Abstract
The symbiosis between legumes and rhizobia is essential for the nitrogen input into the life cycle on our planet. New root organs, the nodules, are established, which house N2-fixing bacteria internalized into the host cell cytoplasm as horizontally acquired organelles, the symbiosomes. The interaction is initiated by bacterial invasion via epidermal root hair curling and cell division in the cortex, both triggered by bacterial nodulation factors. Of the several genes involved in nodule initiation that have been identified, one encodes the leucine-rich repeat-type receptor kinase SymRK. In SymRK mutants of Lotus japonicus or its orthologs in Medicago sp. and Pisum sativum, nodule initiation is arrested at the level of the root hair interaction. Because of the epidermal block, the role of SymRK at later stages of nodule development remained enigmatic. To analyze the role of SymRK downstream of the epidermis, the water-tolerant legume Sesbania rostrata was used that has developed a nodulation strategy to circumvent root hair responses for bacterial invasion. Evidence is provided that SymRK plays an essential role during endosymbiotic uptake in plant cells.
Keywords: bacterial uptake, endosymbiosis, legume nodulation, Sesbania rostrata, infection thread structure
Leguminous plants can engage into a symbiotic interaction with rhizobia and form new root organs, the nodules, in which the bacteria reside and fix atmospheric dinitrogen. Rhizobia belong to diverse phylogenetic taxa but have in common the capacity to induce nodules on the appropriate legume host. The nodulation process is initiated by a complex signal exchange between both partners. The principal signal molecules, the bacterial nodulation factors (NFs), are lipochitooligosaccharides decorated with specific chemical groups at both the reducing and nonreducing end (1). Recognition of compatible NFs triggers developmental and cellular host responses, such as rapid ion fluxes, membrane depolarization, calcium oscillations, cytoskeletal reorganization, root hair deformations and curling, gene expression, and cell division (2, 3).
Rhizobial invasion usually takes place via root hairs. Upon NF perception, developing root hairs deform and eventually curl (4). The bacteria are entrapped in this curl and a microcolony is formed. Local cell wall hydrolysis and invagination of the plasma membrane lead to the formation of a tubular structure: the infection thread (IT) that proceeds through the root hair and underlying cell layers to guide the bacteria toward a nodule primordium, which develops in the cortex (4). An elaborate IT network spreads into the nodule primordium to provide bacteria to plant cells for endosymbiotic release. Uptake is initiated by local IT wall hydrolysis; unwalled infection droplets are formed from which bacteria are pinched off, thus creating symbiosomes (5). Symbiosomes contain one or a few bacteria, now called bacteroids, surrounded by a plant-derived membrane, the peribacteroid or symbiosome membrane (6).
Considerable progress has been made in analyzing the plant mechanisms that regulate initiation and development of nodules and control of bacterial invasion. Several candidate NF receptors have been identified, which are all receptor-like kinases with extracellular lysin motif (LysM) domains (7–9). Bacterial LysM domains bind peptidoglycan or chitin (10, 11); hence, plant LysM-receptor-like kinases are good candidates for NF binding and perception.
In addition, other NF signal transduction elements act downstream from the LysM-receptor-like kinases, because the corresponding mutants still respond to NFs by root hair swelling, but no longer show a root hair curling (RHC) response. In Medicago truncatula, the Dmi1, Dmi2, and Dmi3 (12) genes encode a putative ligand-gated cation channel, a leucine-rich repeat (LRR)-type receptor kinase, and a putative calcium and calmodulin-dependent protein kinase, respectively (13–15). Orthologs of Dmi2 have been described in M. sativa (MsNork), Lotus japonicus (LjSymRK), and Pisum sativum (Pssym19) (13, 16). Remarkably, mutations in these genes also impair arbuscular mycorrhization, which is an ancient and widespread endosymbiosis that facilitates phosphate acquisition and involves 80% of the land plants and fungi belonging to the Glomeromycota (17). Hence, the pathways of nodulation and mycorrhization share common genetic elements for the establishment of endosymbiotic structures (17). Recently, a nonsymbiotic, NF-independent phenotype of enhanced touch sensitivity has been observed for the MsNork, the MtDmi2, and the LjSymRK mutant lines. When care is taken not to disturb root hairs during experimental manipulations, they curl upon application of rhizobia, but the curling is arrested when the root hair tip touches its own shank. Hence SymRK and its relatives are not needed for the curling process itself, although they do remain necessary for downstream, NF-dependent gene expression (18).
We have investigated the role of SrSymRK, the unique Sesbania rostrata ortholog of MsNork/LjSymRK in the establishment of lateral root base (LRB) nodules. The tropical legume S. rostrata is adapted to nodulate in temporarily flooded habitats and has versatile nodulation features. Under well-aerated conditions, nodules form in the zone of developing root hairs via RHC invasion of the microsymbiont Azorhizobium caulinodans (19). However, on hydroponic roots, N2-fixing nodules develop at LRBs (19). Azorhizobia invade the cortex intercellularly via cracks, resulting from lateral root protrusion, thereby circumventing the epidermis (20). Cortical infection pockets (IPs) are formed by NF-dependent local cell death induction and subsequent colonization of bacteria. Presumably, IPs function as signaling centers, similar to rhizobial colonies within 3D root hair curls (19). From IPs, ITs guide bacteria toward the target cells for symbiotic uptake. A very comparable process takes place at the bases of adventitious rootlets that are located on the stem of S. rostrata and that can develop into “stem” nodules upon inoculation. Because LRB nodulation with intercellular invasion strictly depends on NFs (21), it is an excellent tool to analyze the requirement for and consequences of NF-linked perception-transduction systems in cell layers, which are deeper than the epidermis. Here, we demonstrate that RNA interference (RNAi)-mediated knockdown of SrSymRK inhibits release of bacteria from infection droplets into the plant cytoplasm.
Materials and Methods
Plant and Bacterial Material. S. rostrata Brem and A. caulinodans ORS571(pRG960SD-32) were grown as described (22).
Southern Analysis. DNA gel blot analysis was performed as described (23). Labeled probes were generated from the extracellular domain with SrNORKRTF1 (5′-CCAAACAGACGTGGAAGTGA-3′) and SrNORKRTR2 (5′-CTTCGCTCATGTGTTGGTTGC-3′).
Identification of the Full-Length SrSymRK Sequence. Plaques (105)of a λ-fix II S. rostrata genomic library were screened with a probe corresponding to the extracellular and kinase domains of M. truncatula DMI2 (23). The full-length cDNA sequence was obtained by 5′ and 3′ RACE (Smart RACE cDNA amplification kit, Clontech). The fragments were cloned in a pGEMTeasy vector (Promega).
Expression Analysis. In situ hybridization was as described (24). The LRR region was used as template to produce 35S-labeled antisense probe. RT-PCR was performed (25, 26) with specific probes generated by SrNORKrtF1 (5′-CCAAACAGACGTGGAAGTGA-3′) and SrNORKrtR2 (5′-CTTCGCTCATGTGTTGGTTGC-3′) primers.
Transgenic Roots. To produce the knockout constructs (pK7GWIWG27F2-SymRKKO1 and pK7GWIWG27F2-Sym-RKKO2), two nonoverlapping regions of the LRR domain were recombined in the pK7GWIWG27F2 binary GATEWAY vectors (Invitrogen) (27). For the pK7GWIWG27F2-SymRKKO1 and pK7GWIWG27F2-SymRKKO2 constructs, the primers SrSymRKRTF1attB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCAACCAAACAGACGTGGAAGTGA-3′) and SrSymRKR13attB2 (5′-GGGACCACTTTGTACAAGAAAGCTGGGTCACTGGAAGGAATTGGTCCT-3′); and SrSymRKF16attB1 (5′-GGGGACA AGT T TGTACAAAAA AGCAGGCT TGAGCCACA ACAGT-3′) and SrSymRKRTR2attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTCGCTCATGTGTTGGTTGC-3′) were used, respectively. S. rostrata embryonic axes were transformed as described (28).
Light Microscopy and Transmission EM. Staining with β-glucuronidase was as described (21). For semithin sections, the tissues were embedded in Technovit 7100 (Heraeus), cut, and stained with toluidine blue (20, 28). GFP analysis (28) and transmission EM (21) were performed as described.
Results
SrSymRK Transcripts Accumulate in the Infection Zone of LRB Nodules. The full-length cDNA of SrSymRK was obtained by genomic library hybridization and RT-PCR (see Materials and Methods) (GenBank accession no. AY751547). SrSymRK is 86% and 90% similar at the amino acid level to MtDmi2 and LjSymRK, respectively (Fig. 1A). Southern hybridization with the extracellular domain (Fig. 1B) of MtDmi2 as probe revealed one band in each digest, demonstrating that SrSymRK is a unigene.
Fig. 1.
SrSymRK, the SymRK ortholog of S. rostrata.(A) Sequence of the SrSymRK protein and orthologs. SrSymRK shows a 90% and 86% similarity to L. japonicus LjSymRK and M. truncatula MtDMI2, respectively. Conserved amino acids are highlighted. (B) Southern analysis with the extracellular domain of SrSymRK as a probe.
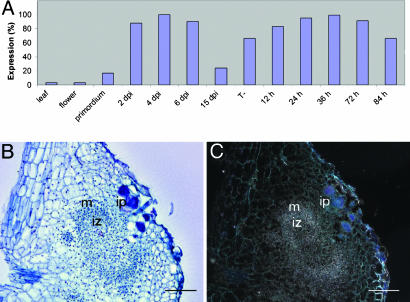
RT-PCR was performed to analyze the expression levels of SrSymRK during LRB nodulation. cDNA was prepared from stem-located adventitious root primordia before and at 2, 4, 6, and 15 d postinoculation (dpi) with A. caulinodans (Fig. 2A). As seen in Fig. 2 A, a basal expression was observed in adventitious root primordia, the expression was rapidly induced between 2 and 4 dpi, and the transcript levels dropped again in mature nodules at 15 dpi. Similarly, but less pronounced, SrSymRK transcripts accumulated during LRB nodulation of hydroponic roots. Uninfected roots and infected LRBs were harvested 12, 24, 36, 72, and 84 h after inoculation. RT-PCR analysis showed that the transcript levels were already slightly elevated 12 h after inoculation. SrSymRK levels were low in flowers and leaves (Fig. 2 A).
Fig. 2.
Expresssion analysis of SrSymRK. (A) RT-PCR analysis. SrSymRK expression levels are shown for leaves, flowers, uninfected adventitious root primordia (primordium), in adventitious root primordia infected with ORS571 after 2, 4, and 6 dpi, in hydroponic roots (T-), and in hydroponic roots 12, 24, 36, 72, and 84 h after inoculation. (B) Bright-field image of an in situ mRNA localization in a semithin section through a nodule primordium 72 h after inoculation. (C) Dark-field image of B; signal is visible as silver grains in the infection zone. ip, infection pocket; iz, infection zone; m, meristem. (Scale bars, 100 μm.)
To analyze the transcript localization during nodule development, developing adventitious root nodules were hybridized in situ at 3 dpi. At this stage, ITs have reached the nodule primordium, which has the shape of an open basket with a meristem and an infection zone where bacteria are released into plant cells. SrSymRK transcripts were detected in the infection zone of the developing nodule (Fig. 2 B and C).
Knockdown of SrSymRK Affects Bacterial Release from ITs. To investigate the role for SrSymRK in LRB nodulation, two independent RNAi constructs were made, each with a 200-bp fragment of the SrSymRK LRR-coding sequence. These constructs were introduced into S. rostrata via Agrobacterium rhizogenes-mediated transformation (see Materials and Methods) with a 35S-enhanced GFP as cotransformation marker (28).
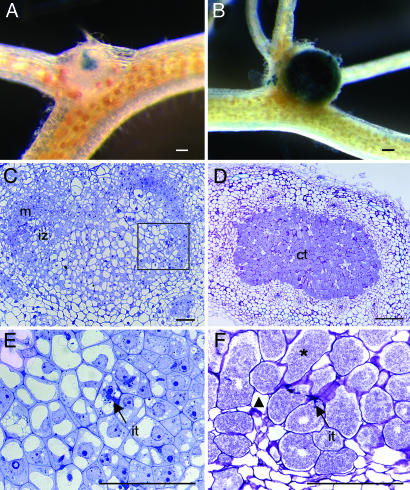
In total, 91 cotransformed roots were selected for analysis. Chimeric plants with transgenic roots were inoculated with A. caulinodans, and nodulation was scored between 8 and 14 dpi. At this stage, control lines transformed with the empty vector, all contained round, N2-fixing nodules (Fig. 3B). Nine lines containing RNAi constructs developed small, white, nodule-like structures at the base of the lateral roots of hydroponically grown roots (Fig. 3A). Infection with ORS571(pRG960–32), which can be visualized by β-glucuronidase staining, revealed that the primary invasion into the infection center was not hampered, but spreading into the cells of the nodule primordium was inhibited. In contrast, the central tissue of the stained control nodules was completely invaded (compare Fig. 3 A with B). RT-PCR analysis of clonally propagated offsprings of the nine lines showed a down-regulation of 65–92% in SrSymRK transcript levels when compared to control lines (data not shown).
Fig. 3.
Knockdown phenotype of SrSymRK.(A) Low magnification view of a SrSymRK knockdown line, 10 dpi. Bacteria are labeled with β-glucuronidase. (B) Low magnification view of a functional WT nodule, 10 dpi. Bacteria are labeled with β-glucuronidase. (C) Semithin section through A. IPs, ITs, and a primordium are formed, but no cells are infected. (D) Semithin section through B, 10 dpi. (E and F) Details of C and D. Uninfected and infected cells are marked by triangles and asterisks, respectively. ct, central tissue; ip, infection pocket; it, infection thread; iz, infection zone; m, meristem. (Scale bars, 100 μm.)
Microscopic analysis of semithin, toluidine blue-stained sections through a nodule-like structure derived from a SrSymRK RNAi line at 10 dpi revealed a nodule primordium in the typical open-basket conformation as is normally seen for WT nodules at 3–6 dpi. IPs were visible, with ITs progressing toward an infection zone adjacent to the meristem (Fig. 3 C and E). Nevertheless, no fixation zone with infected cells was observed but, instead, a zone of differentiated cells that were vacuolated and not filled with bacteria (Fig. 3E). In contrast, the control nodules had a fully differentiated central tissue containing cells with N2-fixing symbiosomes (Fig. 3 D and F).
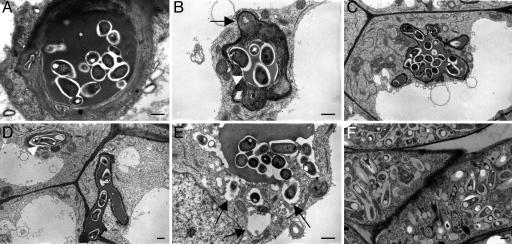
Ultrastructural Analysis of SrSymRK RNAi Lines Reveals Multiple Nodulation Defects. To better investigate eventual morphological abnormalities, in vitro-propagated transgenic roots of a 70% SrSymRK knockdown line were grafted on WT shoots of S. rostrata. This particular line showed nodule primordium and IT formation, but no bacterial uptake, in accordance with the phenotype for SrSymRK knockdown lines described above. The grafts responded to bacterial inoculations in exactly the same manner as the primary transformed roots (data not shown). Ten days after inoculation with A. caulinodans, small, nodule-like structures were harvested for EM analysis (Materials and Methods). High magnifications of ITs in the infection zone showed that, in contrast to WT, a low electron-dense rim was present between the IT matrix and the IT wall (Fig. 4 A, B, and D). The rim was continuous with the low electron-dense bacterial surface polysaccharides. ITs seemed to lose rigidity and became bag-like in appearance, with unusually protruding bulges (Fig. 4 C and D). Extensive cell wall depositions (Fig. 4B) were also obvious that had never been seen in WT ITs (Fig. 4A). Infection droplets were observed, but only a few symbiosome-like structures could be detected in the plant cell cytoplasm. They were different from WT symbiosomes (Fig. 4F) and had an irregular shape (Fig. 4E). The bacteroids were surrounded by a large and irregular peribacteroid space (Fig. 4E).
Fig. 4.
Ultrastructural analysis of the SrSymRK knockdown phenotype. (A) Transverse section through a WT intracellular IT. (B) Transverse section through an intracellular IT in an SrSymRK knockdown background, 10 dpi. Arrowhead and arrow indicate low electron-dense rim and abnormal cell wall depositions in SrSymRK knockdown lines, respectively. (C) Section through a bag-like intracellular IT consisting of numerous branches, 10 dpi. (D) Section through an intracellular IT in an SrSymRK knockdown background. Low electron-dense rim is indicated by an arrowhead. (E) Rare release of bacteria, resulting in abnormal symbiosomes, which are indicated by arrows. (F) WT bacteroids at 6 dpi. (Scale bars, 1 μm.)
Discussion
The search for functions involved in NF perception and signal transduction has revealed a set of genes that are required for nodulation as well as for mycorrhization, the ancient plant-fungi endosymbiosis from which nodulation has been derived (29). We provide more insight into the function of SymRK, a component of the common signal transduction cascade.
LjSymRK and its orthologs MtDmi2, MsNork, and Pssym19 code for LRR-type receptor kinases, and mutations in these genes cause a very early arrest and almost completely block epidermal responses to rhizobia resulting in impaired RHC (13, 16). During mycorrhization, LjSymRK mutants are arrested at the level of intracellular fungal entry in the exodermis/outer cortical cells, indicating that SymRK also functions downstream of the epidermis, in other plant tissues (30). The question of whether during nodulation SymRK plays also a role in tissues other than the epidermis is difficult to answer in legumes that use RHC invasion. However, it can be easily addressed in host plants that offer crack entry invasion, which avoids the epidermis and the root hairs and provides direct intercellular entry in the outer cortex at positions of LRBs. The water-tolerant tropical legume S. rostrata has recruited such a cortical invasion track to nodulate on submergence when accumulating ethylene blocks RHC invasion and root hair development (22) as well as during nodulation at the bases of stem-located adventitious rootlets where root hairs are absent. During LRB nodulation on S. rostrata, the bacteria enter directly in the cortical plant tissues and infection proceeds intercellularly in the outer cortex, without root hair involvement, yet the process depends strictly on NFs (21).
To analyze the role of SymRK downstream of the epidermis, the S. rostrata ortholog was isolated. SrSymRK is ≈85–90% similar to its relatives from other legumes. Southern hybridizations indicated that it is a unigene and expression analysis revealed that it could indeed function downstream of the epidermis. RT-PCR showed that the SrSymRK transcripts were up-regulated during adventitious and lateral root-based nodulation to disappear again in mature nodules. The induction level during adventious root nodulation was more pronounced, reflecting the ease of specific and synchronous sampling of the stem-located adventitious root tissues. In situ hybridization indicated that SrSymRK transcripts were present in cells underlying the nodular meristem, i.e., in the infection zone where bacteria will be released into the plant cell cytoplasm to differentiate into N2-fixing bacteroids.
To obtain an insight into an eventual function of SrSymRK in the infection zone, RNAi knockdown roots were constructed with A. rhizogenes-mediated root transformation. Lines with 4- to >10-fold reduced SrSymRK transcript levels exhibited the same impaired LRB nodulation phenotype. After 2 wk, only small, nonfixing nodule-like structures were detected that were barely infected with A. caulinodans. Light microscopy and EM revealed two main defects in these nodule-like structures. First, ITs were formed, indicating that the underlying mechanism was not hampered, but they had an irregular appearance with protruding bulges containing mainly IT matrix. Also the walls of the ITs were irregular with large cell wall deposits. At the ultrastructural level, the matrix of the ITs was separated from the IT wall by a layer of low electron-dense material that seemed continuous with the low electron-dense surface polysaccharide layer of the bacteria. A flooding of the surface polysaccharides along the edges of the ITs could imply a change in the physico-chemical nature of the IT matrix. Cross-linking of the matrix concomitant with bacterial cell division has been proposed to be the driving force for IT progression (5); hence, changes in the matrix composition might give rise to irregular IT structures.
A second defect is observed at the level of bacterial uptake when rhizobia are released into the plant cell cytoplasm to form symbiosomes. This process is initiated by local hydrolysis of the IT cell wall and formation of an infection droplet, a small bulge surrounded by a plant cell membrane containing a few bacteria and IT matrix (5). From the infection droplets, individual bacteria are released in the cytoplasm (31). As shown by transmission EM analysis, in the SrSymRK RNAi line, infection droplets were formed, but hardly any individual symbiosome was present. The few symbiosomes that were detected in the cytoplasm had an irregular structure, indicating a defect in bacterial uptake into the plant cells, possibly as a consequence of disabled vesicle trafficking. Similar features were observed in M. truncatula (32). Hence, SymRK is involved in control of IT structure and in bacterial uptake for symbiosome formation.
Taking into consideration these observations, the known role of SymRK in rhizobial root hair entry and mycorrhizal invasion, as well as recent findings on the mechanisms of invasion of biotrophic pathogens, we propose that SymRK might be part of a molecular complex for intracellular residence of endosymbionts that incorporates cell wall integrity control and targeted exocytosis (33). Intracellular penetrations of biotrophic pathogens and symbiotic mycorrhiza have much in common and might depend on related programs (31). During arbuscular myccorhizal initiation in LjSymRK mutants, the fungus invades epidermal cells, but is arrested after appressoria have been formed, at the intracellular passage through the exodermis/outer cortex (30), an arrest that might be explained by a defect in a common pathogen/symbiont entry system. Moreover, part of the phenotype observed in SymRK mutants during the initial steps of the RHC nodulation process is related to elevated touch sensitivity (18). A possible role for SymRK in connection to the recruitment of exocytosis-mediated resistance or mechanical stress-sensing mechanisms for symbiont entry could be suppression of touch-related plant responses.
NFs, the main bacterial signals for nodule development, trigger nodulation-related effects in the epidermis, as well as changes in cortex and pericycle cells. The latter effects are provoked from a distance because it is very unlikely that NFs migrate through symplast or apoplast while they are immobilized in the plant cell walls (34). Hence, NFs probably activate secondary signals that, together with signals coming from the stele, determine the landscape for nodule development and preparation for invasion (2). During LRB nodulation in S. rostrata, the distant NF-dependent response of nodule primordium formation is not disturbed in roots with >10-fold SrSymRK silencing, suggesting that it may not be linked to SymRK function. In contrast, the well established coupling of SymRK to NF signal transduction in RHC nodulation (3, 9, 12, 18, 35) would provide an early epidermal checkpoint, which is circumvented in LRB nodulation that uses direct cortical responses.
Besides signaling from a distance, NFs may affect adjacent cells in the epidermis and during the passage of the ITs in the cortex. In the epidermal root hairs, bacterial entry requires both NF perception (presumably via LysM-type receptors) and active SymRK (3, 7–9, 13, 16). In the infection zone, where SymRK transcripts are up-regulated, a cell-autonomous role in uptake is plausible and presumably uncoupled from local bacterial NFs. Although genes responsible for NF production are transcribed in ITs and NFs have been localized in the infection zone (36), a few cases have been described of NF-deficient strains that could reach the interior of a nodule and were internalized to fix nitrogen as long as NFs were added to the root medium (refs. 21 and 37; J. Den Herder and M. Holsters, unpublished results). The NF-deficient strains NGR234 of Rhizobium sp. and USDA110 of Bradyrhizobium japonicum were able to invade and nodulate Vigna unguiculata and Glycine max when NFs of NGR234 were added at a concentration of 10-7 to 10-6 M (37). An analogous observation has been made for the LRB nodulation on hydroponic roots of S. rostrata by coinoculating a bacterial mutant with defective surface polysaccharides (ORS571-X15), but normal NFs, and a mutant without NF production (ORS571-V44; ref. 21). The ORS571-V44 strain cannot enter the host because of the absence of NFs and does not provoke any nodulation-related effect (38), whereas ORS571-X15 induces nodule primordia at LRBs, but is arrested in superficially located IPs. However, ORS571-X15 can complement ORS571-V44 for nodule invasion and functional nodules are formed, which are exclusively occupied by ORS571-V44 bacteria (21). Presumably, NFs trigger developmental gradients from a distance, without being required in situ at the moment of SymRK-dependent internalization.
In summary, SymRK is required at the heart of endosymbiosis in leguminous plants to release bacteria into nodule cells during a process that might testify of common origins of bacterial and fungal endophytic lifestyles and biotrophic pathogen invasion strategies.
Acknowledgments
We thank Christa Verplancke and Annick De Keyser for excellent technical assistance and Martine De Cock for careful preparation of the manuscript. This work was supported by the Interuniversity Poles of Attraction Program-Belgian Science Policy (P5/13) and the Fund for Scientific Research-Flanders (“Krediet aan Navorsers” 1.5.088.99N and 1.5.192.01N). W.C. and K.S. are indebted to the Institute for the Promotion of Innovation by Science and Technology in Flanders for predoctoral fellowships.
Author contributions: W.C., S.G., and M.H. designed research; W.C., R.D.R., and K.S. performed research; W.C., S.G., R.D.R., K.S., and M.H. analyzed data; and W.C., S.G., and M.H. wrote the paper.
Abbreviations: dpi, days postinoculation; IP, infection pocket; IT, infection thread; NF, nodulation factor; LRB, lateral root base; LRR, leucine-rich repeat; RHC, root hair curling; LysM, lysin motif; RNAi, RNA interference.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY751547).
References
- 1.D'Haeze, W. & Holsters, M. (2002) Glycobiology 12, 79R-105R. [DOI] [PubMed] [Google Scholar]
- 2.Geurts, R. & Bisseling, T. (2002) Plant Cell 14, S239-S249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oldroyd, G. E. D. & Downie, J. A. (2004) Nat. Rev. Mol. Cell Biol. 5, 566-576. [DOI] [PubMed] [Google Scholar]
- 4.Gage, D. J. (2004) Microbiol. Mol. Biol. Rev. 68, 280-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewin, N. J. (2004) Crit. Rev. Plant Sci. 23, 293-316. [Google Scholar]
- 6.Vasse, J., de Billy, F., Camut, S. & Truchet, G. (1990) J. Bacteriol. 172, 4295-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Limpens, E., Franken, C., Smit, P., Willemse, J., Bisseling, T. & Geurts, R. (2003) Science 302, 630-633. [DOI] [PubMed] [Google Scholar]
- 8.Madsen, E. B., Madsen, L. H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., Sato, S., Kaneko, T., Tabata, S., Sandal, N., et al. (2003) Nature 425, 637-640. [DOI] [PubMed] [Google Scholar]
- 9.Radutoiu, S., Madsen, L. H., Madsen, E. B., Felle, H. H., Umehara, Y., Grønlund, M., Sato, S., Nakamura, Y., Tabata, S., Sandal, N., et al. (2003) Nature 425, 585-592. [DOI] [PubMed] [Google Scholar]
- 10.Amon, P., Haas, E. & Sumper, M. (1998) Plant Cell 10, 781-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bateman, A. & Bycroft, M. (2000) J. Mol. Biol. 299, 1113-1119. [DOI] [PubMed] [Google Scholar]
- 12.Catoira, R., Galera, C., de Billy, F., Penmetsa, R. V., Journet, E.-P., Maillet, F., Rosenberg, C., Cook, D., Gough, C. & Dénarié, J. (2000) Plant Cell 12, 1647-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kató, P. & Kiss, G. B. (2002) Nature 417, 962-966. [DOI] [PubMed] [Google Scholar]
- 14.Ané, J.-M., Kiss, G. B., Riely, B. K., Penmetsa, R. V., Oldroyd, G. E. D., Ayax, C., Lévy, J., Debellé, F., Baek, J.-M., Kalo, P., et al. (2004) Science 303, 1364-1367. [DOI] [PubMed] [Google Scholar]
- 15.Lévy, J., Bres, C., Geurts, R., Chalhoub, B., Kulikova, O., Duc, G., Journet, E.-P., Ané, J.-M., Lauber, E., Bisseling, T., et al. (2004) Science 303, 1361-1364. [DOI] [PubMed] [Google Scholar]
- 16.Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczyglowski, K., et al. (2002) Nature 417, 959-962. [DOI] [PubMed] [Google Scholar]
- 17.Parniske, M. (2004) Curr. Opin. Plant Biol. 7, 414-421. [DOI] [PubMed] [Google Scholar]
- 18.Esseling, J. J., Lhuissier, F. G. P. & Emons, A. M. C. (2004) Plant Cell 16, 933-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goormachtig, S., Capoen, W. & Holsters, M. (2004) Trends Plant Sci. 9, 518-522. [DOI] [PubMed] [Google Scholar]
- 20.Goormachtig, S., Mergaert, P., Van Montagu, M. & Holsters, M. (1998) Subcell. Biochem. 29, 117-164. [DOI] [PubMed] [Google Scholar]
- 21.D'Haeze, W., Gao, M., De Rycke, R., Van Montagu, M., Engler, G. & Holsters, M. (1998) Mol. Plant–Microbe Interact. 11, 999-1008. [Google Scholar]
- 22.Goormachtig, S., Capoen, W., James, E. K. & Holsters, M. (2004) Proc. Natl. Acad. Sci. USA 101, 6303-6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 24.Goormachtig, S., Alves-Ferreira, M., Van Montagu, M., Engler, G. & Holsters, M. (1997) Mol. Plant–Microbe Interact. 10, 316-325. [DOI] [PubMed] [Google Scholar]
- 25.Corich, V., Goormachtig, S., Lievens, S., Van Montagu, M. & Holsters, M. (1998) Plant Mol. Biol. 37, 67-76. [DOI] [PubMed] [Google Scholar]
- 26.Kiefer, E., Heller, W. & Ernst, D. (2000) Plant Mol. Biol. Rep. 18, 33-39. [Google Scholar]
- 27.Karimi, M., Inzé, D. & Depicker, A. (2002) Trends Plant Sci. 7, 193-195. [DOI] [PubMed] [Google Scholar]
- 28.Van de Velde, W., Mergeay, J., Holsters, M. & Goormachtig, S. (2003) Plant Sci. 165, 1281-1288. [Google Scholar]
- 29.Kistner, C. & Parniske, M. (2002) Trends Plant Sci. 7, 511-518. [DOI] [PubMed] [Google Scholar]
- 30.Demchenko, K., Winzer, T., Stougaard, J., Parniske, M. & Pawlowski, K. (2004) New Phytol. 163, 381-392. [DOI] [PubMed] [Google Scholar]
- 31.Parniske, M. (2000) Curr. Opin. Plant Biol. 3, 320-328. [DOI] [PubMed] [Google Scholar]
- 32.Limpens, E., Mirabella, R., Fedorova, E., Franken, C., Franssen, H., Bisseling, T. & Geurts, R. (2005) Proc. Natl. Acad. Sci. USA 102, 10375-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schulze-Lefert, P. (2004) Curr. Opin. Plant Biol. 7, 377-383. [DOI] [PubMed] [Google Scholar]
- 34.Goedhart, J., Hink, M. A., Visser, A. J. W. G., Bisseling, T. & Gadella, T. W. J., Jr. (2000) Plant J. 21, 109-119. [DOI] [PubMed] [Google Scholar]
- 35.Wais, R. J., Galera, C., Oldroyd, G., Catoira, R., Penmetsa, R. V., Cook, D., Gough, C., Dénarié, J. & Long, S. R. (2000) Proc. Natl. Acad. Sci. USA 97, 13407-13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timmers, A. C. J., Auriac, M.-C., de Billy, F. & Truchet, G. (1998) Development (Cambridge, U.K.) 125, 339-349. [DOI] [PubMed] [Google Scholar]
- 37.Relić, B., Perret, X., Estrada-García, M. T., Kopcinska, J., Golinowski, W., Krishnan, H. B., Pueppke, S. G. & Broughton, W. J. (1994) Mol. Microbiol. 13, 171-178. [DOI] [PubMed] [Google Scholar]
- 38.Goethals, K., Gao, M., Tomekpe, K., Van Montagu, M. & Holsters, M. (1989) Mol. Gen. Genet. 219, 289-298. [DOI] [PubMed] [Google Scholar]