscillation and induction of circadian clock genes are significant in the eye (8), eye nuclear extracts were used for the assay. Fish were kept in dark for 10 d and then given a 2-h light pulse. Nuclear extracts were prepared from fish before and after the light pulse. Light significantly induced AP-1 DNA-binding activity in zebrafish eye (Fig. 1A, lanes 1 and 2). AP-1 binding was efficiently competed by the homologous unlabeled oligonucleotide but not by a mutated AP-1 (TGAAAA). Supershift analysis by using specific Abs confirmed the presence of c-Fos and c-Jun in the AP-1 complex (not shown). Importantly, binding to the other sequences was not modified by light (Fig. 1 A, lanes 3 and 4 and not shown).
Fig. 1.
AP-1 activity is light-inducible in zebrafish. (A) Gel mobility-shift assay of zebrafish eye nuclear extracts binding to consensus AP-1 or CRE oligonucleotides. Extracts were prepared from animals kept in darkness for 3 d (D) or after a light pulse (L). (B) Nuclear extracts from Z3 cells prepared at different times after a 30-min light pulse tested for binding to AP-1 or CRE sites. (C) Lanes 1-3, binding competition assays carried out by addition of excess unlabeled AP-1 oligonucleotides as indicated. Nuclear extract prepared 2 h after start of light pulse was used. comp., competitor. Lanes 4-7, supershift analysis of AP-1 binding. Nuclear extract prepared 2 h after light pulse was incubated with c-Fos, c-ATF-1, or c-Jun Abs for 1 h before the addition of radiolabeled probe. Specific (arrowhead) and supershifted (asterisk) complexes are indicated. (D) Time course of c-Fos and c-Jun protein levels after a light pulse in Z3 cells. Nuclear extracts were prepared at various time points and processed for Western blotting. (E) Effect of CHX on AP-1 and CRE binding. Z3 cells nuclear extracts were prepared at the indicated times after light pulse, as described in C. CHX (final concentration, 50 μg/ml) was added 1 h before light pulse.
Light Inducibility of AP-1 DNA-Binding Activity in Z3 Cells. We have established Z3 cells, a light-inducible cell-based system that recapitulates the circadian characteristics of the zebrafish clock (11, 28). The effect of light on AP-1 DNA binding was thereby analyzed in Z3 cells. After 3 d in darkness, Z3 cells were exposed to a 30-min light pulse, and nuclear extracts were prepared from 0 to 4 h after the light pulse. The intensity of AP-1 binding was significantly increased 1-2 h after light pulse to then decrease (Fig. 1B, lanes 1-7). This binding was specifically competed by excess of homologous unlabeled AP-1 oligonucleotide but not by the mutated form (Fig. 1C, lanes 1-3). No light inducibility in DNA binding was observed for the other sites (Fig. 1B, lanes 8-14 and not shown). Supershift analyses by using anti-c-Fos or anti-c-Jun Abs confirmed the presence of these proteins in the AP-1 complex, whereas an anti-ATF-1 Ab was ineffective (Fig. 1C, lanes 4-7). We then analyzed the timing of light inducibility of c-Fos and c-Jun proteins. c-Fos levels increased significantly at 1-2 h after light pulse and returned to basal levels within 4 h. c-Jun protein levels were also induced by light but to a lower degree (Fig. 1D). Taken together, these data demonstrate that light directly induces AP-1 DNA binding in Z3 cells.
The kinetics of light-induced AP-1 activity (Fig. 1B) suggest the requirement of de novo protein synthesis for the induction. To verify this possibility, EMSA assays were performed by using nuclear extracts prepared from Z3 cells pretreated with cycloheximide (CHX) for 1 h before the light pulse. As expected, inhibition of translation significantly prevented light-inducibility of AP-1 binding (Fig. 1E, lanes 1-8), whereas had not effect on CRE binding (Fig. 1E, lanes 9-16).
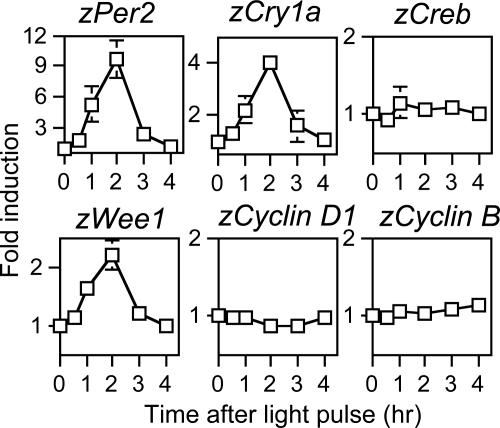
Light Inducibility of Wee1, a Cell Cycle Kinase. We previously have shown that the two circadian clock genes, zPer2 and zCry1a, are light-inducible in Z3 cells (11, 12). We speculated that light-dependent AP-1 activation could be associated with the light responsiveness of circadian genes. We therefore reexamined the kinetics of light induction of zPer2 and zCry1a genes by collecting RNA at several time points from Z3 cells that were kept in darkness for 3 d and then given a 30-min light pulse (Fig. 2). zCreb [CREB, CRE-binding protein] expression is not affected by light (Fig. 2; ref. 25), serving as control.
Fig. 2.
Effect of light on expression of circadian clock and cell cycle genes. Quantitative RT-PCR of zCry1a, zPer2, zCreb, zWee1, zCyclinD1, and zCyclinB mRNA levels. Z3 cells were kept for 3 d under darkness and then given a 30-min light pulse. RNA was harvested at the indicated time points after the light pulse. Expression levels were normalized to that of zActin. Values are mean ± SE of three independent experiments
AP-1 has been involved in the regulation of cell cycle genes, resulting in control of cell proliferation (18, 19). We studied light inducibility in Z3 cells of five zebrafish cell cycle genes, Wee1, Cyclin D1, c-Myc, Cyclin B, and Cdc2. Interestingly, zWee1 inducibility peaked 2 h after light pulse (Fig. 2), thus displaying kinetics similar to zPer2 and zCry1a. We have established that the Wee1 transcript expressed in zebrafish is equivalent to the Wee1b Xenopus isoform (not shown). Light had no effect on the other cell cycle genes (Fig. 2).
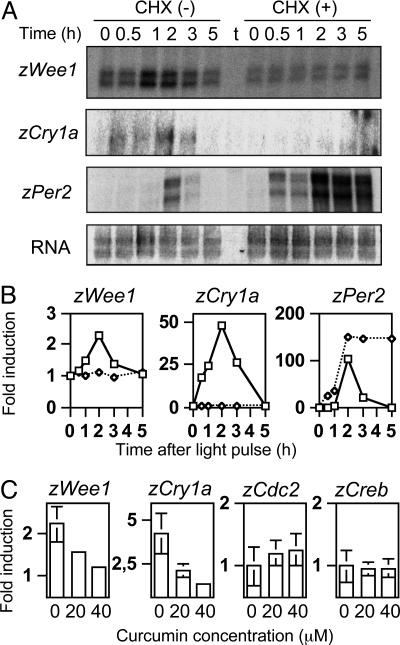
The effect of CHX, which abrogates light-induced AP-1 binding (Fig. 2), was analyzed on the responsiveness of zWee1, zCry1a, and zPer2 by RNA protection assay. The treatment efficiently inhibited light inducibility of zCry1a and zWee1 genes (Fig. 3 A and B). Interestingly, inhibition of translation had no effect on zPer2, whose expression remains sustained after the induction peak (Fig. 3 A and B). Thus, light inducibility follows at least two distinct pathways in Z3 cells, one that is CHX-sensitive and another that is protein synthesis-independent. Importantly, the AP-1 inhibitor curcumin (20) had a drastic effect on light-inducibility of zCry1a and zWee1. Curcumin inhibited inducibility of both genes in a dose-dependent manner, whereas it had no effect on zCreb and zCdc2 (Fig. 3C and not shown).
Fig. 3.
Differential effect of CHX on light-inducible genes. (A) zWee1, zCry1a, and zPer2 gene expression examined by RNA protection assay. RNA was prepared at the indicated times after light pulse, as described in Fig. 2. CHX (final concentration, 50 μg/ml) was added 1 h before light pulse. tRNA is indicated (t). Relative amounts of total RNA for each sample are shown (RNA). (B) Signals obtained in the RNase protection experiments shown in A were quantified by phosphoimaging and normalized with total RNA. The highest values were arbitrarily set as 100 for each mRNA species. (C) Effect of curcumin, an AP-1 inhibitor, on light induction of zWee1, zCry1a, zCreb, and zCdc2. Expression was scored by RT-PCR with two increasing concentrations of curcumin (20 or 40 μM, added 1 h before light pulse). RNA was prepared at the indicated times.
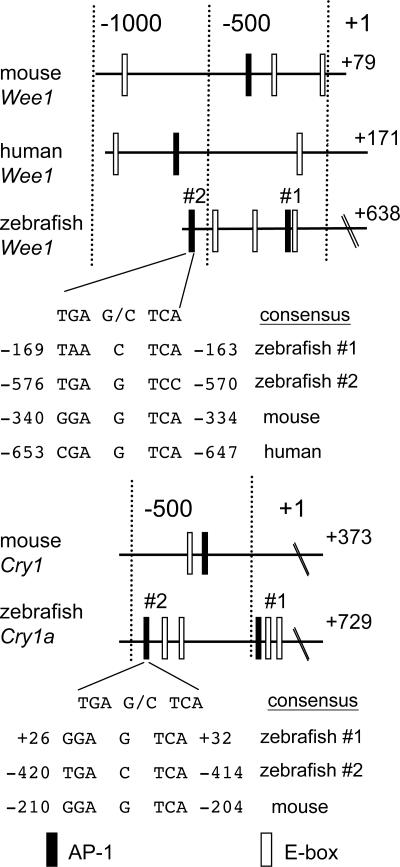
Characterization of zCry1a and zWee1 Promoters. The similar kinetics of zCry1a and zWee1 light inducibility (compare Figs. 1B and 2), the results obtained with curcumin (Fig. 3) and the CHX sensitivity (Fig. 3), suggested that light induction of zCry1a and zWee1 could depend on a light-responsive increase in AP-1 activity. To explore this possibility, we isolated the 5′-flanking regions of the zebrafish Cry1a and Wee1 genes and found two potential AP-1-binding sites within each region, which we indicate as #1 and #2 (Fig. 4). The relative distance between the two AP-1 sites in both promoters is highly similar. Four and three E-boxes are also present within the zebrafish Cry1a and Wee1 regulatory regions, respectively, although their sequence slightly differs from the CACGTG consensus (31). The AP-1 sites in zebrafish Cry1a and Wee1 promoters are intercalated with the E-boxes in both gene promoters, in an organization comparable to the mammalian Cry1 and Wee1 promoters (Fig. 4). By using the 5′ RACE methodology, we mapped the transcription start sites for both genes at 729 nt and 638 nt upstream of the ATG initiation codons of zCry1a and zWee1 genes, respectively.
Fig. 4.
Sequence comparison of zebrafish and mammalian promoters for Wee1 and Cry1a. Numbers represent the position relative to the transcription initiation site. The nucleotide sequences of the potential AP-1 sites are compared with the consensus AP-1-binding sequence. Filled and open boxes indicate AP-1 sites and an E-box, respectively.
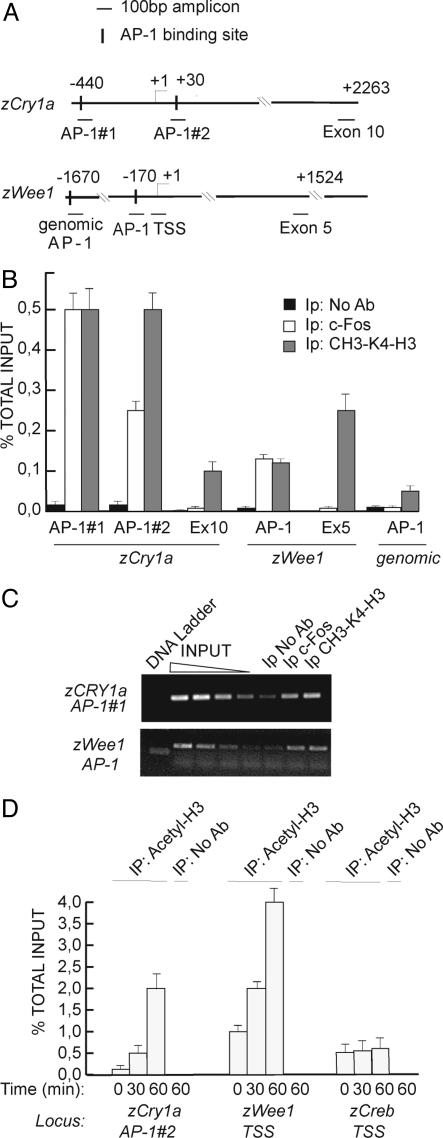
AP-1 Controls zWee1 and zCry1a Promoters in Vivo. To verify whether the AP-1 elements in the zWee1 and zCry1a promoters are functional, we performed ChIP assays. Z3 cells, kept for 3 d under dark, were subjected to a 30-min light pulse and then collected 2 h later. Cross-linked chromatin-protein complexes were extracted and the same amount of sonicated and solubized chromatin-protein complex was than subjected to immunoprecipitation by using either anti-c-Fos Ab or no Ab as negative control. An Ab recognizing dimethyl-K4 histone 3, a mark of transcriptionally active chromatin (32, 33), was also used as positive control. Immunoprecipitated samples were subjected to quantitative real-time PCR by using primers designed to amplify specifically the AP-1 sites in zWee1 and zCry1a promoters (Fig. 5A). As negative control, we amplified a distal exon region of each locus or a random genomic AP-1 consensus site present in an intronic region (referred as “genomic AP-1”). Fig. 5 B and C show that c-Fos specifically and efficiently binds to the AP-1 consensus site in Z3 cells after light pulse in both zCry1a and zWee1 promoters. C-Fos binds to both AP-1 sites of the zCry1a promoter (Fig. 4, #1 and #2).
Fig. 5.
In vivo binding of AP-1 complex to zCry1a and zWee1 promoters analyzed by ChIP. (A) Schematic representation of loci and amplicons used for ChIP assay. PCR primers are available upon request. Amplifications in the zCry1a exon 10 and in zWee1 exon 5 were used as negative controls. Genomic AP-1 indicates an AP-1-binding consensus site (TGAGTCA) in the intron region of zWee1 used as control. (B) Quantification of ChIP by RT-PCR. Z3 cells were subjected to light pulse for 30 min and then collected 2 h later. Cross-linked chromatin-protein complexes were extracted and subjected to immunoprecipitation with anti-cFos and anti-methyl-Lys-4 H3 Abs. ChIP without Ab was used as control. The percentage of immunoprecipitated target sequence relative to that present in total input is indicated. (C) Semiquantitative PCRs were performed on the same samples as described in B. Serial dilutions of input chromatin show the linear range of amplification of PCRs. (D) Light-dependent chromatin modifications in zCry1a and zWee1 promoters. At the indicated time after light pulse, cross-linked chromatin was analyzed by ChIP by using antipan-acetylated histone H3. Precipitated DNA samples were amplified with primers recognizing the transcription start site region of zWee1 and zCry1a genes.
Acetylation of specific lysine residues at the N-terminal tails of histones H3 and H4 marks active transcriptional loci (33). We investigated whether the AP-1 sites in zCry1a and zWee1 promoters are associated with actively transcribing chromatin in response to light. Fig. 5D shows a time-dependent enhancement of H3 pan-acetylation in the transcription start site region of zWee1 gene and zCry1a. One hour after the light pulse, H3 acetylation increases significantly in both promoter regions. The transcription start site amplicon used for zCry1a cover the AP-1 site #2, confirming its light-responsiveness. The zWee1 AP-1 site also shows a significant increase of H3 acetylation after light pulse (not shown). All together these data indicate that light signals to the AP-1 elements in both zWee1 and zCry1a promoters, inducing increased AP-1 binding and chromatin remodeling.
Discussion
The molecular pathways by which light modulates clock functions are of great physiological importance. Here we have shown that light directly activates AP-1 function in zebrafish. We also provide several lines of evidence that light-induced AP-1 activation controls both circadian and cell cycle targets, namely zCry1a and zWee1. These findings underscore AP-1 as a light response transcriptional regulator and reveal that circadian clock and cell cycle share common regulatory pathways.
In many species, the WEE-1 kinase inhibits mitotic cell division by inactivating the M-phase promoting factor (21). In mammals, AP-1 activates Wee1 expression after several stimuli by direct binding to the consensus motif within the Wee1 promoter (19, 20). Consistently with these reports, we have identified AP-1-binding motifs in the zebrafish Wee1 promoter (Figs. 4 and 5). In addition to the AP-1 dependent regulation, the mammalian wee1 gene is driven by circadian transcriptional activators via E-boxes within its promoter region (22). These E-box sites are conserved in the zebrafish zWee1 promoter (Fig. 4). Importantly, zWee1 expression shows a robust rhythmic oscillation in entrained Z3 cells and the oscillation continues after transfer the cells to constant darkness (not shown). Because it is highly likely that zebrafish Wee1 elicits a regulatory function that parallels the mammalian homolog, it is tempting to speculate that light-induced growth suppression of cultured Z3 cells (J.H. and P.S.-C., unpublished data) depends on zWee1 function.
The present study demonstrates that expression of zCry1a is directly light-inducible. We provide evidence that zCry1a transcription is controlled by AP-1 elements within its promoter. This establishes an important link between activation of AP-1 and circadian gene expression. Importantly, seven Cry genes have been cloned in zebrafish (8, 10), and they appear to fall into two groups: one inhibits CLOCK-BMAL1-mediated transcription (repressor-type CRY) and the other does not have transcription inhibitor activity (nonrepressor-type CRY). zCRY1a belongs to the repressor-type class and shows a high degree of homology to mammalian CRY1, an essential negative regulator of the circadian feedback loop (34, 35). Despite the structural and in vitro functional similarities between zCRY1a and mammalian CRY1, their expression profiles are quite different. Unlike zCry1a, mammalian Cry1 expression is not inducible, but shows a robust rhythmic oscillation in constant condition (35). zCry1a exhibits a circadian oscillation in Z3 cells exposed to a light:dark cycle but, similarly to zPer2, this oscillation dampens quickly after transfer of the cells to constant darkness (DD) (12) (J.H. and P.S.-C., unpublished data). It is conceivable that expression of zCry1a is highly regulated so to permit a specific role in clock regulation. Indeed, like mammalian Crys, expression of the other repressor-type zCrys is under circadian clock control and shows no acute inducibility (8, 12). We have previously reported that light-induced zPer2 activation plays a role in the circadian entrainment of Z3 cells (11). Although light inducibility of zCry1a expression is quite similar to that of zPer2 (Fig. 2), a striking difference is apparent. Indeed, Per2 behaves as a bona fide early response gene because its inducibility persists in the absence of protein synthesis (Fig. 3). This effect is characterized by a “superinduction” (Fig. 3A, +CHX), likely due to the block in the synthesis of a transcriptional inhibitor. Thus, the features of zPer2 inducibility are reminiscent of the c-fos and cAMP response element modulator genes, whose early response activation is down-regulated by a de novo synthesized repressor (Fos itself for c-fos and inducible cAMP early repressor for cAMP response element modulator) (36, 37). As zCRY1a strongly interacts with zPER2 (38), the superinduction elicited by treatment with CHX could be due to the absence of newly synthesized CRY proteins. This scenario is in sharp contrast with zCry1a and zWee1 induction profiles (Fig. 3), where CHX treatment causes a block in gene transcription. Thus, Per2 belongs to a different class of early response genes than zCry1a and zWee1. Although the early response of Per2 can be obtained by direct activation of intracellular signaling and phosphorylation of transcription factor CREB (39), thereby in a protein synthesis-independent manner, induction of zCry1a and zWee1 uses the AP-1/Fos complex. These notions show that AP-1-dependent transcription plays a central role in light entrainment of circadian clock.
Acknowledgments
We thank all members of the P.S.-C. laboratory for reagents and discussions. The technical help of C. Birling and N. Fischer is greatly appreciated. J.H. is supported by a fellowship from the Fondation pour la Recherche Médicale. L.C. is supported by a long-term European Molecular Biology Organization fellowship. M.D. is supported by a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists. Work in our laboratory is supported by Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Centre Hospitalier Universitaire Régional, Université Louis Pasteur, and Association pour la Recherche sur le Cancer. The P.S.-C. laboratory is an “Equipe Labelisée” of the Ligue contre le Cancer.
Author contributions: J.H., L.C., and P.S.-C. designed research; J.H., L.C., and M.D. performed research; J.H. and L.C. contributed new reagents/analytic tools; J.H., L.C., and P.S.-C. analyzed data; and J.H., L.C., and P.S.-C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CRE, cyclic AMP-responsive element; CHX, cycloheximide; ChIP, chromatin immunoprecipitation.
References
- 1.Dunlap, J. C. (1999) Cell 96, 271-290. [DOI] [PubMed] [Google Scholar]
- 2.Sancar, A. (2004) J. Biol. Chem. 279, 34079-34082. [DOI] [PubMed] [Google Scholar]
- 3.King, D. P. & Takahashi, J. S. (2000) Annu. Rev. Neurosci. 23, 713-742. [DOI] [PubMed] [Google Scholar]
- 4.Reppert, S. M. & Weaver, D. R. (2002) Nature 418, 935-941. [DOI] [PubMed] [Google Scholar]
- 5.Cahill, G. M. (2002) Cell Tissue Res. 309, 27-34. [DOI] [PubMed] [Google Scholar]
- 6.Pando, M. P. & Sassone-Corsi, P. (2002) BioEssays 24, 419-426. [DOI] [PubMed] [Google Scholar]
- 7.Cermakian, N., Whitmore, D., Foulkes, N. S. & Sassone-Corsi, P. (2000) Proc. Natl. Acad. Sci. USA 97, 4339-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi, Y., Ishikawa, T., Hirayama, J., Daiyasu, H., Kanai, S., Toh, H., Fukuda, I., Tsujimura, T., Terada, N., Kamei, Y., et al. (2000) Genes Cells 5, 725-738. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa, T., Hirayama, J., Kobayashi, Y. & Todo, T. (2002) Genes Cells 7, 1073-1086. [DOI] [PubMed] [Google Scholar]
- 10.Daiyasu, H., Ishikawa, T., Kuma, K., Iwai, S., Todo, T. & Toh, H. (2004) Genes Cells 9, 479-495. [DOI] [PubMed] [Google Scholar]
- 11.Pando, M. P., Pinchak, A. B., Cermakian, N. & Sassone-Corsi, P. (2001) Proc. Natl. Acad. Sci. USA 98, 10178-10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cermakian, N., Pando, M. P., Thompson, C. L., Pinchak, A. B., Selby, C. P., Gutierrez, L., Wells, D. E., Cahill, G. M., Sancar, A. & Sassone-Corsi, P. (2002) Curr. Biol. 12, 844-848. [DOI] [PubMed] [Google Scholar]
- 13.Kornhauser, J. M., Nelson, D. E., Mayo, K. E. & Takahashi, J. S. (1990) Neuron 5, 127-134. [DOI] [PubMed] [Google Scholar]
- 14.Morris, M. E., Viswanathan, N., Kuhlman, S., Davis, F. C. & Weitz, C. J. (1998) Science 279, 1544-1547. [DOI] [PubMed] [Google Scholar]
- 15.Halazonetis, T. D., Georgopoulos, K., Greenberg, M. E. & Leder, P. (1988) Cell 55, 917-924. [DOI] [PubMed] [Google Scholar]
- 16.Kornhauser, J. M., Mayo, K. E. & Takahashi, J. S. (1996) Behav. Genet. 26, 221-240. [DOI] [PubMed] [Google Scholar]
- 17.Karin, M., Liu, Z. & Zandi, E. (1997) Curr. Opin. Cell Biol. 9, 240-246. [DOI] [PubMed] [Google Scholar]
- 18.Shaulian, E. & Karin, M. (2002) Nat. Cell Biol. 4, E131-E136. [DOI] [PubMed] [Google Scholar]
- 19.Kawasaki, H., Komai, K., Ouyang, Z., Murata, M., Hikasa, M., Ohgiri, M. & Shiozawa, S. (2001) EMBO J. 20, 4618-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawasaki, H., Komai, K., Nakamura, M., Yamamoto, E., Ouyang, Z., Nakashima, T., Morisawa, T., Hashiramoto, A., Shiozawa, K., Ishikawa, H., et al. (2003) Oncogene 22, 6839-6844. [DOI] [PubMed] [Google Scholar]
- 21.Russell, P. & Nurse, P. (1987) Cell 49, 559-567. [DOI] [PubMed] [Google Scholar]
- 22.Matsuo, T., Yamaguchi, S., Mitsui, S., Emi, A., Shimoda, F. & Okamura, H. (2003) Science 302, 255-259. [DOI] [PubMed] [Google Scholar]
- 23.Cardone, L. & Sassone-Corsi, P. (2003) Nat. Cell Biol. 5, 859-861. [DOI] [PubMed] [Google Scholar]
- 24.Nagoshi, E., Saini, C., Bauer, C., Laroche, T., Naef, F. & Schibler, U. (2004) Cell 119, 693-705. [DOI] [PubMed] [Google Scholar]
- 25.Whitmore, D., Foulkes, N. S., Strahle, U. & Sassone-Corsi, P. (1998) Nat. Neurosci. 1, 701-707. [DOI] [PubMed] [Google Scholar]
- 26.Whitmore, D., Foulkes, N. S. & Sassone-Corsi, P. (2000) Nature 404, 87-91. [DOI] [PubMed] [Google Scholar]
- 27.Dekens, M. P., Santoriello, C., Vallone, D., Grassi, G., Whitmore, D. & Foulkes, N. S. (2003) Curr. Biol. 13, 2051-2057. [DOI] [PubMed] [Google Scholar]
- 28.Hirayama, J., Kaneko, M., Cardone, L., Cahill, G. M. & Sassone-Corsi, P. (2005) Methods Enzymol. 393, 186-204. [DOI] [PubMed] [Google Scholar]
- 29.Kuo, M. H. & Allis, C. D. (1999) Methods 19, 425-433. [DOI] [PubMed] [Google Scholar]
- 30.Frank, S. R., Schroeder, M., Fernandez, P., Taubert, S. & Amati, B. (2001) Genes Dev. 15, 2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hogenesch, J. B., Gu, Y. Z., Jain, S. & Bradfield, C. A. (1998) Proc. Natl. Acad. Sci. USA 95, 5474-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung, W. L., Briggs, S. D. & Allis, C. D. (2000) Curr. Opin. Cell Biol. 12, 326-333. [DOI] [PubMed] [Google Scholar]
- 33.Rice, J. C. & Allis, C. D. (2001) Curr. Opin. Cell Biol. 13, 263-273. [DOI] [PubMed] [Google Scholar]
- 34.Hirayama, J., Nakamura, H., Ishikawa, T., Kobayashi, Y. & Todo, T. (2003) J. Biol. Chem. 278, 35620-35628. [DOI] [PubMed] [Google Scholar]
- 35.Kume, K., Zylka, M. J., Sriram, S., Shearman, L. P., Weaver, D. R., Jin, X., Maywood, E. S., Hastings, M. H. & Reppert, S. M. (1999) Cell 98, 193-205. [DOI] [PubMed] [Google Scholar]
- 36.Sassone-Corsi, P., Sisson, J. C. & Verma, I. M. (1988) Nature 334, 314-319. [DOI] [PubMed] [Google Scholar]
- 37.Molina, C. A., Foulkes, N. S., Lalli, E. & Sassone-Corsi, P. (1993) Cell 75, 875-886. [DOI] [PubMed] [Google Scholar]
- 38.Hirayama, J., Fukuda, I., Ishikawa, T., Kobayashi, Y. & Todo, T. (2003) Nucleic Acids Res. 31, 935-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travnickova-Bendova, Z., Cermakian, N., Reppert, S. M. & Sassone-Corsi, P. (2002) Proc. Natl. Acad. Sci. USA 99, 7728-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]