Abstract
In higher plants, the circadian clock orchestrates fundamental processes such as light signaling and the transition to flowering. We isolated mutants of the circadian clock from an Arabidopsis thaliana mutagenized reporter line by screening for seedlings with long hypocotyl phenotypes and subsequently assaying for abnormal clock-regulated CAB2::LUC expression. This screen identified five mutant alleles of a clock gene, LUX ARRHYTHMO (LUX), that significantly affect amplitude and robustness of rhythms in both constant white light and dark conditions. In addition, the transition from vegetative to floral development is accelerated and hypocotyl elongation is accentuated in these mutants under light:dark cycles. We genetically mapped the mutations by bulk segregant analysis with high-density oligonucleotide array genotyping to a small putative Myb transcription factor related to other clock components and response regulators in Arabidopsis. The negative arm of the Arabidopsis circadian clock, CIRCADIAN CLOCK ASSOCIATED (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), is repressed in the lux mutants, whereas TIMING OF CAB2 EXPRESSION (TOC1) is activated. We demonstrate that CCA1 and LHY bind to the evening element motif in the LUX promoter, which strongly suggests that these proteins repress LUX expression, as they do TOC1. The data are also consistent with LUX being necessary for activation of CCA1 and LHY expression.
Keywords: Arabidopsis circadian clock, oligonucleotide array mapping
Many biological processes occur in a rhythmic manner within the span of a day. These events, which include mammalian sleep-wake and body temperature cycles, plant leaf movement, and fungal sporulation, coincide with daily environmental cues such as light:dark and temperature cycles and are referred to as circadian rhythms. Circadian rhythms persist with a period close to 24 h in the absence of environmental time cues and are regulated by an internal timing mechanism. Even though the circadian clocks of bacteria, fungi, plants, and animals may have evolved independently, all are comprised of autoregulatory feedback loops based upon transcription and regulated protein turnover (1). An initial model of the Arabidopsis circadian clock consists of a single negative feedback loop comprised of two morning-expressed Myb transcription factors, CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), and the evening-expressed pseudoresponse regulator TIMING OF CAB2 EXPRESSION 1 (TOC1) (2–4). In this model, TOC1 feeds back to either directly or indirectly regulate CCA1 and LHY, which in turn suppress TOC1 expression by binding to its regulatory region (5). The effect of a loss-of-function mutation of any one of them is a pervasive short circadian period, whereas overexpression results in arrhythmicity or dampened amplitude in constant conditions, suggesting that they are essential machinery of the circadian clock (2, 4, 6–10). Their relationship in the model was proposed based on three observations. TOC1 expression is dampened below trough levels when either CCA1 or LHY are overexpressed (5). Reciprocally, the expression of CCA1 and LHY is reduced in the TOC1 loss-of-function mutant, toc1-2 (5). Lastly, both CCA1 and LHY directly bind to a promoter motif (AAATATCT) called the evening element within the TOC1 promoter, likely repressing expression (5, 11). Although intuitively compelling in its first approximation, the single negative feedback loop model is inconsistent with the reduction of CCA1 and LHY observed in TOC1-overexpressing lines as wellasin zeitlupe (ztl) mutants, which are defective in TOC1 protein turnover (12–14). Additionally, the double cca1/lhy mutant is not completely arrhythmic (7, 8). Recently, a mathematical model of the single negative feedback loop predicts a long period for either CCA1 or LHY mutants where, experimentally, they have a short period (15). Thus, the Arabidopsis clock model is very likely to lack essential components and additional circuitry.
Like TOC1, ZTL, CCA1, and LHY, mutations in several other genes including the PSEUDO-RESPONSE REGULATORs(PRR3, PRR5, PRR7, PRR9), GIGANTEA (GI), and TEJ result in a change in circadian period length, but rhythms remain robust (see reviews in refs. 16 and 17). The effect of mutations in some clock-associated genes, such as EARLY FLOWERING 3 (ELF3), EARLY FLOWERING 4 (ELF4), and TIME FOR COFFEE (TIC), results in an erratic period and loss of amplitude up to the point of complete arrhythmicity (18–20). In some cases, the mutant phenotype is conditional. For example, toc1-2 has a short period in CAB2::LUC (≈20 h) under all free run conditions except that it is arrhythmic in monochromatic red light (21). Under constant light (LL), the elf3-1 mutant seems arrhythmic but in constant darkness (DD) maintains a WT period (20, 22). ELF3 has been implicated as a regulator of light input into the oscillator (23); however, expression amplitude in elf3-1, measured by CCR2::LUC, is severely dampened in DD as compared with WT (22). Therefore, normal rhythmicity is disturbed in the mutant, which suggests that the role of ELF3 goes beyond mediating light signaling into the clock (22). In the absence of ELF4 or TIC, plants exhibit erratic rhythms with highly variable period and amplitude in both LL and DD (18, 19). The arrhythmic or erratic behavior caused by mutations in ELF3, ELF4, and TIC falls short of unconditional and unequivocal arrhythmia, leaving their precise role in the clock uncertain.
Even though the organization of the regulatory networks responsible for sustaining circadian oscillations seems similar across eukaryotes, the properties of the individual components are different (1). For example, the functional domains of key mammalian clock proteins include basic/helix–loop–helix (bHLH), basic leucine zipper (bZip), and Per-ARNT-Sim (PAS) (24). On the other hand, key components in the plant clock seem to be Myb DNA-binding proteins, PAS-containing proteins, PRRs, and other proteins with uncharacterized functional domains. In this report, we describe the characterization and cloning of lux arrhythmo (lux, from the Latin word for “light”), an Arabidopsis mutant with a severely compromised circadian clock. Analogous to CCA1 and LHY, the predicted LUX protein contains a single Myb DNA-binding domain (type SHAQKYF) unique to plants. LUX is coregulated with TOC1 and seems to be repressed by CCA1 and LHY by direct binding of these proteins to the evening element in the LUX promoter.
Materials and Methods
Hypocotyl Assay. Hypocotyl assay was conducted as described (25). Values reported are the mean ± 95% confidence interval of 8–16 seedlings.
Bioluminescence Assays. lux arrhythmo mutants were isolated from ethyl-methane sulfonate (EMS) M2 accession Columbia (Col) carrying the CAB2::LUC reporter (26). Seedlings were grown under 12 h light:12 h dark cycles with 60 μmol of photons per m2 per s of cool white fluorescent light at 22°C. Analysis of bioluminescence rhythms under continuous light was performed as described (6). Fast fourier transform nonlinear least square (FFT-NLLS) fit analysis was used to estimate period lengths of individual traces (27). Plants homozygous for the lux-1 mutation and WT were transformed with the CCR2::LUC reporter (3).
Leaf Movement Analysis. Seedlings were grown under 12 h light:12 h dark cycles on MS-agar plates for 6 days and then released to constant 50 μmol·m-2·s-1 white fluorescent light. Time-lapse video recorded cotyledon position every 20 min starting on day 6 for 4–5 days (20). FFT-NLLS analysis was used to estimate period lengths of individual traces (27).
Bulk Segregant Analysis. Bulk segregant analysis was performed as described (28). Mutant and WT pools were comprised of a single leaf from 50 3-week-old F2 seedlings, respectively. The associated array CEL files and R script are available from the authors upon request.
Expression Analysis by RT-PCR. Seedlings were grown for 15–20 days in 12 h light:12 h dark cycles before samples were taken every 3 or 4 h in the light regime indicated. RNA was extracted by using the Qiagen (Valencia, CA) RNeasy Plant Mini Kit. First-strand cDNA was synthesized by using 2 μg of total RNA as template with the Invitrogen SuperScript first-strand synthesis system for RT-PCR kit. The cDNA was diluted 6× with water, and 2 μl were used for PCR amplification by using a Bio-Rad real-time detection system with the following primers and probes: TOC1, forward (F), 5′-TCTTCGCAGAATCCCTGTGAT-3′; reverse (R), 5′-GCTGCACCTAGCTTCAAGCA-3′; probe, 5′-ATGATGTCGAGGCAAGACGAAGTCCC-3′; CCA1, F, 5′-CCAGATAAGAAGTCACGCTCAGAA-3′; R, 5′-GTCTAGCGCTTGACCCATAGCT-3′; probe, 5′-TCTCCAAGGTAGAGAAAGAGGCTGAAGCTAAAGGT-3′; LHY, F, 5′-GACTCAAACACTGCCCAGAAGA-3′; LHY, R, 5′-CGTCACTCCCTGAAGGTGTATTT-3′; probe, 5′-AATCTTGTGGACCGCTCATCCTGTGG-3′. The gene IPP2 (isopentenyl pyrophosphate:dimethyl-allyl pyrophosphate isomerase, At3g02780) and APX3 (ascorbate peroxidase 3, At4g35000) were used as normalization controls, with the following set of primers and probe: IPP2, F, 5′-GTATGAGTTGCTTCTCCAGCAAAG-3′; R, 5′-GAGGATGGCTGCAACAAGTGT-3′; probe, 5′-AAACACAAAGGTTACGTTCCCTCTAGTGTGGACT–3′; APX3, F, 5′-GCCGTGAGCTCCGTTCTCT–3′; R1, 5′-TCGTGCCATGCCAATCG-3′; probe, 5′-CGCGAACAAGAACTGTGCTCCTATCATG-3′. Each PCR was repeated three times.
Expression Analysis Using DNA Microarrays. Seedlings (accession Col) were grown for 7 days in 12 h light:12 h dark cycles under white fluorescents at 120–125 μmol·m-2·s-1 before release to constant light. Samples were collected every 4 h. In a separate experiment, Landsberg erecta (Ler) seedlings were grown in short day (8 h light:16 h dark; 180 μmol·m-2·s-1) or long day (16 h light:8 h dark; 90 μmol·m-2·s-1) conditions for 7 days. Seedlings were collected every 4 h the following 2 days. RNA was extracted as described above. Samples and ATH1 GeneChips were processed according to standard Affymetrix (Santa Clara, CA) protocol. Probe level analysis of the array images was performed collectively disregarding mismatch probes by using a model-based approach (29).
EMSA. EMSA was performed as described (5) with the following probes: LUX EE sense, 5′-TGGTTAGAAGATCTAAAATATCTAGCTAACTCG-3′; LUX EE antisense, 5′-AGGACGAGTTAGCTAGATATTTTAGATCTTCTAA-3′; LUX EE mut sense, 5′-TGGTTAGAAGATCTAAAATGCTCAGCTAACTCG-3′; LUX EE mut antisense, 5′-AGGACGAGTTAGCTGAGCATTTTAGATCTTCTAA-3′. Mutated sequences are indicated by italics.
Results
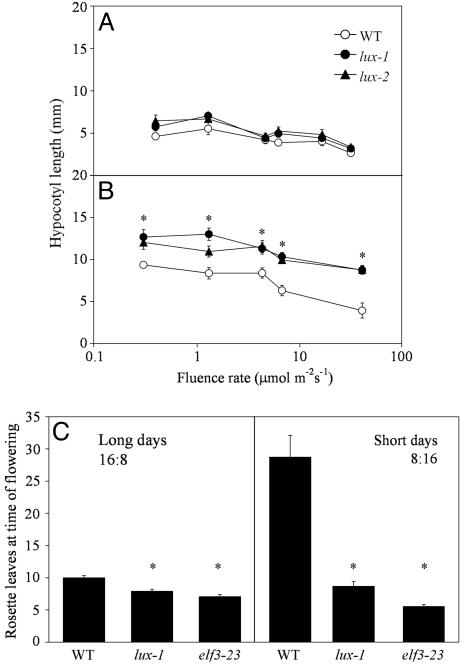
lux Seedlings Exhibit Long Hypocotyl Under Light:Dark Cycles. Mutations affecting the circadian clock go beyond abnormalities in the basic waveform of gene expression; for example, the circadian clock regulates hypocotyl elongation in Arabidopsis, arresting growth at dawn and resuming growth at dusk (30), such that all known clock mutants exhibit aberrant hypocotyl length. In an effort to facilitate the screening for circadian clock mutants, an ethyl-methane sulfonate-mutagenized reporter line was screened for seedlings with long hypocotyl phenotypes and then subsequently assayed for abnormal clock-regulated CAB2::LUC expression (26). Several long hypocotyl mutants were identified and found to have severely compromised rhythms in CAB2::LUC expression under LL. Hypocotyl length was determined for lux seedlings under various fluence rates of LL, 12-h light:12-h dark (LD), and DD. At all fluence rates tested in LD cycles, two different alleles, lux-1 and lux-2, had significantly elongated hypocotyls (Fig. 1B). On the other hand, lux seedlings do not have significantly long hypocotyls in continuous dark (13.44 ± 0.57 mm vs. WT 12.48 ± 1.62 mm) or continuous light (Fig. 1A), regardless of light intensity. These results are similar to that observed for elf3 and elf4 (19) and suggest that differences in hypocotyl length in the lux background are dependent on light:dark cycles.
Fig. 1.
lux seedlings exhibit long hypocotyls and early flowering under light:dark cycles. WT and lux mutants grown for 7 days in either continuous white light (A) or 12 h light:12 h dark cycles (B) at the fluence rate indicated. Each experiment was performed at least twice with similar results. Average ± 95% confidence interval, n = 8–16. *, P < 0.001 (Student's two-tail t test). (C) Average number of rosette leaves (±95% confidence interval) at floral initiation (≈1 cm high inflorescence) measured for WT, lux-1, and elf3-23 in long-day (16 h light:8 h dark) and short-day conditions (8 h light:16 h dark), n = 30–100. The elf3-23 allele has a missense mutation (P667L) that causes a similar mutant phenotype as elf3-1 (28).
lux Is Early Flowering in Long and Short Days. The transition from vegetative to reproductive growth in Arabidopsis is long-dayfacultative. Day length perception is mediated by the circadian clock such that plants grown in long-day conditions (16 h light:8 h dark) flower earlier than plants grown in short days (8 h light: 16 h dark) (31). In addition to a long hypocotyl, lux mutants flower early in short- and long-day conditions (Fig. 1C). These phenotypes were similar to the conditional clock/flowering time mutant elf3, although not quite as severe (Fig. 1C). All three genotypes tested (Col, elf3, and lux) were different for rosette leaves at time of flowering in long days than in short days (P < 0.05).
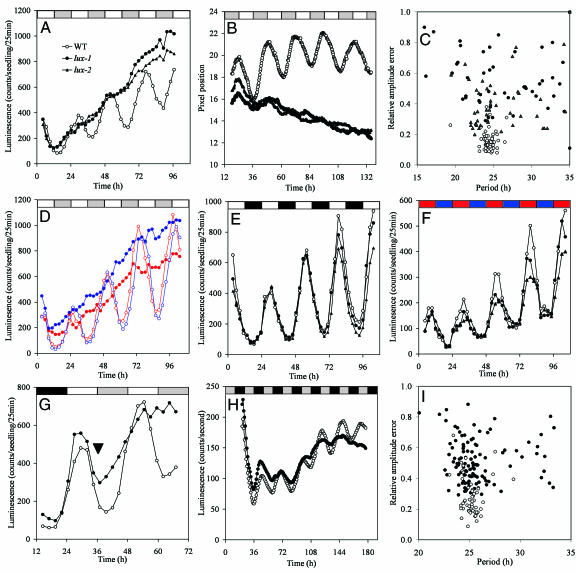
LUX Is Required for Rhythmicity of Multiple Circadian Outputs in Constant Light Conditions. Several mutants were found to have arrhythmic CAB2::LUC expression in LL and were named lux arrhythmo (lux) (Fig. 2A). Analysis of CAB2::LUC traces from lux seedlings using FFT-NLLS (27) indicated the severity of the circadian defect in this mutant: none of the lux seedlings were scored as rhythmic [all relative amplitude error values (RAE) were ≥0.6, n = 48] whereas all of the WT controls were rhythmic (RAE values were <0.6, n = 48). The arrhythmic phenotype was also observed in monochromatic red or blue light (Fig. 2D). Rhythmic leaf movement has been used extensively as a reporter of the circadian clock and is distinct from gene expression rhythms (20). The pervasive effect of the lux mutations on overall circadian rhythms was marked by highly variable period length and RAE for rhythmic leaf movement in lux-1 and lux-2 (Fig. 2 B and C). On the other hand, in the presence of external cues such as light:dark (Fig. 2E) and temperature cycles (Fig. 2F), CAB2::LUC expression in the lux mutants was similar to WT plants, indicating that either clock dysfunction was masked or the external cues are capable of generating and maintaining normal rhythms. To determine the point at which rhythms were disrupted, CAB2::LUC expression was monitored under light:dark cycles before release to constant light. In WT plants, CAB2::LUC expression continued to decrease during the first hour of subjective dark as indicated by a decrease in luminescence between the first two subjective dark time points (Fig. 2G). In lux-1, expression increased during that time, suggesting that clock dysfunction occurred soon after subjective dusk and persisted in the absence of cues from a light:dark transition.
Fig. 2.
LUX is required for normal rhythmicity of multiple circadian outputs in constant light and dark conditions. Transgenic seedlings carrying the CAB2::LUC reporter were entrained under 12 h light:12 h dark cycles for 7 days. Luminescence was monitored in WT (○), lux-1 (•), and lux-2 (▴) under constant white (80 μmol·m-2·s-1) conditions (A) and constant red (55 μmol·m-2·s-1) and blue (62 μmol·m-2·s-1) light conditions (D), average of 24–48 seedlings each. (B) Leaf movement rhythms were monitored under constant white light. (C) Period length versus relative amplitude error (RAE) was calculated by using FFT-NLLS and plotted. WT, lux-1, and lux-2 were similar when entrained and monitored under 12 h light:12 h dark and constant temperature (E) or temperature cycles (12 h 22°C:12 h 12°C) in constant light (F). (G) The transition from light:dark cycles to constant white light conditions. The black arrow indicates the first time point after subjective dusk is where lux-1 and WT CAB2::LUC expression diverges. Transgenic seedlings carrying the CCR2::LUC reporter were entrained under 12 h light:12 h dark cycles for 7 days. Luminescence was monitored in WT (○) and lux-1 (•) under constant darkness (H) and RAE (I). Shown is the average of 12 T3 seedlings from four independent transformation events for a total of 48 WT and 48 lux-1 seedlings.
LUX Is Required for Normal Rhythmicity in Constant Dark Conditions. The plant circadian clock is intimately tied to the regulation of light-signaling pathways. Light also entrains the clock and effects robustness of circadian rhythms (32, 33) while, at the same time, light perception is circadian-regulated (23). The rhythmic expression of CAB2::LUC dampens in constant darkness as does the expression of many other clock-associated genes: CCA1, LHY, TOC1, ELF3, and ELF4 (3, 4, 19, 22). Regardless, circadian rhythms persist in constant darkness and can be measured by using the CCR2::LUC reporter line (3). As with CAB2::LUC and leaf movement, CCR2::LUC expression was severely compromised in lux-1 under constant light (Fig. 5A, which is published as supporting information on the PNAS web site). Only three quarters of the lux-1 plants gave a CCR2::LUC period in LL, and merely 26% of these had RAE values that are <0.6 (Fig. 5B). On the contrary, all but one of the WT seedlings returned a circadian period. As expected, the rhythmicity of CCR2::LUC in lux-1 was normal in the presence of external light:dark cues (Fig. 5C). Although the effects of the lux-1 mutation on rhythmic CCR2::LUC expression in constant dark were not quite as strong as in constant light, the rhythms were still markedly compromised (Fig. 2H). Only 80% of the lux-1 plants gave any period, compared with 98% of the WT seedlings. Among the seedlings with a period estimate, all but one of the RAE values for WT seedlings were <0.6, and considerably more (25%) of RAEs for lux-1 seedlings were greater than 0.6 (Fig. 2I). This observation was similar to the conditionally arrhythmic elf3 mutant, in which amplitude and rhythmicity is also compromised in constant dark but to a lesser extent than in constant light (22).
Genetic Linkage Mapping of the Arabidopsis LUX ARRHYTHMO Locus. lux mutants in the Col background were crossed to the Ler accession to obtain a segregating population for mapping purposes. The segregation of arrhythmic individuals in the F2 generation was consistent with a single recessive mutation (χ2 = 0.000637, P > 0.979, n = 523). The mutation was mapped by bulk segregant analysis (34) by using high-density oligonucleotide arrays as a genotyping tool (35). Genomic DNA hybridization intensities were compared among three replicates of Col and Ler. Eight thousand single-feature polymorphisms were identified, among the 202,806 unique array 25mer features, where Ler alleles had significantly lower hybridization intensity than Col alleles. Single-feature polymorphisms function as molecular markers for genetic linkage mapping and can be simultaneously genotyped in a single assay. The array genotype of a pool of 50 F2 arrhythmic individuals was compared with a pool of 50 F2 WT individuals. The pools were expected to have equal mixtures of Col and Ler genotypes at loci unlinked to lux and therefore exhibit hybridization intensity intermediate to that of the parent genotypes. In the region of the mutation, the mutant pool was enriched for Col alleles, and the WT phenotype pool was enriched for Ler alleles. The peaks of the bulk segregant analysis for three populations of lux mutants placed the mutation at 17.92, 15.96, and 19.59 Mb on chromosome three (Fig. 6A, which is published as supporting information on the PNAS web site). Further recombination analysis was performed in 181 arrhythmic F2s by using single-locus PCR markers (Fig. 6B). In a mapping interval between 17.17 and 17.78 Mb, we identified the circadian-regulated candidate gene At3g46640, a putative Myb transcription factor with a single DNA-binding domain (type SHAQKYF) unique to plants. Sequencing of At3g46640 in independently isolated lux mutants identified five unique alleles with four nucleotide conversions creating premature stop codons and one in the Myb DNA binding domain changing a proline to a leucine (Fig. 6C). lux-3, lux-4, and lux-5 were not characterized beyond their original isolation as long hypocotyl seedlings and segregation analysis of the arrhythmic CAB2::LUC phenotype for genetic linkage mapping. No difference among the five mutant alleles was observed. Based on amino acid sequence similarity, LUX is part of a small family of five genes (At3g46640, At5g59570, At5g05090, At3g10760, and At2g40970) each with a single Myb DNA-binding domain (Fig. 7A, which is published as supporting information on the PNAS web site). A gene on chromosome 5, At5g59570, is most similar (54%) to LUX. In particular, the DNA-binding domain found in this family is very similar to the binding domain found in several genes associated with two-component signal transduction systems (36): the B-type Arabidopsis response regulators (ARRs), GOLDEN2-LIKEs (GLKs), and PRR2 (Fig. 7B).
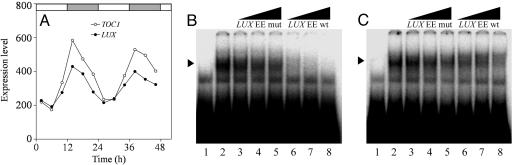
LUX Is a Putative Myb Transcription Factor Whose Expression Is Clock-Regulated. The two key clock genes, CCA1 and LHY, also have a single SHAQKYF-type Myb DNA-binding domain. It is likely that that domain is responsible for binding to the evening element motif in the TOC1 promoter, which in turn suppresses TOC1 expression (5, 11). The LUX promoter contains one evening element 250 bases upstream of the translational start site. Expression analysis revealed that these two genes, TOC1 and LUX, had very similar expression in several conditions. In WT plants released into constant white light (Fig. 3A), long-day and short-day conditions (Fig. 8 A and B, which is published as supporting information on the PNAS web site), the expression of TOC1 and LUX was highly correlated (r = 0.98, 0.96, and 0.96, respectively). The expression of TOC1 and LUX is also remarkably similar in different mutant backgrounds. In 12 h light:12 h dark conditions, LHY accumulation peaks in the early morning; on the other hand, in the LHY overexpressing line (lhy), LHY protein accumulates during the day and decays in the dark (37). As expected, a shift in peak expression of both TOC1 and LUX from dusk (lights off) to morning (lights on) in lhy was observed (Fig. 9 A and B, which is published as supporting information on the PNAS web site). Similarly, the expression of both TOC1 and LUX is shifted in the CCA1-overexpressing line under the same conditions (Fig. 9 C and D). In addition, like TOC1 (5), the expression of LUX was arrhythmic in the elf3-1 mutant (data not shown). These data suggest that TOC1 and LUX share a common regulatory mechanism, which in the case of TOC1 is known to rely on repression elicited by the specific binding of CCA1 and LHY to the evening element present in this promoter. Consistent with this paradigm is the constitutive high expression of TOC1 in the lhy12 cca1-1 double mutant in constant light (7). Similarly, LUX and TOC1 repression is reduced in the cca1-1 lhy-R (8) double mutant under constant conditions (Fig. 9 E and F).
Fig. 3.
The expression of the core clock component TOC1 and LUX are coregulated. (A) Complementary RNA samples were hybridized to high-density oligonucleotide microarrays. Expression of TOC1 (○) and LUX (•) in plants entrained under 12 h light:12 h dark cycles for 7 days and released into constant light. EMSAs revealed that GST-LHY (B) and GST-CCA1 (C) proteins specifically bind the evening element in the LUX promoter. Labeled probe was incubated in the absence (lane 1) or presence (lanes 2–8) of protein extracts. No competitor DNA was added in lane 2. Mutant (lanes 3–5) and WT (lanes 6–8) LUX evening element oligonucleotides (-270/-240) were used as unlabeled competitors as indicated. Black triangles (top) represent increasing amount of competitor, which was present at 50× (lanes 3 and 6), 100× (lanes 4 and 7), and 200× (lanes 5 and 8) molar excess of the labeled probe. The arrowhead indicates the specific protein–DNA complexes.
To further explore the possibility of this regulatory mechanism, EMSAs were performed to determine whether LHY and CCA1 bind to the evening element of the LUX promoter (Fig. 3 B and C). Extracts of Escherichia coli expressing either GST-LHY or GST-CCA1 provided with a radio-labeled probe corresponding to the -270/-240 region of the LUX promoter, which contains the evening element, produced DNA species with retarded mobility. On the other hand, bacterial extracts harboring the empty GST vector did not produce a comparable shifted species (data not shown). Competition experiments showed that GST-LHY and GST-CCA1 specifically bind to a DNA fragment corresponding to this part of the promoter (Fig. 3 B and C). Taken together, these data suggest that repression of LUX expression is mediated by the direct binding of LHY and CCA1 to the evening element present in its promoter.
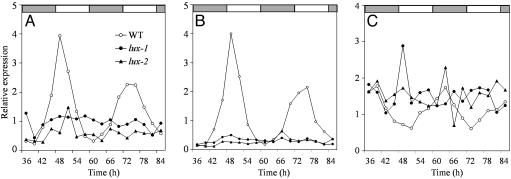
In Constant Light, LUX Is Required for both Activation of CCA1 and LHY and Rhythmic Expression of TOC1. CCA1 and LHY are known to act at the evening element within the TOC1 promoter to repress gene expression as part of the negative arm of the core clock feedback loop (5). LUX seems to be negatively regulated by this same set of proteins, which also contribute to TOC1 expression, suggesting that LUX may participate in the clock in a manner analogous to TOC1. To determine whether LUX contributes to the activation of CCA1 and LHY, the expression of CCA1 and LHY was examined in lux mutants. Consistent with LUX having a positive role in the expression of these two transcription factors, the expression of both CCA1 and LHY was clamped low in lux mutants (Fig. 4 A and B). In addition, TOC1 was arrhythmic and clamped moderately high (Fig. 4C) in lux-1 and lux-2, as would be expected if CCA1 and LHY were expressed at a low and constant level. Although this analysis does not conclusively demonstrate that LUX directly activated LHY and CCA1 expression, it does suggest that at least one function of LUX is to have a positive influence on the expression of these two genes.
Fig. 4.
In constant light, LUX is required for activation of CCA1 and LHY and rhythmic expression of TOC1. Shown is expression of clock-associated genes in WT (○), lux-1 (•), and lux-2 (▴), entrained under 12 h light:12 h dark cycles for 14 days and released to constant light. Shown are data from two independent experiments. mRNA was assayed by real-time RT-PCR for the abundance of CCA1 (A), LHY (B), and TOC1 (C) mRNA relative to internal IPP2 (At3g02780) and APX3 (At4g35000) controls.
Discussion
It is clear from detailed analysis of lux mutants that LUX is required for normal rhythmic expression of multiple clock outputs in both constant light and darkness. The mutant phenotypes are similar to a group of circadian clock mutants that includes elf3, elf4, and tic. Loss of each of these genes produced defects in circadian amplitude and period length (18–20). Measurements of circadian-regulated gene expression suggest that the circadian defects in this group of mutants occurs only in the absence of external cues, and that signals such as light and temperature can drive rhythms in this class of mutants (18, 23). In lux-1, expression of the CAB2::LUC and CCR2::LUC reporters is remarkably similar to WT when seedlings are exposed to either light or temperature cycles. When lux-1 seedlings are released into constant light, CAB2::LUC diverges from WT only after the point of anticipated dark, which correlates with peak LUX expression. On the other hand, in short- and long-day conditions where external cues are provided, lux, elf3, and elf4 exhibit abnormalities in more complex traits such as hypocotyl elongation and flowering time, which are in part circadian clock-regulated (18, 30, 31). Therefore, the clock is either perturbed in a manner that is not detected by measuring circadian-regulated gene expression or these genes act on flowering time and hypocotyl elongation independent of the circadian clock.
There are striking similarities among the circadian clocks of animals, plants, and fungi (1). Each one derives rhythmicity from interlocking autoregulatory feedback loops based upon transcription and regulated protein turnover. In contrast to similarity in design, the components that make up the different clocks have distinct properties. For example, in the mammalian clock, transcriptional activation and repression within the central oscillator are caused by direct binding of bHLH and basic leucine zipper (bZip) transcription factors to the E/E′-box and D-box promoter motifs, respectively (24). Although there are large families of both bZip (38) and bHLH (39) proteins in Arabidopsis, and even though several bHLH proteins of the PHYTOCHROME-INTERACTING FACTOR (PIF) and PIF3-LIKE families seem to interact with TOC1, no circadian defects have been detected in mutants of either family (40). In plants, it is Myb DNA-binding proteins, namely CCA1, LHY, and now LUX, that seem to be important for transcriptional regulation within the circadian clock. These proteins are part of a unique subfamily of putative Myb transcription factors with only a single Myb domain, because it is more common to find two or three Myb domain repeats in a typical member of this super family (41). The single Myb domains found in CCA1, LHY, and LUX are similarly classified as SHAQKYF-type, but LUX is not part of the circadian-associated CCA1/LHY gene family (42, 43). LUX has a distinct gene structure, sequence, and regulation, as well as mutant phenotypes. The LUX DNA-binding domain shares a great deal of similarity to the Myb domain found in a family of genes known as B-type response regulators (ARRs) (36). This family of genes participate in multiprotein signal transduction by means of transfer of a phosphate by a His kinase or His phosphotranfer to the receiver domain of the response regulator (44). Two other subfamilies of response regulators include the A-type, which do not have a DNA-binding domain and the circadian clock-associated PRRs (45). Although the possibility exists, there is currently no evidence relating to circadian rhythms for a direct functional connection between the response regulators (ARRs or PRRs) and LUX/ARR-type Myb DNA-binding domains.
The identification of LUX as a putative Myb transcription factor that is required for CCA1/LHY expression may help to resolve some of the inadequacies of the single CCA1/LHY-TOC1 negative feedback loop model (15). For example, the core clock component TOC1 has been established as an important factor in maintaining a 24-h period in free run conditions as well as up-regulation of CCA1 and LHY expression (5, 7), yet this protein lacks a known DNA-binding motif (3). Detailed analysis of LUX temporal expression patterns indicated that it is tightly coregulated with TOC1. In addition, the LUX promoter contains an evening element, which serves as a binding site for two other key clock components, CCA1 and LHY (11). These two transcription factors are known to act at the evening element within the TOC1 promoter to repress gene expression as part of the negative arm of the core clock feedback loop (5). Thus, LUX seems to be negatively regulated by this same set of proteins in a manner analogous to TOC1.
In lux mutants, the expression of both CCA1 and LHY was clamped low, which suggests that LUX is required for their expression. Reciprocally, TOC1 was arrhythmic and clamped moderately high, the expected results if LUX were in fact necessary for activation of CCA1 and LHY. However, it is important to keep in mind that an increase in CCA1/LHY expression has never been observed in any loss-of-function or gain-of-function clock mutant. On the other hand, in addition to lux, a decrease in CCA1 and/or LHY expression has been observed in elf3, elf4, toc1, gi, and prr5/prr7 (2, 5, 7, 19, 46). Taken together, these results suggest that, while LUX plays an essential role in the circadian clock, possibly contributing to up-regulation of CCA1 and LHY, its addition to the Arabidopsis clock model does not simply “close the loop.” Future experiments will be directed toward determining the direct target(s) of LUX and how it is associated with the circadian clock.
Supplementary Material
Acknowledgments
We thank Todd Michael, Franklin G. Harmon, Ghislain Breton, Takato Imaizumi, and Eva Farre for helpful discussion and critical reading of this manuscript. We thank H. Bird Richardson for expert assistance with figure formatting. We acknowledge members of The Scripps Research Institute DNA Microarray Core Facility and Steve Head, Marcelo Yanovsky, Stacey Harmer, and Mike Covington for expert assistance. This work was supported by National Institutes of Health (NIH) Grants GM56006 and GM67837 (to S.A.K.) and by a Ruth L. Kirschstein NIH Postdoctoral Fellowship (to S.P.H.). This is manuscript 16917-CB of The Scripps Research Institute.
Author contributions: S.P.H., T.F.S., J.O.B., and S.A.K. designed research; S.P.H., T.F.S., and J.L.P.-P. performed research; S.P.H., T.F.S., J.L.P.-P., J.O.B., J.R.E., and S.A.K. contributed new reagents/analytic tools; S.P.H. and J.O.B. analyzed data; and S.P.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CCA1, CIRCADIAN CLOCK ASSOCIATED; LHY, LATE ELONGATED HYPOCOTYL; TOC1, TIMING OF CAB2 EXPRESSION; PRR, PSEUDO-RESPONSE REGULATOR; ELF, EARLY FLOWERING; LL, constant light; DD, constant darkness; bHLH, basic/helix–loop–helix; FFT-NLLS, fast fourier transform nonlinear least square; RAE, relative amplitude error; Ler, Landsberg erecta.
References
- 1.Young, M. W. & Kay, S. A. (2001) Nat. Rev. Genet. 2, 702-715. [DOI] [PubMed] [Google Scholar]
- 2.Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I. A. & Coupland, G. (1998) Cell 93, 1219-1229. [DOI] [PubMed] [Google Scholar]
- 3.Strayer, C., Oyama, T., Schultz, T. F., Raman, R., Somers, D. E., Mas, P., Panda, S., Kreps, J. A. & Kay, S. A. (2000) Science 289, 768-771. [DOI] [PubMed] [Google Scholar]
- 4.Wang, Z. Y. & Tobin, E. M. (1998) Cell 93, 1207-1217. [DOI] [PubMed] [Google Scholar]
- 5.Alabadi, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Mas, P. & Kay, S. A. (2001) Science 293, 880-883. [DOI] [PubMed] [Google Scholar]
- 6.Somers, D. E., Webb, A. A. R., Pearson, M. & Kay, S. A. (1998) Development (Cambridge, U.K.) 125, 485-494. [DOI] [PubMed] [Google Scholar]
- 7.Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H. R., Carre, I. A. & Coupland, G. (2002) Dev. Cell 2, 629-641. [DOI] [PubMed] [Google Scholar]
- 8.Alabadi, D., Yanovsky, M. J., Mas, P., Harmer, S. L. & Kay, S. A. (2002) Curr. Biol. 12, 757-761. [DOI] [PubMed] [Google Scholar]
- 9.Green, R. M. & Tobin, E. M. (1999) Proc. Natl. Acad. Sci. USA 96, 4176-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsushika, A., Makino, S., Kojima, M., Yamashino, T. & Mizuno, T. (2002) Plant Cell Physiol. 43, 118-122. [DOI] [PubMed] [Google Scholar]
- 11.Harmer, S. L., Hogenesch, L. B., Straume, M., Chang, H. S., Han, B., Zhu, T., Wang, X., Kreps, J. A. & Kay, S. A. (2000) Science 290, 2110-2113. [DOI] [PubMed] [Google Scholar]
- 12.Makino, S., Matsushika, A., Kojima, M., Yamashino, T. & Mizuno, T. (2002) Plant Cell Physiol. 43, 58-69. [DOI] [PubMed] [Google Scholar]
- 13.Somers, D. E., Kim, W. Y. & Geng, R. (2004) Plant Cell 16, 769-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mas, P., Kim, W. Y., Somers, D. E. & Kay, S. A. (2003) Nature 426, 567-570. [DOI] [PubMed] [Google Scholar]
- 15.Locke, J. C. W., Millar, A. J. & Turner, M. S. (2005) J. Theor. Biol. 234, 383-393. [DOI] [PubMed] [Google Scholar]
- 16.Eriksson, M. E. & Millar, A. J. (2003) Plant Physiol. 132, 732-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salome, P. A. & McClung, C. R. (2004) J. Biol. Rhythms 19, 425-435. [DOI] [PubMed] [Google Scholar]
- 18.Hall, A., Bastow, R. M., Davis, S. J., Hanano, S., McWatters, H. G., Hibberd, V., Doyle, M. R., Sung, S. B., Halliday, K. J., Amasino, R. M., et al. (2003) Plant Cell 15, 2719-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doyle, M. R., Davis, S. J., Bastow, R. M., McWatters, H. G., Kozma-Bognar, L., Nagy, F., Millar, A. J. & Amasino, R. M. (2002) Nature 419, 74-77. [DOI] [PubMed] [Google Scholar]
- 20.Hicks, K. A., Millar, A. J., Carre, I. A., Somers, D. E., Straume, M., MeeksWagner, D. R. & Kay, S. A. (1996) Science 274, 790-792. [DOI] [PubMed] [Google Scholar]
- 21.Mas, P., Alabadi, D., Yanovsky, M. J., Oyama, T. & Kay, S. A. (2003) Plant Cell 15, 223-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Covington, M. F., Panda, S., Liu, X. L., Strayer, C. A., Wagner, D. R. & Kay, S. A. (2001) Plant Cell 13, 1305-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McWatters, H. G., Bastow, R. M., Hall, A. & Millar, A. J. (2000) Nature 408, 716-720. [DOI] [PubMed] [Google Scholar]
- 24.Ueda, H. R., Hayashi, S., Chen, W., Sano, M., Machida, M., Shigeyoshi, Y., Iino, M. & Hashimoto, S. (2005) Nat. Genet. 37, 187-192. [DOI] [PubMed] [Google Scholar]
- 25.Somers, D. E., Schultz, T. F., Milnamow, M. & Kay, S. A. (2000) Cell 101, 319-329. [DOI] [PubMed] [Google Scholar]
- 26.Millar, A. J., Carre, I. A., Strayer, C. A., Chua, N. H. & Kay, S. A. (1995) Science 267, 1161-1163. [DOI] [PubMed] [Google Scholar]
- 27.Plautz, J. D., Straume, M., Stanewsky, R., Jamison, C. F., Brandes, C., Dowse, H. B., Hall, J. C. & Kay, S. A. (1997) J. Biol. Rhythms 12, 204-217. [DOI] [PubMed] [Google Scholar]
- 28.Hazen, S. P., Borevitz, J. O., Harmon, F. G., Pruneda-Paz, J. L., Schultz, T. F., Yanofsky, M., Liljegren, S. J., Ecker, J. R. & Kay, S. A. Plant Physiol. 138, 990-997. [DOI] [PMC free article] [PubMed]
- 29.Li, C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98, 31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dowson-Day, M. J. & Millar, A. J. (1999) Plant J. 17, 63-71. [DOI] [PubMed] [Google Scholar]
- 31.Yanovsky, M. J. & Kay, S. A. (2003) Nat. Rev. Mol. Cell. Biol. 4, 265-275. [DOI] [PubMed] [Google Scholar]
- 32.Devlin, P. F. & Kay, S. A. (2001) Annu. Rev. Physiol. 63, 677-694. [DOI] [PubMed] [Google Scholar]
- 33.Salome, P. A. & McClung, C. R. (2005) Plant Cell Envviron. 28, 21-38. [Google Scholar]
- 34.Michelmore, R. W., Paran, I. & Kesseli, R. V. (1991) Proc. Natl. Acad. Sci. USA 88, 9828-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borevitz, J. O., Liang, D., Plouffe, D., Chang, H. S., Zhu, T., Weigel, D., Berry, C. C., Winzeler, E. & Chory, J. (2003) Genome Res. 13, 513-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang, I., Chen, H. C. & Sheen, J. (2002) Plant Physiol. 129, 500-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, J. Y., Song, H. R., Taylor, B. L. & Carre, I. A. (2003) EMBO J. 22, 935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T. & Parcy, F. (2002) Trends Plant Sci. 7, 106-111. [DOI] [PubMed] [Google Scholar]
- 39.Bailey, P. C., Martin, C., Toledo-Ortiz, G., Quail, P. H., Huq, E., Heim, M. A., Jakoby, M., Werber, M. & Weisshaar, B. (2003) Plant Cell 15, 2497-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamashino, T., Matsushika, A., Fujimori, T., Sato, S., Kato, T., Tabata, S. & Mizuno, T. (2003) Plant Cell Physiol. 44, 619-629. [DOI] [PubMed] [Google Scholar]
- 41.Stracke, R., Werber, M. & Weisshaar, B. (2001) Curr. Opin. Plant Biol. 4, 447-456. [DOI] [PubMed] [Google Scholar]
- 42.Carre, I. A. & Kim, J. Y. (2002) J. Exp. Bot. 53, 1551-1557. [DOI] [PubMed] [Google Scholar]
- 43.Kuno, N., Moller, S. G., Shinomura, T., Xu, X., Chua, N. H. & Furuya, M. (2003) Plant Cell 15, 2476-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imamura, A., Hanaki, N., Umeda, H., Nakamura, A., Suzuki, T., Ueguchi, C. & Mizuno, T. (1998) Proc. Natl. Acad. Sci. USA 95, 2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizuno, T. (2004) Curr. Opin. Plant Biol. 7, 499-506. [DOI] [PubMed] [Google Scholar]
- 46.Nakamichi, N., Kita, M., Ito, S., Sato, E., Yamashino, T. & Mizuno, T. (2005) Plant Cell Physiol. 46, 609-619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.