Abstract
Cellular imaging techniques based on vibrational spectroscopy have become powerful tools in cell biology because the molecular composition of subcellular compartments can be visualized without the need for labeling. Using high-resolution, nonresonant confocal Raman microscopy on individual cells, we demonstrate here that lipid bodies (LBs) rich in arachidonate as revealed by their Raman spectra associate with latex bead-containing phagosomes in neutrophilic granulocytes. This finding was corroborated in macrophages and in PLB-985 cells, which can be induced to differentiate into neutrophil-like cells, by selective staining of LBs and visualization by confocal fluorescence microscopy. We further show that the accumulation of LBs near phagosomes is mediated at least in part by the flavohemoprotein gp91phox (in which “phox” is phagocyte oxidase), because different LB distributions around phagocytosed latex beads were observed in WT and gp91phox-deficient PLB-985 cells. gp91phox, which accumulates in the phagosomal membrane, is the catalytic subunit of the leukocyte NADPH oxidase, a critical enzyme in the innate immune response. Finally, time-lapse fluorescence microscopy experiments on neutrophils revealed that the LB-phagosome association is transient, similar to the “kiss-and-run” behavior displayed by endosomes involved in phagosome maturation. Because arachidonic acid (AA) has been shown to be involved in NADPH oxidase activation and phagosome maturation in neutrophils and macrophages, respectively, the findings reported here suggest that LBs may provide a reservoir of AA for local activation of these essential leukocyte functions.
Keywords: innate immunity, NADPH oxidase, Raman microscopy, phagocytes
Lipid bodies (LBs), cytoplasmic inclusions composed mainly of triglycerides, sterol esters, and phospholipids, are widespread in prokaryotic and eukaryotic cells (1, 2). They are prominent in adipocytes and certain plant seeds where they serve as lipid storage depots. In recent years, it has become clear that LBs found in a range of other cell types might have very different functions (2). For example, a recent proteomic study by Liu et al. (3) showed that besides structural proteins commonly associated with LBs, other proteins involved in lipid metabolism, signaling, membrane traffic, and caveolae and lipid raft formation were also found on LBs isolated from CHO K2 cells. The surprising presence of different Rab GTPases suggested that LBs may have a role in regulating membrane lipids in various trafficking pathways.
In leukocytes engaged in inflammation, an increased number of LBs is usually found (4, 5). Rapid LB induction by priming agents such as cis-unsaturated fatty acids, protein kinase C activators, or platelet-activating factor has been correlated with an enhanced capacity to generate eicosanoids (6, 7), which are powerful mediators of inflammation (8). Arachidonic acid (AA) is the precursor for the synthesis of eicosanoids, and the enzyme that liberates AA from phospholipids upon stimulation of neutrophils and macrophages, cytosolic phospholipase A2 (cPLA2), has been colocalized with LBs (9). Interestingly, cPLA2 recently was shown also to translocate to phagosomes in phagocytes that had ingested zymosan particles (10, 11). It was found that gp91phox (in which “phox” is phagocyte oxidase) is responsible for binding cPLA2 to the phagosomal membrane (10). This flavohemoprotein is the catalytic subunit of the leukocyte NADPH oxidase enzyme that is critical in innate immunity (12-14). Upon phagocytosis of microorganisms, gp91phox, which is complexed with p22phox into a membrane-bound heterodimer called cytochrome b558, generates large amounts of superoxide (O2-) from oxygen in a process termed the respiratory burst. Superoxide is converted into other reactive oxygen species (ROS) that directly and indirectly contribute to the killing of ingested microbes (15-17). Spatiotemporal regulation of NADPH oxidase activity is very important, because accidental or excessive production of ROS may lead to inflammatory tissue injury, carcinogenesis, atherosclerosis, and rheumatoid arthritis (18). The cytosolic subunits p47phox, p67phox, and p40phox (present as a trimeric complex), and the small GTPase Rac provide intrinsic regulation of NADPH oxidase activity because they interact only with phagosomal or plasma membrane-bound gp91phox and p22phox upon activation of leukocytes (12). These interactions, many of which have been characterized structurally (19), lead to an active assembly that is required for producing superoxide. The importance of NADPH oxidase in the innate immune response is illustrated by patients with chronic granulomatous disorder, where a genetic defect in membrane or cytosolic subunits of NADPH oxidase leads to an enhanced susceptibility to bacterial and fungal infections (20).
It is well known that AA stimulates superoxide production in neutrophils (21, 22), and the fact that cPLA2-deficient phagocytes cannot generate superoxide suggests that AA is necessary for NADPH oxidase activation (23). The exact phospholipid source providing AA for NADPH oxidase activation has remained elusive. It has been proposed (5) that arachidonyl phospholipids present in LBs might provide a source of AA for eicosanoid synthesis that does not require the disturbance of membrane integrity and that can be replenished from the abundant arachidonyl triglycerides in LBs.
We report here, using high-resolution nonresonant Raman microscopy on single neutrophilic granulocytes, that AA-induced LBs rich in esterified arachidonate as identified by their Raman spectra are frequently associated with phagocytosed latex beads. Selective Raman detection of AA in neutrophils was achieved by exposing cells to octadeuterated AA (AA-d8). Confocal fluorescence microscopy experiments on WT and gp91phox-deficient [X-linked chronic granulomatous disorder (X-CGD)] PLB-985 cells, which are myeloid leukemic cells that can be differentiated into neutrophil-like cells, confirmed our Raman microscopy results and showed that the distribution of LBs around phagosomes depends on gp91phox, suggesting that this NADPH oxidase subunit may mediate LB-phagosome interactions. Time-lapse fluorescence microscopy experiments on live neutrophils revealed that the association between phagosomes and LBs is of a transient nature. The accumulation of LBs near ingested latex beads was also demonstrated in macrophages by fluorescence microscopy experiments. The experiments reported here lead us to hypothesize that controlled release of AA from LBs in the vicinity of phagosomes may be an important mechanism in regulating NADPH oxidase activity and/or phagosome maturation. To our knowledge, the only reported case of lipid body association with phagosomes has been a transmission electron microscopy study in which macrophages were incubated with radiolabeled AA and allowed to phagocytose zymosan particles (24). However, the observed association of LBs with and their possible discharge into phagosomes was not the focus of that study, and the possible function of LBs in phagocytosis was not discussed.
Methods
Details about materials, cell cultures, granulocyte isolation, and confocal fluorescence microscopy are provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Induction of LBs and Phagocytosis. Cells suspended in gelatin buffer (PBS/0.1% gelatin/5 mM glucose) were adhered to poly(l-lysine)-coated glass (for fluorescence microscopy) or CaF2 (for Raman imaging) slides for 15 min at 37°C, followed by incubation for 1 h at 37°C with 10 μM AA or AA-d8 in gelatin buffer. After washing with gelatin buffer and phagocytosis buffer (PBS/1 mM CaCl2/0.5 mM MgSO4/5 mM glucose/1% BSA), the samples were incubated in phagocytosis buffer with latex beads (typically 107 beads per 106 cells), which had been serum-opsonized as described in ref. 25 for 30 min at 37°C. For Raman experiments, cells were subsequently washed with PBS and fixed for 30 min in 2% paraformaldehyde in PBS at room temperature. For fluorescence experiments on PLB-985 cells, cells were incubated for 10 min at room temperature with Nile red (0.1 μg/ml) after phagocytosis, washed with PBS, and fixed for 30 min in 2% paraformaldehyde in PBS at room temperature. For time-lapse fluorescence experiments on live neutrophils, cells were incubated with Nile red (0.1 μg/ml), washed with PBS, and subsequently incubated with latex beads as described above.
Confocal Raman Microspectroscopy and Imaging. Nonresonant Raman spectroscopy and imaging experiments were performed on a laser-scanning confocal Raman microspectrometer that is described in ref. 26. Imaging experiments were performed by raster-scanning the laser beam over a cell or an intracellular region of interest and accumulating a full Raman spectrum at each pixel (1 s/pixel at 100 mW 647.1-nm excitation power). Noise in the resulting 3D (spatial × spatial × spectral dimension) data matrix was reduced by singular value decomposition (25, 26). Raman images were constructed by plotting the integrated intensity of the vibrational band of interest as a function of position. Hierarchical cluster analysis (HCA) was performed on Raman imaging data matrices to visualize regions in cells with high Raman spectral similarities. In the cluster analysis routine, principal component analysis scores were taken as input variables, squared Euclidean distances were used as distance measure, and Ward's algorithm was used to partition Raman spectra into clusters. All data manipulations were performed in routines written in matlab 6.5 (MathWorks, Natick, MA).
Results and Discussion
Raman Imaging on Neutrophils. We used confocal Raman microspectroscopy because cellular imaging techniques based on vibrational spectroscopy [e.g., IR (27), Raman (26, 28), and coherent anti-Stokes Raman spectroscopy (29)] do not require labeling of the biomolecules of interest and, more importantly, provide detailed information about the molecular composition of the subcellular volume being probed. These features render vibrational microscopic methods unique tools in biology and medicine, because measurements are conveniently performed on unlabeled intact cells and tissues (30).
Unstimulated neutrophils isolated from peripheral blood contain few LBs (4), which we verified by performing confocal nonresonant Raman microscopy on freshly isolated neutrophils adhered to a poly(l-lysine)-coated CaF2 substrate. Fig. 1A shows a typical Raman cluster image obtained by Raman microscopy on a single neutrophil, followed by HCA of the 3D data matrix that is obtained during Raman imaging (see Methods). The multilobed nucleus, characteristic for neutrophils, is shown in blue. The average Raman spectrum of this cluster (Fig. 1C, spectrum 1) shows diagnostic Raman bands at, for example, 788, 1,095, and 1,577 cm-1 due to several nucleotide and DNA/RNA backbone vibrational modes (31). The Raman spectrum from the green cluster (Fig. 1C, spectrum 2) is typical for the cytoplasm (32) and differs from the red cluster (spectrum not shown) mainly in its higher overall intensity. Because LBs in cells are easily recognized by Raman spectroscopy because their high aliphatic chain density gives rise to very strong Raman signals (33), the absence of a cluster containing strong lipid signals in the image shown in Fig. 1 A confirms previous findings that unstimulated neutrophils contain few or no LBs (4).
Fig. 1.
Confocal Raman image of a quiescent neutrophil. (A) HCA image (12.2 × 12.2 μm2) constructed from Raman imaging data of a single neutrophil. Each color represents a different cluster. (B) Corresponding white-light transmission image. The area enclosed by the black square was used for Raman imaging. (C) Average Raman spectra extracted from the blue (spectrum 1) and green (spectrum 2) clusters displayed in A.
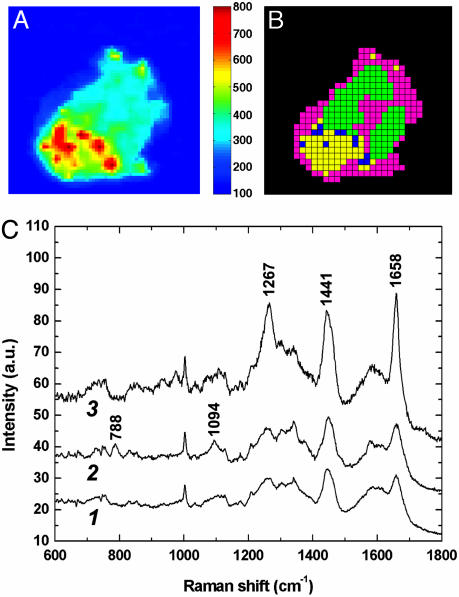
To elicit the formation of LBs in neutrophils, we exposed cells to 10 μM AA for 1 h at 37°C. This concentration is high enough for efficient LB induction (6) but too low for the generation of large amounts of superoxide by NADPH oxidase (21). Nonresonant Raman microscopy on neutrophils after LB induction revealed the presence of intracellular punctate regions with high lipid content, as demonstrated by the yellow/red regions in Fig. 2A and the blue cluster in Fig. 2B. The Raman image in Fig. 2A was constructed in the 1,658-cm-1 band, which has a large contribution from cis- unsaturated C═C stretching vibrations. The presence of unsaturated moieties is evident from spectrum 3 in Fig. 2C, which is the average Raman spectrum from the blue cluster in Fig. 2B. Besides the intense band at 1,658 cm-1, the strong signals at 1,267 cm-1 (HC═ in-plane deformation) and 1,441 cm-1 (CH2 deformations) also point to high levels of unsaturated lipids in the blue cluster. We assign the punctate regions in Fig. 2 A and B to LBs, although the large scanning range of these images (14.1 × 14.1 μm2), in combination with a step size of 440 nm, precludes definite assignment because image pixels that are roughly the same size as LBs are obtained in this way. Other Raman images shown here have much smaller image pixel sizes (vide infra).
Fig. 2.
Formation of LBs in neutrophils after incubation with 10 μM AA for 1 h. (A) Raman image (14.1 × 14.1 μm2) in the 1,635 to 1,680-cm-1 region. (B) Corresponding HCA image. (C) Average Raman spectra extracted from the magenta (spectrum 1), green (spectrum 2), and blue (spectrum 3) clusters displayed in B. Strong bands at 1,267, 1,441, and 1,658 cm-1 in spectrum 3 are assigned to lipids.
Using resonance Raman microscopy, we recently showed that upon phagocytosis of latex beads, gp91phox (the catalytic subunit of NADPH oxidase) translocates from specific granules to the phagosome in neutrophils (25, 34). We now report that non-resonant Raman experiments show the redistribution of LBs upon phagocytosis of latex beads. In neutrophils with internalized beads, several LBs were frequently found in close proximity to the latex particles, as shown by the representative example in Fig. 3. Fig. 3A displays the Raman image in the strong 1,000-cm-1 band of polystyrene, whereas LBs are visualized by the Raman image in the 1,658-cm-1 band of unsaturated lipids (Fig. 3C). Both of these images are constructed from the same data set, exemplifying the abundant chemical information present in Raman spectra. Fig. 3D shows that clusters with distinct average spectra result from HCA on this data set. In addition to two clusters (blue and green) corresponding to the phagocytosed latex bead (spectrum 1 in Fig. 3E) and one cluster from the LBs (yellow, spectrum 3 in Fig. 3E), other clusters displayed in magenta and black correspond to the cytoplasm (spectrum 2 in Fig. 3E) and the nucleus (spectrum not shown), respectively.
Fig. 3.
High-resolution Raman imaging in the cytoplasm of a neutrophil shows that LBs are found in close proximity to a phagocytosed latex bead. (A) Raman image (6.7 × 6.7 μm2) in the 1,000-cm-1 band of polystyrene. (B) White-light transmission image. The area enclosed by the black square was used for Raman imaging. (C) Raman image in the 1,658-cm-1 band of unsaturated lipids. (D) Corresponding HCA image. (E) Average Raman spectra extracted from the blue (spectrum 1), magenta (spectrum 2), and yellow (spectrum 3) clusters displayed in D. Spectra 2 and 3 are shifted along the ordinate for clarity, whereas spectrum 1 has been downscaled.
The Raman spectra shown in Figs. 2C and 3E suggest that LBs are rich in unsaturated lipids, which is most likely due to the exposure of the cells to AA, because it has been reported that the majority of radiolabeled AA added to neutrophils is incorporated into LBs (6). However, from Figs. 2C and 3E, the origin of the lipids present in LBs cannot be established. To clarify this issue, we used AA-d8 to induce LB formation in neutrophils. In AA-d8, the eight vinylic protons have been substituted for deuterium atoms. Due to the higher mass of deuterium, the vinylic ═C─H stretch vibration normally present at 3,013 cm-1 in AA shifts to a spectrally silent region (1,800-2,800 cm-1) when AA-d8 is used. As shown in Fig. 4G, vinylic ═C─D stretching vibrations in AA-d8 are found at 2,220 and 2,249 cm-1, in between the fingerprint (<1,800 cm-1) and high-frequency (>2,800 cm-1) regions of the nonresonant Raman spectrum of neutrophils. The lack of any cellular Raman signal between 1,800 and 2,800 cm-1 allows the presence of AA-d8 anywhere in the cell to be visualized, with ultimate selectivity,§ by Raman imaging in the 2,200- to 2,280-cm-1 region. This selectivity is exemplified by Fig. 4 A, C, and E. A high-resolution Raman image of intracellular LBs, induced in neutrophils by 1 h of exposure to AA-d8, is shown in Fig. 4A. The Raman images in Fig. 4 B and C demonstrate that several LBs rich in AA-d8 (Fig. 4C) associate with an ingested latex bead (Fig. 4B), confirming the results shown in Fig. 3. Another example is provided by Fig. 4D (Raman image of polystyrene) together with Fig. 4E (image in AA-d8) and Fig. 4F (corresponding HCA image). Fig. 4G displays the average Raman spectra corresponding to the blue (spectrum 1), orange (spectrum 2), and green (spectrum 3) clusters displayed in 4F. Spectrum 3 in Fig. 4G shows that the C═C stretch vibration has shifted from 1,658 cm-1 to 1,633 cm-1 upon substituting the vinylic protons in AA for deuteriums in AA-d8. Of further interest in spectrum 3 is the band at 1,740 cm-1, which is due to the carboxyl ester C═O stretch vibration. The relatively high intensity of this band in spectrum 3 suggests that AA-d8 is at least partially esterified in LBs. This finding is in accordance with previous studies that relied on isolation of AA-induced LBs and/or lipid extraction from neutrophils followed by chromatography to demonstrate that AA is mainly present as triglycerides and phospholipids in LBs (9). In contrast to these investigations, the confocal Raman imaging technique presented here allows the composition of LBs to be determined directly inside intact cells without labeling.
Fig. 4.
Raman imaging shows that LBs, enriched in AA-d8, associate with phagocytosed latex beads in neutrophils. (A) Raman image (7.5 × 7.5 μm2) constructed from the 2,200- to 2,280-cm-1 region of the cytoplasm of a neutrophil incubated with AA-d8 for 1 h. (B and C) Neutrophil incubated for 1 h with AA-d8, followed by phagocytosis of latex beads. Raman images (5.2 × 5.2 μm2) are shown in the polystyrene (B) and AA-d8 (C) bands at 1,605 and 2,200-2,280 cm-1, respectively. (D and E) Similar images as in B and C, respectively, but from a different cell. (F) HCA image (5.2 × 5.2 μm2) of the Raman data set used to construct images D and E.(G) Average Raman spectra extracted from the blue (spectrum 1), orange (spectrum 2), and green (spectrum 3) clusters displayed in F. Spectra 2 and 3 are shifted along the ordinate for clarity. Bands marked with an asterisk in spectrum 1 are from polystyrene.
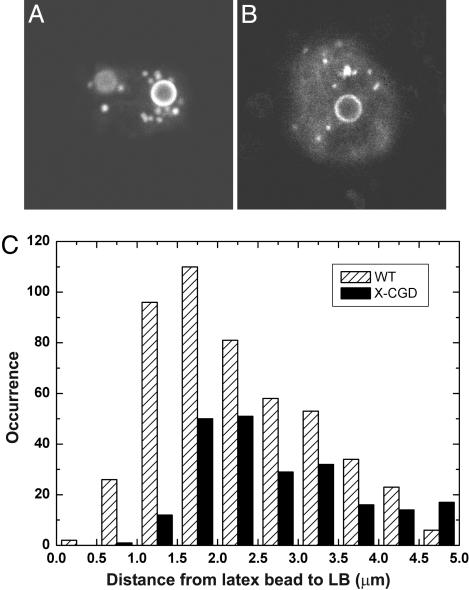
Distribution of LBs Around Phagosomes in WT and gp91phox-Deficient PLB-985 Cells. The superoxide-generating ability of NADPH oxidase is tightly regulated in space and time, and various molecular pathways for regulating NADPH oxidase activity have been unraveled (12, 13). It is well known that AA is essential for generating superoxide (23), and although the exact nature of the interactions between AA and different components of the NADPH oxidase enzyme is unknown, it has been shown that gp91phox (35), p47phox (36), and the Rac2-RhoGDI complex (37) are all influenced by AA. The recent observation that translocation of cPLA2 to the phagosome of phagocytes stimulated with zymosan particles depends on the presence of gp91phox in the phagosomal membrane (10), together with the demonstrated colocalization of cPLA2 with LBs in another study (9), led us to hypothesize that association between LBs and phagocytosed latex beads, as observed in the present study, might be influenced by the interaction of cPLA2 on LBs with gp91phox embedded in the phagosomal membrane. We tested part of this hypothesis by using WT and gp91phox-deficient (X-CGD) PLB-985 cells. These human myeloid leukemic cells can be cultured in vitro and induced to differentiate into cells displaying neutrophil-like behavior, such as phagocytosis and superoxide production upon stimulation (38). After induction of LBs in PLB-985 cells by exposure to AA (10 μM for 1 h), subsequent phagocytosis of fluorescent latex beads, and staining of the LBs with Nile red (39), we examined the association between phagocytosed latex beads and LBs by confocal fluorescence microscopy. Although the fluorescence of both the LBs and the latex beads was observed in one emission channel, Fig. 5A (WT cell) and Fig. 5B (X-CGD cell) show that LBs are easily distinguished from beads by size and appearance (the beads have a fluorescent rim, whereas the fluorescence in the smaller LBs is homogeneously distributed). We analyzed 100 images (of both WT and X-CGD PLB-985 cells) from confocal fluorescence z-stacks of randomly selected cells with clearly internalized latex beads. The distances between the center of latex beads and the center of LBs were measured.¶ Fig. 5C shows that the distribution of these distances is different for WT and X-CGD PLB-985 cells. Taking into account that the average radius of the latex beads is 1.0 μm, we find that in WT cells, 48% of the LBs is found within 1 μm of the phagosomal membrane,∥ whereas in X-CGD cells, this value is 26%. The average distance between the analyzed LBs and the phagosomal membrane is 1.3 ± 1.0 μm and 1.9 ± 1.2 μm for WT and X-CGD cells, respectively. These results indicate that gp91phox is at least in part responsible for the increased accumulation of LBs near the phagosome in WT cells compared with X-CGD cells, suggesting that gp91phox may mediate the interaction between phagosomes and LBs.
Fig. 5.
Association of LBs with latex bead-containing phagosomes in differentiated PLB-985 cells. (A and B) Confocal fluorescence images (10 × 10 μm2) of a WT(A) and X-CGD (B) PLB-985 cell. LBs (small, bright features) are clearly distinguished from the larger latex beads (fluorescent rim). (C) Histograms of distances between the center of latex beads and the center of LBs in WT and X-CGD PLB-985 cells.
Time-Lapse Fluorescence Microscopy on Neutrophils. To study the dynamics of LB association with latex bead-containing phagosomes in living cells, we performed time-lapse confocal fluorescence microscopy on adherent neutrophils incubated with AA, fluorescent latex beads, and Nile red. These experiments revealed frequent transient contacts between LBs and phagosomes. Two of these contacts (with different association times) are shown in Fig. 6 (LBs “1” and “2”), and Movie 1, which is published as supporting information on the PNAS web site, illustrates this LB behavior. The time-lapse experiments also showed that phagocytosed latex beads and LBs are occasionally found moving together through the cytoplasm for a certain period before the LBs dissociate from the beads (Movie 1). This behavior is inconsistent with the possibility that LB-phagosome associations are caused by random independent movements of LBs and phagocytic vacuoles. Interestingly, a “kiss-and-run” hypothesis has been put forward by Desjardins (40) to account for phagosome maturation proceeding by means of multiple transient fusion-fission interactions of phagosomes with endocytic organelles. The spatiotemporal characteristics of these dynamic contacts, and the extensive membrane remodeling that is required for phagosomal maturation, may be regulated by key proteins and/or lipids such as members of the Rab GTPase family, soluble N-ethylmaleimide-sensitive factor attachment protein receptors, and phosphoinositides that have been found inserted into or associated with the phagosomal membrane at different stages of maturation (41). Remarkably, recent independent proteomic analyses have shown that several Rab GTPases (Rab2, Rab5, Rab7, Rab10, Rab11, and Rab14) are present on both purified latex bead-containing phagosomes from J774 macrophage-like cells (42) and on LBs in CHO K2 cells (3). The kiss-and-run behavior of LBs reported here may constitute a mechanism for Rab GTPase transport to and from the phagosome, a process that may be necessary for phagosome maturation.
Fig. 6.
A series of pseudocolor confocal fluorescence still images (12.0 × 12.0 μm2), extracted from Movie 1, showing transient contacts between LBs (marked “1” and “2”) stained with Nile red and fluorescent latex bead-containing phagosomes (marked with asterisks) in an adherent live neutrophil. LB 1 shows contact only in one frame (at 5.2 s), whereas LB 2 is associated with the latex bead-containing phagosome for multiple frames.
Fluorescence Microscopy on Macrophages. Similar to neutrophils, LBs can also be induced in other professional phagocytes such as macrophages (24). We performed confocal fluorescence microscopy experiments on murine RAW 264.7 macrophages (see Supporting Materials and Methods) to establish whether the association of LBs with phagosomes also occurs in these leukocytes. Indeed, we found that LBs are accumulated near ingested particles in both fixed and live macrophages (Fig. 7, which is published as supporting information on the PNAS web site).
A Rationale for the Association of LBs with Phagosomes. Recent work has indicated that LBs in a variety of cell types are not merely lipid reservoirs (2, 3). In phagocytes engaged in inflammation, LBs have been mainly implicated in the synthesis of eicosanoids (4, 5). The AA-releasing enzyme cPLA2, its activating mitogen-activated protein kinases, and the eicosanoid-generating enzymes 5-lipoxygenase and cyclooxygenase all have been found on the surface of LBs in several leukocyte types. Also, phosphatidylinositol 3-kinase (PI3K) was recently found to colocalize with LBs (43). PI3K is a key lipophilic enzyme involved in intracellular signaling of diverse cellular responses, including phagocytosis (44), NADPH oxidase activation (45), and AA release from bacteria-stimulated macrophages (46). The present study reveals that arachidonate-rich LBs associate with latex bead-containing phagosomes in neutrophilic granulocytes, PLB-985 cells differentiated into neutrophil-like cells, and macrophages. This finding suggests that these organelles may be involved in aspects of the innate immune response of phagocytes other than eicosanoid synthesis [for example, phagosome maturation and reactive oxygen species (ROS) formation]. Our microscopy experiments with WT and X-CGD PLB-985 cells, which demonstrate that the accumulation of LBs near phagosomes depends on gp91phox, point to a role for LBs in ROS production through the regulation of superoxide-generating NADPH oxidase. Taking into account the known dependency of several NADPH oxidase subunits (including gp91phox) on AA, we suggest that LBs may be used by phagocytes to locally release AA for NADPH oxidase activation near the phagosome. This hypothesis is strengthened by reports showing the presence of AA-releasing cPLA2 on LBs (9) and the translocation of cPLA2 to the phagosome in PLB-985 cells and macrophages incubated with zymosan particles (10, 11). We expect that future studies with cPLA2-deficient PLB-985 cells, which have become available (23), may provide insight related to this hypothesis.
In conclusion, we have demonstrated here that LBs, cytoplasmic organelles rich in triglycerides and phospholipids, associate with phagosomes in professional phagocytes. We used single-cell confocal Raman microscopy to analyze the chemical composition of these lipid inclusions and found that incubation of phagocytes with AA and latex beads leads to the accumulation of LBs, rich in esterified arachidonate, near the phagocytosed microspheres. These results exemplify the power of vibrational spectroscopy in label-free cellular imaging. We corroborated our Raman microscopy results by fluorescence microscopy experiments, which further revealed that the association of LBs with the phagosome is transient and dependent on gp91phox, the catalytic subunit of the enzyme NADPH oxidase. Together, our findings suggest that LBs play a role in the innate immune response. Their release of AA near the phagosome may be used to locally activate NADPH oxidase and/or to facilitate phagosome maturation, both of which are essential for the degradation of microbes upon infection.
Supplementary Material
Acknowledgments
We thank Drs. G. J. Puppels and T. C. Bakker Schut (Erasmus Medical Center, Rotterdam) for providing the cluster analysis routines and valuable advice and Dr. M. C. Dinauer (Indiana University School of Medicine, Indianapolis) for the generous gift of PLB-985 cells. We gratefully acknowledge the support of The Chronic Granulomatous Disorder Research Trust.
Author contributions: H.-J.v.M., D.R., and C.O. designed research; H.-J.v.M. and Y.M.K. performed research; H.-J.v.M. and C.O. analyzed data; and H.-J.v.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LB, lipid body; AA, arachidonic acid; cPLA2, cytosolic phospholipase A2; AA-d8, octadeuterated arachidonic acid; X-CGD, X-linked chronic granulomatous disorder; HCA, hierarchical cluster analysis; phox, phagocyte oxidase.
Footnotes
The very low acidity of vinylic deuteriums (pKa ≈ 40 for vinylic protons) prevents deuterium exchange inside the cell.
We found that in 93 of 100 investigated WT PLB-985 cells, 1 or more LBs (489 LBs in total and an average of 5.3 LBs per cell) were localized within 4 μm from the surface of phagocytosed beads. In gp91phox-deficient PLB-985 cells, 77 of 100 imaged cells satisfied this criterion (238 LBs in total and an average of 3.1 LBs per cell).
We assume that the phagosomal membrane is tightly wrapped around internalized latex beads. See also ref. 42.
References
- 1.Zweytick, D., Athenstaedt, K. & Daum, G. (2000) Biochim. Biophys. Acta 1469, 101-120. [DOI] [PubMed] [Google Scholar]
- 2.Murphy, D. J. (2001) Prog. Lipid Res. 40, 325-438. [DOI] [PubMed] [Google Scholar]
- 3.Liu, P., Ying, Y., Zhao, Y., Mundy, D. I., Zhu, M. & Anderson, R. G. W. (2004) J. Biol. Chem. 279, 3787-3792. [DOI] [PubMed] [Google Scholar]
- 4.Bozza, P. T., Payne, J. L., Morham, S. G., Langenbach, R., Smithies, O. & Weller, P. F. (1996) Proc. Natl. Acad. Sci. USA 93, 11091-11096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pacheco, P., Bozza, F. A., Gomes, R. N., Bozza, M., Weller, P. F., Castro-Faria-Neto, H. C. & Bozza, P. T. (2002) J. Immunol. 169, 6498-6506. [DOI] [PubMed] [Google Scholar]
- 6.Weller, P. F., Ryeom, S. W., Picard, S. T., Ackerman, S. J. & Dvorak, A. M. (1991) J. Cell Biol. 113, 137-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozza, P. T., Payne, J. L., Goulet, J. L. & Weller, P. F. (1996) J. Exp. Med. 183, 1515-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funk, C. D. (2001) Science 294, 1871-1875. [DOI] [PubMed] [Google Scholar]
- 9.Yu, W., Bozza, P. T., Tzizik, D. M., Gray, J. P., Cassara, J., Dvorak, A. M. & Weller, P. F. (1998) Am. J. Pathol. 152, 759-769. [PMC free article] [PubMed] [Google Scholar]
- 10.Shmelzer, Z., Haddad, N., Admon, E., Pessach, I., Leto, T. L., Eitan-Hazan, Z., Hershfinkel, M. & Levy, R. (2003) J. Cell Biol. 162, 683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girotti, M., Evans, J. H., Burke, D. & Leslie, C. C. (2004) J. Biol. Chem. 279, 19113-19121. [DOI] [PubMed] [Google Scholar]
- 12.Babior, B. M. (1999) Blood 93, 1464-1476. [PubMed] [Google Scholar]
- 13.Roos, D., Van Bruggen, R. & Meischl, C. (2003) Microbes Infect. 5, 1307-1315. [DOI] [PubMed] [Google Scholar]
- 14.Cross, A. R. & Segal, A. W. (2004) Biochim. Biophys. Acta 1657, 1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reeves, E. P., Lu, H., Lortat Jacobs, H., Messina, C. G. M., Bolsover, S., Gabella, G., Potma, E. O., Warley, A., Roes, J. & Segal, A. W. (2002) Nature 416, 291-297. [DOI] [PubMed] [Google Scholar]
- 16.Roos, D. & Winterbourn, C. C. (2002) Science 296, 669-671. [DOI] [PubMed] [Google Scholar]
- 17.Rada, B. K., Geiszt, M., Káldi, K., Timár, C. & Ligeti, E. (2004) Blood 104, 2947-2953. [DOI] [PubMed] [Google Scholar]
- 18.Babior, B. M. (2000) Am. J. Med. 109, 33-44. [DOI] [PubMed] [Google Scholar]
- 19.Groemping, Y. & Rittinger, K. (2005) Biochem. J. 386, 401-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heyworth, P. G., Cross, A. R. & Curnutte, J. T. (2003) Curr. Opin. Immunol. 15, 578-584. [DOI] [PubMed] [Google Scholar]
- 21.Curnutte, J. T., Badwey, J. A., Robinson, J. M., Karnovsky, M. J. & Karnovsky, M. L. (1984) J. Biol. Chem. 259, 11851-11857. [PubMed] [Google Scholar]
- 22.Henderson, L. M. & Chappell, J. B. (1996) Biochim. Biophys. Acta 1273, 87-107. [DOI] [PubMed] [Google Scholar]
- 23.Dana, R., Leto, T. L., Malech, H. L. & Levy, R. (1998) J. Biol. Chem. 273, 441-445. [DOI] [PubMed] [Google Scholar]
- 24.Dvorak, A. M., Dvorak, H. F., Peters, S. P., Shulman, E. S., Macglashin, D. W., Pyne, K., Harvey, S., Galli, S. J. & Lichtenstein, L. M. (1983) J. Immunol. 131, 2965-2976. [PubMed] [Google Scholar]
- 25.van Manen, H.-J., Kraan, Y. M., Roos, D. & Otto, C. (2004) J. Phys. Chem. B 108, 18762-18771. [Google Scholar]
- 26.Uzunbajakava, N., Lenferink, A., Kraan, Y., Volokhina, E., Vrensen, G., Greve, J. & Otto, C. (2003) Biophys. J. 84, 3968-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diem, M., Romeo, M., Boydston-White, S., Miljković & Matthäus, C. (2004) Analyst 129, 880-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puppels, G. J., De Mul, F. F. M., Otto, C., Greve, J., Robert-Nicoud, M., Arndt-Jovin, D. J. & Jovin, T. M. (1990) Nature 347, 301-303. [DOI] [PubMed] [Google Scholar]
- 29.Cheng, J.-X. & Xie, X. S. (2004) J. Phys. Chem. B 108, 827-840. [Google Scholar]
- 30.Hanlon, E. B., Manoharan, R., Koo, T.-W., Shafer, K. E., Motz, J. T., Fitzmaurice, M., Kramer, J. R., Itzkan, I., Dasari, R. R. & Feld, M. S. (2000) Phys. Med. Biol. 45, R1-R59. [DOI] [PubMed] [Google Scholar]
- 31.Peticolas, W. L., Kubasek, W. L., Thomas, G. A. & Tsuboi, M. (1987) Biological Applications of Raman Spectroscopy (Wiley, New York), Vol. 1, pp. 81-134. [Google Scholar]
- 32.Puppels, G. J., Garritsen, H. S. P., Segers-Nolten, G. M. J., De Mul, F. F. M. & Greve, J. (1991) Biophys. J. 60, 1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takai, Y., Masuko, T. & Takeuchi, H. (1997) Biochim. Biophys. Acta 1335, 199-208. [DOI] [PubMed] [Google Scholar]
- 34.van Manen, H.-J., Uzunbajakava, N., Van Bruggen, R., Roos, D. & Otto, C. (2003) J. Am. Chem. Soc. 125, 12112-12113. [DOI] [PubMed] [Google Scholar]
- 35.Doussière, J., Bouzidi, F., Poinas, A., Gaillard, J. & Vignais, P. V. (1999) Biochemistry 38, 16394-16406. [DOI] [PubMed] [Google Scholar]
- 36.Shiose, A. & Sumimoto, H. (2000) J. Biol. Chem. 275, 13793-13801. [DOI] [PubMed] [Google Scholar]
- 37.Chuang, T.-H., Bohl, B. P. & Bokoch, G. M. (1993) J. Biol. Chem. 268, 26206-26211. [PubMed] [Google Scholar]
- 38.Zhen, L., King, A. A. J., Xiao, Y., Chanock, S. J., Orkin, S. H. & Dinauer, M. C. (1993) Proc. Natl. Acad. Sci. USA 90, 9832-9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenspan, P., Mayer, E. P. & Fowler, S. D. (1985) J. Cell Biol. 100, 965-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desjardins, M. (1995) Trends Cell Biol. 5, 183-186. [DOI] [PubMed] [Google Scholar]
- 41.Vieira, O. V., Botelho, R. J. & Grinstein, S. (2002) Biochem. J. 366, 689-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garin, J., Diez, R., Kieffer, S., Dermine, J.-F., Duclos, S., Gagnon, E., Sadoul, R., Rondeau, C. & Desjardins, M. (2001) J. Cell Biol. 152, 165-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, W., Cassara, J. & Weller, P. F. (2000) Blood 95, 1078-1085. [PubMed] [Google Scholar]
- 44.Stephens, L., Ellson, C. & Hawkins, P. (2002) Curr. Opin. Cell Biol. 14, 203-213. [DOI] [PubMed] [Google Scholar]
- 45.Perisic, O., Wilson, M. I., Karathanassis, D., Bravo, J., Pacold, M. E., Ellson, C. D., Hawkins, P. T., Stephens, L. & Williams, R. L. (2004) Adv. Enzyme Regul. 44, 279-298. [DOI] [PubMed] [Google Scholar]
- 46.Hiller, G., Sternby, M., Sundler, R. & Wijkander, J. (2000) Biochim. Biophys. Acta 1485, 163-172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.