Abstract
WNK (with no lysine [K]) kinases are serine-threonine protein kinases with an atypical placement of the catalytic lysine. Intronic deletions increase the expression of WNK1 in humans and cause pseudohypoaldosteronism type II, a form of hypertension. WNKs have been linked to ion carriers, but the underlying regulatory mechanisms are unknown. Here, we report a mechanism for the control of ion permeability by WNK1. We show that WNK1 activates the serum- and glucocorticoid-inducible protein kinase SGK1, leading to activation of the epithelial sodium channel. Increased channel activity induced by WNK1 depends on SGK1 and the E3 ubiquitin ligase Nedd4-2. This finding provides compelling evidence that this molecular mechanism contributes to the pathogenesis of hypertension in pseudohypoaldosteronism type II caused by WNK1 and, possibly, in other forms of hypertension.
Keywords: ion transport, Nedd4-2, pseudohypoaldosteronism type II
WNK (with no lysine [K]) kinases are large protein kinases conserved in multicellular organisms with a unique arrangement of the active site (1, 2). The four human WNK family members display >85% sequence identity within their kinase domains. WNK1, the first identified, is expressed in differentially spliced forms containing >2,100 residues (3, 4). Outside the ≈260-residue N-terminal WNK kinase core, sequence identity among WNKs is much lower, and few homologous regions exist. No other folded domains have yet been identified in WNK1, although numerous potential interaction motifs are present. Thus, nothing further about the function of WNK1 has been deduced from sequence analysis alone.
A positional cloning study identified WNK1 and WNK4 as genes mutated in families of patients with hypertension characterized as pseudohypoaldosteronism type II (PHA II) (5). Mutations in the WNK1 gene are intronic deletions, leading to increased expression, whereas those in the WNK4 gene are missense mutations in the coding sequence. Disruption of the WNK1 gene causes embryonic lethality in mice, indicating that WNK1 is required for development (6). WNK1+/- mice display hypotension, establishing the importance of WNK1 in the normal regulation of blood pressure in rodents. Taken together, these findings are consistent with the idea that blood pressure regulation is sensitive to changes in the amount of WNK1 expressed. WNK1 is widely expressed in tissues and cell lines, indicating ubiquitous functions (1, 3, 5). In contrast, WNK4 is expressed predominantly in kidney and other epithelial tissues (3, 5, 7). Although WNK1 likely modulates the effects of WNK4 on the sodium chloride cotransporter (NCC), no direct effects of WNK1 on transporter activity have yet been validated (8-11). WNK1, but not WNK4, phosphorylates synaptotagmin II, modulating its membrane binding, suggesting that WNK1 is a switch, regulating vesicle trafficking and fusion (12).
The serum- and glucocorticoid-inducible protein kinase SGK1 was originally discovered as a consequence of its induction by the glucocorticoid dexamethasone and serum in epithelial cells (13). A screen to identify candidate regulators of the epithelial sodium channel ENaC also identified SGK1 (14). SGK1 stimulates sodium transport by increasing the amount of the ENaC at the cell surface (15). ENaC is the rate-limiting step for sodium transport across high-resistance epithelia and is regulated by insulin and mineralocorticoid hormones. The activity of SGK1 depends on phosphatidylinositol 3-kinase (PI-3 kinase), an essential mediator of insulin signaling. PI-3 kinase is also required for SGK1-dependent stimulation of ENaC by mineralocorticoids (16). The impact of SGK1 on ENaC, together with its induction by aldosterone and insulin, places this protein kinase in a key position to regulate sodium homeostasis and blood pressure (14).
Ubiquitinylation controls the cell-surface expression of many membrane proteins (17, 18). The surface expression of ENaC is regulated by the HECT domain E3 ubiquitin-protein ligase Nedd4-2 via a direct interaction with a PY motif in two subunits of the channel and a WW proline-binding domain of Nedd4-2 (17). Increased ENaC activity is found in Liddle's disease, an inherited form of human hypertension (19). Mutations in the PY motifs in ENaC subunits required for regulated endocytosis of the protein cause the disease by disrupting ENaC regulation by Nedd4-2. SGK1 interacts with Nedd4-2 and phosphorylates it (20). Phosphorylation by SGK1 reduces the interaction between Nedd4-2 and ENaC, leading to elevated ENaC cell-surface expression. SGK1-/- mice have impaired sodium retention and low blood pressure when stressed (21).
Here, we show that WNK1 stimulates the activity of SGK1 and sodium transport by ENaC. We propose that the ability of WNK1 to regulate ENaC through SGK1/Nedd4-2 contributes to the pathogenesis of PHA II and other forms of hypertension.
Materials and Methods
Constructs, Proteins, and Antibodies. pCMV5-Myc-WNK1 full-length and 1-491 and pBlueScript-WNK1 full-length were as described in refs. 1 and 22. WNK1 1-491, SGK1, and Nedd4-2 were subcloned into pBlueScript for in vitro transcription. To make Flag-SGK1 (full-length and Δ1-60), the DNA was amplified by PCR and subcloned into pCMV7.1-3× Flag (Sigma). Human Nedd4-2 was similarly subcloned into pGEX-KG. Site-directed mutagenesis was performed by using the QuikChange kit (Stratagene) according to the manufacturer's directions. Additional constructs and methods were as described in refs. 1, 22, and 23. DNAs were kindly provided as follows: SGK1 by M. E. Greenberg (Harvard University, Cambridge, MA); Nedd4-2 by P. M. Snyder (University of Iowa, Iowa City); PDK1 by E. N. Olson (University of Texas Southwestern); p70 and S6 by J. Blenis (Harvard University); Akt by M. A. White (University of Texas Southwestern); MEKK2/3 by G. L. Johnson (University of North Carolina, Chapel Hill); ENaC α, β, and γ subunits in Xenopus oocyte expression vector by T. R. Kleyman (University of Pittsburgh, Pittsburgh); and ENaC α, β, and γ subunits in mammalian expression vector by J. D. Stockand (University of Texas Health Sciences Center, San Antonio). GST-Nedd4-2 and GST-S6 proteins were expressed and purified from Escherichia coli strain BL21 by using standard procedures. Crosstide (GRPRTSSFAEGRR) was synthesized by the institutional peptide synthesis facility. Anti-Myc (National Cell Culture Center, Minneapolis), anti-HA (Roche), and anti-Flag (Sigma) were from commercial sources. Anti-WNK1 (Q256) was as described in ref. 1.
Two-Hybrid Screen. A rat brain cDNA library (Clontech) was screened with WNK1 (1-555) in a GAL4 vector (24). Among >106 colonies screened, one interacting clone encoded full-length SGK1.
Tissue Culture and Transfection. HEK293 cells were maintained in Dulbecco-Vogt-modified Eagle's medium with 10% FBS, 2 mM l-glutamine, and 100 units/ml penicillin/steptomycin as described in ref. 25. Chinese hamster ovary (CHO) cells (K1 clone from American Type Culture Collection) were cultured in F12-K medium (Invitrogen), as described in ref. 26. HEK293 cells were transfected by using calcium phosphate at 50-80% confluence and harvested in isotonic lysis buffer containing 1% Triton X-100 and phosphatase and protease inhibitors (25). For coimmunoprecipitation, cells were lysed with a Dounce homogenizer in buffer lacking Triton X-100.
Immunoprecipitation and Blotting. Proteins were immunoprecipitated from cell lysates by using the indicated antibodies at 1:100 dilution and protein A-Sepharose beads. For Western blots, total lysates or immunoprecipitates were resolved by SDS/PAGE, and proteins were transferred onto nitrocellulose membranes. The membranes were incubated with the indicated antibodies and developed by using enhanced chemiluminescence.
Kinase Assays. In vitro kinase assays were performed as described in ref. 22. For assays of SGK1, 30-μl reactions containing 10 mM Hepes (pH 7.5), 50 μM ATP (10-50 cpm/fmol), 10 mM MgCl2, 1 mM DTT, 1 mM benzamidine, and 1 μg of GST-Nedd4-2 were incubated for 60 min at 30°C. Assays were terminated with 7.5 μl of 5× electrophoresis sample buffer and resolved by electrophoresis on 10% polyacrylamide gels (30:0.8, acrylamide:bisacrylamide) in 0.1% SDS. Reactions using 1 μg of Crosstide as the SGK1 substrate were spotted on P81 paper and washed in phosphoric acid to remove nucleotide as described in ref. 27, followed by scintillation counting.
Two-Electrode Voltage-Clamp and Whole-Cell Patch-Clamp Recording of ENaC Channels. cRNAs for mouse α, β, and γ subunits of ENaC (28), rat WNK1, mouse SGK1, and human Nedd4-2 were synthesized by using an in vitro transcription kit (Ambion). Xenopus laevis oocytes were prepared as described in ref. 29. The oocytes were injected with cRNAs for respective cDNAs at the concentrations indicated in the figure legends. After injection, oocytes were incubated in ND96 bath solution containing 96 mM NaCl, 2 mM KCl, 1 mM CaCl2, 1 mM MgCl2, and 5 mM Hepes (pH 7.4). I-V relationships were recorded from oocytes (48-72 h postinjection) by using an OC-725C oocyte clamp amplifier, the program pclamp7, and a Digidata 1200A digitizer (Axon Instruments, Union City, CA). Voltage protocols consist of voltage steps (400 msec) from -100 to 50 mV in 25-mV increments from a 0-mV holding potential. Currents were recorded in ND96 bath solution. In some experiments, ND96 was replaced by a bath solution containing 98 mM KCl, 1 mM CaCl2, 1 mM MgCl2, and 5 mM Hepes (pH 7.4) to verify that inward currents were carried by sodium ions. As indicated, oocytes were incubated in ND96 containing 10 μM amiloride. Similar effects of WNK1 on ENaC were found.
CHO cells were cotransfected with cDNAs (0.5 μg each) for GFP, α, β, and γ subunits of ENaC subunits SGK1, Nedd4-2, and/or WNK1, as indicated. The total amount of DNA for transfection was balanced by using an empty vector. Cells were grown in 10 μM amiloride. At 36-48 h posttransfection, whole-cell currents were recorded by using an Axopatch 200B amplifier (Axon Instruments) as described in ref. 26. Transfected cells were identified by using epifluorescent microscopy. The pipette and bath solution contained 130 mM K-methanesulfonate, 10 mM NaCl, and 10 mM Hepes (pH 7.2) and 140 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, and 10 mM Hepes (pH 7.4), respectively. Capacitance and access resistance were monitored and 75% compensated. The voltage protocol consists of a 0-mV holding potential and 400-ms steps from -100 to 100 mV in 20-mV increments. Statistical comparison was made by using the unpaired t test.
Results
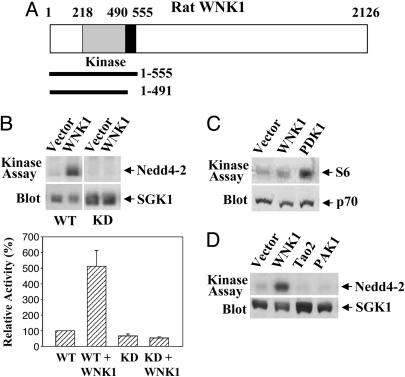
WNK1 Activates SGK1. To identify potential targets of WNK1, we performed a two-hybrid screen with WNK1 (1-555), which includes its protein kinase domain (Fig. 1A), and found SGK1 as an interactor. To evaluate the significance of this interaction, we coexpressed WNK1 (1-491), an active form because of the truncation of its autoinhibitory domain, with SGK1 and found that SGK1 activity was increased with either recombinant Nedd4-2 or the peptide Crosstide as substrate (Fig. 1B). The activity increase was not observed with a kinase-dead (KD) mutant of SGK1 (Fig. 1B). We were unable to detect endogenous SGK1 in 293 or other cell lines. The p70 S6 kinase and Akt (protein kinase B) are similar to SGK1, in that each requires phosphorylation of an activation-loop residue catalyzed by the phosphoinositide-dependent kinase PDK1 and phosphorylation of a C-terminal residue by an as yet unidentified protein kinase (30, 31). WNK1 (1-491) activated neither p70 (Fig. 1C) nor Akt (data not shown), indicating the specificity of SGK1 activation by WNK1. In addition, SGK1 was not activated by other multifunctional protein kinases, TAO2 and PAK1 (Fig. 1D), indicating that related kinases lack the capacity to activate SGK1.
Fig. 1.
WNK1 activates SGK1. (A) Diagram of WNK1 and fragments used in this study. The kinase domain is shaded, and the autoinhibitory domain is in black. (B) Wild-type or KD pCMV7.1-Flag-SGK1 was transfected into HEK293 cells along with empty vector or pCMV5-Myc-WNK1 (1-491). SGK1 activity was assayed with GST-Nedd4-2 (Upper) or Crosstide (Lower) as substrate. Activities are shown as a percentage of basal SGK1 activity. (C) WNK1 does not activate p70. p70 activity was assayed with GST-S6 as substrate. (D) Neither TAO2 (1-451) nor PAK1 activates SGK1. For all experiments, immunoblots of whole-cell lysates are included. Experiments were performed from 3 to 10 times.
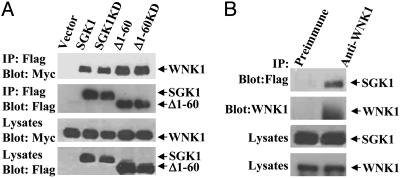
WNK1 Binds to SGK1. To test whether WNK1 and SGK1 interact in cells, Myc-tagged WNK1 (1-491) and Flag-tagged SGK1 were cotransfected into HEK293 cells. WNK1 was detected in SGK1 immunoprecipitates, indicating that these proteins could exist in the same complex (Fig. 2A). The N-terminal 60 residues and the catalytic activity of SGK1 are, apparently, not required for the interaction with WNK1 because SGK1 Δ1-60 (a stable form of SGK1) and a KD mutant coimmunoprecipitated with WNK1. Moreover, endogenous WNK1 also coimmunoprecipitated with SGK1 (Fig. 2B). Because we have been unable to express SGK1 in bacteria, we cannot rule out the possibility that association is through a third protein. These results account for the two-hybrid finding and suggest that WNK1 can associate with SGK1 in cells that express it.
Fig. 2.
WNK1 binds to SGK1. (A) WNK1 coimmunoprecipitates with SGK1. Cells were transfected with Myc-WNK1 (1-491) and either vector or Flag-SGK1 constructs. SGK1 proteins were immunoprecipitated and blotted with anti-Myc and anti-Flag. (B) Endogenous WNK1 coimmunoprecipitates with SGK1. Lysates from cells transfected with Flag-SGK1 (S422D) were immunoprecipitated with anti-WNK1 (Q256) or its preimmune serum, and the precipitates were blotted with anti-Flag and Q256. Immunoblots of whole-cell lysates are included. Experiments were performed four times.
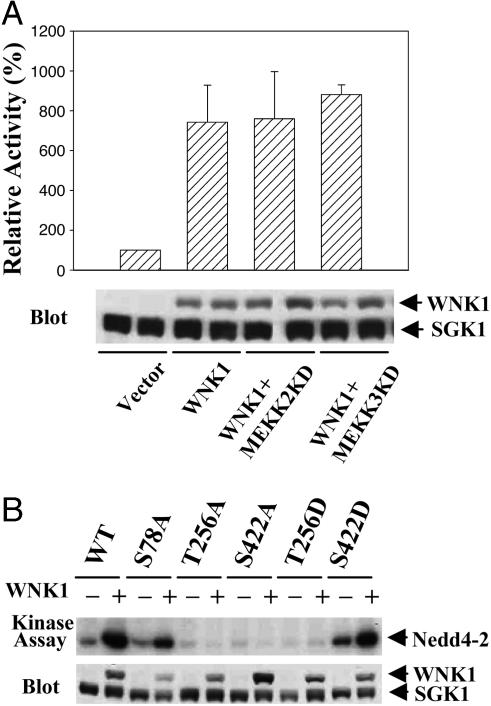
Activation of SGK1 by WNK1 Is Independent of ERK5. We examined the mechanism of SGK1 activation by WNK1. WNK1 activates the MAPK ERK5 via upstream protein kinases MEKK2, MEKK3, and MEK5 (22). ERK5 was reported to phosphorylate and activate SGK1 (32). Inhibitory mutants of MEKK2/3 did not block activation of SGK1 by WNK1 (Fig. 3A). Thus, WNK1 activation of SGK1 is independent of components of ERK5 MAPK pathways.
Fig. 3.
Activation of SGK1 by WNK1 is independent of ERK5 pathway. (A)KD mutants of MEKK2/3 do not block WNK1 activation of SGK1. SGK1 activity was assayed with Crosstide. (B) T256 and S422 on SGK1 are required for WNK1 activation. SGK1 activity was assayed with Nedd4-2. Experiments were performed a minimum of five times in A and twice in B. Immunoblots of whole-cell lysates are included.
Three phosphorylation sites have been identified on SGK1. ERK5 phosphorylates SGK1 on Ser-78, PDK1 phosphorylates the activation loop site Thr-256, and an unknown kinase phosphorylates the C-terminal site Ser-422 (30, 32, 33). Mutating the ERK5 site Ser-78 to alanine had little effect on SGK1 activation by WNK1, consistent with the ERK5-independence of this mechanism (Fig. 3B). Activation of SGK1 by WNK1 was abolished by mutating either Thr-256 or Ser-422 to alanine. SGK1 T256D exhibited low basal activity and was not activated by WNK1, whereas S422D had higher basal activity and was further activated by WNK1 (Fig. 3B). These data indicate that WNK1 promotes SGK1 activation by increasing phosphorylation of Thr-256, its activation loop. However, WNK1 does not phosphorylate SGK1 directly (data not shown). How WNK1 activates SGK1 remains unknown.
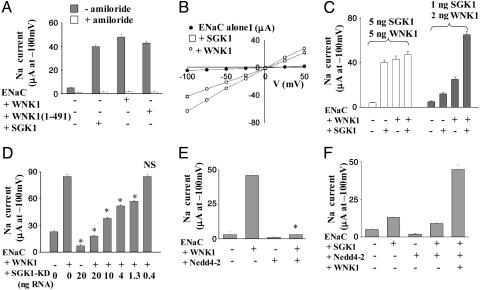
Activation of SGK1 by WNK1 Causes Increased Sodium Transport. Phosphorylation of the ubiquitin E3 ligase Nedd4-2 by SGK1 reduces the interaction between Nedd4-2 and its substrates including ENaC, thus increasing the number of sodium channels in the membrane and, consequently, increasing sodium current (14, 15, 20). We assessed the biological function of WNK1 activation of SGK1 by reconstitution with the α, β, and γ subunits of ENaC in Xenopus oocytes. Xenopus oocytes express endogenous SGK1 (20, 34). Full-length WNK1 activated ENaC current in oocytes (Fig. 4 A and B). The same was true for truncated WNK1 1-491. Expression of SGK1 increased ENaC activity, as shown by others (14, 15, 20, 35). Activation of ENaC by SGK1 and by WNK1 was not additive when the maximal stimulatory concentrations of RNA for SGK1 and WNK1 were used (5 ng of RNA for each) (Fig. 4C, open bars). Coexpression of submaximal (≤50% of the maximal) amounts of WNK1 (2 ng) and SGK1 (1 ng) RNAs caused synergistic activation of ENaC relative to either alone (Fig. 4C, shaded bars). Thus, WNK1 cooperated with SGK1 to activate ENaC. A dominant-negative SGK1 mutant (K127M) inhibited ENaC activation by WNK1 in a dose-dependent manner (Fig. 4D), supporting the conclusion that activation of ENaC by WNK1 requires SGK1 activity. Activation of ENaC by SGK1 in Xenopus oocytes is, at least in part, due to release of inhibition of the channel by the endogenous Nedd4-2 (20). Overexpression of exogenous Nedd4-2 (5 ng of RNA), like KD SGK1, blocked ENaC activation by WNK1 (Fig. 4E), supporting the importance of Nedd4-2 in the actions of WNK1. Moreover, coexpression of WNK1 with an amount of Nedd4-2 RNA (1 ng) less than that required for maximal inhibition resulted in activation of ENaC in the presence of SGK1 (Fig. 4F).
Fig. 4.
WNK1 stimulates ENaC by SGK1 in Xenopus oocytes. (A) WNK1 and SGK1 each increase ENaC activity. Currents were recorded by using two-electrode voltage-clamp from Xenopus oocytes injected with cRNAs for α, β, and γ subunits of ENaC (1 ng each) plus WNK1 (5 ng), or SGK1 (5 ng) in a bath solution containing 96 mM Na+. Inward currents disappeared when Na+ in the bath solution was replaced by K+. Inward Na+ currents (μA) at -100 mV are shown (mean ± SEM, n = 10 oocytes per group). All currents were inhibitable by amiloride (5 μM) (open bars). Background currents (-100 mV) for water-injected oocytes were 0.42 ± 0.03 μA. (B) I-V relationships of currents shown in A. Currents reversed at ≈0 mV, indicating ENaC-mediated intracellular loading of Na+ ions. (C) Relationship between WNK1- and SGK1-activation of ENaC. The experiment paradigm is as in A, except that different amounts of RNA for SGK1 and WNK1, as indicated, were coinjected. (D) Coexpression of SGK1-KD prevents WNK1 stimulation of ENaC in a dose-dependent manner. Experimental paradigm is as in A. *, P < 0.05 vs. ENaC alone. (E) Coexpression of Nedd4-2 (5 ng of RNA) prevents WNK1 stimulation of ENaC. *, P < 0.05 vs. ENaC + WNK1. (F) WNK1 activates ENaC in the presence of a submaximal inhibitory concentration of Nedd4-2 (1 ng of RNA). RNAs encoding α, β, and γ subunits of ENaC and SGK1 were 1 ng each. All experiments were performed at least three times, with similar results.
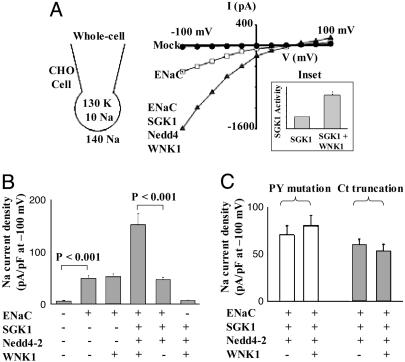
To provide further evidence that WNK1 activates ENaC through an SGK1-Nedd4-2 pathway, we reconstituted ENaC channels in CHO cells that lack endogenous SGK1 by coexpressing ENaC's α, β, and γ subunits (36, 37). Amiloride-sensitive Na+ current was present in ENaC-reconstituted but not in mock-transfected CHO cells (Fig. 5A). Without exogenous SGK1 and Nedd4-2, WNK1 1-491 did not activate ENaC (Fig. 5B). A recent study reported that purified SGK1 can activate ENaC in excised patches, presumably by a direct phosphorylation of the channel (38). However, without exogenous Nedd4-2, we found that SGK1 did not activate ENaC in whole cells (data not shown). WNK1 increased ENaC current by ≈3-fold in the presence of SGK1 and Nedd4-2 (whole-cell current density: 156 ± 13 pA/pF with WNK1 and 47 ± 4 pA/pF without WNK1; Fig. 5B). Consistent with activation of ENaC, WNK1 activated SGK1 activity 3-fold in cotransfected CHO cells (Fig. 5A Inset). As a final experiment to validate the connection between WNK1 and Nedd4-2, the PY motifs in the C termini of the α, β, and γ subunits of ENaC were disrupted by either point mutation or proximal truncation to eliminate the ability of Nedd4-2 to bind to ENaC. These ENaC mutants are no longer susceptible to down-regulation by Nedd4-2 (20, 39). WNK1 did not activate ENaC with point mutations in the PY motifs (white bars) or with C-terminal truncations (gray bars) (Fig. 5C). These findings indicate that WNK1 requires both SGK1 and Nedd4-2 to regulate ENaC and are consistent with the hypothesis that WNK1 activates SGK1, which activates ENaC, at least in part, by preventing ubiquitination and down-regulation by Nedd4-2.
Fig. 5.
WNK1 stimulates ENaC via SGK1-Nedd4-2 pathway in CHO cells. (A) Representative I-V relationships for whole-cell currents in CHO cells. Whole-cell configuration and concentration of the charge carrier are also shown. All currents were inhibitable with amiloride (5 μM). Currents shown in I-V curves are after subtraction of the amiloride-insensitive residual currents. The reversal potential is consistent with Na+ as the charge carrier. The amiloride-sensitive inward currents disappeared when Na+ in the bath solution was replaced by K+. (Inset) WNK1 activated SGK1 by 3-fold in cotransfected CHO cells. (B) SGK1 and Nedd4-2 are required for activation of ENaC by WNK1. Whole-cell amiloride-sensitive current density at -100 mV (pA/pF) is shown (mean ± SEM, n = 5-10 cells per group). Experiments were performed three times, with similar results. (C) The PY motif of ENaC is required for activation by WNK1. Experimental paradigm is as in A. The PY mutation represents Y673A, Y618A, and Y633A point mutations for the α, β, and γ subunits, respectively, of ENaC. Ct truncation represents the truncation at P646, R564, and F611 for α, β, and γ subunits, respectively, of ENaC (mean ± SEM, n = 5-10 cells per group). Experiments were performed twice, with similar results.
Discussion
Sodium reabsorption by the distal nephron of the kidney is critical for blood pressure control (40). ENaC mediates renal sodium excretion and sodium homeostasis and impacts blood pressure. Increased ENaC activity caused by mutations in ENaC subunits, required for properly regulated endocytosis, results in hypertension in Liddle's disease (19). WNK1+/- mice also display low blood pressure, establishing the importance of WNK1 in the normal regulation of blood pressure (6). Interestingly, SGK1-/- mice have impaired sodium retention and low blood pressure when stressed (21). Our study suggests that ENaC is a WNK1 target and that stimulation of ENaC by WNK1 contributes to the pathogenesis of hypertension in PHA II.
The sodium-reabsorptive distal nephron includes the distal convoluted tubule, connecting tubule, and cortical collecting duct. Not only does overexpression of WNK1 leads to PHA II, but missense mutations in the coding sequence of WNK4 also cause PHA II (5). WNK4 reportedly inhibits the activity of the NCC (8-11). WNK4 mutants that cause disease fail to inhibit NCC, suggesting that an increase in NCC activity in the distal convoluted tubule is a cause of hypertension. Consistent with the idea that increased NCC activity is involved, many patients originally characterized with PHA II were highly responsive to thiazide diuretics (41). Indeed, a recent study reported that PHA II patients with the Q565E WNK4 mutation exhibited exquisite sensitivity to thiazides (42). Nevertheless, other transporters are likely involved in the actions of WNK4. WNK4 has been reported to regulate the inward rectifying potassium channel ROMK (Kir1.1), the Na-K-2Cl cotransporter (NKCCl) and the Cl/base exchanger SLC26A6 (CFEX) (7-9, 43). Finally, Yamauchi and coworkers (44) reported that WNK4 phosphorylates claudins 1-4, the tight-junction proteins involved in the regulation of paracellular ion permeability. The paracellular chloride permeability is greater in cells expressing WNK4 mutants than in cells expressing wild-type proteins.
Mutations in the WNK1 gene in PHA II are intronic deletions causing increased expression (5). Because expression of WNK1 abolishes inhibition of NCC caused by WNK4 in Xenopus oocytes (8), it has been suggested that increased expression of WNK1 causes hypertension indirectly by releasing WNK4-mediated inhibition of NCC (8). However, the actions of WNK1 are not mediated solely through WNK4. The cellular and subcellular distributions of WNK1 and WNK4 in kidney and elsewhere in the body are different (3, 5). Unlike PHA II patients with missense mutations of WNK4, patients with intronic deletions of WNK1 are not particularly sensitive to thiazides (45). In a large family with the WNK1 deletion mutation, only one of three affected hypertensive individuals treated with thiazides exhibited blood pressure ≤140/90 mmHg (1 mmHg = 133 Pa) (45). In contrast, two of four affected individuals treated with non-thiazide antihypertensive drugs exhibited normal blood pressure. Responses to other diuretics and antihypertensive drugs in PHA II patients have also been reported (41). Here, we show that WNK1 activates ENaC. We suggest that hypertension in PHA II caused by WNK1 is due, at least in part, to the activation of ENaC. Activation of ENaC, regardless of changes in NCC activity, could account for the reduced response to thiazides in patients with WNK1 mutations.
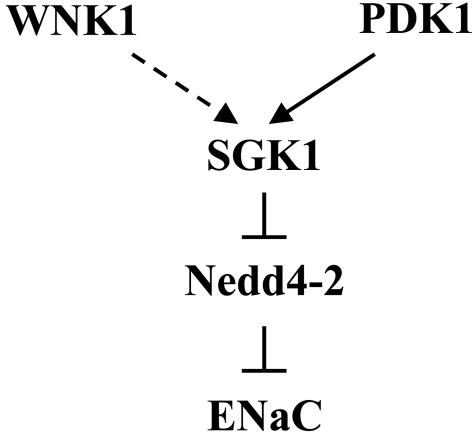
We find that the stimulation of ENaC by WNK1 involves activation of SGK1. Although we have not yet elucidated the activation mechanism, we can propose certain aspects of this process (Fig. 6). We suggest that WNK1 cooperates with PDK1 to activate SGK1 fully; activation of SGK1 leads to inhibitory phosphorylation of Nedd4-2 and the subsequent retention of ENaC on the plasma membrane. WNK1 is ubiquitously expressed (1). At least two transcripts have been detected in kidney by Northern blot analysis: a longer transcript (>10 kb) containing the coding sequence necessary for activating SGK1 and ENaC, and a smaller transcript lacking coding sequence for N-terminal residues (exons I-IV) (4, 46, 47). The longer transcript is present in many tissues, including leukocytes, which were used for analysis of WNK1 expression in PHA II (4, 5, 47). Our results, together with the report of increased WNK1 expression in leukocytes, suggest that increased expression of the longer transcript contributes to hypertension in PHA II. The shorter transcript, lacking the N-terminal domain for activation of SGK1, is expressed most highly in the kidney (4, 47). A recent paper reported that expression of this shorter form of WNK1 in epithelial cells increased amiloride-sensitive short-circuit current (10). Whether the shorter transcript is increased in PHA II is not known. If so, the increase may also contribute to sodium retention and hypertension, perhaps by an SGK1-independent mechanism. Differential expression of WNK1 species lacking the N-terminal regions suggests that WNK1 will display widely variable tissue-specific regulatory properties, depending on the forms that are present. For example, the WNK1 kinase domain is required for modulation of the properties of synaptotagmins by WNK1 (12).
Fig. 6.
Model of ENaC activation by WNK1. See text for details.
Activation of ENaC in the distal nephron would be expected to stimulate K+ and H+ secretion, leading to hypokalemia and alkalosis, as in Liddle's disease (48). PHA II, however, is characterized by hyperkalemia and acidosis (41). It has been proposed that hyperkalemia is due to reduced Na+ delivery to the cortical collecting duct (CCD), consequent to increased NaCl reabsorption in tubules proximal to the CCD (49). However, hyperkalemia in PHA II of WNK1 and WNK4 mutations typically occurs about 10-20 y before development of hypertension, suggesting that other mechanism(s) may also be involved (42, 50). Recently, it has been shown that WNK4 inhibits ROMK K+ channels and the missense mutations increase the inhibition, suggesting that direct inhibition of K+ secretion through ROMK also contributes to hyperkalemia in PHA II (43). The mechanism by which increased expression of WNK1 causes hyperkalemia remains unknown, but may also involve the inhibition of ROMK. Acidosis in PHA II patients may result from the inhibition of ammonia production due to hyperkalemia. Alternatively, WNK1 may have direct effects on renal acid transporters.
SGK1 and Nedd4-2 regulate other ion channels and transporters in addition to ENaC, including the cardiac sodium channel SCN5A (51), the voltage-gated K+ channel KV1.3 (52), the intestinal phosphate transporter NaPi IIb (53), and the neuronal glutamate transporter EAAT1 (54). Both WNK1 and SGK isoforms are widely expressed in tissues, suggesting that WNK1 acts to promote the plasma membrane retention of a select group of proteins involved not only in sodium homeostasis but also in other key processes, such as cardiac and neuronal excitability.
Acknowledgments
We thank Joseph Goldstein, Alfred Gilman, and Michael White for comments about the manuscript; members of the Cobb laboratory for suggestions; and Dionne Ware for administrative assistance. This work was supported by National Institutes of Health Grants GM53032 (to M.H.C.), DK54368 (to C.-L.H.), and DK59530 (to C.-L.H.) and Welch Foundation Grant I1243 (to M.H.C.).
Author contributions: B.-e.X., C.-L.H., and M.H.C. designed research; B.-e.X., S.S., P.-Y.C., A.L., X.-J.L., B.-H.L., J.M.E., and B.O. performed research; B.-e.X., S.S., P.-Y.C., A.L., X.-J.L., B.-H.L., J.M.E., B.O., C.-L.H., and M.H.C. analyzed data; and B.-e.X., C.-L.H., and M.H.C. wrote the paper.
Abbreviations: CHO, Chinese hamster ovary; ENaC, epithelial sodium channel; KD, kinase-dead; NCC, sodium chloride cotransporter; PHA II, pseudohypoaldosteronism type II.
References
- 1.Xu, B., English, J. M., Wilsbacher, J. L., Stippec, S., Goldsmith, E. J. & Cobb, M. H. (2000) J. Biol. Chem. 275, 16795-16801. [DOI] [PubMed] [Google Scholar]
- 2.Min, X., Lee, B. H., Cobb, M. H. & Goldsmith, E. J. (2004) Structure (London) 12, 1303-1311. [DOI] [PubMed] [Google Scholar]
- 3.Verissimo, F. & Jordan, P. (2001) Oncogene 20, 5562-5569. [DOI] [PubMed] [Google Scholar]
- 4.Delaloy, C., Lu, J., Houot, A. M., Disse-Nicodeme, S., Gasc, J. M., Corvol, P. & Jeunemaitre, X. (2003) Mol. Cell. Biol. 23, 9208-9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson, F. H., Disse-Nicodeme, S., Choate, K. A., Ishikawa, K., Nelson-Williams, C., Desitter, I., Gunel, M., Milford, D. V., Lipkin, G. W., Achard, J. M., et al. (2001) Science 293, 1107-1112. [DOI] [PubMed] [Google Scholar]
- 6.Zambrowicz, B. P., Abuin, A., Ramirez-Solis, R., Richter, L. J., Piggott, J., BeltrandelRio, H., Buxton, E. C., Edwards, J., Finch, R. A., Friddle, C. J., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 14109-14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kahle, K. T., Gimenez, I., Hassan, H., Wilson, F. H., Wong, R. D., Forbush, B., Aronson, P. S. & Lifton, R. P. (2004) Proc. Natl. Acad. Sci. USA 101, 2064-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang, C. L., Angell, J., Mitchell, R. & Ellison, D. H. (2003) J. Clin. Invest. 111, 1039-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson, F. H., Kahle, K. T., Sabath, E., Lalioti, M. D., Rapson, A. K., Hoover, R. S., Hebert, S. C., Gamba, G. & Lifton, R. P. (2003) Proc. Natl. Acad. Sci. USA 100, 680-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naray-Fejes-Toth, A., Snyder, P. M. & Fejes-Toth, G. (2004) Proc. Natl. Acad. Sci. USA 101, 17434-17439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang, C. L., Zhu, X., Wang, Z., Subramanya, A. R. & Ellison, D. H. (2005) J. Clin. Invest. 115, 1379-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, B. H., Min, X., Heise, C. J., Xu, B., Chen, S., Shu, H., Luby-Phelps, K., Goldsmith, E. J. & Cobb, M. H. (2004) Mol. Cell 15, 741-751. [DOI] [PubMed] [Google Scholar]
- 13.Webster, M. K., Goya, L. & Firestone, G. L. (1993) J. Biol. Chem. 268, 11482-11485. [PubMed] [Google Scholar]
- 14.Chen, S. Y., Bhargava, A., Mastroberardino, L., Meijer, O. C., Wang, J., Buse, P., Firestone, G. L., Verrey, F. & Pearce, D. (1999) Proc. Natl. Acad. Sci. USA 96, 2514-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarez de la Rosa, D., Zhang, P., Naray-Fejes-Toth, A., Fejes-Toth, G. & Canessa, C. M. (1999) J. Biol. Chem. 274, 37834-37839. [DOI] [PubMed] [Google Scholar]
- 16.Rozansky, D. J., Wang, J., Doan, N., Purdy, T., Faulk, T., Bhargava, A., Dawson, K. & Pearce, D. (2002) Am. J. Physiol. 283, F105-F113. [DOI] [PubMed] [Google Scholar]
- 17.Flores, S. Y., Debonneville, C. & Staub, O. (2003) Pflügers Arch. 446, 334-338. [DOI] [PubMed] [Google Scholar]
- 18.Marmor, M. D. & Yarden, Y. (2004) Oncogene 23, 2057-2070. [DOI] [PubMed] [Google Scholar]
- 19.Toka, H. R. & Luft, F. C. (2002) Semin. Nephrol. 22, 81-88. [DOI] [PubMed] [Google Scholar]
- 20.Debonneville, C., Flores, S. Y., Kamynina, E., Plant, P. J., Tauxe, C., Thomas, M. A., Munster, C., Chraibi, A., Pratt, J. H., Horisberger, J. D., et al. (2001) EMBO J. 20, 7052-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wulff, P., Vallon, V., Huang, D. Y., Volkl, H., Yu, F., Richter, K., Jansen, M., Schlunz, M., Klingel, K., Loffing, J., et al. (2002) J. Clin. Invest. 110, 1263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu, B., Stippec, S., Lenertz, L., Lee, B. H., Zhang, W., Lee, Y. K. & Cobb, M. H. (2004) J. Biol. Chem. 279, 7826-7831. [DOI] [PubMed] [Google Scholar]
- 23.Xu, B., Min, X., Stippec, S., Lee, B. H., Goldsmith, E. J. & Cobb, M. H. (2002) J. Biol. Chem. 277, 48456-48462. [DOI] [PubMed] [Google Scholar]
- 24.Bai, C. & Elledge, S. J. (1996) Methods Enzymol. 273, 331-347. [DOI] [PubMed] [Google Scholar]
- 25.Xu, B., Wilsbacher, J. L., Collisson, T. & Cobb, M. H. (1999) J. Biol. Chem. 274, 34029-34035. [DOI] [PubMed] [Google Scholar]
- 26.Yeh, B. I., Sun, T. J., Lee, J. Z., Chen, H. H. & Huang, C. L. (2003) J. Biol. Chem. 278, 51044-51052. [DOI] [PubMed] [Google Scholar]
- 27.House, C. & Kemp, B. E. (1987) Science 238, 1726-1727. [DOI] [PubMed] [Google Scholar]
- 28.Ahn, Y. J., Brooker, D. R., Kosari, F., Harte, B. J., Li, J., Mackler, S. A. & Kleyman, T. R. (1999) Am. J. Physiol. 277, F121-F129. [DOI] [PubMed] [Google Scholar]
- 29.Zeng, W. Z., Li, X. J., Hilgemann, D. W. & Huang, C. L. (2003) J. Biol. Chem. 278, 16852-16856. [DOI] [PubMed] [Google Scholar]
- 30.Lang, F. & Cohen, P. (2001) Sci. STKE 2001, RE17. [DOI] [PubMed] [Google Scholar]
- 31.Frodin, M., Antal, T. L., Dummler, B. A., Jensen, C. J., Deak, M., Gammeltoft, S. & Biondi, R. M. (2002) EMBO J. 21, 5396-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi, M., Tapping, R. I., Chao, T. H., Lo, J. F., King, C. C., Yang, Y. & Lee, J. D. (2001) J. Biol. Chem. 276, 8631-8634. [DOI] [PubMed] [Google Scholar]
- 33.Park, J., Leong, M. L., Buse, P., Maiyar, A. C., Firestone, G. L. & Hemmings, B. A. (1999) EMBO J. 18, 3024-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loffing, J., Zecevic, M., Feraille, E., Kaissling, B., Asher, C., Rossier, B. C., Firestone, G. L., Pearce, D. & Verrey, F. (2001) Am. J. Physiol. 280, F675-F682. [DOI] [PubMed] [Google Scholar]
- 35.Snyder, P. M., Olson, D. R. & Thomas, B. C. (2002) J. Biol. Chem. 277, 5-8. [DOI] [PubMed] [Google Scholar]
- 36.Coric, T., Hernandez, N., De La Rosa, D. A., Shao, D., Wang, T. & Canessa, C. M. (2004) Am. J. Physiol. 286, G663-G670. [DOI] [PubMed] [Google Scholar]
- 37.Tong, Q., Gamper, N., Medina, J. L., Shapiro, M. S. & Stockand, J. D. (2004) J. Biol. Chem. 279, 22654-22663. [DOI] [PubMed] [Google Scholar]
- 38.Diakov, A. & Korbmacher, C. (2004) J. Biol. Chem. 279, 38134-38142. [DOI] [PubMed] [Google Scholar]
- 39.Staub, O., Dho, S., Henry, P., Correa, J., Ishikawa, T., McGlade, J. & Rotin, D. (1996) EMBO J. 15, 2371-2380. [PMC free article] [PubMed] [Google Scholar]
- 40.Guyton, A. C. (1991) Science 252, 1813-1816. [DOI] [PubMed] [Google Scholar]
- 41.Gordon, R. D. (1986) Aust. N.Z. J. Med. 16, 183-184. [DOI] [PubMed] [Google Scholar]
- 42.Mayan, H., Vered, I., Mouallem, M., Tzadok-Witkon, M., Pauzner, R. & Farfel, Z. (2002) J. Clin. Endocrinol. Metab. 87, 3248-3254. [DOI] [PubMed] [Google Scholar]
- 43.Kahle, K. T., Wilson, F. H., Leng, Q., Lalioti, M. D., O'Connell, A. D., Dong, K., Rapson, A. K., MacGregor, G. G., Giebisch, G., Hebert, S. C., et al. (2003) Nat. Genet. 35, 372-376. [DOI] [PubMed] [Google Scholar]
- 44.Yamauchi, K., Rai, T., Kobayashi, K., Sohara, E., Suzuki, T., Itoh, T., Suda, S., Hayama, A., Sasaki, S. & Uchida, S. (2004) Proc. Natl. Acad. Sci. USA 101, 4690-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Disse-Nicodeme, S., Achard, J. M., Desitter, I., Houot, A. M., Fournier, A., Corvol, P. & Jeunemaitre, X. (2000) Am. J. Hum. Genet. 67, 302-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, Q., Modrek, B. & Lee, C. (2002) Nucleic Acids Res. 30, 3754-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Reilly, M., Marshall, E., Speirs, H. J. & Brown, R. W. (2003) J. Am. Soc. Nephrol. 14, 2447-2456. [DOI] [PubMed] [Google Scholar]
- 48.Lifton, R. P. (1995) Proc. Natl. Acad. Sci. USA 92, 8545-8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schambelan, M., Sebastian, A. & Rector, F. C., Jr. (1981) Kidney Int. 19, 716-727. [DOI] [PubMed] [Google Scholar]
- 50.Achard, J. M., Warnock, D. G., Disse-Nicodeme, S., Fiquet-Kempf, B., Corvol, P., Fournier, A. & Jeunemaitre, X. (2003) Am. J. Med. 114, 495-498. [DOI] [PubMed] [Google Scholar]
- 51.Boehmer, C., Wilhelm, V., Palmada, M., Wallisch, S., Henke, G., Brinkmeier, H., Cohen, P., Pieske, B. & Lang, F. (2003) Cardiovasc. Res. 57, 1079-1084. [DOI] [PubMed] [Google Scholar]
- 52.Gamper, N., Fillon, S., Huber, S. M., Feng, Y., Kobayashi, T., Cohen, P. & Lang, F. (2002) Pflügers Arch. 443, 625-634. [DOI] [PubMed] [Google Scholar]
- 53.Palmada, M., Dieter, M., Speil, A., Embark, H., Bohmer, C., Mack, A. F., Wagner, H. J., Klingel, K., Kandolf, R., Murer, H., et al. (2004) Am. J. Physiol. 287, G143-G150. [DOI] [PubMed] [Google Scholar]
- 54.Boehmer, C., Henke, G., Schniepp, R., Palmada, M., Rothstein, J. D., Broer, S. & Lang, F. (2003) J. Neurochem. 86, 1181-1188. [DOI] [PubMed] [Google Scholar]