Abstract
Teichoic acids (TAs), and especially lipoteichoic acids (LTAs), are one of the main immunostimulatory components of pathogenic Gram-positive bacteria. Their contribution to the immunomodulatory properties of commensal bacteria and especially of lactic acid bacteria has not yet been investigated in detail. To evaluate the role of TAs in the interaction between lactic acid bacteria and the immune system, we analyzed the antiinflammatory properties of a mutant of Lactobacillus plantarum NCIMB8826 affected in the TA biosynthesis pathway both in vitro (mononuclear cells stimulation) and in vivo (murine model of colitis). This Dlt- mutant was found to incorporate much less d-Ala in its TAs than the WT strain. This defect significantly impacted the immunomodulation reactions induced by the bacterium, as shown by a dramatically reduced secretion of proinflammatory cytokines by peripheral blood mononuclear cells and monocytes stimulated by the Dlt- mutant as compared with the parental strain. Concomitantly, a significant increase in IL-10 production was stimulated by the Dlt- mutant in comparison with the WT strain. Moreover, the proinflammatory capacity of L. plantarum-purified LTA was found to be Toll-like receptor 2-dependent. Consistent with the in vitro results, the Dlt- mutant was significantly more protective in a murine colitis model than its WT counterpart. The results indicated that composition of LTA within the whole-cell context of L. plantarum can modulate proinflammatory or antiinflammatory immune responses.
Keywords: lactic acid bacteria, probiotic, immunonodulation, inflammation, cell-wall mutant
The human intestinal lumen houses a complex bacterial microflora constituted of >400 cultivable species (1). The microbiota established after birth is considered to be essential in priming the immune system during ontogeny, to limit dysfunctional responses (2). Recent evidence clearly demonstrated that commensal bacteria regulate intestinal development and function (3), and interruption of these interactions results in pathological features (4). Different factors have been reported to contribute to the protective function of gut microflora such as maintaining a physical barrier against colonization or invasion by pathogens, facilitating nutrient digestion and assimilation, and providing immunological surveillance signals at the gut mucosa-lumen interface (5). It is widely accepted that the intestinal microflora plays an important role in maintaining the health of the host and possesses immunomodulating capacities (6-10). Lactic acid bacteria (LAB) are normal inhabitants of the human gastrointestinal tract and are major components of the dominant flora in the small bowel. They are considered beneficial to the host and as such are being developed for probiotic applications (11, 12). The health-promoting properties attributed to LAB strains are multiple and include their capacity to interact with the host immune system. Notably, their successful use in the prevention of inflammatory bowel disease (IBD) has been described (13). Recent studies have reported the positive effects of probiotics not only in the prevention of colitis in animal models (14-16) but also in the maintenance of remission after surgery in IBD (17-19). We established that the in vivo protective effect of LAB in a murine 2,4,6-trinitrobenzenesulfonic acid (TNBS)-induced model of colitis is strain-specific (20, 21). Several LAB strains were reported to exert in vitro stimulatory properties on cells of the innate immune system, including macrophages and natural killer cells, and to induce the production of both proinflammatory cytokines, such as TNF-α and IL-12, and the antiinflammatory IL-10 cytokine (6, 7, 10, 22). Previous reports have suggested that Gram-positive bacteria tend to preferentially stimulate IL-12 secretion, whereas Gram-negative bacteria predominantly induce IL-10 from stimulated monocytes and macrophages (23). Nevertheless, substantial differences were found among LAB strains in their capacity to induce a specific cytokine profile (6-10). Because IL-10 plays a central role in down-regulating inflammatory cascades, strains capable of inducing this cytokine would likely be good candidates for use in IBD therapeutic intervention studies.
Little is known about the precise mechanisms by which lactobacilli may exert their beneficial effects. The increasing availability of genome sequences from intestinal species of LAB (24) opens new possibilities for studying their functionality in the gut. In particular, the complete genome sequence of Lactobacillus plantarum WCFS1 (a single colony of the L. plantarum NCIMB8826 strain) has been established (25), which allows detailed studies of the bacterial compounds involved in the interaction with the host immune system. Components of the Gram-positive bacterial cell wall, such as peptidoglycan and teichoic acid (TA) (especially lipoteichoic acid, LTA), have been shown to be involved in cytokine induction. For LTA, conflicting results have been reported, which might be explained by differences in the preparation protocols leading to more or less endotoxin contamination and loss of residues, especially d-Ala (26, 27). The quality and level of d-Ala substitution on TA were shown to be key factors for cytokine induction, as demonstrated by the synthesis of LTA substituted with l-Ala instead of d-Ala (28). In the same manner, a number of controversial reports on the involvement of Toll-like receptors (TLRs) (especially TLR-2 and TLR-4) in LTA-induced cell activation have been published. Most of these studies have been performed with endotoxin-contaminated LTAs. Nevertheless, the use of highly purified LTAs clearly demonstrated TLR-2 signaling in NF-κB activation, resulting in cytokine production (29, 30). The majority of the studies conducted so far focused on pathogenic streptococci and staphylococci. Therefore, further studies are needed to elucidate the active bacterial components responsible for the variable immunomodulating capacities of LAB. LTAs have been proposed to mediate adhesion of lactobacilli to epithelial cells (31, 32). LTAs were also recently shown to be a major mediator of the capacity of Lactobacillus strains to stimulate TNF-α from macrophages (33). To our knowledge, the importance of the specific composition of LTAs in the immunomodulating capacity of lactobacilli has not been analyzed in detail. In the present work, we compared a mutant of L. plantarum NCIMB8826 affected in its TA composition with its WT counterpart. A defect in the LTA d-Ala content profoundly affected the immunomodulation profile of the mutant. The mutant was considerably more antiinflammatory than the parental strain, both in vitro and in a murine colitis model.
Materials and Methods
Bacterial Strains and Growth Conditions. L. plantarum NCIMB8826 (National Collection of Industrial and Marine Bacteria, Aberdeen, U.K.) used in this study was isolated from human saliva. A mutant deficient in the d-alanylation of TAs was constructed by disrupting the dlt operon by single-step homologous recombination using a suicide knockout vector. To this end, an internal 596-bp fragment of dltB gene [lp_2018 (24)] was amplified by PCR from NCIMB8826 chromosomal DNA using primers NCIDLTB1 (5′-TTGGTACCGCTGTCGTTAAGATCACACC-3′) and NCIDLTBR2 (5′-TCGCCCGGGCAACCGCATGTAGATGAAATCACG-3′). The amplicon was cloned into pCR2.1 plasmid (Invitrogen and Life Technologies, Leek, The Netherlands), recovered as a BamHI-XhoI fragment, and subcloned into the BamHI-SalI-digested pJDC9 suicide plasmid (eryR, ref. 34) to generate pGIE007. The pGIE007 plasmid was electroporated into L. plantarum NCIMB8826, and six candidate mutants were selected on Man, Rogosa, and Sharpe (MRS) (Difco) plates containing both erythromycin (5 μg/ml) and lincomycin (10 μg/ml) (Sigma). One candidate mutant, designated EP007, was chosen for further characterization. Correct integration of pGIE007 at the dlt locus was confirmed by PCR. The expected decrease in mRNA size and absence of a dltB-specific transcript were verified by Northern blot analyses that confirmed the mutant genotype (data not shown). Lactobacilli were grown at 37°C in MRS broth in the presence of 5 μg/ml erythromycin for the Dlt- mutant. Streptococcus gordonii LMG 17843 strain (35) and Escherichia coli TG1 strain (36) were grown at 37°C in brain heart infusion (Difco) and Luria broth (36), respectively. For immune cell stimulation, bacteria were grown until the mentioned times, and the number of colony-forming units (CFU) was estimated by measuring the optical density at 600 nm. Cells were harvested by centrifugation, washed twice in sterile PBS (pH 7.2), resuspended at 109 CFU per ml in PBS containing 20% glycerol, and stored at -80°C until used for stimulation assays. For in vivo mouse colitis experiments, bacteria were grown for 48 h, washed twice, and resuspended at 108 CFU per ml in 0.2 M NaHCO3 buffer containing 2% glucose.
Cell Purification. Peripheral blood mononuclear cells (PBMC) were isolated from peripheral blood of healthy donors as described (37). Briefly, after a Ficoll gradient centrifugation (Amersham Pharmacia), mononuclear cells were collected, washed twice with RPMI medium 1640 (Life Technologies, Paisley, Scotland), and adjusted to 2 × 106 cells per ml in RPMI medium 1640 supplemented with 50 μg/ml gentamycin, 2 mM l-glutamine, and 10% FCS (GIBCO/BRL). Monocytes (CD14+ cells) were purified by using CD14 antibodies coupled to magnetic microbeads (Miltenyi Biotec), according to the manufacturer's recommendations. Briefly, PBMC (3 × 108 cells) were incubated on ice for 30 min with 300 μl of CD14 antibodies coupled to magnetic microbeads. After a wash, they were applied onto a column placed in a magnetic field of a MidiMACS separation unit (Miltenyi Biotec). After elution of the column, cells obtained by positive selection were washed twice and adjusted at 0.5 × 106 cells per ml in complete RPMI medium 1640.
Induction of Cytokine Release. PBMC (2 × 106 cells per ml) or monocytes (0.5 × 106 cells per ml) were seeded in 24-well tissue culture plates (Corning). Twenty microliters of a thawed bacterial suspension at 109 CFU per ml or purified LTA was then added. PBS containing 20% glycerol was used as negative (nonstimulated) control. After 24-h stimulation at 37°C in 5% CO2, culture supernatants were collected, clarified by centrifugation, and stored at -20°C until cytokine analysis. Cytokines were measured by ELISA using commercial kits according to the manufacturers' recommendations.
Purification of LTAs. Highly purified LTAs were prepared from L. plantarum NCIMB8826 and its isogenic Dlt- mutant as described (26). Briefly, defrosted aliquots of bacteria were mixed with an equal volume of 1-butanol (Merck) under stirring for 30 min at room temperature. After centrifugation at 13,000 × g for 20 min, the aqueous phase was lyophilized, resuspended in chromatography start buffer (15% n-propanol in 0.1 M ammonium acetate, pH 4.7), and centrifuged at 45,000 × g for 15 min. The supernatant was subjected to hydrophobic interaction chromatography on octyl-Sepharose. The purity and structure of LTA were analyzed by NMR and MS as described (26).
TLR Knockout Cells. Bone marrow cells were prepared from C3H/HeN (TLR-4+/+) and C3H/HeJ (TLR-4-/-) mice (purchased from Charles River Laboratories) and from TLR-2 WT (TLR-2+/+) and TLR-2 knockout (TLR-2-/-) mice, generated by homologous recombination by Deltagen (Menlo Park, CA) and kindly provided by Tularik (South San Francisco). The genetic background of the TLR-2 mice was C57BL/6. All TLR-2 mice were bred in the animal facility at the University of Konstanz. Mice were fed ad libitum with Altromin 1314 (Altromin, Lage an der Lippe, Germany) and kept at 20-22°C, 50-60% air humidity at a 12-h day/night cycle. Bone marrow cells were isolated from the femurs by rinsing with 10 ml of ice-cold PBS and transferred to siliconized glass tubes (Vacutainer, BD Biosciences, Heidelberg). After centrifugation, cells were resuspended in RPMI 1640 medium (PAA, Linz, Austria) containing 10% FCS (GIBCO/Invitrogen) and transferred to 96-well cell culture plates (5 × 105 cells per well, Greiner, Frickenhausen, Germany). Murine bone marrow cells were then stimulated with LTA from Staphylococcus aureus, prepared in house according to Morath et al. (26), L. plantarum NCIMB8826, the Dlt- mutant (for preparation see above), or whole bacteria, and then incubated for 24 h at 37°C and 5% CO2. LPS from Salmonella abortus equi (Sigma) served as a positive stimulation control for TLR-2-/- cells and also to confirm the defect of TLR-4 of cells from the C3H/HeJ mice.
Induction of Colitis. Animal experiments were performed at the animal facility of the Institut Pasteur de Lille (French-accredited establishment A59107) according to French government guidelines 86/609/CEE. Adult BALB/c mice purchased from Iffa Credo, St Germain sur l'Arbresle, France were group-housed (8-10 per cage) and had free access to regular rodent chow and tap water. For colitis induction, anesthesized mice received an intrarectal administration of 40 μl of a solution of TNBS (100 mg/kg, Fluka) dissolved in 0.9% NaCl/ethanol (60:40 vol/vol). Control mice received 40% ethanol. Animals were killed by cervical dislocation 2 days after TNBS administration. To study the effect of selected LAB on colitis, 107 CFU of L. plantarum NCIMB8826 WT or Dlt- mutant were administered once daily by intragastric gavage, starting 5 days before and until 1 day after TNBS administration. Mice were weighed before colitis induction and at death. Suboptimal bacterial doses (leading to partial or no protection in the case of the WT) were used in these experiments to highlight the functional differences between the WT and the mutant strains. Macroscopic lesions of the colon were evaluated according to the Wallace criteria (38) that reflect both the intensity of inflammation and the extent of the lesions. A colon sample located in the most injured area was fixed in 4% paraformaldehyde and embedded in paraffin. Four-micrometer sections were stained with May Grünwald-Giemsa and blindly scored according to the Ameho criteria (39). This grading on a scale from 0 to 6 takes into account the degree of inflammatory infiltrate, the presence of erosion, ulceration, or necrosis, and the depth and surface extension of the lesion.
Statistics. Comparisons between the different animal groups were analyzed by the nonparametric Wilcoxon ANOVA test. Differences were considered significant if P was <0.05.
Results
Dlt- Mutant of L. plantarum NCIMB8826 Presents a Defect in d-Alanylation of TAs. L. plantarum is described in the literature as producing LTA substituted with d-Ala and d-glucose (40). The dlt operon, responsible for d-alanylation of TAs (41), was disrupted in the L. plantarum NCIMB8826 strain by single-step chromosomal integration of a suicide vector carrying dltB, generating the Dlt- mutant (see Materials and Methods). The recently published protocol for the preparation of highly purified biologically active LTA (26) was applied to L. plantarum, and the molecular composition of the WT and mutant LTAs was determined by NMR spectroscopy. This analysis confirmed that the LTA from L. plantarum NCIMB8826 is constituted of polyglycerol phosphate (polygroP) polymers with d-Ala as major substituent. LTA from the WT strain displayed 41.7% of groP units substituted with d-Ala, whereas only 1.1% of groP of the major LTA fraction from the Dlt- mutant carried d-Ala residues. This strong reduction in d-alanylation of the Dlt- LTA was expected because the Dlt machinery necessary for d-Ala incorporation was inactivated. Surprisingly, the LTA from the Dlt- mutant contained a large amount of glucose substitutions on groP (24.2%), whereas glucose substitutions were nearly undetectable in WT LTA (data not shown).
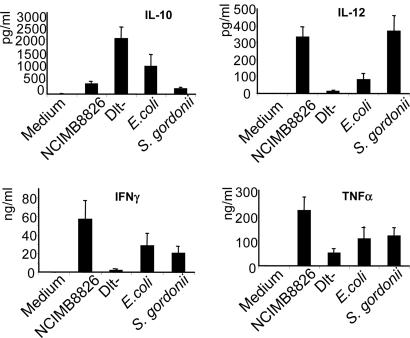
L. plantarum NCIMB8826 WT and Dlt- Mutant Differentially Induce IL-10, IL-12, TNF-α, and IFN-γ Secretion by PBMC and Monocytes. We first compared the cytokine profiles induced by the L. plantarum WT and Dlt- mutant strains collected at different times of bacterial growth (8, 24, 48, and 72 h). Both strains were able to induce IL-10 secretion from stimulated PBMC, but the induction of this cytokine was substantially enhanced when immune cells were stimulated by the Dlt- mutant. Concomitantly, the Dlt- mutant showed a significant decrease of proinflammatory cytokines, and this affect was more pronounced when bacteria were grown for 48 h (data not shown). This time point of 48 h was, therefore, selected for further experiments performed on a larger number of blood donors (n = 11). In addition, a Gram-positive strain (Streptococcus gordonii LMG 17843) and a Gram-negative (E. coli TG1) strain were included as internal controls. Consistent with the previous experiment, the Dlt- mutant induced increased levels of IL-10 secretion from PBMC in comparison to the WT strain (Fig. 1) together with decreased production of T helper 1/proinflammatory cytokines (IL-12, TNF-α, and IFN-γ). As described (23), the Gram-negative bacterium (E. coli) induced much higher levels of IL-10 than the Gram-positive bacteria (L. plantarum NCIMB8826 and Streptococcus gordonii). Conversely, the L. plantarum WT induced more IL-12 than E. coli. Similar results were obtained for IL-10, IL-12, and TNF-α secretion after stimulation of purified monocytes (data not shown). After stimulation of both PBMC and monocytes, the ratio of IL-10 to IL-12 production (Table 1) was similarly low for L. plantarum and Streptococcus gordonii (1.1 and 0.5 for PBMC and 3.4 and 2.7 for monocytes, respectively). In contrast, the IL-10/IL-12 ratio was higher for E. coli (12.9 for PBMC and 26.8 for monocytes) and dramatically increased for the Dlt- mutant (160.8 for PBMC and 122.0 for monocytes).
Fig. 1.
Cytokine response of human PBMC to stimulation with L. plantarum NCIMB8826 WT, the isogenic Dlt- mutant, E. coli TG1, and Streptococcus gordonii LMG 17843 strains. Bacteria were collected after 48 h (lactobacilli) or 24 h (E. coli and Streptococcus gordonii) of growth. Results represent the mean ± SEM for the response of 11 blood samples (individual donors).
Table 1. IL-10/IL-12 ratio secreted by human PBMC and monocytes stimulated by selected bacteria.
| Bacteria | PBMC | Monocytes |
|---|---|---|
| NCIMB8826 | 1.1 | 3.5 |
| Dlt- mutant | 160.8 | 122.0 |
| E. coli | 12.9 | 26.8 |
| Streptococcus gordonii | 0.5 | 2.7 |
Human PBMC and monocytes from 11 donors were purified and stimulated with each of the bacterial strains at a concentration of 1 × 107 CFU per ml. IL-12 and IL-10 concentrations were measured by ELISA in the cell-free supernatant after 24 h. Results are expressed as IL-10/IL-12 ratio.
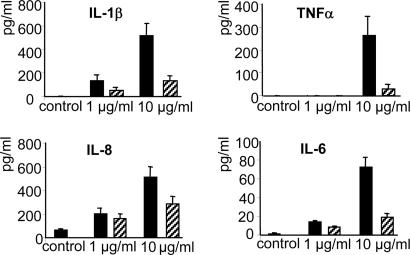
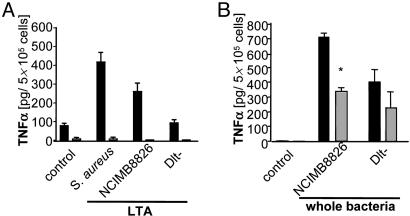
The in Vitro Immunomodulating Properties of the Dlt- Mutant Are Linked to Its LTA Composition. Because a defect in d-alanylation of TAs in the Dlt- mutant dramatically altered the immune modulating properties of L. plantarum NCIMB8826, the immunostimulatory properties of purified LTAs were evaluated. The LTA from the NCIMB8826 WT strain was able to stimulate the production of TNF-α, IL-1β, IL-6, and IL-8 by PBMC especially at a dose of 10 μg/ml, whereas LTA isolated from the Dlt- mutant induced very low levels of IL-1β and IL-6 and virtually no TNF-α even at this highest dose (Fig. 2). Previous reports have shown that cytokine induction by highly purified LTAs from pathogenic Gram-positive bacteria is TLR-2-dependent. TNF-α induction by L. plantarum LTAs was therefore examined by using bone marrow cells isolated from TLR-2+/+ and TLR-2-/- mice. The stimulating capacity of LTAs purified from the L. plantarum WT and Dlt- strains was compared with that of Staphylococcus aureus LTA. Although being a weaker inducer of cytokine release than the latter, the LTA from L. plantarum WT was able to induce TNF-α release from bone marrow cells of TLR-2+/+ mice (Fig. 3A). In line with the results obtained for PBMC, the TNF-α-inducing capacity of the LTA isolated from the Dlt- mutant was strongly reduced. It was notable that all three LTAs tested were unable to stimulate TNF-α release from TLR-2-/- mice-derived cells, suggesting a strong dependence of TLR-2 for the induction of cytokines by LTAs. We next investigated the TLR-2 dependence of whole bacteria (Fig. 3B). As expected, the L. plantarum WT strain was more potent in inducing TNF-α from the cells derived from TLR-2+/+ mice than its isogenic Dlt- mutant. Cytokine induction by whole WT L. plantarum bacteria from cells of TLR-2-/- mice was significantly but not totally inhibited, showing only a partial TLR-2 dependence. In contrast, the Dlt- mutant displayed no significant TLR-2 dependence. The same experiments were performed with bone marrow cells from TLR-4 mutated (C3H/HeJ) mice and compared with TLR-4 WT (C3H/HeN) mice. The results demonstrated no TLR-4 dependence for either purified LTAs or whole bacteria (data not shown), highlighting the specific role of TLR-2 in the observed effect.
Fig. 2.
Cytokine response of human PBMC to stimulation with 1 or 10 μg/ml of LTAs purified from the L. plantarum NCIMB8826 WT (filled bars) or the isogenic Dlt- mutant (hatched bars). Results represent the mean ± SEM for the response of six blood samples (individual donors).
Fig. 3.
TNF-α response of bone marrow cells derived from TLR-2+/+ (black bars) and TLR-2-/- (gray bars) mice to stimulation by 10 μg/ml purified bacterial LTA (A) or whole bacteria (5 × 104 CFU) (B). Results represent the mean ± SEM (n = 4). *, P < 0.05 represents significant differences.
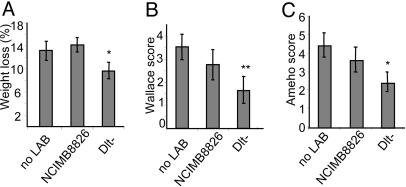
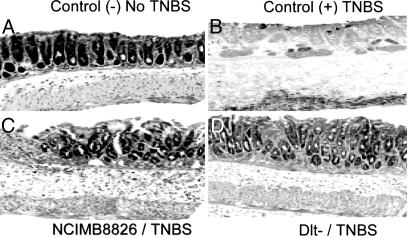
In Vitro Antiinflammatory Properties of the Dlt- Mutant Were Confirmed in Vivo in a TNBS-Induced Murine Colitis Model. To evaluate the in vivo immunomodulatory potential of the two L. plantarum strains, we compared the development of TNBS-induced colitis in mice that were treated, or not, with L. plantarum WT and Dlt- mutant strains. Notably, the percentage of mice displaying diarrhea after TNBS administration was much lower in the group of animals gavaged with the Dlt- mutant (11.1%) than in the group of mice receiving the L. plantarum WT strain or no bacteria treatment (55.5% in both cases) (data not shown). We observed that weight loss was significantly reduced in mice receiving the Dlt- mutant as compared with mice receiving no bacteria or receiving the L. plantarum WT strain (Fig. 4A). Characteristic features of colitis were observed 2 days after administration of TNBS in the mice receiving no bacteria, leading to a Wallace score of 3.5 ± 0.5 (Fig. 4B), which corresponds to several areas of ulceration accompanied by intestinal wall thickening. Histological analysis performed on these mice revealed large areas of ulceration with important inflammatory infiltrates together with necrosis, leading to an Ameho score of 4.4 ± 0.6 (Figs. 4C and 5B). More precisely, four of nine mice in this group displayed large areas of necrosis extending through the muscularis mucosae (Ameho score of 6). Even though the results obtained with mice treated with L. plantarum WT strain in these experimental conditions were not significantly different (macroscopic and histological analyses) from the group of mice receiving no bacteria, mice gavaged with the WT bacteria had a tendency to display milder colitis as assessed macroscopically (Fig. 4B) and histologically (Figs. 4C and 5C). In particular, only one of eight mice displayed histological inflammation, leading to an Ameho score graded to 6. In contrast, mice that had received the Dlt- mutant displayed significantly (P < 0.05) less severe lesions. On a macroscopic level, necrotic lesions were observed in only one mouse of eight, whereas the others suffered solely from hyperemia and thickening of the intestinal wall, leading to a Wallace score of 1.6 ± 0.6 (Fig. 4B). These results were confirmed by histological observations (Ameho score of 2.4 ± 0.5) (Fig. 4C) that evidenced only mild inflammatory infiltrates and limited edema (Fig. 5D). Control mice receiving 40% ethanol intrarectally displayed neither macroscopic (data not shown) nor histological lesions (Fig. 5A).
Fig. 4.
Effect of preventive administration of L. plantarum NCIMB8826 WT and Dlt- mutant strains on acute colitis induced in BALB/c mice by intrarectal administration of TNBS. (A) Weight variation between day 5 (TNBS administration) and day 7 (death). (B and C) Wallace (B) and Ameho (C) inflammation scores. Results are means ± SEM of one representative experiment (eight or nine mice per group). Significant P < 0.05 (**) or < 0.1 (*), as compared with the TNBS control group.
Fig. 5.
Representative histological sections of colonic tissues of BALB/c mice obtained 2 days after intrarectal TNBS/ethanol administration. Shown are colon sections of mice receiving no LAB and ethanol 40% (A), TNBS-treated mice receiving no LAB (B), TNBS-treated mice gavaged with L. plantarum NCIMB8826 WT strain (C), or TNBS-treated mice gavaged with the Dlt- mutant (D). (Magnification: ×100.)
Discussion
The use of probiotics has been proposed as a way to stimulate or regulate the mucosal immune system and thereby assist in maintaining the gut immunological barrier. Their positive effects have been studied in the treatment and prevention of various intestinal disorders, including colitis in animal models and IBD in humans (14-19). However, the mechanisms underlying these effects and especially the role of bacterial cell-wall components are currently not well understood. The capacity of lactobacilli to variably induce proinflammatory or antiinflammatory cytokines indicates that different strains of Lactobacillus may differentially stimulate immune cells and could therefore exert opposite immunoregulatory effects (6-8, 10).
In this study, we established the importance of TA (especially LTA) composition in the ability of lactobacilli to differentially impact on the host immune system. By analyzing the properties of a cell-wall mutant affected in d-alanylation of TA (Dlt- mutant) in vitro, we observed that a defect in d-Ala substitution of the TA and/or the replacement of d-Ala by glucose led to a strong increase in the capacity of the strain to induce IL-10. The IL-10/IL-12 ratios obtained after PBMC or monocyte stimulation indicated a significant change in the immunomodulation properties of the Dlt- mutant in comparison to the parental strain, leading to a substantial reduction in the inflammatory potential. d-Ala substitution in TA was further linked with the in vitro induction of T helper 1/proinflammatory cytokines by the WT L. plantarum strain. Because traditional methods used to prepare LTAs were reported to compromise their purity and quality and may lead to a loss of d-Ala residues, a purification method was used in this study that allowed preparation of highly purified and biologically active LTA from both strains (26). We confirmed that LTA from L. plantarum NCIMB8826 was constituted of polyglycerol phosphate (polygroP) polymers with d-Ala as major constituent (41% of substitution). Strikingly, LTA from this WT strain was a more potent inducer of TNF-α, IL-1β, IL-6, and IL-8 secretion by PBMC than was LTA from the Dlt- mutant, which displayed only a 1.1% d-Ala substitution. It is noted that Staphylococcus aureus LTA, which carries >70% of d-Ala substitution (26), induced even higher levels of TNF-α. Collectively, these observations provide further evidence of the proinflammatory properties of d-Ala as a constituent of LTA, as was demonstrated by the synthesis of LTA substituted with l-Ala instead of d-Ala (28).
It is becoming evident that the recognition of specific microbial/pathogen-associated patterns, such as LTA, is mediated by pattern recognition receptors (PRRs) that signal the presence of specific microorganisms to the host. The TLR family is the best-characterized class of PRRs in mammalian species. Rakoff-Nahoum et al. (42) recently reported that signaling of commensal bacteria through TLRs under normal steady-state conditions plays a crucial role in the maintenance of intestinal epithelial homeostasis. Previous studies have shown that highly purified LTA from Staphylococcus aureus was able to activate cellular responses via TLR-2 (29, 30). Matsuguchi et al. (33) provided similar results for LTA-enriched protoplast fractions and purified LTA from two Lactobacillus strains (L. fermentum and L. casei). However, as mentioned by those authors, the identification of the responsible TLR in the recognition of the LTA of different Gram-positive bacteria remains controversial. Here, we established that the proinflammatory capacity of highly purified and biologically active LTAs from the L. plantarum WT and Dlt- mutant strains is clearly TLR-2-dependent. Nevertheless, a partial but significant TLR-2 dependence of the immunostimulating capacity of L. plantarum WT whole cells was observed. Interestingly, the Dlt- mutant cells were less potent in inducing the proinflammatory cytokine TNF-α and showed no significant TLR-2 dependence. This finding indicates that TLR-2 dependence is linked to the presence of biological active LTA, but that other receptors might also be involved in Gram-positive bacteria recognition depending on cell envelope structural differences among species or even strains (33). In our case, we have demonstrated the impact of LTA substitution on the immunomodulating properties of L. plantarum. The increased antiinflammatory capacity of the L. plantatum Dlt- mutant might actually be an indirect consequence of the reduction in d-Ala substitution and could be linked to other cell envelope components that are less effective or masked when TAs are correctly substituted and exert full biological activity. Travasso et al. (43) recently demonstrated that highly purified peptidoglycan is preferentially detected through intracellular receptors (Nod1/Nod2). Nod2/Card15 signaling was shown to inhibit TLR-2-driven activation of NF-κB (44), and Card15 mutations have been associated with Crohn's disease (45). Therefore, perhaps in the absence of biologically active LTA, unmasked peptidoglycan of the Dlt- mutant strain may exert an inhibitory effect on the proinflammatory NF-κB cascade through Nod2 signaling. The protective effect of beneficial LAB could also be mediated by the release of soluble factors that alter epithelial permeability (46) or exert inhibitory effects on the inflammatory cascade (47). These observations raised the question of whether soluble factors or cytoplasmic bacterial components, such as DNA, play a major role in the immune effect of probiotic bacteria. Recent reports indeed described the role of probiotic genomic DNA, notably in IL-10 induction (48). Moreover, Rachmilewitz et al. (49) showed that TLR-9 signaling is essential in mediating the antiinflammatory effect of the bacteria they studied, supporting the key role of genomic DNA carrying specific unmethylated CpG motifs. In a recent article (50), it was argued that such findings do not rule out the effect of other bacterial components in different signaling pathways, such as IL-10 synthesis and secretion. IL-10 plays a central role in down-regulating inflammatory cascades. A genetically engineered Lactococcus lactis strain has been used successfully for local administration of this cytokine, leading to protection against colitis in two different mouse models (51). The Dlt- mutant was shown to be a strong inducer of IL-10, and in vivo studies in a murine TNBS-induced colitis model demonstrated a significant antiinflammatory effect. This result would suggest that endogenous production of IL-10 was as beneficial as local delivery of this cytokine.
Taken together, these results emphasize the importance of LTA composition in the proinflammatory or antiinflammatory properties of Lactobacillus cells, but also point to the potential for use of specific Lactobacillus cell wall mutants for treatment of IBD. Moreover, the data provide a rational explanation for the sometimes-conflicting evidence reported in the literature on the immunomodulation effect of different closely related species or strains, owing to variations in cell wall composition. Last, this study emphasizes the usefulness of targeted isogenic mutants in assessing the role of specific bacterial components in the interaction with the host immune system, an approach that is fully supported by the recent availability of full genome sequences for a number of probiotic LAB.
Acknowledgments
We thank Benoit Foligné and Véronique Dennin for their skillful help with animal experiments, Stephanie Traub and Susanne Deiniger for their expert technical support, and Jean Delcour for fruitful collaboration. This work was supported by European Union Grant QLK1-CT2000-0146, the Institut Pasteur de Lille, Konstanz University, and the Fondation Louvain from Université Catholique de Louvain. P.H. is research associate at Fonds National de la Recherche Scientifique.
Author contributions: C.G., P.H., and A.M. designed research; C.G., S.N., S.M., C.H., and J.D. performed research; E.P., S.M., T.H., and P.H. contributed new reagents/analytic tools; C.G., S.N., C.H., T.H., and A.M. analyzed data; C.G., S.N., C.H., B.P., P.H., and A.M. wrote the paper; and C.G., T.H., P.H., and A.M. provided collaboration management.
Abbreviations: CFU, colony-forming units; IBD, inflammatory bowel disease; LAB, lactic acid bacteria; LTA, lipoteichoic acid; PBMC, peripheral blood mononuclear cells; TA, teichoic acid; TLR, toll-like receptor; TNBS, 2,4,6-trinitrobenzenesulfonic acid.
References
- 1.Berg, R. D. (1996) Trends Microbiol. 4, 430-435. [DOI] [PubMed] [Google Scholar]
- 2.Bjorksten, B. (1999) Proc. Nutr. Soc. 58, 729-732. [DOI] [PubMed] [Google Scholar]
- 3.Hooper, L. V. & Gordon, J. I. (2001) Science 292, 1115-1118. [DOI] [PubMed] [Google Scholar]
- 4.Duchmann, R., Kaiser, I., Hermann, E., Mayet, W., Ewe, K. & Meyer zum Buschenfelde, K. H. (1995) Clin. Exp. Immunol. 102, 448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercenier, A., Pavan, S. & Pot, B. (2003) Curr. Pharm. Des. 9, 175-191. [DOI] [PubMed] [Google Scholar]
- 6.Miettinen, M., Matikainen, S., Vuopio-varkila, J., Pirhonen, J., Varsila, K., Kurimoto, M. & Julkunen, I. (1998) Infect. Immun. 66, 6058-6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hessle, C., Hanson, L. A. & Wold, A. E. (1999) Clin. Exp. Immunol. 116, 276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maassen, C. B. M., van Holten-Neelen, C., Blak, F., Heijne den Bak-Glashouwer, J., Leer, R. J., Laman, J. D., Boersma, W. J. A. & Claassen, E. (2000) Vaccine 18, 2613-2623. [DOI] [PubMed] [Google Scholar]
- 9.Vitini, E., Alvarez, S., Medina, M., Medici, M., de Budeguer, M. V. & Perdigon, G. (2000) Biocell 24, 223-232. [PubMed] [Google Scholar]
- 10.Christensen, H. R., Frokiaer, H. & Pestka, J. J. (2002) J. Immunol. 168, 171-178. [DOI] [PubMed] [Google Scholar]
- 11.Fuller, R. (1989) J. Appl. Bacteriol. 66, 365-378. [PubMed] [Google Scholar]
- 12.Guarner, F. & Schaafsma, G. J. (1998) Int. J. Food Microbiol. 39, 237-238. [DOI] [PubMed] [Google Scholar]
- 13.Fedorak, R. N. & Madsen, K. L. (2004) Inflamm. Bowel Dis. 10, 286-299. [DOI] [PubMed] [Google Scholar]
- 14.Fabia, R., Ar'Rajab, A., Johansson, M. L., Willen, R., Anderson, R., Molin, G. & Bengmark, S. (1993) Scand. J. Gastroenterol. 28, 155-162. [DOI] [PubMed] [Google Scholar]
- 15.Mao, Y., Nobaek, S., Kasravi, B., Adawi, D., Stenram, U., Molin, G. & Jeppsson, B. (1996) Gastroenterology 111, 334-344. [DOI] [PubMed] [Google Scholar]
- 16.Madsen, K. L., Doyle, J. S., Jewell, D., Tavernini, M. M. & Fedorak, N. (1999) Gastroenterology 116, 1107-1114. [DOI] [PubMed] [Google Scholar]
- 17.Rembacken, B. J., Snelling, A. M., Hawkey, P. M., Chalmers, D. M. & Axon, A. T. (1999) Lancet 354, 635-639. [DOI] [PubMed] [Google Scholar]
- 18.Gionchetti, P., Rizzello, F., Venturi, A., Brigidi, P., Matteuzzi, D., Bazzocchi, G., Poggioli, G., Miglioli, M. & Campieri, M. (2000) Gastroenterology 119, 305-309. [DOI] [PubMed] [Google Scholar]
- 19.Mimura, T., Rizzello, F., Helwig, U., Poggioli, G., Schreiber, S., Talbot, I. C., Nicholls, R. J., Gionchetti, P., Campieri, M. & Kamm, M. A. (2004) Gut 53, 108-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foligne, B., Grangette, C. & Pot, B. (2005) Gut 54, 727-728. [PMC free article] [PubMed] [Google Scholar]
- 21.Mercenier, A., Hols, P., Roussel, Y., Perez-Martinez, G., Buesa, J., Wilks, M., Pozzi, G., Remaut, E., Morelli, L., Grangette, C., et al. (2004) Microb. Ecol. Health Dis. 16, Suppl., 86-95. [Google Scholar]
- 22.Schiffrin, E. J., Rochat, F., Link-Amster, H., Aeschlimann, J. M. & Donnet-Hughes, A. (1995) J. Dairy Sci. 78, 491-497. [DOI] [PubMed] [Google Scholar]
- 23.Hessle, C., Andersson, B. & Wold, A. E. (2000) Infect. Immun. 68, 3581-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klaenhammer, T., Altermann, E., Arigoni, F., Bolotin, A., Breidt, F., Broadbent, J., Cano, R., Chaillou, S., Deutscher, J., Gasson, M., et al. (2002) Antonie Van Leeuwenhoek 82, 29-58. [DOI] [PubMed] [Google Scholar]
- 25.Kleerebezem, M., Boekhorst, J., van Kranenburg, R., Molenaar, D., Kuipers, O. P., Leer, R., Tarchini, R., Peters, S. A., Sandbrink, H. M., Fiers, M. W. E. J., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morath, S., Geyer, A. & Hartung, T. (2001) J. Exp. Med. 193, 393-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morath, S., Geyer, A., Spreitzer, I., Hermann, C. & Hartung, T. (2002) Infect. Immun. 70, 938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morath, S., Stadelmaier, A., Geyer, A., Schmidt, R. R. & Hartung, T. (2002) J. Exp. Med. 195, 1635-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Opitz, B., Schroder, N. W., Spreitzer, I., Michelsen, K. S., Kirschning, C. J., Hallatschek, W., Zahringer, U., Hartung, T., Gobel, U. B. & Schumann, R. R. (2001) J. Biol. Chem. 276, 22041-22047. [DOI] [PubMed] [Google Scholar]
- 30.Schroder, N. W., Morath, S., Alexander, C., Hamann, L., Hartung, T., Zahringer, U., Gobel, U. B., Weber, J. R. & Schumann, R. R. (2003) J. Biol. Chem. 278, 15587-15594. [DOI] [PubMed] [Google Scholar]
- 31.Sherman, L. A. & Savage, D. C. (1986) Appl. Environ. Microbiol. 52, 302-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granato, D., Perotti, F., Masserey, I., Rouvet, M., Golliard, M., Servin, A. & Brassart, D. (1999) Appl. Environ. Microbiol. 65, 1071-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuguchi, T., Takagi, A., Matsuzaki, T., Nagaoka, M., Ishikawa, K., Yokokura, T. & Yoshikai, Y. (2003) Clin. Diagn. Lab. Immunol. 10, 259-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen, J. D. & Morrison, D. A. (1988) Gene 64, 155-164. [DOI] [PubMed] [Google Scholar]
- 35.Carlsson, J. (1967) Odontol. Rev. 18, 173-178. [PubMed] [Google Scholar]
- 36.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 37.Muller-Alouf, H., Proft, T., Zollner, T. M., Gerlach, D., Champagne, E., Desreumaux, P., Fitting, C., Geoffroy-Fauvet, C., Alouf, J. E. & Cavaillon, J. M. (2001) Infect. Immun. 69, 4141-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallace, J. L., MacNaughton, W. K., Morris, G. P. & Beck, P. L. (1989) Gastroenterology 96, 29-36. [DOI] [PubMed] [Google Scholar]
- 39.Ameho, C. K., Adjei, A. A., Harrison, E. K., Takeshita, K., Morioka, T., Arakaki, Y., Ito, E., Suzuki, I., Kulkarni, A. D., Kawajiri, A. & Yamamoto, S. (1997) Gut 41, 487-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Archibald, A. R. & Baddiley, J. (1966) Adv. Carbohydr. Chem. Biochem. 21, 323-375. [DOI] [PubMed] [Google Scholar]
- 41.Neuhaus, F. C., Heaton, M. P., Debabov, D. V. & Zhang, Q. (1996) Microb. Drug Resist. 2, 77-84. [DOI] [PubMed] [Google Scholar]
- 42.Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. (2004) Cell 118, 229-241. [DOI] [PubMed] [Google Scholar]
- 43.Travassos, L. H., Girardin, S. E., Philpott, D. J., Blanot, D., Nahori, M. A., Werts, C. & Boneca, I. G. (2004) EMBO J. 5, 1000-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe, T., Kitani, A., Murray, P. J. & Strober, W. (2004) Nat. Immunol. 5, 800-808. [DOI] [PubMed] [Google Scholar]
- 45.Hugot, J. P., Chamaillard, M., Zouali, H., Lesage, S., Cezard, J. P., Belaiche, J., Almer, S., Tysk, C., O'Morain, C. A., Gassull, M., et al. (2001) Nature 411, 599-603. [DOI] [PubMed] [Google Scholar]
- 46.Madsen, K., Cornish, A., Soper, P., McKaigney, C., Jijon, H., Yachimec, C., Doyle, J., Jewell, L. & De Simone, C. (2001) Gastroenterology 121, 580-591. [DOI] [PubMed] [Google Scholar]
- 47.Menard, S., Candalh, C., Bambou, J. C., Terpend, K., Cerf-Bensussan, N. & Heyman, M. (2004) Gut 53, 821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lammers, K. M., Brigidi, P., Vitali, B., Gionchetti, P., Rizzello, F., Caramelli, E., Matteuzzi, D. & Campieri, M. (2003) FEMS Immunol. Med. Microbiol. 38, 165-172. [DOI] [PubMed] [Google Scholar]
- 49.Rachmilewitz, D., Katakura, K., Karmeli, F., Hayashi, T., Reinus, C., Rudensky, B., Akira, S., Takeda, K., Lee, J., Takabayashi, K. & Raz, E. (2004) Gastroenterology 126, 520-528. [DOI] [PubMed] [Google Scholar]
- 50.Reid, G., Guarner, F., Gibson, G., Tompkins, T., Gill, H., Rowland, I., Rastall, B., Pot, B. & Sanders, M. E. (2004) Gastroenterology 127, 366-367. [DOI] [PubMed] [Google Scholar]
- 51.Steidler, L., Hans, W., Schotte, L., Neirynck, S., Obermeier, F., Falk, W., Fiers, W. & Remaut, E. (2000) Science 289, 1352-1355. [DOI] [PubMed] [Google Scholar]