Abstract
Soluble Hsp70 homologs cotranslationally interact with nascent polypeptides in all kingdoms of life. In addition, fungi possess a specialized Hsp70 system attached to ribosomes, which in Saccharomyces cerevisiae consists of the Hsp70 homologs Ssb1/2p, Ssz1p, and the Hsp40 homolog zuotin. Ssz1p and zuotin are assembled into a unique heterodimeric complex termed ribosome-associated complex. So far, no such specialized chaperones have been identified on ribosomes of higher eukaryotes. However, a family of proteins characterized by an N-terminal zuotin-homology domain fused to a C-terminal two-repeat Myb domain is present in animals and plants. Members of this family, like human MPP11 and mouse MIDA1, have been implicated in the regulation of cell growth. Specific targets of MPP11/MIDA1, however, have remained elusive. Here, we report that MPP11 is localized to the cytosol and associates with ribosomes. Purification of MPP11 revealed that it forms a stable complex with Hsp70L1, a distantly related homolog of Ssz1p. Complementation experiments indicate that mammalian ribosome-associated complex is functional in yeast. We conclude that despite a low degree of homology on the amino acid level cooperation of ribosome-associated chaperones with the translational apparatus is well conserved in eukaryotic cells.
Keywords: myb domain, NatA, protein folding, translation
Newly synthesized polypeptides leave the ribosome through a tunnel in the large ribosomal subunit. At the tunnel exit a number of ribosome-associated factors interact with the nascent polypeptide and initiate folding, covalent modifications, and translocation to the various cellular compartments (1). Yeast Ssb1/2p was the first Hsp70 homolog identified as a component associated with ribosomes (2). Ssb1/2p closely contacts nascent polypeptides right after they emerge from the tunnel (3, 4). Later, it was found that the major fraction of the Hsp40 homolog zuotin is bound to ribosomes of yeast. Based on localization and concordant phenotypes of Δssb1Δssb2 and Δzuo1 strains, it was proposed that Ssb1/2p and zuotin function together in the same cellular process (5). In vitro, however, zuotin does not stimulate Ssb1/2p's ATPase as would be expected for a classical J partner (6). This puzzle was solved with the identification of Ssz1p, a third chaperone component on yeast ribosomes. The Hsp70 homolog Ssz1p together with zuotin forms an unusual dimeric ribosome-associated complex (RAC) (7). It is now established that the two subunits of RAC and Ssb1/2p form a chaperone triad, and that each of the chaperones is essential for functionality in vivo and in vitro (6, 8, 9). The mechanism of the yeast chaperone triad, however, remains poorly defined. Ssb1/2p and RAC might separate newly synthesized polypeptides from the surface of the ribosome or postpone folding until a full domain has emerged from the tunnel (10). Recently, it was discovered that the absence of functional RAC or Ssb1/2p reduces translational fidelity (11).
Because the translational apparatus of eukaryotic cells displays a high degree of conservation, one might expect the yeast chaperone triad to be a general component of eukaryotic ribosomes. However, no member of the mammalian Hsp70 family stands out as an obvious homolog of Ssz1p or Ssb1/2p. In contrast, cytosolic Hsp70s with classical chaperone properties like human Hsc70 and yeast Ssa1p are highly conserved. Southern hybridization experiments using zuotin DNA to probe plant and animal DNA were negative (12), and so far no biochemical evidence for the existence of a Ssb1/2p, Ssz1p, or zuotin homolog has been reported. Hence it was speculated that yeast and fungi contain a specialized group of chaperones that is not conserved in higher eukaryotes (10, 13, 14). Zuotin contains an unusual N-terminal domain found only in a small group of eukaryotic Hsp40 homologs, such as mouse MIDA1 (15) and human MPP11 (16). MPP11 was identified with an antibody recognizing a specific set of phosphopeptides, and it was suggested that it functions during cell division (17). MIDA1 was identified as an Id1-interacting protein in a GST pull-down assay (18). Id1 is a member of a family of helix-loop-helix proteins involved in cell type-specific transcription and cell lineage commitment (19). In addition, it was found that MIDA1 displays DNA-binding activity (18, 20). According to the current model, MPP11/MIDA1 is a multifunctional protein involved in transcriptional control through interaction with multiple factors (21). These data provided no evidence for a function of MPP11/MIDA1 during translation and ribosome association was never tested directly. We speculated that if MPP11/MIDA1 was a functional homolog of zuotin the protein should predominantly localize to cytosolic ribosomes and interact with an Hsp70 partner. Here, we show that indeed MPP11/MIDA1 forms a stable complex with the Hsp70 homolog Hsp70L1, and that this mammalian complex rescues growth defects in yeast strains lacking functional RAC. These results indicate that ribosome-associated chaperones are conserved from yeast to human and have important implications for our understanding of MPP11's role in the cell.
Materials and Methods
Cloning, Expression, and Generation of Polyclonal Antibodies. The genes encoding MPP11 (GenBank accession no. CAA66913), Hsp70L1 (GenBank accession no. NP_057383), and HYPK (GenBank accession no. NP_057484) were cloned from human cDNA. Each gene was fused to an N-terminal hexahistidine tag by using the Escherichia coli expression vector pET28a (Novagen). His-6-MPP11 and His-6-HYPK were soluble, and His-6-Hsp70L1 was contained in inclusion bodies. Upon coexpression of His-6-MPP11 and His-6-Hsp70L1 a significant fraction of His-6-Hsp70L1 was detected in the soluble fraction. The proteins were purified by using Ni-NTA according to the manufacturer's protocol for native or denatured protein purification, respectively (Qiagen, Valencia, CA) and used to generate polyclonal rabbit antibodies (Eurogentec, Brussels). α-HYPK recognizes HYPK only when highly overexpressed in E. coli.
Yeast Strains and Plasmids for Expression in Yeast. Standard yeast genetic techniques were used (22). MH272-3f a/α (ura3/ura3, leu2/leu2, his3/his3, trp1/trp1, ade2/ade2) is the parental wild-type strain of all haploid derivatives used in this study. Deletion strains lacking SSZ1 (Δssz1: ssz1::LEU2), ZUO1 (Δzuo1: zuo1::TRP1), or both genes (Δzuo1Δssz1: zuo1::TRP1 ssz1::LEU2) were as described (7). Construction of the J-domain mutant of zuotin was as described (8). SSA1 was expressed from the 2-μ plasmid pRS423. For expression in yeast MPP11, MPP11Δ362-568, which lacks the C-terminal Myb domain, and Hsp70L1 were cloned into plasmid pESC-Ura (GAL1/GAL10 promoter, Stratagene). MPP11 and MPP11Δ362-568 were fused to an N-terminal flag tag, and Hsp70L1 was fused to an N-terminal myc tag. Coexpression vectors were generated by cloning two genes into a single vector. Resulting plasmids are: pESC-flagMPP11, pESC-flagMPP11ΔC, pESC-mycHsp70L1, pESC-f lagMPP11-mycHsp70L1, and pESC-flagMPP11ΔC-mycHsp70L1.
Immunofluorescence Microscopy. HeLa cells were fixed in 3.7% formaldehyde and subsequently permeabilized with 0.1% Triton X-100. Cells were treated with α-MPP11 followed by incubation with a secondary Cy3-conjugated donkey anti-rabbit antibody (Jackson ImmunoResearch). Samples were analyzed with a Nikon Eclipse TE300 microscope (23).
Preparation of Ribosomes from Rat Liver. Purification of free ribosomes was performed as described (24) with minor modifications. Fifty grams of liver from male wistar rats was homogenized in 200 ml of S1 buffer (20 mM Hepes-KOH, pH 7.4/5 mM MgAcetate/50 mM KAcetate/1 mM DTT/1 mM PMSF) containing 250 mM sucrose. Postmitochondrial supernatant (PMS) was prepared by centrifugation at 18,000 × g for 15 min followed by filtration through glass wool. PMS (4.2 ml) was layered on top of a two-step sucrose cushion (3.3 ml of 0.5 M sucrose in S1 buffer, 2.5 ml of 2 M sucrose in S1 buffer) and was centrifuged for 1.5 h at 200,000 × g to separate ribosomes from cytosolic proteins.
Partial Purification of MPP11 from Reticulocyte Lysate. Ribosomes were isolated from 100 ml of rabbit reticulocyte lysate (Green Hectares, Oregon, WI) by centrifugation at 200,000 × g for 1.5 h. The ribosomal pellet was washed once in 10 ml of S1 buffer and resuspended in S1 buffer containing KAcetate concentrations as indicated. For purification of MPP11, ribosome-associated proteins were released by 400 mM KAcetate. High-salt washed ribosomes (hs-pel) were separated from released proteins (hs-sup) by centrifugation at 200,000 × g for 1.5 h. The hs-sup was diluted with 3 volumes of buffer S1 lacking KAcetate, MgAcetate, and DTT and was loaded onto a MonoS HR5/5 (Amersham Pharmacia). Bound proteins were eluted with a 50-1,200-mM, 25-ml linear KAcetate gradient in 40 mM Hepes-KOH, pH 7.4. MPP11 eluted at a concentration of 330-400 mM KAcetate. MPP11-containing fractions were pooled, diluted with 40 mM Hepes-KOH, pH 7.4 to a final salt concentration of 100 mM KAcetate, and loaded onto a MonoQ HR5/5 (Amersham Pharmacia). Bound proteins were eluted with a 50- to 1,000-mM, 18-ml linear KAcetate gradient in 40 mM Hepes-KOH, pH 7.4. MPP11 eluted at a concentration of 330-450 mM KAcetate.
Blue Native PAGE (BN-PAGE). BN-PAGE was performed as described on 6-16.5% gradient gels at 4°C (25, 26). Native protein samples were applied to BN-PAGE in S2 buffer (10% glycerol/10 mM bis-Tris, pH 7.0/50 mM ε-aminocaproic acid). Denatured protein samples were prepared by resuspending trichloroacetic acid-precipitated samples in S2 buffer containing 0.5% SDS. Proteins from BN-PAGE were either transferred onto poly(vinylidene difluoride) membranes or lanes were cut, incubated in 0.1% SDS/1% β-mercaptoethanol for 15 min, and applied to a second-dimension 10% Tris-Tricine gel (27). Spots from second-dimension gels were digested with trypsin essentially as described (28). Mass spectra were recorded on a Bruker Reflex III instrument (Bruker Daltonics, Bremen, Germany). Data bank searches with the peptide masses obtained were performed with mascot software (www.matrixscience.com).
Ribosome Profiles. HeLa cells at 90% confluency were incubated with 100 μg/ml cycloheximide for 1 h, trypsinated, collected, and lysed with 0.5% Triton X-100 in S3 buffer (50 mM Hepes-KOH, pH 7.4/1 mM MgAcetate/1 mM PMSF) containing 80 mM KAcetate, 700 mM KAcetate, or 80 mM KAcetate plus 0.25 mg/ml RNase A as indicated. After a clarifying spin for 20 min at 20,000 × g 10 OD260 units were loaded on top of a 10.8-ml 15-55% sucrose gradient in S3 buffer containing either 80 or 700 mM KAcetate. After centrifugation at 150,000 × g for 5 h at 4°C gradients were fractionated with a density gradient fractionator monitoring A260 (ISCO).
Miscellaneous. 125I-labled protein A (29) or ECL (PerkinElmer) was used to develop immunoblots.
Results
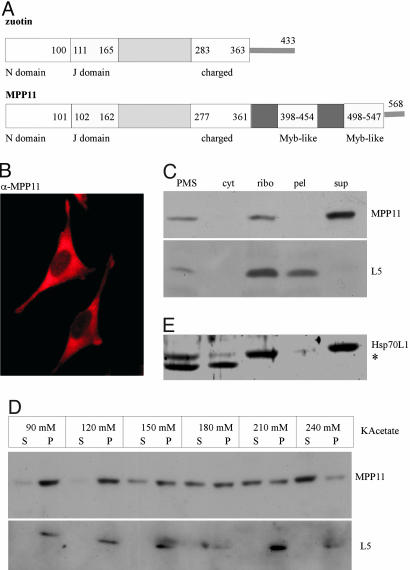
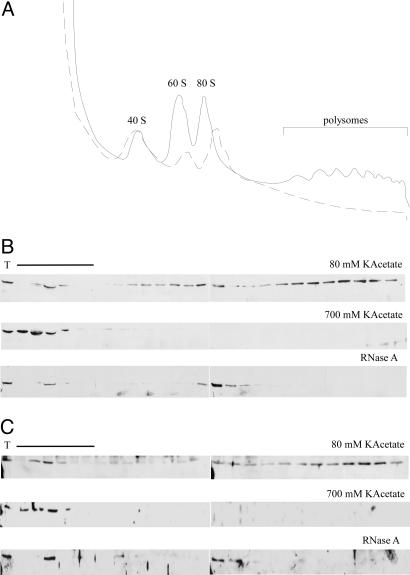
The Mammalian Zuotin Homolog MPP11/MIDA1 Is Bound to Cytosolic Ribosomes. Yeast zuotin belongs to the type III subfamily of J proteins (30). Mutational analysis revealed that the N domain, the J domain, and the charged region are essential for zuotin's in vivo function (5). Zuotin's N domain is conserved only in close homologs from higher eukaryotes, which possess an overall similar domain structure plus an extra C-terminal domain containing two Myb DNA-binding motifs (Fig. 1A). As a tool to investigate the function of the mammalian zuotin homologs we raised a polyclonal antibody (α-MPP11). On immunoblots of HeLa cell extracts α-MPP11 recognized a single band indicating specificity of the antibody (data not shown). Immunofluorescence of HeLa cells revealed that MPP11 was localized to the cytosol (Fig. 1B). Localization of MPP11 was more precisely assessed by fractionation of a rat liver homogenate. The bulk of rat liver MPP11 was contained in the cytosolic fraction and cosedimented with a ribosomal marker (Fig. 1C and data not shown). Binding of rat liver MPP11 to ribosomes was salt-sensitive, and the protein was released at a concentration of 200-250 mM KAcetate (Fig. 1D). When HeLa cell extracts were analyzed on sucrose density gradients the bulk of MPP11 colocalized with monosomes and polysomes. Upon high-salt treatment MPP11 was released to the cytosolic fraction. RNase A treatment destroyed polysomes and concomitantly MPP11 was shifted to monosomes (Fig. 2 A and B). The combination of data indicates that the bulk of MPP11 is associated with cytosolic mammalian ribosomes in a salt-sensitive manner.
Fig. 1.
MPP11 and Hsp70L1 localize to the mammalian cytosol and are bound to ribosomes in a salt-sensitive manner. (A) Domain structure of zuotin and MPP11. Yeast zuotin and human MPP11 share 37% identity within the zuotin-homology region (30). MPP11 possesses an additional two-repeat Myb domain (37). (B) Cytosolic distribution of MPP11. HeLa cells were immunostained with α-MPP11 as described in Materials and Methods. (C) Rat liver MPP11 is bound to cytosolic ribosomes. Postmitochondrial supernatant (PMS) was separated into a cytosolic fraction (cyt) and a ribosomal pellet (ribo) by centrifugation at low-salt concentration (see Materials and Methods). The ribosomal pellet was resuspended, adjusted to 700 mM KAcetate, and separated into a ribosomal pellet (pel) and the corresponding supernatant (sup). (D) MPP11 from rabbit reticulocytes is released from ribosomes by KAcetate. Ribosomes isolated at 50 mM KAcetate were adjusted to the indicated concentrations of KAcetate and subsequently separated into a ribosomal pellet (P) and supernatant (S). (E) Rat liver Hsp70L1 is bound to cytosolic ribosomes. Localization of Hsp70L1 was analyzed as described in C. Aliquots of the samples in C-E were analyzed by immunoblotting using α-MPP11, α-Hsp70L1, and α-L5 (ribosomal protein of the mammalian large subunit). In rat liver extracts α-Hsp70L1 detected an additional band (*), which is either a degradation product of Hsp70L1 or a cross-reaction.
Fig. 2.
MPP11 and Hsp70L1 comigrate with ribosomes. (A) HeLa cell extracts were run through sucrose gradients, and A260 of the gradient fractions was traced. Solid line indicates untreated, and dashed line indicates RNase A-treated. (B) In a low-salt sucrose gradient (80 mM KAcetate) the major fraction of MPP11 colocalizes with polysomes; in a high-salt sucrose gradient (700 mM KAcetate) MPP11 is released from ribosomes and polysomes and colocalizes with cytosolic proteins; after RNase A treatment polysomes are converted to monosomes and MPP11 colocalizes with the monosomes. (C) As in B but analyzed for Hsp70L1. T indicates total extract. Because of the high protein content in the cytosol, when compared with ribosomal fractions, loading in some areas (labeled with the bar) was reduced to 25%. Fractions were analyzed by immunoblotting using α-MPP11 and α-Hsp70L1.
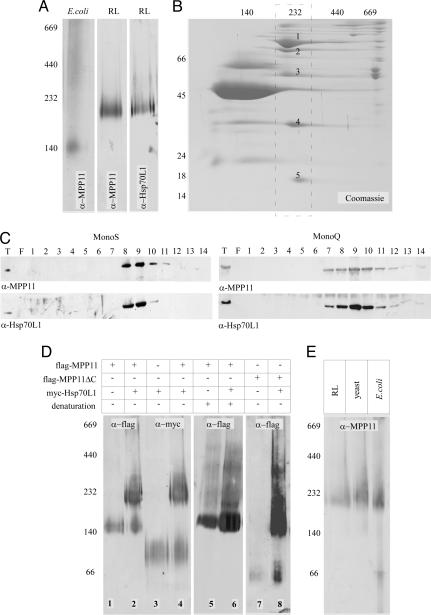
The Hsp40 Homolog MPP11 Forms a Complex with Hsp70L1. To assess whether MPP11 was part of an oligomeric complex, we performed BN-PAGE analysis. MPP11 from reticulocytes was detected at ≈200 kDa, whereas His-6-MPP11 expressed in E. coli was detected at ≈140 kDa (Fig. 3A). Apparent size on BN-PAGE often differs from the actual molecular mass of proteins and complexes. However, the difference between reticulocyte lysate MPP11 and heterologously expressed MPP11 suggested that the authentic mammalian protein was part of a larger complex.
Fig. 3.
MPP11 and Hsp70L1 form a heterodimeric complex. (A) A lysate derived from E. coli expressing His-6-MPP11 and proteins released by high-salt treatment from rabbit reticulocyte ribosomes (RL) were run on BN-PAGE and analyzed by immunoblotting using α-MPP11 or α-Hsp70L1 as indicated. (B) Partly purified MPP11 was loaded onto BN-PAGE followed by a second-dimension 10% Tris-Tricine gel. The 200-kDa band (dashed rectangle) contained five major proteins, which were identified as the rabbit homologs of human NMDA (spot 1), MPP11 (spot 2), Hsp70L1 (spot 3), ARD1 (spot 4), and HYPK (spot 5). (C) MPP11 and Hsp70L1 coelute on cation (MonoS) and anion (MonoQ) exchange columns. Proteins released from rabbit reticulocyte ribosomes were loaded onto MonoS and eluted with a KAcetate gradient. Fractions 8 and 9 were pooled and loaded onto a MonoQ. Immunoblots were developed with α-MPP11 and α-Hsp70L1. (D) MPP11 and Hsp70L1 form a complex in the yeast cytosol. Extracts from wild-type yeast expressing flag-MPP11, flag-MPP11ΔC, or myc-Hsp70L1 as indicated were loaded onto BN-PAGE. Samples in lanes 5 and 6 were denatured with SDS before analysis. BN gels were analyzed by immunoblotting using α-flag and α-myc antibodies as indicated. (E) Rabbit reticulocyte lysate (RL), extracts from yeast coexpressing flag-MPP11 and myc-Hsp70L1, and extracts from E. coli coexpressing His-6-MPP11 and His-6-Hsp70L1 were analyzed on BN-PAGE followed by immunoblotting using α-MPP11 (for details see Materials and Methods).
Reticulocyte lysate was used for the identification of possible complex partner(s) (see Materials and Methods). As a final step, fractions enriched for MPP11 were loaded onto a preparative BN-PAGE followed by a second-dimension SDS/PAGE (Fig. 3B). Five major proteins were contained in the 200-kDa region and were identified as the rabbit homologs of human NMDA (101.2 kDa), MPP11 (67.4 kDa), Hsp70L1 (54.7 kDa), ARD1 (26.4 kDa), and HYPK (14.6 kDa). A homolog of Id1 was not detected, suggesting that the ribosome-associated fraction of MPP11 was not bound to this transcription factor (see Introduction). NMDA and ARD1 are homologs of yeast Nat1p and Ard1p, respectively. In yeast and mammals these two proteins are known components of the NatA complex (31, 32). Using a crosslink approach we recently found that Nat1p is close to nascent polypeptides in yeast (4). The fact that NMDA and ARD1 were contained in the ribosomal salt wash suggests that mammalian NatA is also ribosome-associated. HYPK is a poorly characterized protein originally identified in a screen for proteins interacting with huntingtin (33). The reason for its copurification with the NMDA/ARD1 and MPP11/Hsp70L1 complexes awaits further investigation.
The cellular localization of the poorly characterized Hsp70 homolog Hsp70L1 has previously not clearly been established (34). On the sequence level neither peptide binding domain nor ATPase domains of Hsp70L1 are particularly similar to Ssz1p (data not shown). To determine whether MPP11 and Hsp70L1 behaved like components of a stable complex, a polyclonal antibody recognizing Hsp70L1 (α-Hsp70L1) was raised. Consistent with complex formation MPP11 and Hsp70L1 were contained in the same fractions of a rat liver homogenate (Fig. 1 C and E) and comigrated on BN-PAGE (Fig. 3A). In addition, MPP11 and Hsp70L1 coeluted on cation (MonoS) as well as on anion exchange columns (MonoQ) (Fig. 3C). Ribosome binding of Hsp70L1 was confirmed on sucrose density gradients. Hsp70L1 comigrated with polysomes and ribosomes and was released by high-salt treatment (Fig. 2 A and C).
To test complex formation more rigorously, MPP11 and Hsp70L1 were expressed in yeast (Fig. 4 Upper). Under native conditions yeast-expressed MPP11 migrated with an apparent molecular mass ≈140 kDa on BN-PAGE (Fig. 3D, lane 1). The size corresponded well to the size of E. coli-expressed MPP11 (Fig. 3A). When Hsp70L1 was coexpressed in yeast, a large fraction of MPP11 was shifted to a band of ≈200 kDa, corresponding to the apparent size of authentic MPP11 in reticulocyte lysate (Fig. 3 D, lane 2, and E). Conversely, when Hsp70L1 was expressed in yeast, the protein migrated with an apparent size of ≈80 kDa, and only upon coexpression of MPP11 a large fraction of Hsp70L1 was detected in a 200-kDa complex (Fig. 3D, lanes 3 and 4). Yeast RAC is a heterodimeric complex (7). The apparent molecular mass of mammalian RAC (mRAC) (≈200 kDa), MPP11 (≈140 kDa), and Hsp70L1 (≈80 kDa) on BN-PAGE was more compatible with a heterotrimeric composition (Fig. 3D, lanes 1-4). To clarify whether mRAC contained two subunits of MPP11 per Hsp70L1 the complex was denatured before analysis on BN-PAGE. The 200-kDa band disappeared upon denaturation, whereas the 140-kDa band persisted (Fig. 3D, lanes 5 and 6). The result suggested that, in the absence of Hsp70L1, MPP11 was a monomer migrating aberrantly on BN-PAGE. We generated a truncated version of MPP11, lacking the C-terminal Myb-like domain (amino acids 362-568, Fig. 1A). MPP11ΔC migrated with an apparent molecular mass of ≈70 kDa, suggesting that the C-terminal domain of MPP11 was responsible for the unusual migration of full-length MPP11 on BN-PAGE. MPP11ΔC was capable of forming a complex with Hsp70L1, which migrated with an apparent molecular mass of 140 kDa (Fig. 3D, lanes 7 and 8). The combined data suggest that mRAC contains one subunit of MPP11 and one subunit of Hsp70L1 like its counterpart from yeast. Because of the poor quality of our HYPK antibody we were unable to directly test whether HYPK was contained in the MPP11-Hsp70L1 complex from authentic tissues. However, the complex recognized by α-MPP11 was of similar size independent of whether reticulocyte lysate or extracts from yeast or E. coli coexpressing MPP11 and Hsp70L1 were analyzed on BN-PAGE (Fig. 3E).
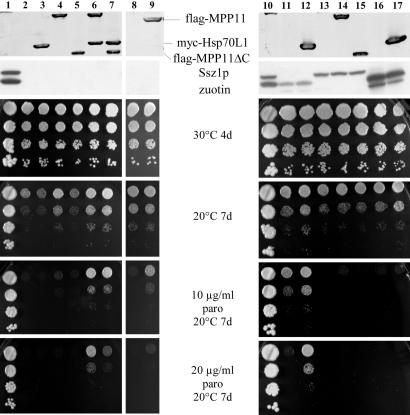
Fig. 4.
mRAC complements growth defects of yeast strains lacking functional yeast RAC. (Upper) Immunoblot of total yeast extracts decorated with α-flag (recognizing flag-MPP11 and flag-MPP11ΔC), α-myc (recognizing myc-Hsp70L1), α-zuotin, and α-Ssz1p antibodies. Note that in the absence of Ssz1p zuotin is destabilized and a degradation product is generated (7). (Lower) Haploid yeast strains were grown to early log phase at 30°C on galactose containing minimal medium. Serial 10-fold dilutions containing the same number of cells were spotted onto rich galactose plates containing paromomycin (paro) as indicated. Lane 1, wild type. Lane 2, Δzuo1Δssz1. Lane 3, Δzuo1Δssz1+pESC-mycHsp70L1. Lane 4, Δzuo1Δssz1+pESC-flagMPP11. Lane 5, Δzuo1Δssz1+pESC-flagMPP11ΔC. Lane 6, Δzuo1Δssz1+pESC-flagMPP11-mycHsp70L1. Lane 7, Δzuo1Δssz1+pESC-flagMPP11ΔC-mycHsp70L1. Lane 8, Δzuo1Δssz1+pRS-SSA1. Lane 9, Δzuo1Δssz1+pESC-flagMPP11+pRS-SSA1. Lane 10, wild type. Lane 11, Δssz1. Lane 12, Δssz1+pESC-mycHsp70L1. Lane 13, Δzuo1. Lane 14, Δzuo1+pESC-flagMPP11. Lane 15, Δzuo1+pESC-flagMPP11ΔC. Lane 16, Δzuo1Δssz1+2 μzuo1-H128Q. Lane 17, Δzuo1Δssz1+2μzuo1-H128Q+pESC-mycHsp70L1.
mRAC Complements Defects Caused by the Absence of Ssz1p and Zuotin in Yeast. Yeast strains lacking functional RAC suffer from severe growth defects. Δzuo1, Δssz1, and Δzuo1Δssz1 strains grow slowly and are cold-sensitive and hypersensitive toward aminoglycosides, such as paromomycin (5, 7). To test whether mRAC was functional in yeast, MPP11 and Hsp70L1 were expressed in the respective deletion strains (Fig. 4 Upper). In a first set of experiments expression of the mammalian homologs was analyzed in Δzuo1Δssz1 (Fig. 4, lanes 2-9). MPP11 was able to partly complement cold sensitivity and hypersensitivity toward paromomycin at 30°C, but failed to support growth in the presence of paromomycin at 20°C (Fig. 4, lane 4 and data not shown). Hsp70L1 by itself failed to complement any of the growth defects displayed by Δzuo1Δssz1 (Fig. 4, lane 3). Most efficient complementation was obtained when MPP11 and Hsp70L1 were coexpressed. In the presence of both mRAC subunits, Δzuo1Δssz1 tolerated paromomycin even at 20°C (Fig. 4, lane 6). The synergistic effect of MPP11 and Hsp70L1 indicated that mRAC was not only assembled (Fig. 3D) but also functional in yeast. To test the specificity of the MPP11/Hsp70 interaction the cytosolic Hsp70 homolog Ssa1p was overexpressed in the absence or presence of MPP11. An increased level of Ssa1p alone did not affect growth of Δzuo1Δssz1 (Fig. 4, lane 8 and data not shown). Combined expression of MPP11 and Ssa1p failed to improve growth of Δzuo1Δssz1 at 20°C; however, it increased tolerance toward paromomycin (Fig. 4, lanes 8 and 9). A direct comparison of Δzuo1Δssz1 coexpressing either MPP11/Hsp70L1 or MPP11/Ssa1p revealed that complementation was significantly more efficient in the presence of Hsp70L1 (Fig. 4, lanes 6 and 9, and data not shown).
Next, we asked whether the Myb domain of MPP11 had any effect on the function of the protein in yeast. To that end, MPP11ΔC lacking the Myb domain was expressed in Δzuo1Δssz1 (Fig. 4 Upper, lanes 5 and 7). Suppression by MPP11ΔC was less efficient than by the full-length protein (Fig. 4, lane 5). Coexpression of Hsp70L1 improved suppression by MPP11ΔC, however, not to the level obtained by full-length MPP11 (Fig. 4, lane 7).
Mammalian Hsp70L1 Functionally Interacts with Yeast Zuotin, Whereas Mammalian MPP11 Is Not Supported by Yeast Ssz1p. To address whether functional complexes could be formed by a combination of yeast and mRAC subunits, MPP11 and MPP11ΔC were expressed in a Δzuo1 strain, whereas Hsp70L1 was expressed in a Δssz1 background. A direct comparison revealed that the presence of Ssz1p did not improve complementation by MPP11 or MPP11ΔC (Fig. 4, lanes 14 and 15). Hsp70L1, which was unable to complement in a Δzuo1Δssz1 background, improved growth in the presence of yeast zuotin, suggesting that the two proteins interact (Fig. 4, lane 12). Finally, we used a nonfunctional J-domain mutant of zuotin (zuo1-H128Q), which is known to associate with ribosomes and Ssz1p but fails to functionally interact with Ssb1/2p (6, 8). Expression of Hsp70L1 in the presence of zuo1-H128Q failed to complement growth defects of Δzuo1Δssz1 (Fig. 4, lane 17). The finding suggested that it was not sufficient to anchor Hsp70L1 to the ribosome, but that complementation by Hsp70L1 required functional interaction with the J domain of zuotin.
Discussion
Human MPP11 and mouse MIDA1 are widely expressed in different tissues (16, 35). In the course of this study we have detected MPP11, or a close homolog, in rabbit reticulocyte lysate, rat liver, and the HeLa cell line. In all three samples MPP11 was found together with Hsp70L1 in a ribosome-associated complex that we subsequently termed mRAC. Complementation in yeast suggested that mRAC can productively interact with the yeast ribosome. However, the individual mRAC subunits differ in their ability to interact with the yeast translation machinery. Hsp70L1 was able to functionally interact with zuotin. In this context it is important to recall that only the complex of zuotin and Ssz1p (RAC) is able to stimulate the ATPase of the yeast ribosome-bound Hsp70 Ssb1/2p (6, 8). As complementation by zuotin/Hsp70L1 depended on an intact J domain of zuotin, Hsp70L1 most likely can replace Ssz1p and support zuotin's interaction with Ssb1/2p (see Introduction). MPP11 was unable to functionally interact with Ssz1p. Rather, suppression by MPP11 was decreased in the presence of Ssz1p. Although the reason for this observation is unclear, one possible explanation is that MPP11 and Ssz1p interact; however, the complex is nonfunctional and might either trap MPP11 or block ribosomal binding sites. When Hsp70L1 is absent, MPP11 most likely functions in combination with Ssa1p and/or other cytosolic Hsp70 homolog(s). After all, overexpression of Ssa1p improves MPP11-mediated suppression, although not to the same extent as Hsp70L1. Possibly, some properties of Hsp70L1 are more similar to Ssa1p than to Ssz1p. Alternatively, the complex of MPP11 and Hsp70L1 might function in combination with an additional Hsp70 homolog. This situation would resemble the yeast ribosome where RAC and Ssb1/2p form a functional triad (6, 8). One should keep in mind that, as for Ssz1p and Hsp70L1, the primary sequence of Ssb1/2p's functional mammalian homolog might not be well conserved between fungi and mammals.
MPP11/MIDA1 is an unusual multidomain protein with an N-terminal J domain plus a C-terminal two-repeat Myb domain (Fig. 1A). The domain structure of MPP11 is of specific interest as the J domain is an ancient module that is conserved throughout all kingdoms of life, whereas the Myb domain is confined to eukaryotes. J proteins as well as Myb proteins comprise large and diverse protein families with multiple cellular functions. J proteins stimulate the rate of ATP hydrolysis in partner chaperones of the Hsp70 family, and together they function in folding, translocation, and degradation of protein substrates. Proteins containing a Myb domain bind to DNA and are involved in transcriptional regulation and cell-cycle progression (30, 36, 37). Interestingly, the mammalian endoplasmic reticulum contains a membrane-bound J protein that interacts with ribosomes and contains a C-terminal domain with homology to the Myb domain (38). Phylogenetic analysis suggests that fungal zuotin homologs originate from MPP11-like ancestors, which have lost the Myb domain during evolution (39). Our in vivo complementation data demonstrate that the C-terminal domain of MPP11 is not required for complex formation with Hsp70L1 but contributes to complementation. Whether or not the Myb domain of MPP11 has a function beyond the ribosome remains to be established. Although the mouse homolog MIDA1 can bind to DNA the current picture is somewhat inconsistent, and the significance of DNA binding has remained elusive. First, the entire zuotin-homology region binds to the left-handed DNA conformation, termed Z-DNA in vitro (18). Second, the Myb domain of MIDA1 recognizes a specific hairpin structure consisting of seven nucleotides surrounded by a short palindrome (20). Third, DNA binding in vivo was suggested based on the ability of MIDA1 to act as a transcriptional activator (21). Surprisingly, however, MIDA1 in vivo affected only heterologously expressed genes in a promoter-independent manner and the J domain rather than the Myb domain was required for transcriptional activation. In this context it is interesting to recall that yeast zuotin was originally identified as a protein interacting with Z-DNA (12), has affinity to tRNA (40), and, last but not least, binds to ribosomal RNA (5). Based on this evidence, it was suggested that zuotin interacts with a variety of nucleic acids and its specificity for ribosomes in vivo is achieved via additional interactions with ribosomal proteins (5). Association of MPP11/MIDA1's zuotin-homology domain with DNA might reflect a similar kind of general affinity for nucleic acids.
Whether the function of MPP11 is confined to translation or additionally involves transcriptional regulation is also of medical relevance. Increased levels of MPP11 were detected in a high percentage of head and neck squamous cell tumors (16) and in patients with acute myeloid leukemia (41). MPP11 was also found as a leukemia-associated antigen in patients with leukemias (42). MIDA1 was identified as a tumor-associated antigen in a mouse model, and significant suppression of tumor growth in animals immunized with MIDA1 was demonstrated (43). A role of MPP11/MIDA1 in transcriptional regulation is consistent with these results and was put forward in the context of MPP11/MIDA1's role in tumorogenesis. However, transcriptional regulation is not the only possible function that could connect MPP11/MIDA1 to cancer progression. In the past few years it has become clear that translation is an important control point, particularly during embryonic development and regulation of cell growth and differentiation. Several initiation factors, elongation factors, and ribosomal proteins are overexpressed in human tumors (44, 45), and important tumor suppressors and protooncogenes affect ribosome biogenesis or regulate the activity of translation factors (46). Our findings raise the possibility that MPP11 might be involved in cellular transformation via its connection to the translational apparatus.
Acknowledgments
We thank Yves Dubaquié, Matthias Gautschi, and members of the Rospert laboratory for discussion and critical reading of the manuscript. This work was supported by the Fonds der Chemischen Industrie (S.R.) and Sonderforschungsbereich Grants 388 and 610 (to S.R).
Author contributions: S.R. designed research; H.O., C.C., P.M., T.W., C.K.S., P.J., P.R., J.S., and S.R. performed research; H.O., C.C., P.M., C.K.S., P.J., P.R., J.S., and S.R. analyzed data; C.K.S. contributed new reagents/analytic tools; and S.R. wrote the paper.
Abbreviations: RAC, ribosome-associated complex; mRAC, mammalian RAC; BN-PAGE, blue native PAGE.
References
- 1.Rospert, S., Gautschi, M., Rakwalska, M. & Raue, U. (2005) in Protein Folding Handbook, eds. Buchner, J. & Kiefhaber, T. (Wiley, Weinheim, Germany), Vol. II, pp. 429-458. [Google Scholar]
- 2.Nelson, R. J., Ziegelhoffer, T., Nicolet, C., Werner-Washburne, M. & Craig, E. A. (1992) Cell 71, 97-105. [DOI] [PubMed] [Google Scholar]
- 3.Pfund, C., Lopez-Hoyo, N., Ziegelhoffer, T., Schilke, B. A., Lopez-Buesa, P., Walter, W. A., Wiedmann, M. & Craig, E. A. (1998) EMBO J. 17, 3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gautschi, M., Just, S., Mun, A., Ross, S., Rücknagel, P., Dubaquié, Y., Ehrenhofer-Murray, A. & Rospert, S. (2003) Mol. Cell. Biol. 23, 7403-7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan, W., Schilke, B., Pfund, C., Walter, W., Kim, S. & Craig, E. A. (1998) EMBO J. 17, 4809-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang, P., Gautschi, M., Walter, W., Rospert, S. & Craig, E. A. (2005) Nat. Struct. Mol. Biol. 12, 497-504. [DOI] [PubMed] [Google Scholar]
- 7.Gautschi, M., Lilie, H., Fünfschilling, U., Mun, A., Ross, S., Lithgow, T., Rücknagel, P. & Rospert, S. (2001) Proc. Natl. Acad. Sci. USA 98, 3762-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gautschi, M., Mun, A., Ross, S. & Rospert, S. (2002) Proc. Natl. Acad. Sci. USA 99, 4209-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hundley, H., Eisenman, H., Walter, W., Evans, T., Hotokezaka, Y., Wiedmann, M. & Craig, E. (2002) Proc. Natl. Acad. Sci. USA 99, 4203-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukau, B., Deuerling, E., Pfund, C. & Craig, E. A. (2000) Cell 101, 119-122. [DOI] [PubMed] [Google Scholar]
- 11.Rakwalska, M. & Rospert, S. (2004) Mol. Cell. Biol. 24, 9186-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, S., Lockshin, C., Herbert, A., Winter, E. & Rich, A. (1992) EMBO J. 11, 3787-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartl, F. U. & Hayer-Hartl, M. (2002) Science 295, 1852-1858. [DOI] [PubMed] [Google Scholar]
- 14.Craig, E. A., Eisenman, H. C. & Hundley, H. A. (2003) Curr. Opin. Microbiol. 6, 157-162. [DOI] [PubMed] [Google Scholar]
- 15.Shoji, W., Inoue, T., Yamamoto, T. & Obinata, M. (1995) J. Biol. Chem. 270, 24818-24825. [DOI] [PubMed] [Google Scholar]
- 16.Resto, V. A., Caballero, O. L., Buta, M. R., Westra, W. H., Wu, L., Westendorf, J. M., Jen, J., Hieter, P. & Sidransky, D. (2000) Cancer Res. 60, 5529-5535. [PubMed] [Google Scholar]
- 17.Matsumoto-Taniura, N., Pirollet, F., Monroe, R., Gerace, L. & Westendorf, J. M. (1996) Mol. Biol. Cell 7, 1455-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue, T., Shoji, W. & Obinata, M. (1999) Biochem. Biophys. Res. Commun. 266, 147-151. [DOI] [PubMed] [Google Scholar]
- 19.Sikder, H. A., Devlin, M. K., Dunlap, S., Ryu, B. & Alani, R. M. (2003) Cancer Cell 3, 525-530. [DOI] [PubMed] [Google Scholar]
- 20.Inoue, T., Shoji, W. & Obinata, M. (2000) Genes Cells 5, 699-709. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida, M., Inoue, T., Shoji, W., Ikawa, S. & Obinata, M. (2004) Biochem. Biophys. Res. Commun. 324, 326-332. [DOI] [PubMed] [Google Scholar]
- 22.Sherman, F., Fink, G. R. & Hicks, J. B. (1986) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 23.Wang, K., Feramisco, J. R. & Ash, J. F. (1982) Methods Enzymol. 85, 514-562. [DOI] [PubMed] [Google Scholar]
- 24.Blobel, G. & Potter, V. R. (1967) J. Mol. Biol. 26, 279-292. [DOI] [PubMed] [Google Scholar]
- 25.Schägger, H., Cramer, W. A. & von Jagow, G. (1994) Anal. Biochem. 217, 220-230. [DOI] [PubMed] [Google Scholar]
- 26.Schägger, H. & von Jagow, G. (1991) Anal. Biochem. 199, 223-231. [DOI] [PubMed] [Google Scholar]
- 27.Schägger, H. & von Jagow, G. (1987) Anal. Biochem. 166, 368-379. [DOI] [PubMed] [Google Scholar]
- 28.Perrot, M., Sagliocco, F., Mini, T., Monribot, C., Schneider, U., Shevchenko, A., Mann, M., Jenö, P. & Boucherie, H. (1999) Electrophoresis 20, 2280-2298. [DOI] [PubMed] [Google Scholar]
- 29.Haid, A. & Suissa, M. (1983) Methods Enzymol. 96, 192-205. [DOI] [PubMed] [Google Scholar]
- 30.Walsh, P., Bursac, D., Law, Y. C., Cyr, D. & Lithgow, T. (2004) EMBO Rep. 5, 567-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polevoda, B. & Sherman, F. (2003) Biochem. Biophys. Res. Commun. 308, 1-11. [DOI] [PubMed] [Google Scholar]
- 32.Sugiura, N., Adams, S. M. & Corriveau, R. A. (2003) J. Biol. Chem. 278, 40113-40120. [DOI] [PubMed] [Google Scholar]
- 33.Faber, P., Barnes, G., Srinidhi, J., Chen, J., Gusella, J. & MacDonald, M. (1998) Hum. Mol. Genet. 7, 1463-1474. [DOI] [PubMed] [Google Scholar]
- 34.Wan, T., Zhou, X., Chen, G., An, H., Chen, T., Zhang, W., Liu, S., Jiang, Y., Yang, F., Wu, Y. & Cao, X. (2004) Blood 103, 1747-1754. [DOI] [PubMed] [Google Scholar]
- 35.Hughes, R., Chan, F. Y., White, R. A. & Zon, L. I. (1995) Genomics 29, 546-550. [DOI] [PubMed] [Google Scholar]
- 36.Jin, H. & Martin, C. (1999) Plant Mol. Biol. 41, 577-585. [DOI] [PubMed] [Google Scholar]
- 37.Ito, M. (2005) J. Plant Res. 118, 61-69. [DOI] [PubMed] [Google Scholar]
- 38.Dudek, J., Volkmer, J., Bies, C., Guth, S., Müller, A., Lerner, M., Feick, P., Schäfer, K. H., Morgenstern, E., Hennessy, F., et al. (2002) EMBO J. 21, 2958-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braun, E. L. & Grotewold, E. (2001) Mol. Biol. Evol. 18, 1401-1412. [DOI] [PubMed] [Google Scholar]
- 40.Wilhelm, M. L., Reinbolt, J., Gangloff, J., Dirheimer, G. & Wilhelm, F. X. (1994) FEBS Lett. 349, 260-264. [DOI] [PubMed] [Google Scholar]
- 41.Greiner, J., Ringhoffer, M., Taniguchi, M., Li, L., Schmitt, A., Shiku, H., Dohner, H. & Schmitt, M. (2004) Int. J. Cancer 108, 704-711. [DOI] [PubMed] [Google Scholar]
- 42.Greiner, J., Ringhoffer, M., Taniguchi, M., Hauser, T., Schmitt, A., Dohner, H. & Schmitt, M. (2003) Int. J. Cancer 106, 224-231. [DOI] [PubMed] [Google Scholar]
- 43.Okada, H., Attanucci, J., Giezeman-Smits, K. M., Brissette-Storkus, C., Fellows, W. K., Gambotto, A., Pollack, L. F., Pogue-Geile, K., Lotze, M. T., Bozik, M. E. & Chambers, W. H. (2001) Cancer Res. 61, 2625-2631. [PubMed] [Google Scholar]
- 44.Clemens, M. J. & Bommer, U. A. (1999) Int. J. Biochem. Cell Biol. 31, 1-23. [DOI] [PubMed] [Google Scholar]
- 45.Abbott, C. M. & Proud, C. G. (2004) Trends Biochem. Sci. 29, 25-31. [DOI] [PubMed] [Google Scholar]
- 46.Ruggero, D. & Pandolfi, P. P. (2003) Nat. Rev. Cancer 3, 179-192. [DOI] [PubMed] [Google Scholar]